Abstract
Background
High-risk human papillomavirus type 16 (HPV16) is a risk factor for cervical cancer. The progression from initial infection to cervical cancer has been linked to properties of the viral sequences. However, the distribution of HPV16 variants among Chinese women has not been extensively addressed and the role of HPV16 variants in the risk of cervical carcinogenesis remains poorly understood.
Methods
HPV16 positive cervical exfoliated cell samples were collected from 249 women living in Beijing, China. PCR products from two fragments of E6-E7 and LCR of HPV16 in these samples were sequenced and analyzed.
Results
Lineage A was found in the subjects, including A1, A2, A3 and A4 sublineages. Based on the HPV16 reference sequences, 26 nucleotide mutations of A4 sublineage and 39 nucleotide mutations of A1-3 sublineages were found in the E6, E7 and LCR of HPV16 isolates. Point mutations T843C, A7287C and A7872G of A4 sublineage were significantly associated with high-grade cervical lesions. The high-frequency sites in HPV16 LCR located at regions that can bind to multiple transcription factors.
Conclusions
This study contributes to the identification of unique variants in the E6, E7 and LCR of HPV16 isolates infected in Chinese women. Mutations of T843C, A7287C and A7872G in A4 sublineages were significantly associated with high-grade cervical lesions, suggesting that mutations in the E7 and LCR region have potential effects on viral replication and progression of cervical cancer.
Keywords: HPV16, E6, E7, LCR, Polymorphism, Phylogeny
Highlights
-
•
Lineage A is the predominant HPV16 variant in Chinese women.
-
•
Based on the HPV16 prototype, 25 nucleotide mutations in E6 and E7 were found and 41 nucleotide mutations in LCR were found.
-
•
The mutations of C843T, A7287C and A7872G were significantly correlated with the cervical lesions.
1. Introduction
Cervical cancer is the most common malignant tumor in the female genital tract system. Approximately 569,847 new cases were diagnosed and 311,365 deaths occurred worldwide in 2018, mainly in developing countries [1]. Persistent infection with high-risk human papillomavirus (HR-HPV) is a major risk factor for cervical cancer [2]. HPV type 16 (HPV16) is the most prevalent HR-HPV which has been found in the majority of cervical cancer cases [3].
HPV16 genome is ca. 7,906 bp in length and is divided into three functional regions: an early region encoding core viral proteins (E1, E2, E4, E5, E6 and E7), long control region (LCR) and a late region. E6 and E7, which are involved in tumorigenesis of cervical cancer, are highly expressed in tumors. The LCR, adjacent to the upstream of E6 gene, contains regulatory elements for viral DNA replication and transcription. The late region encodes the L1 and L2 capsid proteins responsible for the viral DNA packaging and assembling in an icosahedral structure [4].
HPV variants can be classified into lineages and sublineages based on the complete genome differences of 1.0–10.0% and 0.5–1.0%, respectively [5,6]. HPV16 has been grouped into four phylogenetic lineages: A, B, C and D, and each lineage was further divided into four sublineages (A1-4, B1-4, C1-4, and D1-4) respectively [7,8]. The previously named Asian (As) and North American 1 (NA1) variants are designated sublineages A4 and D1, respectively [9,10]. Sequencing and Epidemiological evidence suggested that nucleotide polymorphism of HPV genome could contribute to viral pathogenicity and cervical carcinogenicity [6,[11], [12], [13], [14], [15]]. In addition, a few reports suggest that co-evolution of HPV16 and humans is strongly associated with risk of cervical cancer risk [12,16,17].
In this study, samples from cervical cancer patients were obtained from Han women in Beijing, China, and polymorphic sites within the genes of E6, E7 and LCR were analyzed. The aims of this study were to investigate the genetic diversity of HPV16 E6, E7 and LCR in isolates from Chinese Han women and correlation of the E6, E7 and LCR polymorphisms with disease status of infected patients. Finally, functional prediction of HPV16 LCR variants in cervical specimens was performed to determine the polymorphisms of transcription factor binding sites of the HPV16 LCR variants in Beijing, China.
2. Materials and methods
2.1. Collection of clinical specimens
This study collected cervical exfoliated cells samples from 392 HPV-positive patients who visited the gynecological outpatient department of Chinese PLA general hospital in 2018. According to the cytological and histological evaluations of fresh specimens, the cytological results were categorized as normal, cervicitis, atypical squamous cells of undetermined significance (ASCUS), cervical intraepithelial neoplasia grade 1, 2 and 3 (CIN1, CIN2 and CIN3) and cervical cancer (CC). Histopathological diagnosis was completed by a pathologist who was unaware of the HPV detection results. Informed consent was obtained from participation in the study and this study was approved by the ethics committee of National Institute for Viral Disease Control and Prevention, China.
2.2. PCR amplification and sequencing of HPV16
PCR was performed as that previously described [18]. According to the HPV16 reference sequence (Accession Number: K02718), the primer pairs were used for amplifications of both fragments of E6-E7 and LCR as well. The primers were synthesized by Sangon Biotech (Shanghai, China). The amplification of the LCR fragments was performed in 50 μL reaction volumes containing one unit of ExTaq® DNA Polymerase (TaKaRa), 4 mM of MgCl2, 0.25 mM of dNTPs, 0.25 μM of each primer and 1 μL of extracted DNA. The thermal profiling was as: 95 °C for 5min, followed by 5 cycles of 95 °C for 1min, 55 °C for 1min, 72 °C for 5min, and then 35 cycles of 95 °C for 20s, 55 °C for 30s, 72 °C for 5min, and final elongation at 72 °C for 5min. Purification and sequencing of PCR products were completed by Sangon Biotech (Shanghai, China). The following primers were used:
HPV16 E6E7-F: GTAACCGAAATCGGTTGAACCG;
HPV16 E6E7-R: CATAAAACCATCCATTACATCCCG;
HPV16 LCR-F: CTACAACTGCTAAACGCAAAAAACG;
HPV16 LCR-R: AACATTGCAGTTCTCTTTTGGTGC.
