Abstract
Human metapneumovirus (hMPV) has emerged as a frequent cause of acute respiratory infections (ARI) among young children. The prevalence and genetic diversity of hMPV circulating in Chennai, Southern India, has not been studied yet. Hence, this study was aimed to investigate the prevalence, co-infection with other respiratory viruses like HRSV A and B, influenza A and B, hRV and HPIV 1–4 viruses, socio-demographic associations, and genotypes of hMPV among children in Chennai. A total of 350 nasal swab specimens were collected from children with ARI during April 2016 to August 2018 and tested for hMPV by real time PCR method. In this study, hMPV was detected in 4% (14/350) of the samples. One hMPV positive sample was found to be co-infected with influenza B virus. The mean and median ages of the children with hMPV infection were 61.5 months (5.1 years) and 83 months (6.9 years), respectively. Phylogenetic analysis of the partial F gene revealed the presence of A2c subcluster among the study strains as well as with B1 and B2 lineages. The prevalence data obtained in this study is important in evaluating the role of hMPV in childhood ARI and emphasizes the importance of routine viral diagnosis in hospitals. To the best of our knowledge, this is the first study to report the prevalence, seasonality, and genetic diversity of hMPV in Chennai as well as the first study to report A2c subcluster of hMPV among children in India.
Supplementary Information
The online version contains supplementary material available at 10.1007/s42770-022-00689-2.
Keywords: hMPV, ARI, Real time PCR, Fusion gene, Phylogeny, A2c subcluster
Introduction
Human metapneumovirus (hMPV) is an enveloped, single-stranded RNA virus belonging to the family Pneumoviridae and Metapneumovirus genus [1]. It was first isolated in 2001 in Netherlands [2]. Since then, it has been documented as a frequent cause of both upper respiratory tract infection and lower respiratory tract infection among people of all age groups, predominantly affecting the children, elderly, and immunocompromised individuals [3–5].The epidemiological scenario of hMPV is understudied in India. The first case of hMPV in India was reported in 2004 from Pune [6]. Other studies have been conducted in India at Lucknow, New Delhi, and Vellore, where children were the study population [7–9]. Premature birth, pre-existing nosocomial infection, young age, and underlying chronic heart, pulmonary, or neural disorders are some of the risk factors associated with severe hMPV infection [10]. hMPV circulates as two distinct genotypes — hMPV A and B [3, 11]. Based on the phylogenetic analysis of the nucleocapsid (N) and F gene sequences, Genotype A is further classified into A1 and A2 and Genotype B is divided into B1 and B2 lineages [12]. Lineage A2 is divided into A2a and A2b [12, 13], A2b1 and A2b2 subclusters [14], while lineage B2 consists of B2a and B2b [15]. Recently, an additional A2c subcluster was revealed in Japan, Malaysia, and Croatia [16–18]. It is important to determine the lineages of circulating hMPV strains which could aid in hMPV preventive measures. However, there is limited data on the prevalence and genetic diversity of hMPV in Chennai, South India. This study describes the prevalence of hMPV and its association with demographic factors like age, gender, and seasonality during April 2016–August 2018 in Chennai, South India. The circulating lineages of hMPV strains were determined, and mutational analysis was performed.
Materials and methods
Study population and sample collection
Children below 18 years of age and visiting the pediatric outpatient department of three tertiary care hospitals (ESI hospital, K.K. Nagar, Chennai, Institute of Child Health, Egmore, Chennai and Rabindran’s Health Care Centre, Ambattur, Chennai) and one pediatric clinic (Triplicane, Chennai) with symptoms of acute respiratory infection such as fever, cough, nasal congestion, rhinorrhea, headache, sore throat, myalgia, and dyspnea were enrolled in the study during April 2016–August 2018. Children with co-morbidities including cystic fibrosis, asthma, bronchiectasis, pulmonary hypertension, congenital heart disease, congestive heart failure, neutropenia, and babies born prematurely were excluded from the study. The samples were collected from children experiencing symptoms for ≤ 21 days.
The children were grouped [19] as follows: infants (1 month–2 years), 121/350, 34.6%; young children (2–6 years), 148/350, 42.3%; older children (6–12 years), 68/350, 19.4%; and adolescents (12–18 years) 13/350, 3.7%.
Seven children had repeated infection and hence, three hundred and fifty nasal swabs were collected from three hundred and forty-three children. The nasal swabs were immediately transported to laboratory in HiViral Transport medium (HiMedia, Mumbai). Informed consent was obtained from their parents/guardian.
