SUMMARY
Maximum entropy ecological niche modelling and spatial scan statistic were utilized to predict the geographical range and to investigate clusters of infections for Neospora caninum and Coxiella burnetii in dairy cattle farms in Catalonia, northeastern Spain, using the Maxent and SaTScan programs, respectively. The geographical distribution of Neospora and Coxiella with the highest level of probability (P > 0·60) covers central Catalonia and spreads towards northeastern Catalonia which contains a high concentration of dairy cattle farms. The most important environmental factor that contributed to the ecological niche modelling was precipitation of driest month followed by elevation. Significant clusters (P < 0·001) were detected for Neospora and Coxiella infections in the western and eastern regions of Catalonia, respectively.
Key words: Cattle, Coxiella burnetii, ecological niche modelling, dairy farms, Neospora caninum
INTRODUCTION
Neospora caninum is an obligate intracellular protozoan and Coxiella burnetii is an intracellular Gram-negative bacterium. Both pathogens cause abortions in cattle worldwide [1–3]. Cattle infection with N. caninum is facilitated through the ingestion of sporulated oocysts in the environment passed in the faeces of the definitive carnivore host [4]. Cattle infection with C. burnetii is facilitated through contact with birth products (birth fluids, placenta), vaginal mucus, milk, faeces, urine and semen of C. burnetii-shedding cattle [1, 5]. A common characteristic of Neospora and Coxiella infections is that in both cases cattle are infected by the respective pathogens present in the environment. Therefore, the presence of these pathogens in the environment makes their spread and geographical distribution susceptible to environmental conditions.
Ecological niche modelling offers the opportunity to derive predictive distribution maps from species occurrence and environmental data [6, 7]. In the present study a cross-sectional survey was conducted in dairy cattle farms in Catalonia, northeastern Spain to collect information on the spatial distribution of N. caninum-infected and C. burnetii-infected dairy farms in order to construct predictive maps of Catalonia for the two pathogens, based on ecological niche modelling, and to investigate clusters of infections.
MATERIALS AND METHODS
Farms and sampled areas
Bulk tank milk (BTM) samples were collected on the same day in October 2010 from dairy cattle farms in Catalonia, northeastern Spain during daily quality BTM control of all Catalan dairy farms. Catalonia is divided into 41 regions and at the time of the study there were 814 dairy farms which were located in 15 regions [8]. Farms were randomly sampled within each region (10–12%) in order to be representative of all areas and the coordinates of the location of each farm were recorded in a database. The association between the prevalence of Neospora-positive and Coxiella-positive farms was estimated using Fisher's exact test (P < 0·05) and Cramer's V2 test. Cramer's V2 test measures the association between two categorical variables. A value of zero indicates that there is no association. A value of 1 indicates that there is a perfect association.
Anti-N. caninum antibody detection in BTM
A commercial indirect enzyme-linked immunosorbent assay (ELISA) kit (Civtest Bovis Neospora, Spain), based on the whole tachyzoite lysate of N. caninum NC-1 was used to determine antibodies against N. caninum. The skimmed milk samples were centrifuged at 1000 g for 15 min at 4 °C. Absorbance was measured as optical density (OD) values at 405 nm using a microplate reader (Multiskan FC, Finland). A relative index percent (RIPC) per BTM sample was obtained according the manufacture's procedure:
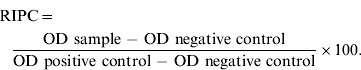 |
The kit manufacturer considers individual cow prevalence of antibodies against N. caninum to be less than 5–10% when BTM RIPC is ⩽3·0 and greater than 10% when BTM RIPC is >3·0. Comparing results of BTM with individual serology, it was found that at the cut-off point of 3·0 the sensitivity and specificity of the test were 60% and 91%, respectively. For analysis of data, BTM was considered negative when RIPC ⩽3·0 and positive when RIPC >3·0.
Anti-C. burnetii antibody detection in BTM
A commercial indirect ELISA LSIVET Ruminant Milk/Serum Q Fever (CoxLS kit; Laboratoire Service International, France) was used to determine antibodies to C. burnetii in the BTM samples. The milk samples were skimmed and the test was performed according to the manufacturer's instructions. The antigen of the ELISA CoxLS kit has been isolated from domestic ruminants by INRA (France). Absorbance was measured as OD values at 450 nm using a microplate reader (Multiskan FC). The results were expressed as a S/P ratio calculated as follows:
The BTM results were expressed as titres (titre = S/P × 100). BTM was considered negative when the titre was ⩽30 and positive when the titre was >30 [9].
Environmental data
The environmental datasets obtained for this study were climate, elevation, and land cover data. These datasets were converted to a common projection, map extent and resolution prior to use in the modelling program. WorldClim version 1.4 climate data [10] was obtained from the WorldClim website (http://www.worldclim.org). WorldClim is a set of global climate layers (climate grids) with a spatial resolution of 1 km2. They can be used for mapping and spatial modelling in a geographical information system (GIS) or other computer programs. The data layers were generated through interpolation of average monthly climate data from weather stations on a 30 arc-second (1 km2) resolution grid. Variables included are monthly total precipitation, and monthly mean, minimum and maximum temperature. The data are further processed into a series of bioclimatic variables (Table 1). Input data were gathered from a variety of sources and, where possible, were restricted to records from the period 1950–2000. The thin-plate smoothing spline algorithm implemented in the ANUSPLIN package (Centre for Resources and Environmental Studies at the Australian National University, Australia) was used for interpolation, using latitude, longitude, and elevation as independent variables. The database is documented in detail in Hijmans et al. [10]. The bioclimatic variables with an approximate spatial resolution of 1 km2 were used for this project.