2.3. Phylogenetic analysis of HPV16 E6-E7 and LCR
The phylogenetic diagrams of either HPV16 E6-E7 or LCR sequences were constructed using the Neighbor-Joining method (NJ) with MEGA Suite version 7.0. The reference sequences of each variant used in the analysis were NC_001526.4, K02718 (A1), AF536179 (A2), HQ644236 (A3), AF534061 (A4), AF536180 (B1), AF472509 (C1) and AF402678 (D3). The mutation points were manually interpreted according to the actual phenogram. After the sequence alignment, integrated analysis of alignment results and the clinical data was performed to find the relationship between mutation and clinicopathology.
2.4. Prediction of transcription factor binding sites
The online database JASPAR (http://jaspar.genereg.net/) was used to investigate the transcription factor binding sites (TFBS) within HPV-16 LCR [19,20]. The transcription factors include CEBPB, ETS1, FOS, FOXA1, HSF1, HOXC11, JUN, IRF7, IRF1, IRF2, MAFK, NFE2L2, NFKB1, NFIL3, PHOX2A, RUNX1, RAX, SP1, SRY, SOX9, SPIB, STAT1, STAT3, SRF, VAX1, and YY1. The relative profile score threshold was set at 85%.
2.5. Statistical analysis
Contingency table (R × C) data analysis, Chi-square test (CMH-χ2) were used for the difference of HPV16 mutations and tumorigenicity of cervical cancer, a P-value <0.05 was considered statistically significant. Furthermore, the magnitude of the associations of HPV16 mutations with ≥CIN2,3 was assessed by calculating odds ratios (OR) and respective 95% confidence intervals (CI). Statistical analyses were performed with SPSS 26.0.
3. Results
3.1. Characteristics of the study populations and distribution of HPV16
Information about the studied population is shown in Table 1. Among the 392 HPV-positive samples, 249 were identified HPV16 positive, accounting for 63.5%. The average age of the subjects was 36.32 ± 8.12 years old. Among the populations, 99 women were Normal (39.7%), 44 were CIN1 (17.7%), 44 were CIN2 (17.7%), 41 were CIN3 (16.5%) and 21 were cervical cancer (8.4%).
Table 1.
Information about HPV16 infected women.
| HPV16 positive women (%) n = 249 | |
|---|---|
| Mean ± SD | 36.32 ± 8.12 |
| Range | 20–60 |
| Cervical disease (no. of women) | |
| Normal | 99(39.76) |
| CIN1 | 44(17.67) |
| CIN2 | 44(17.67) |
| CIN3 | 41(16.47) |
| CC | 21(8.43) |
3.2. Phylogenetic analysis of nucleotide sequence HPV16
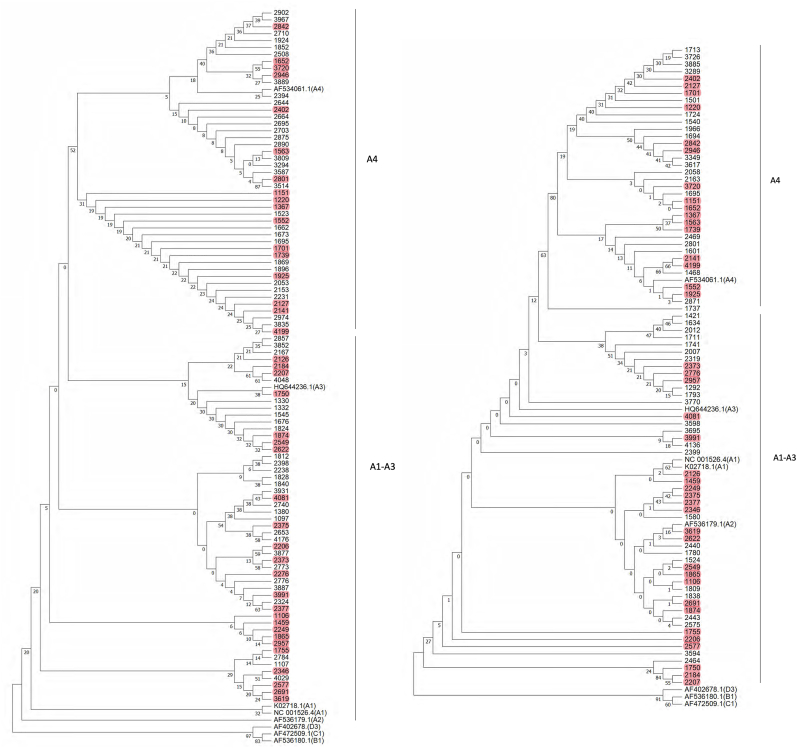
Among the 249 HPV16 isolates, the E6E7 or LCR sequences from 141 isolates were analyzed. The 98 E6-E7 sequences and 84 LCR sequences were obtained, respectively. Both E6-E7 and LCR sequences were obtained from 41 samples. The phylogenetic trees of HPV16 E6-E7 and LCR were constructed, respectively (Fig. 1). Lineage A alone was observed in the subjects. 98 E6-E7 sequences were subjected to phylogenetic tree analysis (Figs. 1A), 52 samples were A1-3 sublineages, and 46 samples were A4 sublineages. According to the phylogenetic analysis based on 84 LCR sequences, 49 were A1-3 sublineages and 35 were A4 sublineages (Fig. 1B).
Fig. 1.