Detection of human metapneumovirus
RNA was extracted from 150 μl of the sample using NucleoSpin RNA virus kit (Macherey Nagel, Germany) according to the manufacturer’s instructions. The extracted RNA was quantified using NanoDrop Spectrophotometer ND-1000 (Thermo Fisher Scientific, USA) and immediately subjected to reverse transcription. Reverse transcription was carried out using RevertAid first strand cDNA synthesis kit (Thermo Fisher Scientific, USA) according to manufacturer’s protocol and stored at − 20 °C until use.
Real time PCR was performed using previously published primers targeting the highly conserved nucleoprotein (N) gene and capable of identifying all the four lineages [20]. The 10-µL real time PCR master mix consisted of 5 µL of 5 × FastStart Universal Probe Master (ROX) (Roche Diagnostic, Germany), 0.2 µL each of forward and reverse primers (10 µM), 0.2 µL of probe (25 µM), 2 µL of cDNA, and 2.4 µL of nuclease free water. The cycling conditions were standardized at 95 °C for 10 min followed by 45 cycles of at 95 °C for 15 s and 60 °C for 1 min. Extension was carried out for 10 s at 72 °C.
Real time PCR was carried out in StepOnePlus Real Time instrument (Thermo Fisher Scientific, USA).
Analysis of co-infection and seasonality
The samples were further screened for the presence of other respiratory viruses namely, influenza viruses A and B [21], human respiratory syncytial virus [22], human parainfluenza viruses types 1 to 4, and human rhinovirus. The details of primers, PCR reaction mixture, and cycling conditions for hPIV and hRV are given in Supplement Table 1. Temporal distribution of hMPV was determined by plotting number of hMPV detected against total number of samples collected.
Sequencing and phylogeny
For phylogenetic analysis, the partial F gene of hMPV strains was amplified by heminested RT-PCR using primers previously described [23]. For the first round PCR, 2 μL of cDNA was added to 18 μL master mix containing 2 μL of 10 × PCR buffer, 0.5 μL each of forward and reverse primer (HRSV common primers) (10 μM), 0.3 μL of 10 mM dNTPs, and 0.025 U of Taq polymerase (5U/μL). Thermal cycling conditions for the first round were as follows: initial denaturation of 94 °C for 3 min, followed by 40 cycles of denaturation at 94 °C for 30 s, annealing at 55 °C for 30 s, extension at 72 °C for 30 s, and a final extension at 72 °C for 10 min.
The second round was performed with 1 μL of the first-round product as template with same reaction mixture and thermal cycling conditions except that the annealing temperature was set at 58 °C, and the PCR was carried out for 20 cycles.
PCR products were purified using the QIAquick Gel Extraction Kit (QIAGEN, Hilden, Germany) and sequenced using ABI 3731 × L DNA sequencer (Applied Biosystems, USA) at AgriGenome Labs Pvt Ltd, Kerala, India. Nucleotide sequences of partial F gene of hMPV strains representative of A1 (isolate 00–1) [2], A2a (CAN97-83) [13], A2b (JPS03-240) [24], A2c (hMPV/OkinawaJPN/137/10) [16], B1 (NL/1/99 and CAN97-82) [25, 26], and B2 (NL/94/01) [27] lineages were retrieved from the GenBank for reference. All the sequences were contig aligned using ClustalW and phylogenetic analysis was done using the MEGA 6 software (Pennsylvania, USA) [28]. Approximately 291 bp of the partial F gene was used in the phylogenetic analysis. The best model for constructing phylogenetic tree was estimated using the Find Best DNA/Protein Models (ML). Using this method, it was found that the K2 + I (Kimura-2-parameter + evolutionarily invariable sites) contained the lowest BIC (Bayesian Information Criterion) score and hence, was the appropriate model (Supplement Fig. 1). The statistical robustness and reliability of the tree topology were analyzed using 1000 bootstrapping replicates. The evolutionary distances were computed using the p-distance method, which was given in the units of the number of base differences per site.
Mutational analysis of the hMPV strains
RNA viruses can accrue genetic differences when outbreaks occur, which leads to modulations in virulence and evolvability [29]. Hence, the deduced amino acid sequences from partial F gene of hMPV strains were compared with reference strains CLZ0741 for A2c lineage, NL/94/01 for B2 lineage and NL/1/99 and CAN97-82 for B1 lineage for mutational analysis. Synonymous and nonsynonymous mutations were analyzed by the method of Nei and Gojobori using SNAP (Synonymous/Nonsynonymous Analysis Program) provided by the HIV database website. The O-glycosylated residues and potential N-glycosylation sites (Asn–X–Ser/Thr, where X is any amino acid except Proline) were predicted using the NetOGlyc version 4.0 server (http://www.cbs.dtu.dk/services/NetOGlyc/) and NetNGlyc version 1.0 server (http://www.cbs.dtu.dk/services/NetNGlyc/) software, respectively.