Table 1.
List of WorldClim bioclimatic variables used in the model [10]
| Bioclimatic variable | Description |
|---|---|
| BIO1 | Annual mean temperature |
| BIO2 | Mean diurnal range [mean of monthly (max temp – min temp)] |
| BIO3 | Isothermality (BIO2/BIO7) (×100) |
| BIO4 | Temperature seasonality (standard deviation × 100) |
| BIO5 | Max temperature of warmest month |
| BIO6 | Min temperature of coldest month |
| BIO7 | Temperature annual range (BIO5–BIO6) |
| BIO8 | Mean temperature of wettest quarter |
| BIO9 | Mean temperature of driest quarter |
| BIO10 | Mean temperature of warmest quarter |
| BIO11 | Mean temperature of coldest quarter |
| BIO12 | Annual precipitation |
| BIO13 | Precipitation of wettest month |
| BIO14 | Precipitation of driest month |
| BIO15 | Precipitation seasonality (coefficient of variation) |
| BIO16 | Precipitation of wettest quarter |
| BIO17 | Precipitation of driest quarter |
| BIO18 | Precipitation of warmest quarter |
| BIO19 | Precipitation of coldest quarter |
Elevation data were also downloaded from the WorldClim website. WorldClim processed this dataset from NASA Shuttle Radar Topography Mission (SRTM) data to have the same projection and resolution as the other WorldClim layers. SRTM obtained elevation data on a near-global scale to generate the most complete high-resolution digital topographic database of Earth. SRTM consisted of a specially modified radar system that flew onboard the Space Shuttle Endeavour during an 11-day mission in February 2000. Land-cover data were downloaded from the United States Geological Survey's (USGS) Global Land Cover Characteristics (GLCC) database, version 2 Global (http://www.edsns17.crusgs.gov/glcc/). It is a 1 km resolution global land-cover characteristics database generated for use in a wide range of environmental research and modelling applications.
The land-cover data were developed from 1 km advanced very high-resolution radiometer (AVHRR) satellite data from April 1992 to March 1993. More precisely, the 1 km AVHRR normalized differential vegetation index (NDVI) composites are the core dataset used in land-cover characterization. In addition, other key geographical data include digital elevation data, ecoregion interpretations and country-level or regional-level vegetation and land-cover maps. Land-cover data are available in multiple classification schemes; the Global Ecosystems land cover classification, which contains 100 classes, was used for the modelling work. This global database was used because it is not geographically limited and is available on the USGS data portal for most of the countries in the world. In this way, the presented application can be easily reproduced worldwide, producing comparable results. Moreover, the GLCC database is compatible with the WorldClim global climate layers used in this study having the same spatial resolution (1 km2).
Modelling algorithm
Model building was performed using the maximum entropy method implemented in the Maxent program [6]. The Maxent program develops models of species' distributions using species' presence data and environmental data [6, 11, 12]. Maxent is based on the idea of estimating a target probability distribution by finding the probability distribution of maximum entropy (i.e. the most spread out, or closest to uniform), subject to a set of constraints that represent the incomplete presence-only data information about the target distribution. The constraints are that the expected value of each environmental variable should match its empirical average. A detailed description of the Maxent algorithm is given in Philips et al. [6]. Maxent has been used for a number of studies and has been shown to be a high performing modelling program [7, 13].
Model performance evaluation and contribution of environmental factors to the models
The model performance was evaluated using the threshold independent method based on the area under the curve (AUC) of receiver-operating characteristics curve (ROC). The AUC is computed through a ROC plot of sensitivity (the true-positive fraction) against 1 – specificity (the false-positive fraction) [14]. The AUC value ranges from 0·5 (random accuracy) to a maximum value of 1 (perfect discrimination). In order to determine which variables contribute most to the model's development, the Maxent program was set to perform the jackknife tests in which the method is run multiple times: (i) using all variables, (ii) dropping one variable at a time, and (iii) running the model using only one variable. Variables which produced the highest training gains or reduced the training gain when omitted from the model are considered to be the most important variables. Strong correlation between variables can be located in curves showing how each environmental variable affects the Maxent prediction disregarding all other variables. The GIS software ArcGIS v. 9.2 (ESRI Inc., USA) was used to display the prediction results and represent the sampled localities.
Spatial cluster analysis
The spatial scan statistic implemented in SaTScan software (version 8.0; SaTScan™, USA) was used to investigate geographical clusters of infection. The concept of a spatial scan statistic is based on the generalization of a test probability [15–17]. The spatial scan statistic uses a circular window of variable radius that moves across the map to represent potential geographical clusters. The radius of the cluster varies from zero up to a specified maximum value. By gradually changing the circle centre and radius, the window scans the geographical areas for potential localized clusters without incorporating prior assumptions about their size and location and notes the number of observed and expected observations inside the window at each location. The test of significance is based on the likelihood ratio test for which the window with the maximum likelihood is the most likely cluster [16]. The assessment of a cluster is performed by comparing the number of cases (infection) within the circle with the number of expected cases under the assumption that cases are randomly distributed in the space. The P value is obtained through Monte Carlo hypothesis testing [18]. The spatial scan statistic adjusts for spatial variations in the density of the population in the study area [17]. The detection of clusters in the present study was performed under the Bernoulli probability model using the maximum cluster size of 50% of the total population for Neopora and Coxiella infections separately. Test-positive farms were considered as cases while test-negative farms were regarded as controls. The number of simulations for Monte Carlo testing was set to 9999. For each window of varying position and size, the SaTScan program tested the risk of Neospora and Coxiella infection each within and outside the window, with the null hypothesis of equal risk.