Phylogenetic tree of the HPV16 variants based on E6E7 and LCR sequences. A. Phylogenetic tree based on 98 E6E7 sequences. B. Phylogenetic tree based on 84 LCR sequences. The Neighbor-Joining method and the Tamura 3-Parameter model were used to construct the phylogenetic tree by MEGA version 7.0. The standard sequences included NC_001526.4, K02718 (A1), AF536179 (A2), HQ644236 (A3), AF534061 (A4), AF536180 (B1), AF472509 (C1) and AF402678 (D3). Numbers closest to the branch points are bootstrap values (1,000 replicates). Both E6E7 and LCR sequences of 41 samples were marked in red. . (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.3. Single nucleotide polymorphisms of HPV16 isolates
22 nucleotide mutations were found in E6 and E7 sequences of 98 HPV16 isolates, including 10 missense mutations in E6 (A95G, G176A, T178A, T185G, G267A, A276G, C335T, T350G, A442C and C516G) and 2 missense mutations in E7 (A646C and C790T). T350G mutation accounted for 8.2% (8/98), followed by T178A mutation, accounting for 4.1% (4/98), and A442C mutation accounted for 3.1% (3/98). Both T350G and T178A mutations occurred in A1-3 variants, and the mutation rates were 15.4% and 7.7% respectively. The sporadic mutation sites of A1-3 variants and A4 variants are shown in Table 2, Table 3.
Table 2.
Sporadic mutations in E6 and E7 of A1-3 variants.
| ORF |
E6 |
Number of variation samples | E7 |
Number of variation samples | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 |
0 |
1 |
1 |
1 |
2 |
2 |
2 |
3 |
3 |
4 |
6 |
6 |
7 |
7 |
7 |
|||
| 9 |
9 |
6 |
7 |
7 |
4 |
5 |
7 |
3 |
5 |
4 |
4 |
6 |
2 |
6 |
9 |
|||
| Variants | 4 | 5 | 9 | 6 | 8 | 1 | 6 | 6 | 5 | 0 | 2 | 6 | 6 | 3 | 0 | 0 | ||
| K02178.1 (A1) | G | A | T | G | T | T | C | A | C | T | A | A | G | T | T | C | ||
| #01 | . | . | . | . | G | . | . | . | . | . | 3 | . | A | . | . | . | 20 | |
| #02 | A | . | . | . | . | . | . | . | . | . | . | 6 | . | . | . | . | T | 1 |
| #03 | . | . | . | . | A | . | . | . | . | . | . | 1 | . | . | . | C | . | 6 |
| #04 | . | . | . | . | . | . | . | . | . | G | . | 6 | C | . | . | . | . | 2 |
| #05 | . | . | . | A | . | . | . | . | . | . | . | 1 | . | . | C | . | . | 1 |
| #06 | . | . | . | . | . | . | . | G | . | . | . | 1 | ||||||
| #07 | . | . | . | . | . | . | T | . | . | G | . | 1 | ||||||
| #08 | A | . | . | . | . | . | . | . | . | . | C | 1 | ||||||
| #09 | A | . | . | . | A | . | . | . | . | . | . | 3 | ||||||
| #10 | A | . | . | . | . | . | T | . | . | . | . | 1 | ||||||
| #11 | . | . | C | . | . | . | . | . | . | . | C | 1 | ||||||
| #12 | A | . | . | . | . | . | . | . | T | . | . | 1 | ||||||
| #13 | A | . | . | . | . | . | . | . | . | G | . | 1 | ||||||
| #14 | A | G | . | . | . | . | . | . | . | . | . | 1 | ||||||
| Mutation | A | G | C | A | A | G | T | G | T | G | C | C | A | C | C | T | ||
| Amino acid change | – | – | – | D25 N | D25E | – | – | N58S | H78Y | L83V | E113D | N29H | – | – | – | R77C | ||
| Number of variation samples (n = 52) | 14 | 1 | 1 | 1 | 4 | 3 | 2 | 1 | 1 | 8 | 2 | 2 | 20 | 1 | 6 | 1 | ||
| Variation frequency (%) | 26.9 | 1.9 | 1.9 | 1.9 | 7.7 | 5.8 | 3.8 | 1.9 | 1.9 | 15.4 | 3.8 | 3.8 | 38.5 | 1.9 | 11.5 | 1.9 |
Table 3.
Sporadic mutations in E6 and E7 of A4 variants.
| ORF |
E6 |
Number of variation samples | E7 |
Number of variation samples | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
1 |
3 |
5 |
6 |
6 |
8 |
8 |
|||
| 0 |
8 |
6 |
1 |
0 |
6 |
4 |
4 |
|||
| Variants | 3 | 5 | 0 | 6 | 6 | 3 | 0 | 3 | ||
| AF534061 (A4) | T | G | A | C | G | G | C | T | ||
| #01 | G | . | . | . | 2 | . | . | . | C | 10 |
| #02 | . | . | . | G | 1 | A | . | . | . | 1 |
| #03 | . | A | . | . | 1 | . | A | . | . | 1 |
| #04 | . | . | C | . | 1 | . | . | T | C | 1 |
| Mutation | G | A | C | G | A | A | T | C | ||
| Amino acid change | L38V | R55K | E113D | S138C | – | – | – | – | ||
| Number of variation samples(n = 46) | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 11 | ||
| Variation frequency (%) | 4.3 | 2.2 | 2.2 | 2.2 | 2.2 | 2.2 | 2.2 | 23.9 |
In addition, compared with the reference sequences of sublineages A1 and A4 in LCR region, 41 different point mutations were detected in within the entire LCR fragment (Table 4, Table 5). Sequence analysis of 98 samples showed that the mutations T843C, G666A, A7287C, A7418G, C7780T and A7872G in the A4 isolates and T7711G in the A1 isolates were clustered and the relationship between them and cervical lesions was shown in Table 6. In the χ2 statistical test, we first normalized the grouping. As shown in Table 6, we combined the groups of normal and CIN1 into one category, meanwhile, combined CIN2 and above lesions into another category. The statistical results suggested that A4 sublineage T843C, A7287C and A7872G were significantly related to CIN2 and above grade lesions.
