SUMMARY
This paper shows that control of foodborne disease outbreaks may be challenging even after establishing the source of infection. An outbreak of Salmonella Typhimurium U323 infections occurred in Denmark from March to September 2010, involving 172 cases. Before the detection of human cases, several positive isolates of the outbreak strain had been found in a particular pig slaughterhouse and thus early traceback, investigation and control measures were possible. Several batches of pork and pork products were recalled and the slaughterhouse was closed twice for disinfection. No single common food item was identified as the outbreak source, but repeated isolation of the outbreak strain from the slaughterhouse environment and in pork and products as well as patient interviews strongly suggested different pork products as the source of infection. Furthermore, a matched case-control study identified a specific ready-to-eat spreadable pork sausage (teewurst) as the source of a sub-outbreak (matched odds ratio 17, 95% confidence interval 2·1–130).
Key words: Epidemiology, foodborne infections, outbreaks, Salmonella (Typhimurium), Salmonella typing
INTRODUCTION
In 2009, S. Typhimurium was the most common type of Salmonella in Denmark with an incidence of 14/100 000 population [1]. During the same year, this serotype caused 5/13 salmonellosis outbreaks and 409/678 (60%) laboratory-confirmed outbreak-associated cases. From 2005 to 2010, S. Typhimurium was responsible for 29/63 salmonellosis outbreaks in the country. In 2009, due to two large outbreaks, the most common S. Typhimurium phage types were U292 and DT135 (28% and 13%, respectively) [2]. The phage type DT120 was detected in 8% of the human cases.
The majority of foodborne infections in Denmark are detected by the laboratory-based surveillance system or by direct reports from citizens to the Danish Veterinary and Food Administration (DVFA) or to the local medical officer. Cross-regional and national foodborne outbreaks are typically investigated in collaboration by the National Food Institute at the Technical University of Denmark (DTU Food), Statens Serum Institut (SSI) and DVFA in the Central Outbreak Management Group (COMG) which conducts weekly meetings for outbreak preparedness, response and investigation.
Isolates from the national Salmonella surveillance of food and animals in Denmark are serotyped and tested for antimicrobial resistance. Relevant isolates are further subtyped using methods such as phage-typing, multiple-locus variable-number tandem repeat analysis (MLVA) and pulsed-field gel electrophoresis (PFGE). S. Typhimurium strains from human patients are routinely MLVA-subtyped [3] and tested for antimicrobial resistance at SSI and phage-typed at DTU Food. Molecular typing of isolates from humans, food, animals and feed are compared via a shared database. Clusters of more than five patient isolates in a 2-week period with identical MLVA types are considered as potential outbreaks [4]. When a cluster is identified, the COMG is notified and an outbreak investigation is initiated. The core of an outbreak investigation consists of comparative typing of isolates from patients, food and animals, coupled with patient interviews, and descriptive and analytical patient, food and veterinary epidemiology. If relevant, traceback investigations and intensive investigation on farms, in restaurants, and grocery chains, etc. are conducted.
On 23 March 2010, DTU Food reported a steady occurrence of positive isolates of S. Typhimurium, of unknown phage type and resistant to ampicillin, streptomycin and sulphamethoxazole, in samples from mainly pork meat and products. Several isolates originated from a specific pig slaughterhouse A or a closely associated cutting plant. At this point, although no human cases had yet been identified, an investigation was initiated with the focus on food contamination. On 20 April, a total of 14 human cases with the outbreak type had been confirmed and an outbreak investigation began with the aim of describing the outbreak and identifying a common food exposure in order to guide control measures. A case-control study was undertaken to confirm an association between illness in a subgroup of patients and consumption of teewurst or ‘tea sausage’, a spreadable sausage made from fresh salted and smoked pork and beef which is fermented but not heat-treated. The producer of the teewurst had received pork from slaughterhouse A during the period that the outbreak strain had been isolated.
This paper reports the investigations of the entire outbreak.
METHODS
Microbiology
For human and veterinary isolates, MLVA was performed and allele numbers assigned using a modified version [5] of the method described by Lindstedt et al. [6]. Susceptibility to a standard panel of 15 antimicrobial agents was performed using Sensititre (TREK Diagnostic Systems Ltd, UK) as described in DANMAP [7].