RESULTS
A total of 90 (11%) dairy cattle farms were sampled. The prevalence of Neospora-positive and Coxiella-positive farms in the examined farms was 90% and 74%, respectively. No association between the prevalence of Neospora-positive and Coxiella-positive farms was found (Fisher's exact test: P = 0·22, Cramer's V2 = 0·02). Figures 1 and 2 display the potential geographical distribution of Neospora-positive and Coxiella-positive farms predicted by the Maxent program, respectively. The geographical distribution of Neospora-positive and Coxiella-positive farms with the highest level of probability (P > 0·60) covers central Catalonia and spreads towards northeastern Catalonia.
Fig. 1.
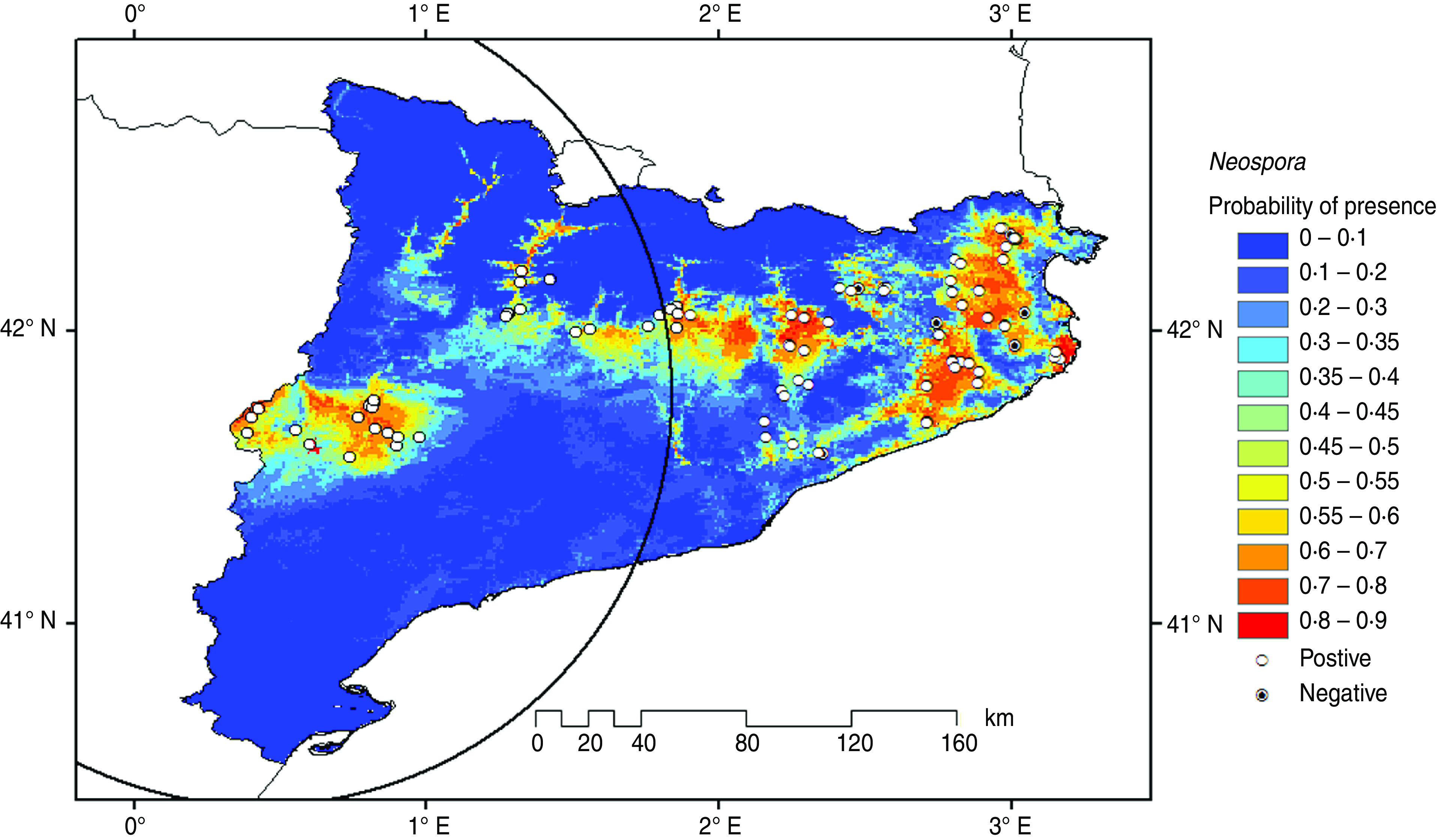
[colour online]. Predicted geographical distribution ranges for Neospora caninum. The circle encloses the location identified within the most likely cluster.
Fig. 2.