Table 4.
Sporadic mutations in LCR of A1-3 variants.
| ORF |
7 |
7 |
7 |
7 |
7 |
7 |
7 |
7 |
7 |
7 |
7 |
7 |
7 |
7 |
7 |
7 |
7 |
7 |
7 |
7 |
7 |
7 |
0 |
Number of variation samples |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
1 |
1 |
1 |
2 |
3 |
3 |
3 |
3 |
3 |
3 |
4 |
4 |
6 |
4 |
5 |
6 |
7 |
7 |
7 |
7 |
8 |
0 |
||
| 6 |
7 |
7 |
9 |
3 |
0 |
1 |
1 |
8 |
9 |
9 |
2 |
4 |
3 |
9 |
9 |
2 |
1 |
2 |
6 |
8 |
6 |
1 |
||
| Variants | 6 | 0 | 2 | 5 | 1 | 8 | 2 | 9 | 0 | 2 | 3 | 8 | 7 | 7 | 6 | 7 | 8 | 1 | 7 | 0 | 9 | 6 | 3 | |
| K02178.1 (A1) | A | T | A | A | A | C | C | A | T | C | C | C | T | C | A | C | A | T | A | T | C | G | C | |
| #01 | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | . | 1 |
| #02 | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | 1 |
| #03 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | 9 |
| #04 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | G | . | . | . | . | . | . | 1 |
| #05 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | . | 5 |
| #06 | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | 1 |
| #07 | . | . | . | . | . | . | . | . | C | . | . | . | . | . | C | . | . | . | . | . | . | . | . | 1 |
| #08 | . | . | . | . | T | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | 1 | |
| #09 | G | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 6 |
| #10 | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | . | 1 |
| #11 | . | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | . | 3 |
| #12 | G | . | . | C | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | |
| #13 | G | . | . | . | . | T | . | C | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 |
| #14 | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | T | . | . | 1 |
| #15 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | T | . | . | 1 |
| #16 | . | . | . | C | . | A | . | . | . | . | . | . | . | . | . | . | . | G | . | A | . | . | . | 1 |
| #17 | G | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 |
| #18 | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | 1 |
| #19 | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | 1 |
| #20 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | T | 1 |
| Mutation | G | G | C | C | C | T/A | T | C | C | T | T | T | C | T | C | T | G | G | C | A | T | A | T | |
| Number of variation samples (n = 49) | 11 | 3 | 6 | 1 | 1 | 4 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 12 | 1 | 1 | 2 | 10 | 1 | |
| Variation frequency (%) | 22 | 6 | 12 | 2 | 2 | 8 | 2 | 2 | 2 | 4 | 2 | 2 | 2 | 4 | 2 | 2 | 2 | 25 | 2 | 2 | 4 | 20 | 2 |
Table 5.
Sporadic mutations in LCR of A4 variants.
| ORF |
7 |
7 |
7 |
7 |
7 |
7 |
7 |
7 |
7 |
7 |
7 |
7 |
7 |
7 |
7 |
7 |
7 |
0 |
Number of variation samples |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 |
2 |
2 |
2 |
3 |
3 |
4 |
4 |
4 |
4 |
6 |
7 |
7 |
8 |
8 |
8 |
8 |
0 |
||
| 0 |
1 |
3 |
8 |
6 |
9 |
0 |
1 |
2 |
2 |
5 |
6 |
8 |
2 |
4 |
7 |
7 |
7 |
||
| Variants | 9 | 8 | 2 | 7 | 3 | 6 | 4 | 8 | 7 | 8 | 9 | 3 | 0 | 5 | 3 | 2 | 4 | 9 | |
| AF534061 (A4) | C | G | A | C | T | G | T | A | C | G | A | C | C | G | A | A | C | T | |
| #01 | . | . | . | . | . | . | . | . | . | . | . | . | T | A | . | . | . | . | 1 |
| #02 | . | . | . | . | . | . | . | G | . | . | . | . | T | . | . | . | . | . | 6 |
| #03 | . | . | . | . | . | . | . | . | . | A | . | . | . | . | . | . | . | . | 2 |
| #04 | A | . | G | . | G | . | . | . | . | . | . | . | T | . | . | . | . | . | 1 |
| #05 | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | 2 |
| #06 | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | 2 |
| #07 | . | . | . | A | . | . | . | . | . | . | G | . | . | . | . | . | . | . | 1 |
| #08 | . | . | . | A | . | . | . | . | . | . | . | . | T | . | G | . | . | 5 | |
| #09 | . | . | . | . | . | A | . | G | . | . | . | . | . | . | . | . | . | . | 1 |
| #10 | . | . | . | . | . | . | . | G | . | . | . | T | . | . | . | . | . | . | 1 |
| #11 | . | . | . | . | . | . | . | . | . | A | . | . | . | . | . | . | . | G | 1 |
| #12 | . | . | . | A | . | . | . | . | . | . | . | . | T | . | . | . | . | . | 3 |
| #13 | A | . | G | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | 1 |
| #14 | . | . | . | . | . | . | . | . | . | . | . | . | T | . | G | . | 1 | ||
| #15 | . | C | . | . | . | . | . | . | A | . | . | . | T | . | . | . | . | . | 1 |
| #16 | . | . | . | A | . | . | . | . | . | . | . | . | T | A | . | G | . | . | 1 |
| #17 | . | . | . | . | . | . | G | G | . | . | . | . | T | . | . | . | . | . | 1 |
| #18 | A | . | G | . | . | . | . | . | . | . | . | . | T | . | C | . | . | . | 1 |
| Mutation | A | C | G | A | G | A | G | G | A | A | G | T | T | A | C | G | G | G | |
| Number of variation samples (n = 35) | 3 | 1 | 3 | 10 | 1 | 1 | 1 | 11 | 1 | 3 | 1 | 1 | 24 | 2 | 1 | 6 | 1 | 1 | |
| Variation frequency (%) | 8.6 | 2.9 | 8.6 | 29 | 2.9 | 2.9 | 2.9 | 31 | 2.9 | 8.6 | 2.9 | 2.9 | 69 | 5.7 | 2.9 | 17 | 2.9 | 2.9 |
Table 6.