Sequence data were registered under accession numbers MK080266, MK080267, MK080268, MK080269, MK080270, MK080271, MK080272, MK080273, MK080274, MK080275, MK080276, MK080277, and MK080278 at the NCBI GenBank.
Results and discussion
Among three hundred and fifty samples collected, fourteen samples (14/350, 4%) were positive for hMPV by real time PCR (Table 1). The 4% positivity rate was comparable with other studies from India; 5% in Pondicherry; 3% in Kolkata; and 3.6% in Lucknow [30–32]. A high prevalence rate of 12.7% was observed in a study from Vellore, South India [9]. Prevalence rates of as low as 1.7% and as high as 27% have been reported globally [33, 34]. This variation in the prevalence rates could be attributed to the different study population, ununified sampling methods, different techniques employed for viral detection, and the study period. hMPV is reported to be responsible for 4–16% of ARI-hospitalizations in children [35]; hence, the prevalence data obtained in this study is valuable in evaluating the role of hMPV in childhood ARI and emphasizes the importance of routine viral diagnosis in hospitals.
Table 1.
Socio-demographic and molecular data of hMPV positive patients
| S. no | Age/sex | Location | Year of detection | Lineage | Accession number |
|---|---|---|---|---|---|
| 1 | 3.5Y/F | Ammayathoppu | July 2016 | A2c | MK080266 |
| 2 | 3Y 8 m/F | Manali Mathur | July 2016 | A2c | MK080267 |
| 3 | 2Y 1 m/M | Anna Nagar | September 2016 | A2c | MK080268 |
| 4 | 9Y/F | Triplicane | March 2018 | A2c | MK080269 |
| 5 | 6Y/M | Triplicane | March 2018 | B1 | MK080270 |
| 6 | 2Y/M | Egmore | June 2018 | A2c | MK080271 |
| 7 | 7 m/F | Thirumullaivoyal | July 2018 | Not Applicable* | - |
| 8 | 13Y/F | Ambattur | July 2018 | A2c | MK080272 |
| 9 | 3Y/F | Annanur | July 2018 | B2 | MK080273 |
| 10 | 10Y/M | Menambedu | July 2018 | B2 | MK080274 |
| 11 | 11 m/F | Thirumullaivoyal | August 2018 | A2c | MK080275 |
| 12 | 6Y/M | Senthil Nagar | August 2018 | B1 | MK080276 |
| 13 | 6Y/M | Annanur | August 2018 | B2 | MK080277 |
| 14 | 6Y/F | Ambattur | August 2018 | B2 | MK080278 |
*Sequencing could not be performed
Among the fourteen samples positive for hMPV, only one sample (1/14, 7.1%) was found to be co-infected with influenza B virus. In a study from India, hRSV and human rhino virus were frequently detected with hMPV [36]. The association between such dual infections and disease severity is an ongoing debate. Nandhini et al. found that two hMPV patients co-infected with HRSV presented with severe infection requiring hospitalization [30]. While some studies have documented increased risk of severe infection in cases of co-infection [37, 38], other studies have shown no impact on severity of infection [39, 40].
A male:female ratio of 3:4 was observed among the patients with hMPV infection. The mean and median ages were 61.5 months (5.1 years) and 83 months (6.9 years), respectively. Similar studies from other countries have reported the median of children with hMPV infection as 17.2 months [41] and 24 months [42]. In these studies, a male preponderance was observed which was in contrast to the results of present study. Studies have documented higher prevalence of hMPV in children less than 5 years of age [31, 41]. In the current study, hMPV was more prevalent in young children (57.1%, 8/14) which could be attributed to a more immature immune system when compared to older children and exposure to infections in the school environment.
With respect to the annual distribution of hMPV in our study, three cases were detected in 2016 (21.4%) and eleven cases in 2018 (78.6%). No hMPV was detected among samples collected in 2017. This data is in accordance with earlier observations documenting high and low incidence of hMPV in alternating years [30, 43]. This biennial pattern has also been reported globally [18, 41, 44, 45].
Most of the hMPV strains were detected during monsoon season (July–August) in this study. In contrast to these results, the peak incidence of hMPV occurred in the month of February in North-Western India [46]. Narayanan et al. found that the hMPV-positive ARIs were more common during the cooler and wetter months of July to January [9]. Similarly, Agrawal et al. reported that majority of the hMPV positive samples were detected during July–November, although low frequency of hMPV was observed throughout the year [47].