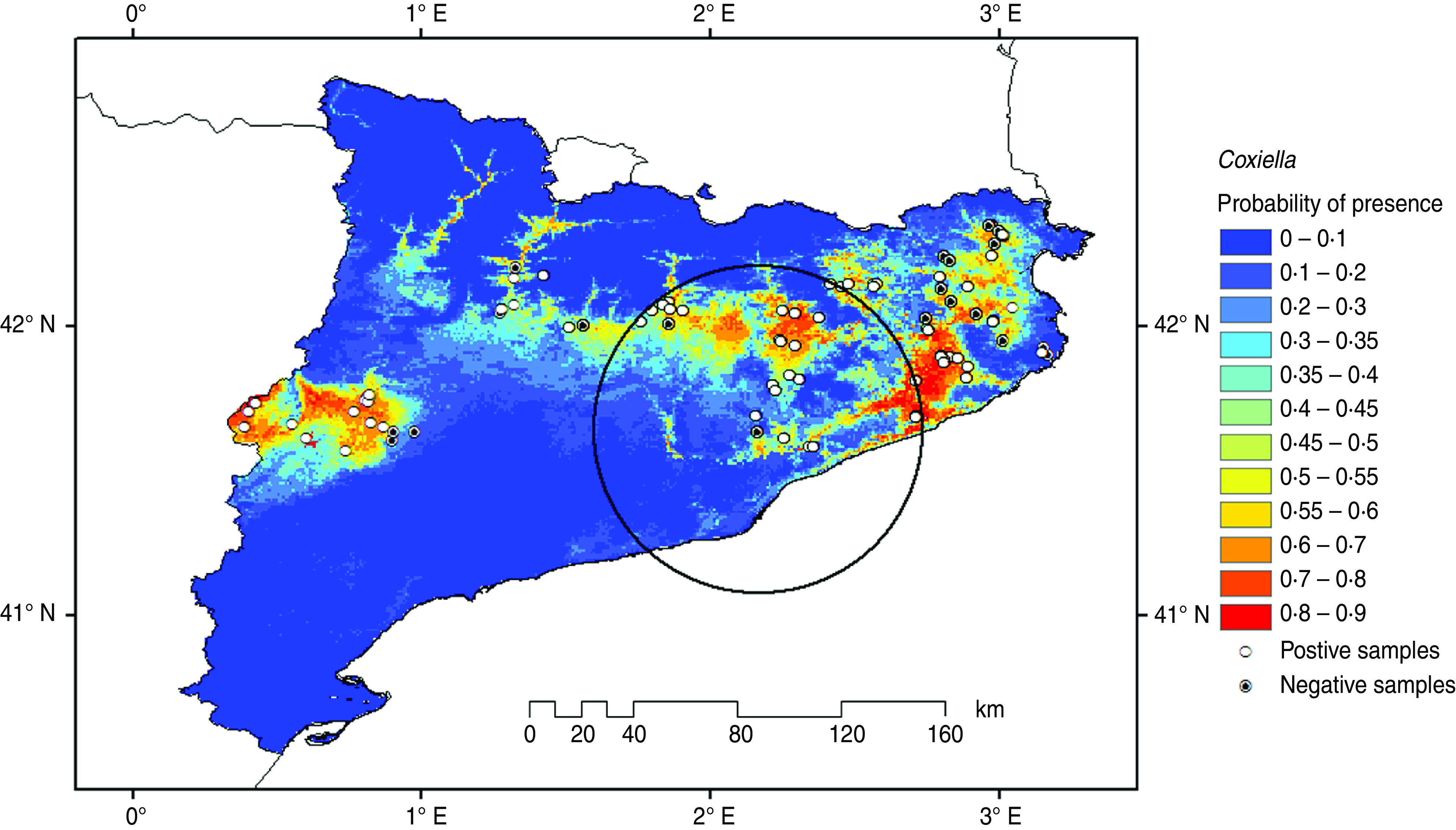
[colour online]. Predicted geographical distribution ranges for Coxiella burnetii. The circle encloses the location identified within the most likely cluster.
Model validation
The AUC values for the models range from 0·886 to 0·926, indicating that the models are good to very good (Table 2). Both models have low P values (Table 2) using minimum training presence as the threshold with Neospora-positive farms having the lowest (P = 0·0001) indicating the best prediction.
Table 2.
Statistical evaluation of the Maxent model
| Species | Number of presence records* (training/testing) | AUC for training data | AUC for test data | P value using minimum training presence as threshold |
|---|---|---|---|---|
| Neospora caninum | 60/20 | 0·914 | 0·886 | 0·0001 |
| Coxiella burnetii | 50/16 | 0·926 | 0·910 | 0·001 |
AUC, Area under the curve.
Duplicate presence records for the same localities were removed from the model.
