SUMMARY
Surveillance is integral for the monitoring and control of infectious diseases. We conducted prospective laboratory surveillance of methicillin-resistant Staphylococcus aureus (MRSA) in five Singaporean public-sector hospitals from 2006 to 2010, using WHONET 5.6 for data compilation and analysis. Molecular profiling using multilocus variable-number tandem-repeat analysis, staphylococcal cassette chromosome mec classification and multilocus sequence typing was performed for a random selection of isolates. Our results showed overall stable rates of infection and bacteraemia, although there was significant variance among the individual hospitals, with MRSA rates increasing in two smaller hospitals and showing a trend towards decreasing in the two largest hospitals. The proportion of blood isolates that are EMRSA-15 (ST22-IV) continued to increase over time, slowly replacing the multi-resistant ST239-III. A new MRSA clone – ST45-IV – is now responsible for a small subset of hospital infections locally. More effort is required in Singaporean hospitals in order to reduce the rates of MRSA infection significantly.
Key words: Antibiotic resistance, bacterial typing, methicillin-resistant S. aureus (MRSA)
Methicillin-resistant Staphylococcus aureus (MRSA) remains a major global cause of healthcare-associated infections despite advances in rapid diagnostics and infection control practices [1]. This is partially attributable to the sustained need for intravascular devices in hospitals, as well as the continuing emergence and dissemination of new clones of community-associated MRSA (CA-MRSA) and healthcare-associated MRSA (HA-MRSA) in healthcare settings [2]. UK-EMRSA-15 (ST22-IV) – first observed in Singaporean hospitals in the early 2000s – competed successfully against the endemic ST239-III clone, replacing >40% of infections caused by the latter by 2006 [3]. We report on the results of MRSA surveillance in five of six Singaporean public-sector hospitals between 2006 and 2010. These comprised a tertiary hospital (hospital 4:1600 beds), a dedicated maternity and child hospital (hospital 3:900 beds), and three secondary hospitals (hospital 1:400 beds; hospital 2:900 beds; hospital 5:1450 beds).
Microbiological data were extracted from the laboratory information system of each hospital and analysed quarterly using WHONET 5.6 (WHO, Switzerland), with duplicates eliminated monthly according to the Clinical and Laboratory Standards Institute (CLSI) guidelines [4]. All laboratories predominantly used disk susceptibility testing following CLSI guidelines [5]. Denominator data in the form of hospital inpatient days were obtained from hospitals' administrative records. Combined and individual hospital MRSA incidence densities were explored independently for trend over time by linear regression, corrected for first-level autocorrelation using the Cochrane–Orcutt estimation.
There were two separate prospectively defined collections of clinical MRSA isolates that were typed. First, MRSA isolates from all routine clinical (but not surveillance) samples cultured within a pre-defined 1-month period were collected from all hospitals in 2007 and 2008. Lack of funding precluded similar sampling in other years. Second, 20 MRSA blood isolates each year were randomly obtained from hospitals 2, 4 and 5 outside of the collection periods for 2007 and 2008 listed above. Minimum inhibitory concentration (MIC) testing via Sensititre broth microdilution plates (TREK Diagnostic Systems, USA) was performed for vancomycin, ciprofloxacin, erythromycin, clindamycin, gentamicin, and sulfamethoxazole. All isolates were typed using multilocus variable-number tandem-repeat (VNTR) analysis (MLVA) [6], with two representative isolates from clonal clusters from the first routine clinical sample collection in addition to all 100 blood isolates from the second collection further typed via multilocus sequence typing (MLST) [7]. Staphylococcal cassette chromosome mec (SCCmec) type was classified using a published multiplex PCR strategy [8]. The major antibiotic resistance profiles of ST22 strains identified on MLST were then used to classify MRSA blood isolates within the WHONET files into ST22 and non-ST22 MRSA groups, respectively.