Phage-typing of human and veterinary isolates was performed at DTU Food in accordance with international standards [8, 9]. The phage-typing lysis pattern of the outbreak strain is not described in the scheme from the World Health Organization Collaborative Centre for phage-typing of Salmonella –Health Protection Agency (HPA), Colindale, UK [10] and thus the phage type was first assigned as RDNC. Confirmatory phage-typing of a few isolates was performed at HPA in Colindale in order to assign a phage type.
The outbreak strain was defined by phage-type profile and the MLVA profile JPX.0007.DK (3-12-9-NA-211), allowing for variation at loci STTR6 and STTR10 [5]. As the MLVA profile is common among other S. Typhimurium phage types (e.g. DT120), phage-typing was important for the definition of cases.
Case definition and hypothesis generation
For this investigation, a case was defined as the outbreak strain (defined by the MLVA profile and phage type) being isolated from a specimen in a person residing in Denmark, who had not travelled abroad in the 7 days before disease onset.
Cases were interviewed by telephone using an S. Typhimurium specific hypothesis-generating questionnaire.
Case-control study
A matched case-control study was undertaken to test the hypothesis that human cases diagnosed between 20 July and 14 September belonged to a sub-outbreak caused by teewurst. Controls were selected from the Danish Population Registry [11], matched to cases by municipality, gender, and date of birth. Two controls were interviewed for each case. Controls who reported foreign travel or who had experienced symptoms of gastrointestinal illness in the 7 days prior to interview were excluded. Participants were interviewed by phone between 24 August and 17 September using a tailored questionnaire focusing on consumption of various types of meats, cold cuts and a range of other exposures in the week prior to onset of disease (cases) or interview (controls).
Data were entered into an EpiData database [12]. Statistical analyses were conducted in Stata v. 10 (StataCorp, USA). In order to examine relationships between each exposure and disease, matched odds ratios (mORs) and 95% confidence intervals (CIs) were calculated. Conditional logistic regression analysis was performed to investigate possible interaction between different exposure types.
Environmental and herd investigations
As part of the national Salmonella control programme, all Danish slaughterhouses are required to take swab samples from slaughtered carcases and, following the ‘own-check’ programme, analyse these for the presence of Salmonella. At larger slaughterhouses (the size of slaughterhouse A), five samples are taken daily. All Salmonella isolates deriving from the own-check programme and export control must be sent for subtyping at DTU Food. These isolates are included in the surveillance of foodborne outbreaks. Further, as part of the case-by-case control programme, spot checks are performed where Danish and imported meat is tested for Salmonella. If a sample is found positive, the whole batch of meat is subjected to a risk analysis at DTU Food and results are forwarded to DVFA, where it is determined if the affected batch of meat must be withdrawn from the market. Withdrawn batches are traced using the traceability systems implemented in all food establishments according to EU legislation. Fresh meat is traceable to farm-level, but the likelihood of cross-contamination at the slaughterhouse should also be considered. During foodborne disease outbreaks, all meat and products found positive for the outbreak strain will be traced. Environmental investigations including swabs from slaughterhouse equipment are undertaken when in-house contamination is suspected. For this outbreak, upon detection of S. Typhimurium in meat samples, traceback investigations were performed in each instance. Because the majority of contaminated meat was traced back to slaughterhouse A and the associated cutting plant, further environmental investigation was initiated at slaughterhouse A in early May.
Herds delivering pigs to slaughterhouse A were identified by a unique herd identification number given to all animal herds in Denmark. Special attention was given to herds which had delivered pigs to the slaughterhouse on dates immediately prior to detection of the outbreak strain in food or at the slaughterhouse. For each of these herds, the likelihood of the herd being the source of contamination was assessed based on delivery size and pattern to slaughterhouse A as well as herd mortality and previous Salmonella incidence (based on serological monitoring and Salmonella culture follow-up in herds with moderate and high seroprevalence). Generally in Denmark, serological monitoring of Salmonella antibody levels in meat juice samples from slaughtered pigs is used to classify farms into three Salmonella levels [13]. At the time of the outbreak, farms placed in the two highest levels were followed up by bacteriological sampling and culturing. A herd from which the outbreak strain had previously been isolated was subjected to a full mapping of trade patterns. Furthermore, control visits were conducted by the veterinary authorities to confirm register data and monitor clinical signs of salmonellosis.