Effect of environmental variables on the model
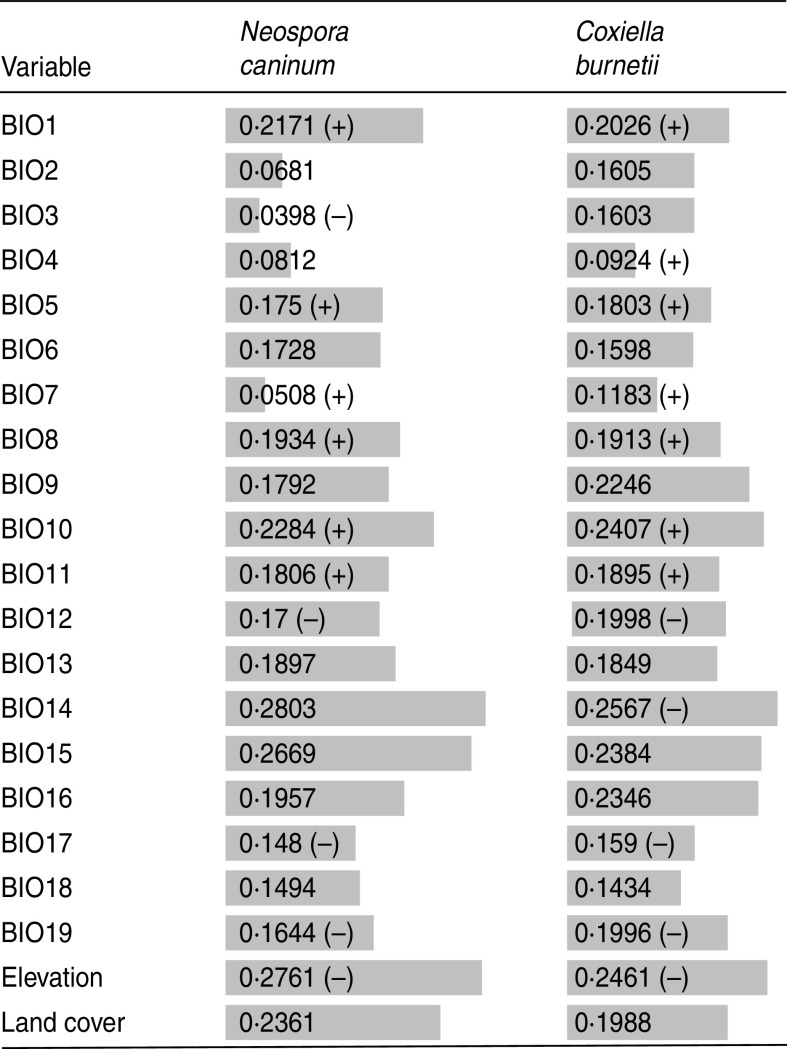
For Neospora-positive and Coxiella-positive farms, ‘precipitation of driest month’ (BIO14) followed by elevation were the variables that achieved the highest training gain when used to build a model with no other variables (Fig. 3).
Fig. 3.
Training gain achieved by models using single variables. Length of bar ( ) represents the training gain value. A longer bar represents a higher training gain. Positive or negative signs, if present, indicate linear correlation between predicted suitability and environmental variable.
) represents the training gain value. A longer bar represents a higher training gain. Positive or negative signs, if present, indicate linear correlation between predicted suitability and environmental variable.
Other environmental variables that show training gain when modelling with only a single variable included: ‘precipitation seasonality’ (BIO15), land cover, ‘mean temperature of warmest quarter’ (BIO10), ‘annual mean temperature’ (BIO1), ‘precipitation of wettest quarter’ (BIO16), ‘mean temperature of wettest quarter’ (BIO8), and ‘mean temperature of coldest quarter’ (BIO11), for Neospora-positive farms, and ‘precipitation of driest month’ (BIO14), elevation, ‘mean temperature of warmest quarter’ (BIO10), ‘precipitation seasonality’ (BIO15), ‘precipitation of wettest quarter’ (BIO16), ‘mean temperature of driest quarter’ (BIO9), ‘precipitation of coldest quarter’ (BIO19), ‘mean temperature of wettest quarter’ (BIO8), and land cover for Coxiella-positive farms (Fig. 3).
Clear or almost clear linear (positive or negative) or no linear correlation between predicted suitability and environmental variable are indicated in Figure 3.
Land-cover types that occurred at the species sampling areas are shown in Table 3. The primary land covers are similar for both Neospora-positive and Coxiella-positive farms and include mostly Mediterranean scrub followed by cool crops and towns.
Table 3.
Numbers of sample localities within different land-cover categories of the USGS Global Ecosystems land cover classification
| Land cover (class number) | Neospora caninum | Coxiella burnetii |
|---|---|---|
| Mixed forest (24) | 8 | 6 |
| Cool crops and towns (30) | 10 | 8 |
| Crops and town (31) | 9 | 7 |
| Mediterranean scrub (46) | 39 | 33 |
| Small leaf mixed woods (60) | 5 | 5 |
| Woody savanna (91) | 5 | 4 |
| Grass crops (93) | 5 | 4 |
USGS, United States Geological Survey.
Spatial cluster analysis
The results of the spatial scan statistic analyses showed one most likely cluster of infected farms (P < 0·05) for each pathogen. The relative risk (RR) of the most likely cluster was 1·25 and 1·57 for Neospora-positive and Coxiella-positive farms, respectively (Table 4).
Table 4.