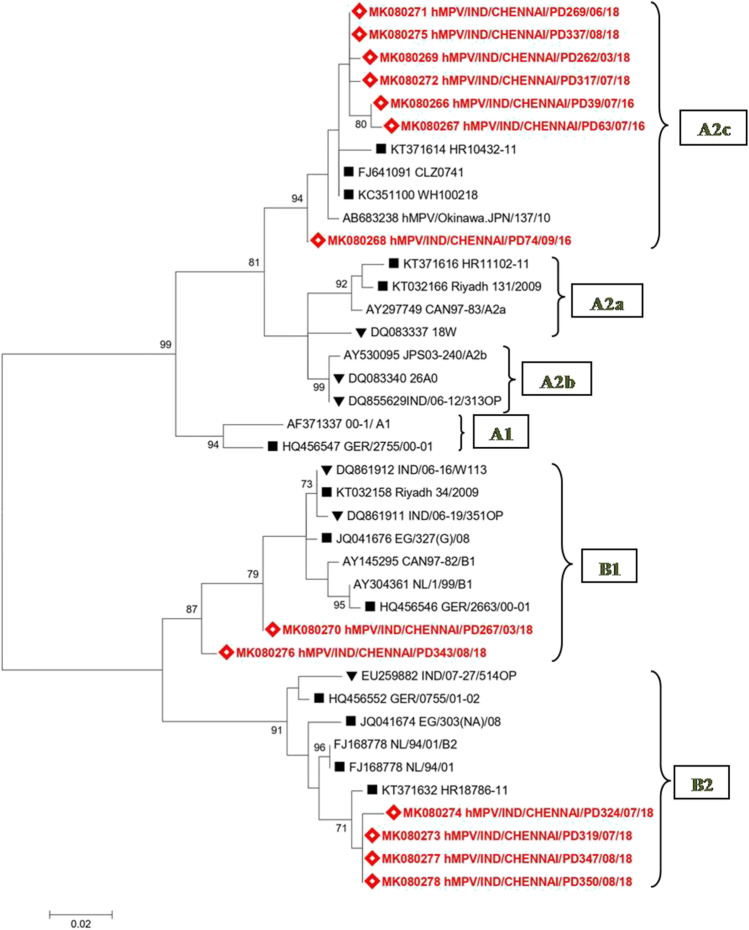
The partial F gene sequencing (219–342 aa) was carried out for 13/14 (92.9%) hMPV positive strains. Phylogenetic analysis of the thirteen strains revealed that three different lineages of hMPV circulated in our region during the study period (Fig. 1); B1 (2/13, 15.4%), B2 (4/13, 30.8%), and A2c (7/13, 53.8%). Three hMPV samples detected in 2016 were found to belong to the A2c subcluster. In 2018, hMPV strains belonging to all the three lineages (B1, B2, and A2c subcluster) were co-circulating. Two out of the ten samples (20%) belonged to the B1 lineage; four (40%) samples each were of B2 lineage and A2c subcluster. Co-circulation of hMPV lineages is a familiar trend which has been documented from countries all over the world [18]. None of our hMPV strains belonged to the A1 lineage, A2a, and A2b subclusters, though they have been previously reported from India [43, 48, 49]. However, this is the first report of the A2c subcluster in India. This subcluster was earlier reported from epidemiologic studies in Japan, Malaysia, Croatia, and recently in Bangladesh [16–18, 50] and our A2c strains closely matched with strains from these countries (Fig. 1).
Fig. 1.
Phylogenetic analysis of the human metapneumovirus strains obtained in this study. Phylogenetic tree of hMPV based on the partial F gene was constructed using maximum likelihood method using 1000 bootstrapping replicates. Values greater than 70% are shown. Sequences of reference strains and strains from other countries were obtained from NCBI GenBank. Sequences obtained in this study are indicated with a lozenge symbol. Other Indian strains are indicated with a black down-pointing triangle symbol. Strains from other countries are represented with a black square symbol
The strains belonging to the A2c subcluster in our study clustered with a significant bootstrap value of 94%. The intragenotypic p distance was found to be less than 0.07 between all the members of the same cluster (Supplement Table 2) suggesting very little divergence among the strains. This finding is similar to previous studies which suggest minimal progressive drift and considerable stability of genetic lineages over a period of time [51].
On comparing the A2c, B1, and B2 strains obtained from our study with their respective reference strains, it was found that the A2c strains in the present study had 97.1–97.9% and 100% identity at nucleotide and amino acid level, respectively. The two B1 strains in this study had 95.8% and 97.6% identity at nucleotide level and 100% identity at amino acid level. The identity of the B2 strains in this study ranged from 97.4 to 98.2% at nucleotide level and 99.2% at amino acid level due to the D280N mutation observed among all our B2 strains.
The F protein of all the analyzed strains had a length of 127 amino acids. The deduced amino acid sequence alignment of the groups A and B hMPV strains revealed that they were highly conserved with very few lineage specific mutations (Supplement Figs. 2, 3, and 4). Two mutations were observed among all the strains when compared with the hMPV reference isolate 00–1; F258I and G294E. The N233Y, V286I, and Q312K mutations were observed among all the B1 (Supplement Fig. 3) and B2 lineage strains (Supplement Fig. 4); they were however absent among the A2c strains (Supplement Fig. 2). The K296N and K296D point mutations were present only in B1 and B2 lineage strains, respectively. We also observed the D280N mutation in our B2 strains which was previously reported from other countries. Selection pressure analysis revealed that this region of F gene was under high negative selection with a dS/dN ratio of 49.13.