RESULTS
Microbiology
The S. Typhimurium outbreak strain was characterized as phage type U323 at HPA, Colindale; a phage type not previously found in veterinary, food or human isolates in Denmark. The majority of human isolates (n = 132) were resistant to ampicillin (A), streptomycin (S) and sulphamethoxazole (Su). The resistance profile of human isolates changed during the outbreak, with more SSu-resistant and fully sensitive isolates being observed in the second half of the outbreak period. A similar change in resistance pattern over time was also observed in food isolates.
Descriptive epidemiology and hypothesis generation
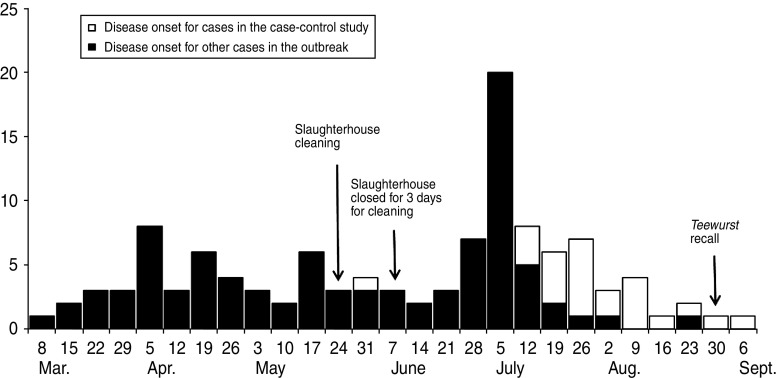
The outbreak comprised of a total of 172 cases. Patient disease onset dates (n = 116) ranged from 13 March to 10 September (Fig. 1). Sixty-one percent (n = 107) of patients were female. Ages ranged from 8 months to 92 years (median 51 years). Nineteen percent (n = 33) were children aged <15 years. Twenty-six (15%) cases were hospitalized. Eight (5%) patients in the cohort died.
Fig. 1.
Confirmed cases of S. Typhimurium U323 by week of onset of illness, Denmark, 13 March to 10 September 2010 (n=116). Date of onset of illness missing for 56 cases.
Patients' addresses were distributed throughout Denmark. However, higher incidences were observed in eastern and southern parts of the country with the highest incidence (7 cases/100 000 population) observed in the same region (one of five administrative regions in Denmark) where slaughterhouse A is located.
A total of 133 patients were interviewed by telephone. None of the interviewed patients reported that they were vegetarians or that they did not eat pork for religious or other reasons. Ten (8%) patients had eaten takeaway food in the 7 days preceding illness and 11 (8%) had eaten in cafes or restaurants. No particular catering premise or chain of premises was mentioned by more than one patient and there was no evident link between illness and attending events. Ninety-one percent (121/133) of patients had eaten fresh pork in the 7 days prior to illness; primarily minced meat (42%) or pork chops (20%). Sixty-two percent (83/133) and 42% (56/133) had eaten fresh beef and chicken, respectively, in the week before disease onset. Ninety-two percent (122/133) had eaten some type of ready-to-eat delicatessen meat or paté, containing pork, in the 7 days before they became ill. The questionnaire section on ready-to-eat delicatessen meat included a question specifically concerning the consumption of teewurst. None of the initially interviewed patients (excluding those from the case-control study and onwards) had eaten teewurst. In July, it was noted that an increased number of patients began mentioning consumption of teewurst, which led to the initiation of a case-control study (see below).
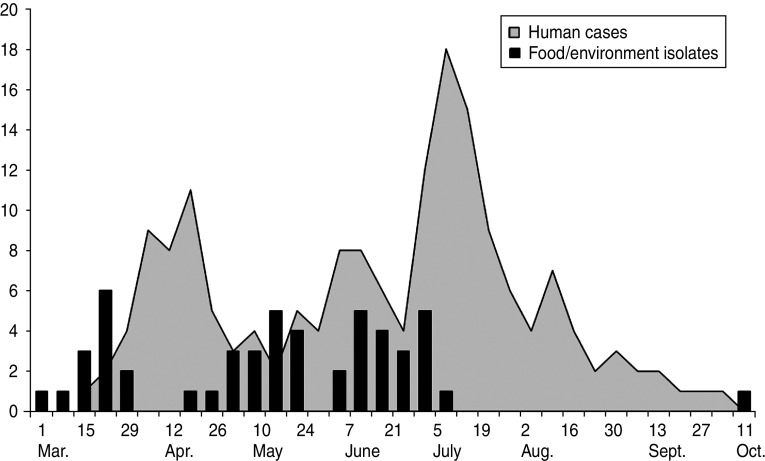
An increased identification of human isolates of S. Typhimurium U323 corresponded in time (with an approximate 3-week delay) with an increased finding of veterinary/environmental isolates (Fig. 2).