Significant clusters of Neospora caninum-infected and Coxiella burnetii-infected dairy farms
| Species | Cluster | Location (coordinates) [radius] | No. of cases | Expected cases | Relative risk | P value |
|---|---|---|---|---|---|---|
| Neospora caninum | Most likely | Almacelles (41·738712° N, 0·424497° E) [158·08 km] | 45 | 40·50 | 1·25 | 0·01 |
| Coxiella burnetii | Most likely | Caldes de Montbui (41·636959° N, 2·158792° E) [64·16 km] | 35 | 27·54 | 1·57 | 0·01 |
DISCUSSION
Dairy cattle farms are an important source of income for the economy of Catalonia region in northeastern Spain. Despite the low number of farms, Catalonia is one of the most dynamic dairy regions of Spain [19]. The samples for testing were chosen randomly within each region of Catalonia (10–12%) in order to be representative of the overall area. Abortions due to N. caninum and C. burnetii have a grave impact on the reproductive performance of dairy cattle with major consequences on the profitability of cow production [20, 21]. The seroprevalence of infections due to N. caninum and C. burnetii has been reported to be 83% in Spanish farms [22] and 61% in Catalan farms [23], respectively. In Catalonia, the risk of abortion was found to be 12–19 times higher in Neospora-seropositive dairy cows than in their seronegative partners [21, 24]. No similar data is available for Coxiella abortions.
Even though N. caninum can be transmitted vertically when tachyzoites are transmitted from an infected dam to her foetus during pregnancy [4, 25], the ingestion of oocysts is the only demonstrable mode for horizontal transmission in herbivores [4] and the mode of first introduction of the disease to the farm.
N. caninum [26, 27] and C. burnetii [9] analysis of antibodies using ELISA in BTM has been demonstrated to be very useful in estimating the prevalence of these pathogens in dairy farms. However, as the volume of milk that each lactating cow provides to the tank is variable throughout the year, the prevalence value obtained from BTM has to be considered as a guiding result. The BTM samples used in the present study were the same routinely obtained by the Catalan dairy industry for milk quality testing and enabled detection of Neospora and Coxiella infection at an affordable price. This available, reliable, and simple to use diagnostic technique for screening and monitoring large number of cows can be very useful for bovine neosporosis and coxiellosis implementation control programmes [26, 27], but individual milk or serum analysis should be performed in order to better control the positive farms.
The present study reports the potential geographical range of N. caninum and C. burnetii in northeastern Spain using the Maxent program. Maxent is a mathematical framework that can be applied for several species at once and/or for spatially structured communities [28]. There have been no previous attempts to model Neospora and Coxiella distribution using GIS.
Papes & Baubert [29] found that the minimum number of records needed for accurate modelling with Maxent is around 15. Pearson et al. [30] found that models built with as few as five records showed some statistical significance using Maxent. It is important to mention that by modelling the ecological niche for the two pathogens in the present study, it does not mean that the models represent their actual or realized distribution. These models should be considered as an initial effort to approximate their potential geographical range.
The results of the present study indicate that various bioclimatic variables, and most importantly, precipitation of driest month (BIO14), precipitation seasonality (BIO15) and elevation correlate similarly with the geographical distribution of Neospora and Coxiella. In two previous studies on climate effects on the risk of Neospora in cattle in Germany [31] and Italy [32] it was found that ‘mean temperature in July in the municipality where the herd is localized’ and ‘mean temperature in spring in a buffer zone around farm location’ were identified as risk factors, respectively. In addition, two previous studies on climate effects on abortions due to Neospora in The Netherlands [33] and California [34] indicated that mild temperatures and humidity increase the risk of infection. Other studies on climate effects on the risk of Coxiella in cattle showed that the number of antibody-positive cows and their antibody titre in Japan were significantly elevated in winter and decreased in summer [35], while in Italy all the C. burnetii-related abortions were recorded between October and April [36]. These observations for Neospora and Coxiella may be attributed to the effect of climate on the rate of sporulation or survival of Neospora oocysts and the preservation of Coxiella bacteria in the environment.
Another finding of the present study is that Mediterranean scrub followed by cool crops and towns are the primary land covers for the geographical distribution of both Neospora-infected and Coxiella-infected farms. Farm location or proximity to a city, town, or village has been found to be a risk factor for N. caninum-associated abortion in Switzerland [37] and N. caninum positivity in bulk milk ELISA [31]. The observed land covers that favour the geographical distribution of Neospora may be associated with high human population density which is correlated with a high density of dogs [32] that serve as reservoirs of Neospora. In the case of Coxiella, the observed favourable land covers may be associated with the presence of ticks that have an active role in maintaining the disease in wild and peridomestic cycles [38] or other wild ungulates [39].
The areas where significant clusters of Neospora-positive and Coxiella-positive farms were observed are centred on Almacelles and Caldes de Montbuy, respectively. Almacelles is an irrigated flat area with fruit trees and forage crop production with a medium density of big dairy farms and a high density of beef feedlots. Dogs and other small wild animals are present in this area facilitating the spread of neosporosis to cattle [40, 41]. The area of Caldes de Montbuy is surrounded by mountains and is very close to two natural parks (Parc natural del Montseny and Parc natural de Sant Llorenç del Munt i l'Obac). This area is associated with wild animals and ticks that can carry coxiellosis [38].
The most likely cluster for Neospora-positive farms did not coincide with that for Coxiella-positive farms, suggesting that different risk factors might be at play for clusters. Since both Neospora and Coxiella occur in the same hosts, a possible discriminating factor might be the presence of reservoir hosts such as wild animals and ticks. The distribution range of suitable reservoir hosts may vary between high-risk areas associated with both significant clusters as a result of the heterogeneity of climates and landscapes. For example it is known that the habitat of ticks is more set by abiotic factors such as vegetation and climate (which determine tick development and survival rates on the ground) rather than by host-related factors [42].