Nucleotide mutations of HPV16 and tumor progression.
| Mutations | <CIN2 | ≥CIN2 | χ2 | P | OR | 95% CI | P |
|---|---|---|---|---|---|---|---|
| A4 843T | 8 | 27 | 9.197 | 0.004* | 0.111 | 0.024–0.520 | 0.005* |
| A4 843C | 8 | 3 | |||||
| A1 666G | 15 | 17 | 1.456 | 0.180 | 2.059 | 0.631–6.712 | 0.722 |
| A1 666A | 6 | 14 | |||||
| A4 7287A | 7 | 18 | 7.887 | 0.07* | 0.097 | 0.016–0.576 | 0.010* |
| A4 7287C | 8 | 2 | |||||
| A4 7418A | 12 | 12 | 1.591 | 0.281 | 2.667 | 0.566–12.557 | 0.215 |
| A4 7418G | 3 | 8 | |||||
| A4 7780C | 3 | 8 | 1.591 | 0.281 | 0.375 | 0.08–1.766 | 0.215 |
| A4 7780T | 12 | 12 | |||||
| A4 7872A | 10 | 19 | 4.844 | 0.040* | 0.105 | 0.011–1.029 | 0.053 |
| A4 7872G | 5 | 1 | |||||
| A1 7711T | 17 | 20 | 3.272 | 0.095 | 4.25 | 0.816–22.132 | 0.086 |
| A1 7711G | 2 | 10 |
Note: *P < 0.05.
3.4. Prediction of transcription factor binding sites
We utilized JASPAR to predict the influences of SNP in LCR for binding to transcription factors and effects of the variants in HPV16 LCR on binding sites of cellular transcription factors. The results indicated that 14 (34.1%) of the 41 variants were located in the HPV16 LCR binding sites of transcription factors, which potentially influenced the binding of transcription factors and resultant gene expression. The potential transcription factors FOXA1, VAX1, STAT3, CEBPB, SOX9, FOS, RAX1, NFIL3, HOXC11 and PHOX2A were detected on the mutation sites of A1 and A4 variant strains, respectively (Table 7). The point mutation of A7166G, T7170G or A7172C resulted in loss of a putative binding site for oncogenic transcription factor FOXA1. Furthermore, co-variations of C7308T and C7312T resulted in a new binding site for the transcription factors CEBPB and NFIL3, while G7218C resulted in loss of two binding site for the transcription factors STAT3 and HOXC11. The mutations of C7312T, A7496C, A7727C, G7218C, T7363G, A7659G and G7825A resulted in loss of transcription factors, while the mutations of C7597T, C13T and G7825A resulted in addition of binding sites for transcription factors.
Table 7.
Nucleotide mutations of HPV-16 LCR and the predicted binding sites for transcription factors.
| Sublineage | Nucleotide mutation site | Transcription factors |
|---|---|---|
| A1 | A7166G | △FOXA1 |
| A1 | T7170G | △FOXA1 |
| A1 | A7172C | △FOXA1 |
| A1 | C7308T, C7312T | NFIL3 |
| A1 | C7312T | △ETS1 |
| A1 | A7496C | △VAX1 |
| A1 | C7597T | SOX9 |
| A1 | A7727C | △PHOX2A |
| A1 | C0013T | RAX |
| A4 | G7218C | STAT3, HOXC11 |
| A4 | T7363G | △VAX1 |
| A4 | A7659G | △ETS1 |
| A4 | G7825A | △SOX9, FOS |
Note: △ mutation site resulted in loss the transcription factor.
4. Discussion
Cervical cancer is the second most common cancer in women in the world, with high malignancy and high mortality. Persistent infection of HR-HPV is a high-risk factor for cervical cancer. With the development of molecular biology and epidemiology, the research on the relationship between HPV infection and lower genital tract diseases has been deepened, and it has been found that HPV16 infection is closely related to the occurrence of cervical cancer together with high-grade cervical lesions. In our study, HPV16 was still the most prevalent HR-HPV in China. Among the 392 HPV-positive samples, 249 samples (63.5%) were confirmed HPV16 positive.
In 1993, Ho et al. conducted an evolutionary analysis on the HPV LCR partial sequences (7478–7841 nt) of HPV16-infected persons from 25 different races and regions around the world, and the results divided the HPV16 variant strains into 5 main sublineages [11]. Occurrence groups and are distributed in different regions. D2 and D3 sublineages are mainly found in Central and South America and Spain, lineage B and C lineages are mainly found in Africa, A4 sublineage is mainly found in Southeast Asia, A1 and A2 sublineages are found all over the world, except for Africa. Among cervical cancer patients in China, HPV16 lineage A is the most common variant, and the distribution of HPV16 isolates in different regions [21,22]. In this study, the evolution analysis of HPV E6E7 and LCR sequence showed that HPV16 isolates in Beijing women was lineage A according to lineage distribution mentioned above.
Large of studies have conducted in-depth research on the relationship between HPV16 variation and cervical lesions. The nucleotide difference between HPV16 mutants was less than 1%, but there was significant difference in carcinogenicity. Among the point mutations, a few mutations in E6 gene can affect the diversity of mutant strains, and L83V mutation is the most common E6 mutation [23]. Previous studies proved that the L83V mutation of HPV16 lineage D (D2, D3) has stronger immortalization, transformation and tumorigenicity than lineage A variant through sequencing of HPV16 E6 [[24], [25], [26], [27], [28]]. In this study, the point mutation of E6 T350G (L83V) accounted for only 8.2% (8/98), L83V mutation accounted for 6.6% (4/61) of CIN2 and above lesions. There was no significant difference between the L83V mutation and the incidence of CIN2 and above lesions (the results were not shown). Therefore, we suggested that L83V mutation in E6 gene did not affect the high carcinogenicity.