In agreement with previous studies, the partial F gene sequences from our study were highly conserved, especially at the amino acid level [18, 51]. This could be due to the involvement of this gene in host immunity and viral functions. These sequences also lacked potential O-linked and N-linked glycosylation sites, which have been hypothesized to be involved in escaping the host immune response in other viruses [52].
In India, in spite of the fact that a number of children die due to ARI every year, community-based studies documenting the contribution of circulating respiratory viruses are very sparse. Our study is the first epidemiologic study on hMPV from Chennai in South India. This study underlines the importance of hMPV as an etiology of ARI in children and emphasizes the need for routine laboratory diagnosis of this virus. This study also provides significant understanding of the circulating hMPV strains in Chennai. This is also the first report on the presence of the subcluster A2c in India.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary file3 (jpg 418 kb) Supplement Fig. 1 Maximum Likelihood fits of 24 different nucleotide substitution models
Supplementary file4 (jpeg 365 kb) Supplement Fig. 2 Deduced amino acid alignment of partial F gene of hMPV strains belonging to the A2c subcluster obtained in this study
Supplementary file5 (jpg 370 kb) Supplement Fig. 3 Deduced amino acid alignment of partial F gene of hMPV strains belonging to the B1 lineage obtained in this study
Supplementary file6 (jpg 393 kb) Supplement Fig. 4 Deduced amino acid alignment of partial F gene of hMPV strains belonging to the B2 lineage obtained in this study
Acknowledgements
We thank Dr. Elilarasi, Dr. Kalpana, and Dr. Sarathbalaji — Department of Pulmonology, Institute of Child Health, Egmore, Chennai; Dr. Indumathi — Pediatric Clinic, Triplicane, Chennai; and Dr. Rohit Bharadwaj — Rabindran’s Health Care Centre, Ambattur, Chennai for providing the clinical samples used in this study.
Funding
This work was supported by the Intramural Research funds, University of Madras, Chennai, India.
Declarations
Ethics approval
The study was approved by the Institutional Human Ethics Committee of Dr. ALM Post Graduate Institute of Basic Medical Sciences, University of Madras (Approval No: UM/IHEC/12–2014-II and UM/IHEC/07–2017-I).
Conflict of interest
The authors declare no competing interests.
Footnotes
Responsible Editor: Mauricio Nogueira
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Afonso CL, Amarasinghe GK, Bányai K, Bào Y, Basler CF, Bavari S, Bejerman N, Blasdell KR, Briand FX, Briese T, Bukreyev A. Taxonomy of the order Mononegavirales: update. Arch Virol. 2016;161:2351–2360. doi: 10.1007/s00705-016-2880-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van den Hoogen BG, de Jong JC, Groen J, Kuiken T, de Groot R, Fouchier RA, Osterhaus AD. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat Med. 2001;7:719–724. doi: 10.1038/89098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boivin G, Abed Y, Pelletier G, Ruel L, Moisan D, Côté S, Peret TC, Erdman DD, Anderson LJ. Virological features and clinical manifestations associated with human metapneumovirus: a new paramyxovirus responsible for acute respiratory-tract infections in all age groups. J Infect Dis. 2002;186:1330–1334. doi: 10.1086/344319. [DOI] [PubMed] [Google Scholar]
- 4.Falsey AR, Walsh EE. Viral pneumonia in older adults. Clin Infect Dis. 2006;42:518–524. doi: 10.1086/499955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kahn JS. Epidemiology of human metapneumovirus. Clin Microbiol Rev. 2006;19:546–557. doi: 10.1128/CMR.00014-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rao BL, Gandhe SS, Pawar SD, Arankalle VA, Shah SC, Kinikar AA. First detection of human metapneumovirus in children with acute respiratory infection in India: a preliminary report. J Clin Microbiol. 2004;42:5961–5962. doi: 10.1128/JCM.42.12.5961-5962.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singh AK, Jain B, Verma AK, Kumar A, Dangi T, Dwivedi M, Singh KP, Jain A. Hospital outbreak of human respiratory syncytial virus (HRSV) illness in immunocompromised hospitalized children during summer. Clin Respir J. 2015;9:180–184. doi: 10.1111/crj.12121. [DOI] [PubMed] [Google Scholar]