Another interesting observation in the present study is that the identified clusters encompass areas with high and low probability of presence for both Neospora-positive and Coxiella-positive farms predicted by Maxent program. In addition, even though the bioclimatic variables were found in the present study to affect similarly the geographical distribution of both Neospora-positive and Coxiella-positive farms, no association between the prevalence of these pathogens was found. These observations suggest that in addition to environmental factors, other factors such as animal and farmer as well as farm and pasture management status affect infection with these two pathogens.
The environmental factors identified in the present study are expected to be valid to model the geographical distribution of Neospora-positive and Coxiella-positive farms in other regions of the world, but other enzootic areas in the world should be located to test this assumption. In addition, it seems that the distribution of Neospora-positive and Coxiella-positive farms is also influenced by other variables not analysed in our study.
In conclusion, the highest level of probability for the geographical distribution of Neospora-positive and Coxiella-positive farms covers central Catalonia and spreads towards northeastern Catalonia which contains a high concentration of farms with potential dairy cattle hosts. Maximum entropy ecological niche modelling has proved to be a useful tool in mapping Neospora and Coxiella in northeastern Spain, while the significant clusters detected might be helpful in investigating the underlying causes of increased risk in the identified areas.
ACKNOWLEDGEMENTS
The authors thank the students of the School of Agricultural Engineering of the University of Lleida Rosa Colome and Montse Navarro and the staff members of the Interprofessional Dairy Laboratory of Catalonia (ALLIC) for their help with the collection of samples for this study. This work was supported by Spanish CICYT grant GL2010-21273-C03-01/GAN.
DECLARATION OF INTEREST
None.
REFERENCES
- 1.Maurin M, Raoult D. Q fever. Clinical Microbiology Reviews 1999; 12: 518–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dubey JP. Neosporosis in cattle. Journal of Parasitology 2003; 89 (Suppl.): 42–56. [Google Scholar]
- 3.García-Ispierto I, et al. Neospora caninum and Coxiella burnetii seropositivity are related to endocrine pattern changes during gestation. Theriogenology 2010; 74: 212–220. [DOI] [PubMed] [Google Scholar]
- 4.Dubey JP, Schares G, Ortega-Mora LM. Epidemiology and control of neosporosis and Neospora caninum. Clinical Microbiology Reviews 2007; 20: 323–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guatteo R, et al. Coxiella burnetii shedding by dairy cows. Veterinary Research 2007; 38: 849–860. [DOI] [PubMed] [Google Scholar]
- 6.Phillips SJ, Anderson RP, Schapire RE. Maximum entropy modelling of species geographical distributions. Ecological Modelling 2006; 190: 231–259. [Google Scholar]
- 7.Kouam MK, et al. Geographic distribution modeling and spatial cluster analysis for equine piroplasms in Greece. Infection, Genetics and Evolution 2010; 10: 1013–1018. [DOI] [PubMed] [Google Scholar]
- 8.Milk Monitoring Service Evolution of the number of farms in Catalonia by regions, 2000–2010. Annual Report, November 2010.
- 9.Muskens J, et al. Prevalence of Coxiella burnetii infection in Dutch dairy herds based on testing bulk tank milk and individual samples by PCR and ELISA. Veterinary Record 2012; 168: 79. [DOI] [PubMed] [Google Scholar]
- 10.Hijmans RJ, et al. Very high resolution interpolated climate surfaces for global land areas. International Journal of Climatology 2005; 25: 1965–1978. [Google Scholar]
- 11.Phillips SJ, Dudik M, Schapire RE. A maximum entropy approach to species distribution modeling. In: Proceedings of the Twenty-First International Conference on Machine Learning. New York: ACM Press, 2004, pp. 472–486. [Google Scholar]
- 12.Phillips SJ, Dudik M. Modeling of species distributions with Maxent: new extensions and a comprehensive evaluation. Ecography 2008; 31: 161–175. [Google Scholar]
- 13.Kantzoura V, et al. Geographic distribution modelling for ruminant liver flukes (Fasciola hepatica) in south-eastern Europe. International Journal for Parasitology 2011; 41: 747–753. [DOI] [PubMed] [Google Scholar]
- 14.Swets JA. Measuring the accuracy of diagnostic systems. Science 1988; 240: 1285–1293. [DOI] [PubMed] [Google Scholar]
- 15.Turnbull BW, et al. Monitoring for clusters of disease: application to leukemia incidence in upstate New York. American Journal of Epidemiology 1990; 132 (Suppl.): 136–143. [DOI] [PubMed] [Google Scholar]
- 16.Kulldorff M, Nagarwalla N. Spatial disease clusters: detection and inference. Statistics in Medicine 1995; 15: 707–715. [DOI] [PubMed] [Google Scholar]
- 17.Kulldorff M. A spatial scan statistic. Communications in Statistics – Theory and Methods 1997; 26: 1481–1496. [Google Scholar]
- 18.Dwass M. Modified randomization tests for nonparametric hypotheses. Annals of Mathematical Statistics 1957; 28: 181–187. [Google Scholar]
- 19.Estany J, Nogareda C.A note on Animal Agriculture in Spain and Catalonia in: Adapting Animal Production to Changes for a Growing Human Population, International Conference, Lleida, Spain, 19–21 May 2010, pp. 145–151 (http://www.aap2010.udl.cat/Book-Adapting%20animal%20production.pdf).