Some studies believed that HPV16 D2 and D3 sublineages are much more carcinogenic than that of HPV16 A1-3 sublineages [29,30]. Wu et al. found that the HPV16 A4 variants may be related to the high incidence of cervical cancer in China [31]. However, Xi et al. showed that the two variants of A1 and A4 had similar risks of high-grade cervical lesions and cervical cancer [17]. This study found that the variants of HPV16 in Chinese women was mainly lineage A. There was no statistical correlation between A4 sublineage and cervical lesions. However, T843C, A7287C and A7872G on the E7 and LCR sequences of A4 variants were significantly correlated with the grade of cervical lesions. We hypothesized that the analysis of the E2, E4, E5, L1 and L2 sequences of A4 sublineage can further reveal the evolutionary information of the A4 variants, which may play an important role in the study of its pathogenic mechanism.
The LCR contains the early promoter and various transcriptional regulatory sites of involved in viral DNA replication and transcription [32,33], such as E2, YY1, NF1, FOS, NFKB1, and others. Several studies have suggested that certain LCR variants of HPV16 may play an important role in viral persistence and the development of cervical cancer [[34], [35], [36]]. For example, Dong et al. suggested that the mutation of the LCR sequence of HPV16-positive cervical cancer patients is mainly located in the YY1 binding region [37,38]. This study found that 14 mutation sites may located in the 11 binding regions, including FOXA2, CEBPB, STAT3, FOS, SOX9 and others. However, we did not find that the loss or addition of transcription factors caused by nucleotide mutations of HPV16 LCR were associated with disease progression.
In recent years, research data have shown that HPV16 variants are associated with different ethnicities, A1-3 variants are more likely to appear in Caucasian women, and D2 and D3 variants are more likely to appear in African Americans [39]. The relationship between race related HPV16 variants and cervical cancer has also been studied. Xi et al. confirmed that certain mutants have different risks of CIN3 in different races [17]. This study only focused on the distribution of HPV16 variants in the Han population. In contrast to previous studies, the D2 and D3 variants, which are more common in cervical cancer in Uyghur women, are not common in Chinese Han women. Therefore, there is sample selection bias in this study, it is still necessary to further expand the sample size. Han and minority women in China need to be also included in the study.
In conclusion, this study has shown that lineage A (including A1-3 and A4 sublineages) of HPV16 is the predominant variants in Beijing, China. 22 mutations in E6, E7 and 43 different point mutations in LCR were detected in HPV16 isolates. In addition, the mutations of T843C, A7287C and A7872G were significantly correlated with the cervical lesions, and the 11 potential transcription factor binding sites were in 14 point mutations. Although our study showed some limitations on sample capacity and source, it provided more data on the distribution of HPV variants in Chinese Han women, the complicated relation among HPV16 E6, E7 and LCR mutations, transcription factors and carcinogenesis.
Author contributions
Jiao WANG designed the experiments, analyzed the data, performed the experiments, and wrote the paper; Hongtu LIU designed the experiments, analyzed the data, and wrote the paper; Yifan GUO, Hui WANG, Ying LI, Li ZHANG, Zhan WANG and Lei SONG review and revise the paper.
Declaration of competing interest
The authors declare no conflicts of interest.
Data availability
Data will be made available on request.
References
- 1.Cohen P.A., Jhingran A., Oaknin A., Denny L. Cervical cancer. Lancet. 2019;393:169–182. doi: 10.1016/s0140-6736(18)32470-x. [DOI] [PubMed] [Google Scholar]
- 2.Ferenczy A., Franco E. Persistent human papillomavirus infection and cervical neoplasia. Lancet Oncol. 2002;3:11–16. doi: 10.1016/s1470-2045(01)00617-9. [DOI] [PubMed] [Google Scholar]
- 3.Li N., Franceschi S., Howell-Jones R., Snijders P.J., Clifford G.M. Human papillomavirus type distribution in 30,848 invasive cervical cancers worldwide: variation by geographical region, histological type and year of publication. Int. J. Cancer. 2011;128:927–935. doi: 10.1002/ijc.25396. [DOI] [PubMed] [Google Scholar]
- 4.Smith B., Chen Z., Reimers L., van Doorslaer K., Schiffman M., Desalle R., Herrero R., Yu K., Wacholder S., Wang T., Burk R.D. Sequence imputation of HPV16 genomes for genetic association studies. PLoS One. 2011;6 doi: 10.1371/journal.pone.0021375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cornet I., Gheit T., Franceschi S., Vignat J., Burk R.D., Sylla B.S., Tommasino M., Clifford G.M. Human papillomavirus type 16 genetic variants: phylogeny and classification based on E6 and LCR. J. Virol. 2012;86:6855–6861. doi: 10.1128/jvi.00483-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burk R.D., Harari A., Chen Z. Human papillomavirus genome variants. Virology. 2013;445:232–243. doi: 10.1016/j.virol.2013.