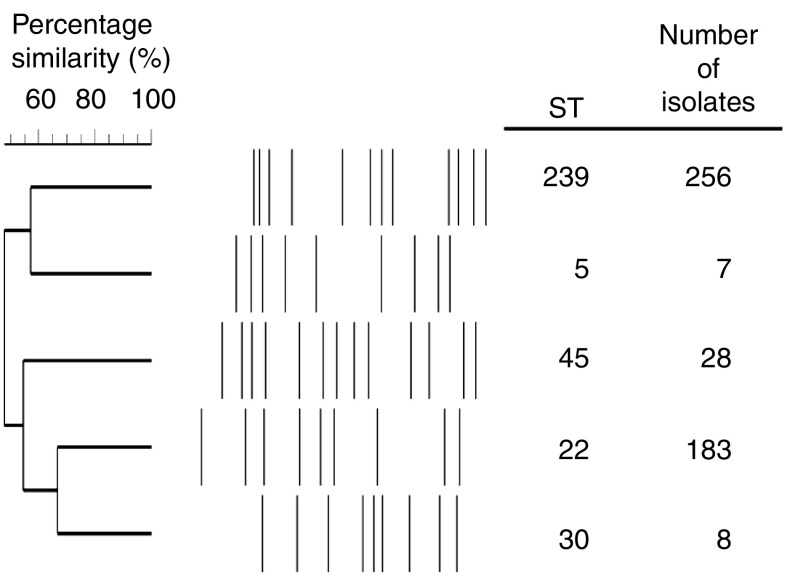
There were 190 and 204 MRSA isolates collected in 2007 and 2008, respectively. MLVA and MLST combined results are displayed in Figure 1, and the antibiotic susceptibility results are shown in Table 1. The two major MRSA clones remain ST239-III and ST22-IV, comprising 88·9% of all typed isolates. Minor clones were ST45-IV, ST5-IV and ST30-IV, the latter probably representing CA-MRSA. Ten isolates with unique MLVA profiles were not further typed. For the separate MRSA blood culture collection, 44 (44·0%) and 51 (51·0%) isolates were ST239-III and ST22-IV, respectively, with one ST5-IV and four ST45-IV isolates. Using the typical ST22-IV antibiogram (resistant to ciprofloxacin, sensitive to gentamicin and cotrimoxazole, variable susceptibility to erythromycin but all erythromycin-resistant isolates also possessing inducible resistance to clindamycin) to classify MRSA blood isolates from the WHONET database revealed that the proportion of MRSA that is ST22 had increased from 39·1% in 2006 to 49·7% by 2010, although this was not universal in all hospitals, and appeared to be a consequence of decline in numbers of non-ST22 isolates (Fig. 2).
Fig. 1.
Multilocus variable-number tandem-repeat analysis fingerprints of major methicillin-resistant Staphylococcus aureus clones in Singaporean hospitals, 2006–2010. ST, Multilocus sequence type.
Table 1.