- 8.Banerjee S, Bharaj P, Sullender W, Kabra SK, Broor S. Human metapneumovirus infections among children with acute respiratory infections seen in a large referral hospital in India. J Clin Virol. 2007;38:70–72. doi: 10.1016/j.jcv.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 9.Narayanan H, Sankar S, Simoes EA, Nandagopal B, Sridharan G. Molecular detection of human metapneumovirus and human bocavirus on oropharyngeal swabs collected from young children with acute respiratory tract infections from rural and peri-urban communities in South India. Mol Diagn Ther. 2013;17:107–115. doi: 10.1007/s40291-013-0030-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Principi N, Esposito S. Paediatric human metapneumovirus infection: epidemiology, prevention and therapy. J Clin Virol. 2014;59(3):141–147. doi: 10.1016/j.jcv.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 11.Van den Hoogen BG, Bestebroer TM, Osterhaus AD, Fouchier RA. Analysis of the genomic sequence of a human metapneumovirus. Virol. 2002;295:119–132. doi: 10.1006/viro.2001.1355. [DOI] [PubMed] [Google Scholar]
- 12.Huck B, Scharf G, Neumann-Haefelin D, Puppe W, Weigl J, Falcone V. Novel human metapneumovirus sublineage. Emerg Infect Dis. 2006;12:147–150. doi: 10.3201/eid1201.050772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Biacchesi S, Skiadopoulos MH, Boivin G, Hanson CT, Murphy BR, Collins PL, Buchholz UJ. Genetic diversity between human metapneumovirus subgroups. Virol. 2003;315:1–9. doi: 10.1016/S0042-6822(03)00528-2. [DOI] [PubMed] [Google Scholar]
- 14.Regev L, Meningher T, Hindiyeh M, Mendelson E, Mandelboim M. Increase human metapneumovirus mediated morbidity following pandemic influenza infection. PLoS ONE. 2012;7:e34750. doi: 10.1371/journal.pone.0034750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carr MJ, Waters A, Fenwick F, Toms GL, Hall WW, O’Kelly E. Molecular epidemiology of human metapneumovirus in Ireland. J Med Virol. 2008;80:510–516. doi: 10.1002/jmv.21081. [DOI] [PubMed] [Google Scholar]
- 16.Nidaira M, Taira K, Hamabata H, Kawaki T, Gushi K, Mahoe Y, Maeshiro N, Azama Y, Okano S, Kyan H, Kudaka J. Molecular epidemiology of human metapneumovirus from 2009 to 2011 in Okinawa, Japan. Jpn J Infect Dis. 2012;65:337–340. doi: 10.7883/yoken.65.337. [DOI] [PubMed] [Google Scholar]
- 17.Chow WZ, Chan YF, Oong XY, Ng LJ, Nor’E SS, Ng KT, Chan KG, Hanafi NS, Pang YK, Kamarulzaman A, Tee KK. Genetic diversity, seasonality and transmission network of human metapneumovirus: identification of a unique sub-lineage of the fusion and attachment genes. Sci Rep. 2016;6:27730–27739. doi: 10.1038/srep27730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jagušić M, Slović A, Ljubin-Sternak S, Mlinarić-Galinović G, Forčić D. Genetic diversity of human metapneumovirus in hospitalized children with acute respiratory infections in Croatia. J Med Virol. 2017;89:1885–1893. doi: 10.1002/jmv.24884. [DOI] [PubMed] [Google Scholar]
- 19.Gauthier P, Cardot JM. Developing drugs for children and the adjustment of medication—is it a new challenge or an adaptation of past ideas? J Pers Med. 2011;1(1):5–16. doi: 10.3390/jpm1010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bonroy C, Vankeerberghen A, Boel A, De Beenhouwer H. Use of a multiplex real-time PCR to study the incidence of human metapneumovirus and human respiratory syncytial virus infections during two winter seasons in a Belgian paediatric hospital. Clin Microbiol Infect. 2007;13:504–509. doi: 10.1111/j.1469-0691.2007.01682.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hindupur A, Dhandapani P, Menon T. Influenza virus among children with acute respiratory infections in Chennai. India Indian Pediatr. 2019;56(1):74–75. [PubMed] [Google Scholar]
- 22.Hindupur A, Menon T, Dhandapani P. Genetic diversity of human respiratory syncytial virus in children with acute respiratory infections in Chennai. South India Indian J Med Microbiol. 2019;37(2):248–254. doi: 10.4103/ijmm.IJMM_19_193. [DOI] [PubMed] [Google Scholar]