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mandal P., Bhattacharjee B., Sen S., Bhattacharya A., Saha S.S., Chowdhury R.R., Mondal N.R., Chakrabarty B., Chatterjee T., Roy S., Sengupta S. Predominance of genomically defined A lineage of HPV16 over D lineage in Indian patients from eastern India with squamous cell carcinoma of the cervix in association with distinct oncogenic phenotypes. Transl. Oncol. 2022;15:101256. doi: 10.1016/j.tranon.2021.101256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Z., DeSalle R., Schiffman M., Herrero R., Wood C.E., Ruiz J.C., Clifford G.M., Chan P.K.S., Burk R.D. Niche adaptation and viral transmission of human papillomaviruses from archaic hominins to modern humans. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1007352. e1007352-e1007352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamada T., Wheeler C.M., Halpern A.L., Stewart A.C., Hildesheim A., Jenison S.A. Human papillomavirus type 16 variant lineages in United States populations characterized by nucleotide sequence analysis of the E6, L2, and L1 coding segments. J. Virol. 1995;69:7743–7753. doi: 10.1128/jvi.69.12.7743-7753.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kukimoto I., Muramatsu M. Genetic variations of human papillomavirus type 16: implications for cervical carcinogenesis. Jpn. J. Infect. Dis. 2015;68:169–175. doi: 10.7883/yoken.JJID.2014.584. [DOI] [PubMed] [Google Scholar]
- 11.Ho L., Chan S.Y., Burk R.D., Das B.C., Fujinaga K., Icenogle J.P., Kahn T., Kiviat N., Lancaster W., Mavromara-Nazos P., et al. The genetic drift of human papillomavirus type 16 is a means of reconstructing prehistoric viral spread and the movement of ancient human populations. J. Virol. 1993;67:6413–6423. doi: 10.1128/jvi.67.11.6413-6423.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mirabello L., Yeager M., Cullen M., Boland J.F., Chen Z., Wentzensen N., Zhang X., Yu K., Yang Q., Mitchell J., Roberson D., Bass S., Xiao Y., Burdett L., Raine-Bennett T., Lorey T., Castle P.E., Burk R.D., Schiffman M. HPV16 sublineage associations with histology-specific cancer risk using HPV whole-genome sequences in 3200 women. J. Natl. Cancer Inst. 2016;108:djw100. doi: 10.1093/jnci/djw100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pientong C., Wongwarissara P., Ekalaksananan T., Swangphon P., Kleebkaow P., Kongyingyoes B., Siriaunkgul S., Tungsinmunkong K., Suthipintawong C. Association of human papillomavirus type 16 long control region mutation and cervical cancer. Virol. J. 2013;10 doi: 10.1186/1743-422X-10-30. 30-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cornet I., Gheit T., Iannacone M.R., Vignat J., Sylla B.S., Del Mistro A., Franceschi S., Tommasino M., Clifford G.M. HPV16 genetic variation and the development of cervical cancer worldwide. Br. J. Cancer. 2013;108:240–244. doi: 10.1038/bjc.2012.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tan G., Duan M., Li Y., Zhang N., Zhang W., Li B., Qu P. Distribution of HPV 16 E6 gene variants in screening women and its associations with cervical lesions progression. Virus Res. 2019;273 doi: 10.1016/j.virusres.2019.197740. [DOI] [PubMed] [Google Scholar]
- 16.Clifford G.M., Tenet V., Georges D., Alemany L., Pavón M.A., Chen Z., Yeager M., Cullen M., Boland J.F., Bass S., Steinberg M., Raine-Bennett T., Lorey T., Wentzensen N., Walker J., Zuna R., Schiffman M., Mirabello L. Human papillomavirus 16 sub-lineage dispersal and cervical cancer risk worldwide: whole viral genome sequences from 7116 HPV16-positive women. Papillomavirus Res. 2019;7:67–74. doi: 10.1016/j.pvr.2019.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xi L.F., Koutsky L.A., Hildesheim A., Galloway D.A., Wheeler C.M., Winer R.L., Ho J., Kiviat N.B. Risk for high-grade cervical intraepithelial neoplasia associated with variants of human papillomavirus types 16 and 18. Cancer Epidemiol. Biomarkers Prev. 2007;16:4–10. doi: 10.1158/1055-9965.Epi-06-0670. [DOI] [PubMed] [Google Scholar]
- 18.Sun Z., Lu Z., Liu J., Wang G., Zhou W., Yang L., Liu C., Ruan Q. Genomic polymorphism of human papillomavirus type 52 in women from Northeast China. Int. J. Mol. Sci. 2012;13:14962–14972. doi: 10.3390/ijms131114962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sandelin A., Alkema W., Engström P., Wasserman W.W., Lenhard B. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004;32:D91–D94. doi: 10.1093/nar/gkh012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Castro-Mondragon J.A., Riudavets-Puig R., Rauluseviciute I., Berhanu Lemma R., Turchi L., Blanc-Mathieu R., Lucas J., Boddie P., Khan A., Manosalva Pérez N., Fornes O., Leung T.Y., Aguirre A., Hammal F., Schmelter D., Baranasic D., Ballester B., Sandelin A., Lenhard B., Vandepoele K., Wasserman W.W., Parcy F., Mathelier A. JASPAR 2022: the 9th release of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2022;50:D165–d173. doi: 10.1093/nar/gkab1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hang D., Yin Y., Han J., Jiang J., Ma H., Xie S., Feng X., Zhang K., Hu Z., Shen H., Clifford G.M., Dai M., Li N. Analysis of human papillomavirus 16 variants and risk for cervical cancer in Chinese population. Virology. 2016;488:156–161. doi: 10.1016/j.virol.2015.11.016. [DOI] [PubMed] [Google Scholar]