- 20.Lee JI, Kim IH. Pregnancy loss in dairy cows: the contributing factors, the effects on reproductive performance and the economic impact. Journal of Veterinary Science 2007; 8: 283–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.López-Gatius F, et al. Neospora associated abortion episode over 1-year period in a dairy herd in north-east Spain. Journal of Veterinary Medicine Series B: Infectious Diseases and Veterinary Public Health 2004. a; 51: 348–352. [DOI] [PubMed] [Google Scholar]
- 22.Quintanilla-Gozalo A, et al. Seroprevalence of Neospora caninum infection in dairy and beef cattle in Spain. International Journal for Parasitology 1999; 29: 1201–1208. [DOI] [PubMed] [Google Scholar]
- 23.Pascual-Velasco F.Q Fever [in Spanish]. Junta de Castilla y León: Heraldo de Zamora Press, 1996, pp. 118. [Google Scholar]
- 24.López-Gatius F, Pabon M, Almería S. Neospora caninum infection does not affect early pregnancy in dairy cattle. Theriogenology 2004; 62: 606–613. [DOI] [PubMed] [Google Scholar]
- 25.Trees AJ, Williams DJL. Endogenous and exogenous transplacental infection in Neospora caninum and Toxoplasma gondii. Trends in Parasitology 2005; 21; 558–561. [DOI] [PubMed] [Google Scholar]
- 26.Wapenaar W, et al. Use of an enzyme-linked immunosorbent assay in bulk milk to estimate the prevalence of Neospora caninum on dairy farms in Prince Edward Island, Canada. Canadian Veterinary Journal 2007; 48: 493–499. [PMC free article] [PubMed] [Google Scholar]
- 27.Gonzalez-Warleta M, et al. Anti-Neospora caninum antibodies in milk in relation to production losses in dairy cattle. Preventive Veterinary Medicine 2011; 101: 58–64. [DOI] [PubMed] [Google Scholar]
- 28.Haegeman B, Etienne RS. Entropy maximization and the spatial distribution of species. American Naturalist 2010; 74–90. [DOI] [PubMed] [Google Scholar]
- 29.Papes M, Baubert P. Modeling ecological niches from low numbers of occurrences: assessment of the conservation status of poorly known viverrids (Mammalia, Carnivora) across two continents. Diversity and Distributions 2007; 13: 890–902. [Google Scholar]
- 30.Pearson RG, et al. Predicting species' distributions from small numbers of occurrence records: a test case using cryptic geckos in Madagascar. Journal of Biogeography 2007; 34: 102–117. [Google Scholar]
- 31.Schares G, et al. Regional distribution of bovine Neospora caninum infection in the German state of Rhineland-Palatinate modelled by logistic regression. International Journal for Parasitology 2003; 33: 1631–1640. [DOI] [PubMed] [Google Scholar]
- 32.Rinaldi L, et al. Neospora caninum in pastured cattle: determination of climatic, environmental, farm management and individual animal risk factors using remote sensing and geographic information systems. Veterinary Parasitology 2005; 128: 219–230. [DOI] [PubMed] [Google Scholar]
- 33.Wouda W, Bartels CJM, Moen AR. Characteristics of Neospora caninum-associated abortion storms in dairy herds in the Netherlands (1995–1997). Theriogenology 1999; 52: 233–245. [DOI] [PubMed] [Google Scholar]
- 34.Thurmond MC, Anderson ML, Blanchard PC. Secular and seasonal trends of Neospora abortion in California dairy cows. Journal of Parasitology 1995; 81: 364–367. [PubMed] [Google Scholar]
- 35.Yanase T, et al. Seasonal variations in the presence of antibodies against Coxiella burnetii in dairy cattle in Hokkaido, Japan. Microbiology and Immunology 1997; 41: 73–75. [DOI] [PubMed] [Google Scholar]
- 36.Parisi A, et al. Diagnosis of Coxiella burnetii-related abortion in Italian domestic ruminants using single-tube nested PCR. Veterinary Microbiology 2006; 118: 101–116. [DOI] [PubMed] [Google Scholar]
- 37.Hassig M, Gottstein B. Epidemiological investigations of abortions due to Neospora caninum on Swiss dairy farms. Veterinary Record 2002; 150: 538–542. [DOI] [PubMed] [Google Scholar]
- 38.Toledo A, et al. Vector-borne and zoonotic diseases detection of Coxiella burnetii in ticks collected from Central Spain. Vector-Borne and Zoonotic Diseases 2009; 9: 465–468. [DOI] [PubMed] [Google Scholar]
- 39.Ruiz-Fons F, et al. Prevalence of Coxiella burnetti infection in wild and farmed ungulates. Veterinary Microbiology 2008; 126: 282–286. [DOI] [PubMed] [Google Scholar]
- 40.Almería S, et al. Red foxes (Vulpes vulpes) are a natural intermediate host of Neospora caninum. Veterinary Parasitolology 2002; 107: 287–294. [DOI] [PubMed] [Google Scholar]
- 41.Hughes JM, et al. Neospora caninum: detection in wild rabbits and investigation of co-infection with Toxoplasma gondii by PCR analysis. Experimental Parasitology 2008; 120: 255–260. [DOI] [PubMed] [Google Scholar]
- 42.Randolph SE. Tick and tick-borne disease systems in space and from space. Advances in Parasitology 2000; 47: 217–243. [DOI] [PubMed] [Google Scholar]