Fig. 2.
The relationship between discovery of positive food/environmental and human isolates of the outbreak strain by week of laboratory date (received), Denmark, 2010.
An urgent enquiry through the Epidemic Intelligence Information System (EPIS), an outbreak notification system hosted by the European Centre for Disease Control (ECDC), identified 12 possible cases in Sweden, suspected of being infected by food from a specific restaurant. A potential connection between the restaurant and slaughterhouse A was investigated, but a direct link could not be established even though meat from the slaughterhouse had been exported to Sweden during the relevant period.
Case-control study
Nineteen cases with a diagnosis date between 20 July and 14 September 2010 and 41 controls were interviewed for the case-control study. A total of three controls were excluded from the analysis; two due to symptoms of gastrointestinal illness and one because of lack of knowledge about foods consumed.
The median age of patients included in the case-control study was 43 years (range 8 months to 87 years). Ten (53%) lived in the southern Jutland region where a frequent distribution of patients was observed in the last months of the outbreak. Interviewed patients' disease onset dates ranged from 1 June to 10 September (Fig. 1).
Results of the case-control interviews (Table 1) showed that 9/19 patients and no controls had eaten brand X teewurst in the 7 days prior to illness or interview, respectively. Univariate analyses revealed a statistically significant association between illness and having bought brand X teewurst in the 7 days before disease onset or interview, respectively. A statistically significant association between illness and ever having eaten brand X teewurst was also observed. Consumption of fresh chicken, pork and beef as well as a range of ready-to-eat products and vegetables was not associated with illness. Consumption of salami and pork tenderloin in the week prior to disease onset was shown to be significantly related to illness, although in a conditional logistic regression analysis, only having bought brand X teewurst was independently associated with being a case (OR 25, 95% CI 2·0–310, P < 0·000).
Table 1.
Risk exposures for cases of Salmonella Typhimurium U323 sub-outbreak and matched controls (single risk variable analysis)
| Risk factor | No. cases exposed/ total | No. controls exposed/total | mOR | 95% CI |
|---|---|---|---|---|
| Chicken | 10/19 | 20/38 | 1·0 | 0·4–2·8 |
| Pork | 18/19 | 35/38 | 1·5 | 0·2–14·4 |
| Beef | 17/19 | 27/38 | 2·6 | 0·7–15·4 |
| Bacon | 8/19 | 16/38 | 1·0 | 0·4–2·8 |
| Pork mince | 11/19 | 17/38 | 1·6 | 0·6–4·7 |
| Pork chops | 4/19 | 7/38 | 1·2 | 0·3–4·2 |
| Pork sausages | 12/19 | 17/38 | 2·0 | 0·7–6·1 |
| Sliced ham | 4/19 | 8/38 | 1·0 | 0·2–4·3 |
| Salami* | 13/19 | 12/38 | 6·8 | 1·5–31·9 |
| Liver paté | 15/19 | 22/38 | 4·0 | 0·8–20·0 |
| Pork tenderloin* | 6/19 | 3/35 | 5·3 | 1·1–26·8 |
| Brand X teewurst | 9/19 | 0/38 | –† | – |
| Buying brand X teewurst * | 9/19 | 2/38 | 16·5 | 2·1–131·4 |
| Ever having eaten brand X teewurst * | 11/19 | 7/38 | 14·2 | 1·8–113·6 |
| Carrots | 11/19 | 29/38 | 0·4 | 0·1–1·6 |
| Tomatoes | 12/19 | 29/38 | 0·5 | 0·1–2·0 |
| Lettuce | 11/19 | 26/38 | 0·6 | 0·2–2·4 |
mOR, Matched odds ratio; CI, confidence interval.
Significant association between illness and exposure to variable.
Unable to estimate as none of the controls in the matched pairs had eaten this food.