- 22.Zhe X., Xin H., Pan Z., Jin F., Zheng W., Li H., Li D., Cao D., Li Y., Zhang C., Fu S., Shao R., Pan Z. Genetic variations in E6, E7 and the long control region of human papillomavirus type 16 among patients with cervical lesions in Xinjiang, China. Cancer Cell Int. 2019;19:65. doi: 10.1186/s12935-019-0774-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gokhale P.S., Sonawani A., Idicula-Thomas S., Kerkar S., Tongaonkar H., Mania-Pramanik J. HPV16 E6 variants: frequency, association with HPV types and in silico analysis of the identified novel variants. J. Med. Virol. 2014;86:968–974. doi: 10.1002/jmv.23924. [DOI] [PubMed] [Google Scholar]
- 24.Niccoli S., Abraham S., Richard C., Zehbe I. The Asian-American E6 variant protein of human papillomavirus 16 alone is sufficient to promote immortalization, transformation, and migration of primary human foreskin keratinocytes. J. Virol. 2012;86:12384–12396. doi: 10.1128/jvi.01512-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richard C., Lanner C., Naryzhny S.N., Sherman L., Lee H., Lambert P.F., Zehbe I. The immortalizing and transforming ability of two common human papillomavirus 16 E6 variants with different prevalences in cervical cancer. Oncogene. 2010;29:3435–3445. doi: 10.1038/onc.2010.93. [DOI] [PubMed] [Google Scholar]
- 26.Hudelist G., Manavi M., Pischinger K.I., Watkins-Riedel T., Singer C.F., Kubista E., Czerwenka K.F. Physical state and expression of HPV DNA in benign and dysplastic cervical tissue: different levels of viral integration are correlated with lesion grade. Gynecol. Oncol. 2004;92:873–880. doi: 10.1016/j.ygyno.2003.11.035. [DOI] [PubMed] [Google Scholar]
- 27.Lu X., Lin Q., Lin M., Duan P., Ye L., Chen J., Chen X., Zhang L., Xue X. Multiple-integrations of HPV16 genome and altered transcription of viral oncogenes and cellular genes are associated with the development of cervical cancer. PLoS One. 2014;9 doi: 10.1371/journal.pone.0097588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andersson S., Alemi M., Rylander E., Strand A., Larsson B., Sallstrom J., Wilander E. Uneven distribution of HPV 16 E6 prototype and variant (L83V) oncoprotein in cervical neoplastic lesions. Br. J. Cancer. 2000;83:307–310. doi: 10.1054/bjoc.2000.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berumen J., Ordoñez R.M., Lazcano E., Salmeron J., Galvan S.C., Estrada R.A., Yunes E., Garcia-Carranca A., Gonzalez-Lira G., Madrigal-de la Campa A. Asian-American variants of human papillomavirus 16 and risk for cervical cancer: a case-control study. J. Natl. Cancer Inst. 2001;93:1325–1330. doi: 10.1093/jnci/93.17.1325. [DOI] [PubMed] [Google Scholar]
- 30.Kang S., Jeon Y.T., Kim J.W., Park N.H., Song Y.S., Kang S.B., Lee H.P. Polymorphism in the E6 gene of human papillomavirus type 16 in the cervical tissues of Korean women. Int. J. Gynecol. Cancer. 2005;15:107–112. doi: 10.1111/j.1048-891x.2005.15010.x. [DOI] [PubMed] [Google Scholar]
- 31.Picconi M.A., Alonio L.V., Sichero L., Mbayed V., Villa L.L., Gronda J., Campos R., Teyssié A. Human papillomavirus type-16 variants in Quechua aboriginals from Argentina. J. Med. Virol. 2003;69:546–552. doi: 10.1002/jmv.10343. [DOI] [PubMed] [Google Scholar]
- 32.Mosmann J.P., Monetti M.S., Frutos M.C., Kiguen A.X., Venezuela R.F., Cuffini C.G. Mutation detection of E6 and LCR genes from HPV 16 associated with carcinogenesis. Asian Pac. J. Cancer Prev. APJCP. 2015;16:1151–1157. doi: 10.7314/apjcp.2015.16.3.1151. [DOI] [PubMed] [Google Scholar]
- 33.Xi J., Chen J., Xu M., Yang H., Luo J., Pan Y., Wang X., Qiu L., Yang J., Sun Q. Genetic variability and functional implication of the long control region in HPV-16 variants in Southwest China. PLoS One. 2017;12 doi: 10.1371/journal.pone.0182388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soeda E., Ferran M.C., Baker C.C., McBride A.A. Repression of HPV16 early region transcription by the E2 protein. Virology. 2006;351:29–41. doi: 10.1016/j.virol.2006.03.016. [DOI] [PubMed] [Google Scholar]
- 35.Gheit T., Cornet I., Clifford G.M., Iftner T., Munk C., Tommasino M., Kjaer S.K. Risks for persistence and progression by human papillomavirus type 16 variant lineages among a population-based sample of Danish women. Cancer Epidemiol. Biomarkers Prev. 2011;20:1315–1321. doi: 10.1158/1055-9965.Epi-10-1187. [DOI] [PubMed] [Google Scholar]
- 36.Bletsa G., Zagouri F., Amoutzias G.D., Nikolaidis M., Zografos E., Markoulatos P., Tsakogiannis D. Genetic variability of the HPV16 early genes and LCR. Present and future perspectives. Expet Rev. Mol. Med. 2021;23:e19. doi: 10.1017/erm.2021.18. [DOI] [PubMed] [Google Scholar]
- 37.Dong X.P., Stubenrauch F., Beyer-Finkler E., Pfister H. Prevalence of deletions of YY1-binding sites in episomal HPV 16 DNA from cervical cancers. Int. J. Cancer. 1994;58:803–808. doi: 10.1002/ijc.2910580609. [DOI] [PubMed] [Google Scholar]
- 38.Bhattacharjee B., Sengupta S. HPV16 E2 gene disruption and polymorphisms of E2 and LCR: some significant associations with cervical cancer in Indian women. Gynecol. Oncol. 2006;100:372–378. doi: 10.1016/j.ygyno.2005.09.016. [DOI] [PubMed] [Google Scholar]
- 39.Xi L.F., Kiviat N.B., Hildesheim A., Galloway D.A., Wheeler C.M., Ho J., Koutsky L.A. Human papillomavirus type 16 and 18 variants: race-related distribution and persistence. J. Natl. Cancer Inst. 2006;98:1045–1052. doi: 10.1093/jnci/djj297. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.