Antibiotic susceptibility profile of major Singaporean hospital MRSA clones
| Sequence type | SCCmec | No. of isolates tested | Vancomycin (MIC50/90 mg/l) | Gentamicin (MIC50/90 mg/l) | Erythromycin (MIC50/90 mg/l) | Clindamycin (MIC50/90 mg/l) | Ciprofloxacin (MIC50/90 mg/l) | Cotrimoxazole (MIC50/90 mg/l) |
|---|---|---|---|---|---|---|---|---|
| 5 | IV | 7 | Susceptible (1/−) | Susceptible (0·5/−) | Variable (⩾32/−) | Susceptible* (0·125/−) | Resistant (⩾16/−) | Resistant (⩾32/−) |
| 22 | IV | 183 | Susceptible (0·5/1) | Susceptible (0·25/0·5) | Variable (⩾32/ ⩾ 32) | Susceptible* (0·125/0·25) | Resistant (⩾16/ ⩾ 16) | Susceptible (⩽0·25/ ⩽ 0·25) |
| 30 | IV | 8 | Susceptible (0·5/−) | Susceptible (0·5/−) | Susceptible (0·25/−) | Susceptible (0·125/−) | Susceptible (0·5/−) | Susceptible (⩽0·25/−) |
| 45 | IV | 28 | Susceptible (0·5/1) | Variable (⩾32/ ⩾ 32) | Variable (⩾32/ ⩾ 32) | Variable* (0·125/ ⩾ 16) | Resistant (⩾16/ ⩾ 16) | Variable (⩽0·25/16) |
| 239 | III | 256 | Susceptible (1/1) | Resistant (⩾32/ ⩾ 32) | Resistant (⩾32/ ⩾ 32) | Variable* (⩾16/ ⩾ 16) | Resistant (⩾16/ ⩾ 16) | Resistant (⩾32/ ⩾ 32) |
Isolates susceptible to clindamycin via minimum inhibitory concentration (MIC) criteria possess inducible resistance to the drug on disk diffusion testing.
Fig. 2.
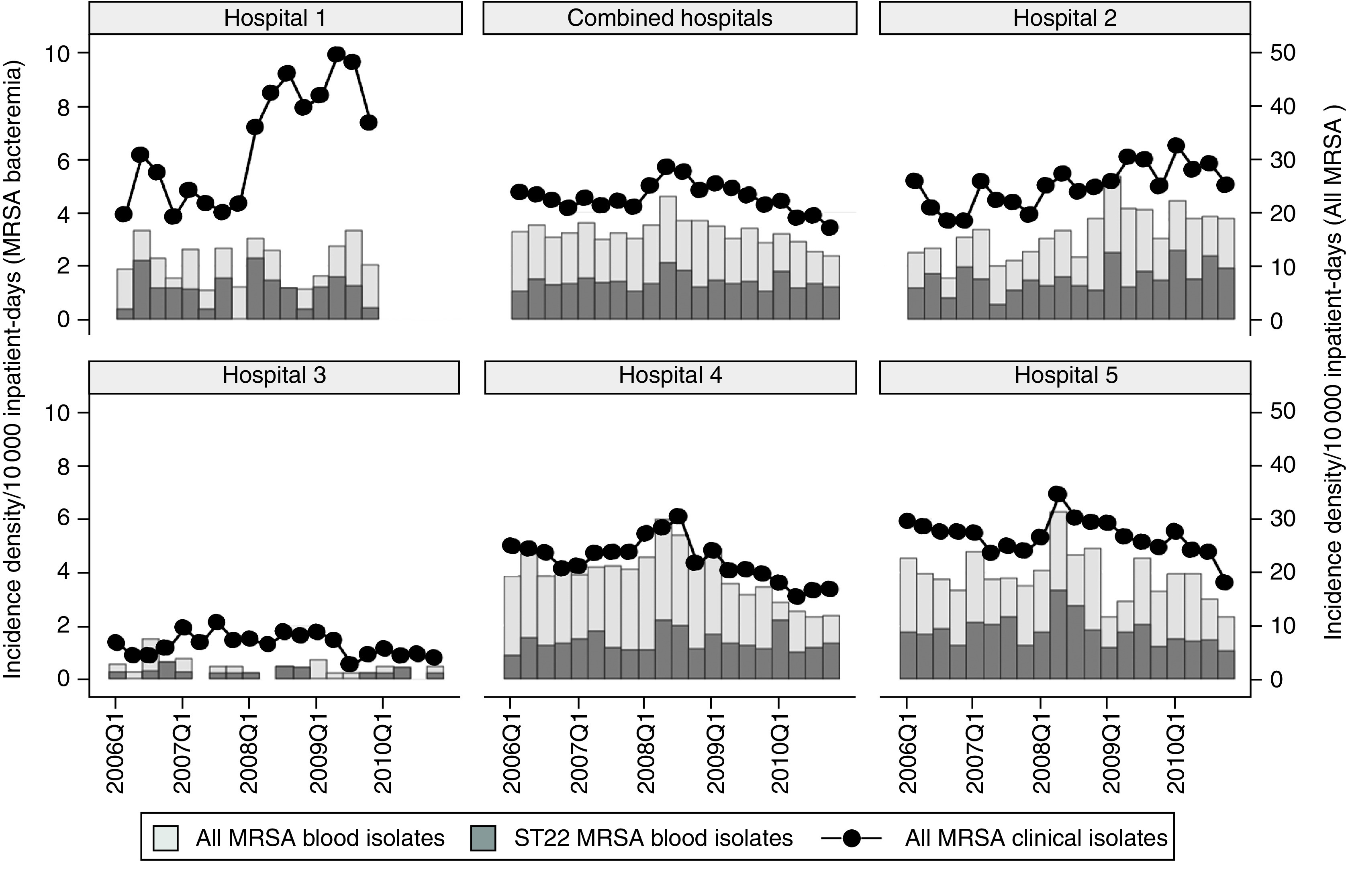
Incidence density of all clinical methicillin-resistant Staphylococcus aureus (MRSA) isolates, MRSA blood isolates and EMRSA-15 blood isolates in Singaporean hospitals. No data was available for hospital 1 in 2010 as it had closed in early 2010.
Overall clinical MRSA incidence density had decreased since 2008, although over the longer 5-year period, this was not significant (Fig. 2; combined clinical isolates: coefficient −0·05, 95% CI −0·11 to 0·02, P = 0·133). This phenomenon was mainly driven by changes within the much larger hospital 4 (coefficient −0·042, 95% CI −0·091 to 0·007, P = 0·086) and hospital 5 (coefficient −0·034, 95% CI −0·076 to 0·008, P = 0·103); MRSA rates remained stable at hospital 3 and increased in hospital 1 (coefficient 0·163, 95% CI 0·043–0·283, P = 0·011) and hospital 2 (coefficient 0·046, 95% CI 0·023–0·69, P = 0·001).