- 23.Kaida A, Iritani N, Kubo H, Shiomi M, Kohdera U, Murakami T. Seasonal distribution and phylogenetic analysis of human metapneumovirus among children in Osaka City, Japan. J Clin Virol. 2006;35:394–399. doi: 10.1016/j.jcv.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 24.Ishiguro N, Ebihara T, Endo R, Ma X, Kikuta H, Ishiko H, Kobayashi K. High genetic diversity of the attachment (G) protein of human metapneumovirus. J Clin Microbiol. 2004;42(8):3406–3414. doi: 10.1128/JCM.42.8.3406-3414.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van den Hoogen BG, Herfst S, Sprong L, Cane PA, Forleo-Neto E, De Swart RL, Osterhaus AD, Fouchier RA. Antigenic and genetic variability of human metapneumoviruses. Emerg Infect Dis. 2004;10(4):658. doi: 10.3201/eid1004.030393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Graaf M, Osterhaus AD, Fouchier RA, Holmes EC. Evolutionary dynamics of human and avian metapneumoviruses. J Gen Virol. 2008;89(12):2933–2942. doi: 10.1099/vir.0.2008/006957-0. [DOI] [PubMed] [Google Scholar]
- 27.Bastien N, Normand S, Taylor T, Ward D, Peret TC, Boivin G, Anderson LJ, Li Y. Sequence analysis of the N, P, M and F genes of Canadian human metapneumovirus strains. Virus Res. 2003;93(1):51–62. doi: 10.1016/S0168-1702(03)00065-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pybus OG, Tatem AJ, Lemey P. Virus evolution and transmission in an ever more connected world. Proc R Soc B: Biol Sci. 2015;282(1821):20142878. doi: 10.1098/rspb.2014.2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nandhini G, Sujatha S, Jain N, Dhodapkar R, Tamilarasu K, Krishnamurthy S, Biswal N. Prevalence of human metapneumovirus infection among patients with influenza-like illness: report from a tertiary care centre, Southern India. Indian J Med Microbiol. 2016;34:27–32. doi: 10.4103/0255-0857.174117. [DOI] [PubMed] [Google Scholar]
- 31.Roy Mukherjee T, Chanda S, Mullick S, De P, Dey-Sarkar M, Chawla-Sarkar M. Spectrum of respiratory viruses circulating in eastern India: prospective surveillance among patients with influenza-like illness during 2010–2011. J Med Virol. 2013;85:1459–1465. doi: 10.1002/jmv.23607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jain B, Singh AK, Dangi T, Agarwal A, Verma AK, Dwivedi M, Singh KP, Jain A. High prevalence of human metapneumovirus subtype B in cases presenting as severe acute respiratory illness: an experience at tertiary care hospital. Clin Respir J. 2014;8:225–233. doi: 10.1111/crj.12064. [DOI] [PubMed] [Google Scholar]
- 33.Arnott A, Vong S, Sek M, Naughtin M, Beauté J, Rith S, Guillard B, Deubel V, Buchy P. Genetic variability of Human metapneumovirus amongst an all ages population in Cambodia between 2007 and 2009. Infect Genet Evol. 2013;15:43–52. doi: 10.1016/j.meegid.2011.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sung CC, Chi H, Chiu NC, Huang DT, Weng LC, Wang NY, Huang FY. Viral etiology of acute lower respiratory tract infections in hospitalized young children in Northern Taiwan. J Microbiol Immunol Infect. 2011;44:184–219. doi: 10.1016/j.jmii.2011.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Panda S, Mohakud NK, Pena L, Kumar S. Human metapneumovirus: review of an important respiratory pathogen. Int J Infect Dis. 2014;25:45–52. doi: 10.1016/j.ijid.2014.03.1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sarkar S, Ratho RK, Singh M, Singh MP, Singh A, Sharma M (2022) Comparative analysis of epidemiology, clinical features, and cytokine response of respiratory syncytial and human metapneumovirus infected children with acute lower respiratory infections. Jpn J Infect Dis 75:56–62 [DOI] [PubMed]
- 37.Foulongne V, Guyon G, Rodière M, Segondy M. Human metapneumovirus infection in young children hospitalized with respiratory tract disease. Pediatr Infect Dis J. 2006;25:354–359. doi: 10.1097/01.inf.0000207480.55201.f6. [DOI] [PubMed] [Google Scholar]
- 38.König B, König W, Arnold R, Werchau H, Ihorst G, Forster J. Prospective study of human metapneumovirus infection in children less than 3 years of age. J Clin Microbiol. 2004;42:4632–4635. doi: 10.1128/JCM.42.10.4632-4635.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Woensel JB, Bos AP, Lutter R, Rossen JW, Schuurman R. Absence of human metapneumovirus co-infection in cases of severe respiratory syncytial virus infection. Pediatr Pulmonol. 2006;41:872–874. doi: 10.1002/ppul.20459. [DOI] [PubMed] [Google Scholar]