Environmental investigations and interventions
From March 2010 and onwards, the outbreak strain was identified in a total of 113 samples; four environmental samples from slaughterhouse A and 109 meat samples, mainly pork, of which 96 were sampled directly at slaughterhouse A or could be traced back there. Positive meat types included mainly minced pork, pork belly, pork loin and loin back ribs. A small number of positive beef meat samples were also identified, although all of these were shown to have a potential cross-contamination route with pork from slaughterhouse A. Investigation of slaughterhouse A showed positive discovery of the outbreak strain in swabs from equipment and meat samples, even after closing down production for thorough cleaning and disinfection. It was concluded that the establishment was most likely contaminated. Repeated cleaning and disinfection was performed and alterations in equipment and procedures were implemented (Fig. 1). From the beginning of July, no further positive samples of the outbreak strain were found at slaughterhouse A.
On 8 July, a press statement was issued jointly by the DVFA and SSI, notifying the public about the salmonellosis outbreak and the link to consumption of pork meat from slaughterhouse A. In addition to describing the outbreak investigation and the action taken to control the outbreak, the statement also contained detailed guidelines on how to prevent infection with Salmonella.
Several samples of the teewurst, produced between 24 August and 3 September 2010, were analysed for the presence of Salmonella, but none were found positive.
Herd investigation
How the S. Typhimurium strain was introduced to slaughterhouse A could not be established with certainty. No herds delivering pigs to the contaminated slaughterhouse were found to be an obvious source of contamination. Among herds with frequent delivery to the slaughterhouse, one particular pig herd had several delivery dates correlating with isolation of the outbreak strain in meat or the slaughterhouse environment. However, this herd had a low Salmonella seroprevalence and a veterinary inspection did not reveal any signs of clinical salmonellosis. An additional investigation found no direct link between the slaughterhouse contamination and a pig farm from which the outbreak strain was isolated in February 2010 as no pigs from this herd had been slaughtered at slaughterhouse A in the 3 months prior to the dates of positive Salmonella findings. However, slaughter of pigs from the culture-positive herd was associated with detection of a one-locus MLVA variation of the outbreak strain in meat which could be traced back to other slaughterhouses. No patients with a Salmonella infection fitting this exact one-locus variation of the strain were identified.
DISCUSSION
We report a relatively large outbreak of salmonellosis in Denmark in a situation where the outbreak strain was found in several samples of pork meat and pork products before the first human cases appeared. Because of the surveillance in Danish meat-handling establishments, combined with an intensive typing effort of isolates, the COMG considered an outbreak to be likely and was able to prepare the investigatory action and possible control measures at an early stage. In essence, the outbreak was detected and resolved even before the first patients were detected. However, in spite of this, controlling the outbreak proved difficult and it ended up lasting for almost 7 months.
A total of 172 cases of S. Typhimurium U323 were reported between March and September 2010. Due to previous positive findings in the surveillance system, human cases were a priori expected to have an association with consumption of pork meat or pork products. Results from patient interviews were compatible with the pork/pork product hypothesis: none of the 133 patients interviewed excluded pork from their diet and almost all had eaten some type of pork in the week before disease onset; either fresh, cooked meat or ready-to-eat products. The demographic characteristics of patients also corresponded to what has been learned from previous pork-associated Salmonella outbreaks in Denmark, i.e. geographically widespread distribution (with a cluster of cases in the region near slaughterhouse A), median age >35 years, and slightly more women than men [14]. Results of the hypothesis-generating interviews were probably subject to mistaken recall as people were asked to remember food they had consumed up to a month before the interview. In this case, however, patients were also asked for their usual shopping and eating habits, possibly reducing this effect.
During the investigation, it was not possible to pinpoint a single specific food product, brand or grocery chain as the source of infection which supports the most likely theory that the outbreak was caused by slaughterhouse contamination, and subsequent contamination of a broad range of fresh pork and pork products as well as some ready-to-eat pork products further processed in other establishments before reaching the consumer. A case-control study, focusing on pork consumption in general was not undertaken, as results from previous outbreaks indicate that such studies are likely to be inconclusive because of the frequent consumption of pork products in Denmark [15]. For the current outbreak, the epidemic curve showed a marked peak of illness in week 26 (i.e. 28 June to 4 July) which we consider to be caused by exposure to an infected ready-to-eat pork product as described below.
In July, it was noted that several cases reported eating a specific ready-to-eat spreadable pork sausage called teewurst. Teewurst is a mixture of fresh pork, beef and fat which is salted, seasoned, fermented and then smoked. The finished product is not heat-treated and has a low acid content. The association between teewurst and foodborne diseases is well documented and it has been linked to cases of both Shiga-toxin producing E. coli [16] and haemolytic uraemic syndrome [17]. Because teewurst contains fresh pork, it appears to be a general risk food item, although a study by Dourou et al. [18] failed to show that teewurst provides a favourable environment for the survival of S. Typhimurium.
Our case-control study showed that infections of S. Typhimurium U323 in July and August were most likely caused by consumption of brand X teewurst. The company that produced the teewurst had frozen pork meat supplied by slaughterhouse A, without knowledge of the potential Salmonella contamination. Patients who submitted faecal samples in July and August had a different geographical distribution and were also younger than the average of the main outbreak, although this was not statistically significant. We concluded that consumption of teewurst caused a sub-outbreak in a younger and more geographically distinct group of people. Following the preliminary results of the case-control study, the factory producing the teewurst was notified and the product was recalled from consumers on 3 September. A notice was published on the DVFA website, informing consumers about the symptoms of salmonellosis and advising them to discard any unopened or remaining brand X teewurst or return the packages to the place of purchase. Batches of teewurst produced in the period before the recall were negative for Salmonella; however, due to the short shelf-life of the product, none of the presumed contaminated batches from July and early August were available for analysis. Interestingly, only two further cases belonging to the outbreak were reported following the last patient known to have eaten teewurst. These cases had not eaten teewurst, although one patient did report having eaten another type of sausage from the same producer. We therefore believe that not only did the spreadable pork sausage cause a sub-outbreak but also that this sub-outbreak constituted the end of the main outbreak.
Intensive cleaning and disinfection of slaughterhouse A as well as recall of several batches of meat and products were important contributing factors in controlling the outbreak. Since the beginning of July, no positive isolates were found at slaughterhouse A and following this, it was concluded that it was safe to resume normal production.
Although the origin of the outbreak was known with near certainty to be contamination of slaughterhouse A, it was still not possible to prevent the occurrence of human cases. This is partly due to the fact that contaminated meat was already on the market at the time when the first positive isolates were identified and partly because there are no EU or national legislative restrictions concerning the presence of Salmonella in fresh meat (unlike those for meat preparations and some meat products, including minced meat). Another factor contributing to the length of the outbreak is the fact that the pork meat had been sold through a variety of different retail outlets. Consumers would have been unable to identify the meat or product as originating from slaughterhouse A or the associated cutting plant due to further cutting and processing in other establishments. From an environmental perspective, the equipment in some slaughterhouses is very difficult to clean thoroughly, which may lead to recurrent problems with contamination. As no single product was identified as the source of infection, the original theory of an in-house infection contaminating a range of fresh and ready-to-eat products has strong support. We note that this outbreak in many ways bears resemblance to a previous S. Typhimurium outbreak, described by Bruun et al. [19], which was also caused by pork or pork products in different forms, originating from the same slaughterhouse, as occurred in the current outbreak. In this case, food traceback and comparative microbiological typing were also instrumental in solving the outbreak.
In conclusion, this national outbreak of S. Typhimurium U323 in Denmark has demonstrated how a combination of typing Salmonella isolates from farm-to-fork and from the human population can provide early warning of a salmonellosis outbreak. It also highlights the importance of national Salmonella surveillance which allowed identification of the slaughterhouse contamination and provided the COMG with valuable information to initiate investigative measures. In spite of the existence of these systems, tracing pork meat that has entered the production chain still poses a significant challenge. If feasible, adoption of a standardized automated system across the EU, with detailed product and distribution information, for tracing products might prove worthwhile. Currently, this is not possible in the EU and such systems are only as good as the data provided by the operators. At present, by the time enough evidence has been gathered to issue a product recall, products with a short shelf-life (such as fresh meat) are most likely to have been consumed.
In this outbreak, early warnings from the Salmonella surveillance system were not sufficient to prevent the outbreak from lasting almost 7 months. State-of-the-art surveillance, typing, epidemiology and food traceback allowed us to firmly establish the source of the outbreak and, in essence, solve it almost before it became evident; however, legislative measures and some delays in traceback did not allow for sufficient control, resulting in one of the longest lasting Salmonella outbreaks in Denmark.