- 40.Zhang C, Zhu N, Xie Z, Lu R, He B, Liu C, et al. Viral etiology and clinical profiles of children with severe acute respiratory infections in China. PLoS One. 2013;8:e72606. doi: 10.1371/journal.pone.0072606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moe N, Krokstad S, Stenseng IH, Christensen A, Skanke LH, Risnes KR, Nordbø SA, Døllner H. Comparing human metapneumovirus and respiratory syncytial virus: viral co-detections, genotypes and risk factors for severe disease. PLoS One. 2017;12(1):e0170200. doi: 10.1371/journal.pone.0170200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu R, Guo C, Zhao L, Deng J, Wang F, Sun Y, Qian Y. Epidemiological and genetic characteristics of human metapneumovirus in pediatric patients across six consecutive seasons in Beijing, China. Int J Infect Dis. 2020;91:137–142. doi: 10.1016/j.ijid.2019.11.012. [DOI] [PubMed] [Google Scholar]
- 43.Choudhary ML, Anand SP, Sonawane NS, Chadha MS. Development of real-time RT-PCR for detection of human metapneumovirus and genetic analysis of circulating strains (2009–2011) in Pune, India. Arch Virol. 2014;159:217–225. doi: 10.1007/s00705-013-1812-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rueda AJV, Mistchenko AS, Viegas M. Phylogenetic and phylodynamic analyses of human metapneumovirus in Buenos Aires (Argentina) for a three-year period (2009–2011) PloS One. 2013;8:e63070. doi: 10.1371/journal.pone.0063070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ljubin-Sternak S, Mlinaric-Galinovic G, Buntic AM, Tabain I, Vilibic-Cavlek T, Cepin-Bogovic J, Tesovic G. Seasonal occurrence of human metapneumovirus infections in Croatia. Pediatr Infect Dis. 2014;J33:165–167. doi: 10.1097/INF.0000000000000026. [DOI] [PubMed] [Google Scholar]
- 46.Malhotra B, Swamy MA, Reddy PJ, Gupta ML. Viruses causing severe acute respiratory infections (SARI) in children ≤ 5 years of age at a tertiary care hospital in Rajasthan, India. Indian J Med Res. 2016;144:877–885. doi: 10.4103/ijmr.IJMR_22_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Agrawal AS, Roy T, Ghosh S, Chawla-Sarkar M. Genetic variability of attachment (G) and fusion (F) protein genes of human metapneumovirus strains circulating during 2006–2009 in Kolkata, Eastern India. Virol J. 2011;8:1–8. doi: 10.1186/1743-422X-8-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Banerjee S, Sullender WM, Choudekar A, John C, Tyagi V, Fowler K, Lefkowitz EJ, Broor S. Detection and genetic diversity of human metapneumovirus in hospitalized children with acute respiratory infections in India. J Med Virol. 2011;83:1799–1810. doi: 10.1002/jmv.22176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Narayanan H, Sankar S, Simoes EA, Nandagopal B, Sridharan G. Analysis of sequence diversity of human metapneumovirus collected from young children with acute respiratory tract infections in South India. Mol diagn Ther. 2013;17:247–255. doi: 10.1007/s40291-013-0032-9. [DOI] [PubMed] [Google Scholar]
- 50.Rahman MZ, Sumiya M, Sahabuddin M, Pell LG, Gubbay JB, Rahman R, Momtaz F, Azmuda N, Shanta SS, Jahan I, Rahman M. Genetic characterization of human metapneumovirus identified through community and facility-based surveillance of infants in Dhaka, Bangladesh. J Med Virol. 2019;91:549–554. doi: 10.1002/jmv.25351. [DOI] [PubMed] [Google Scholar]
- 51.Yang CF, Wang CK, Tollefson SJ, Piyaratna R, Lintao LD, Chu M, Liem A, Mark M, Spaete RR, Crowe JE, Williams JV. Genetic diversity and evolution of human metapneumovirus fusion protein over twenty years. Virol J. 2009;6:138–147. doi: 10.1186/1743-422X-6-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hansen JE, Lund O, Tolstrup N, Gooley AA, Williams KL, Brunak S. NetOglyc: prediction of mucin type O-glycosylation sites based on sequence context and surface accessibility. Glycoconj J. 1998;15:115–130. doi: 10.1023/A:1006960004440. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary file3 (jpg 418 kb) Supplement Fig. 1 Maximum Likelihood fits of 24 different nucleotide substitution models
Supplementary file4 (jpeg 365 kb) Supplement Fig. 2 Deduced amino acid alignment of partial F gene of hMPV strains belonging to the A2c subcluster obtained in this study
Supplementary file5 (jpg 370 kb) Supplement Fig. 3 Deduced amino acid alignment of partial F gene of hMPV strains belonging to the B1 lineage obtained in this study
Supplementary file6 (jpg 393 kb) Supplement Fig. 4 Deduced amino acid alignment of partial F gene of hMPV strains belonging to the B2 lineage obtained in this study