ACKNOWLEDGEMENTS
We thank the regional clinical microbiological laboratories for submitting clinical Salmonella isolates to Statens Serum Institut for national surveillance. The authors are also grateful to the regional veterinary and food administration centres and meat inspection units for general assistance and to Charlotte Kjelsø, Jeppe Nørgaard Rasmussen, Henriette Juel Larsen, Dominik Wessely, Lærke Navntoft and Susanne Hansen at the Outbreak Unit, Statens Serum Institut, for conducting telephone interviews.
DECLARATION OF INTEREST
None.
REFERENCES
- 1.Anon. Annual report on zoonoses in Denmark 2009. National Food Institute, Technical University of Denmark, 2010. [Google Scholar]
- 2.Ethelberg S, et al. Large outbreaks of Salmonella Typhimurium infection in Denmark in 2008. Eurosurveillance 2008; 13: 1–2. [PubMed] [Google Scholar]
- 3.Torpdahl M, et al. Tandem repeat analysis for surveillance of human Salmonella Typhimurium infections. Emerging Infectious Diseases 2007; 13: 388–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ethelberg S, et al. Outbreak with multi-resistant Salmonella Typhimurium DT104 linked to carpaccio, Denmark, 2005. Epidemiology and Infection 2007; 135: 900–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Larsson JT, et al. Development of a new nomenclature for Salmonella typhimurium multilocus variable number of tandem repeats analysis (MLVA). Eurosurveillance 2009; 14: 19174. [PubMed] [Google Scholar]
- 6.Lindstedt BA, et al. Multiple-locus variable-number tandem-repeat analysis of Salmonella enterica subsp enterica serovar Typhimurium using PCR multiplexing and multicolour capillary electrophoresis. Journal of Microbiological Methods 2004; 59: 163–172. [DOI] [PubMed] [Google Scholar]
- 7.DANMAP 2009. Use of antimicrobial agents and occurrence of antimicrobial resistance in bacteria from animals, foods and humans in Denmark, 2009. 2010, ISSN 1600-2032.
- 8.Anderson ES, et al. Bacteriophage-typing designations of Salmonella typhimurium. Journal of Hygiene (London) 1977; 78: 297–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Callow BR. A new phage-typing scheme for Salmonella typhimurium. Journal of Hygiene (London) 1959; 57: 346–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baggesen DL, et al. Phage typing of Salmonella Typhimurium – is it still a useful tool for surveillance and outbreak investigation? Eurosurveillance 2010; 15: 19474. [PubMed] [Google Scholar]
- 11.Pedersen CB, et al. The Danish Civil Registration System. A cohort of eight million. Danish Medical Bulletin 2006; 53: 441–449. [PubMed] [Google Scholar]
- 12.Lauritsen JM (ed.). EpiData Data Entry, Data Management and Basic Statistical Analysis System. Odense, Denmark: EpiData Association, 2000–2008. [Google Scholar]
- 13.Wegener HC, et al. Salmonella control programs in Denmark. Emerging Infectious Diseases 2003; 9: 774–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anon. Dietary habits in Denmark 2003–2008. Main results. National Food Institute, Technical University of Denmark, 2010. [Google Scholar]
- 15.Torpdahl M, et al. A regional outbreak of S. Typhimurium in Denmark and identification of the source using MLVA typing. Eurosurveillance 2006; 11: 134–136. [PubMed] [Google Scholar]
- 16.Werber D, et al. Shiga toxin-producing Escherichia coli infection in Germany – different risk factors for different ages. American Journal of Epidemiology 2006; 165: 425–434. [DOI] [PubMed] [Google Scholar]
- 17.Ammon A, Petersen LR, Karch H. A large outbreak of haemolytic uremic syndrome caused by an unusual sorbitol-fermenting strain of Escherichia coli O157:H. Journal of Infectious Diseases 1999; 179: 1274–1277. [DOI] [PubMed] [Google Scholar]
- 18.Dourou D, et al. Behavior of Escherichia coli 0157:H7, Listeria monocytogenes and Salmonella Typhimurium in teewurst; a raw spreadable sausage. International Journal of Food Microbiology 2009; 130: 245–250. [DOI] [PubMed] [Google Scholar]
- 19.Bruun T, et al. An outbreak of Salmonella Typhimurium infections in Denmark, Norway and Sweden, 2008. Eurosurveillance 2009; 12: 19147. [PubMed] [Google Scholar]