Our surveillance demonstrated stable overall MRSA rates in Singaporean hospitals, with mixed success in MRSA control in individual hospitals. Hospitals 4 and 5 had initiated specific MRSA control programmes since July 2007 and January 2008, respectively, with separate public launches of hospital-wide hand hygiene campaigns in April 2009 as well as focused active surveillance for MRSA in patients transferred into and out of the intensive care units. These appeared to correlate with decreasing trends of MRSA clinical isolates and bacteraemia since mid-2008. In the other hospitals, no specific MRSA control programmes were launched during this period, with resultant MRSA trends that were stable or increasing over time. The reason for the steep increase in MRSA incidence density in hospital 1 is unknown. It is clear that MRSA remains a significant problem locally, with bacteraemia rates that are more than three times higher than comparable UK National Health System acute trusts over the same period [9].
Although the relative proportion of UK-EMRSA-15 has increased since 2006, the rate of replacement of other local clones has slowed considerably. The reasons for this are unclear at present, although it is plausible that other MRSA clones had evolved in the face of this competitive threat. Interestingly, the incidence density of UK-EMRSA-15 from blood isolates had either increased or remained stable in all hospitals, even in hospitals 4 and 5 where progress in terms of reduction of all MRSA bacteraemia cases were seen. This suggests that UK-EMRSA-15 is harder to control compared to the multiresistant ST239 clone, a factor that underlies its success vis-à-vis other local endemic MRSA clones.
A new MRSA clone, ST45-IV, is now present in a relatively small proportion of isolates that had been typed via molecular methods. This clone was probably imported, as there are reports of regional spread of ST45 HA-MRSA [10]. Unfortunately, the actual dynamics of spread of this clone cannot be ascertained with the current data.
Local MRSA remained sensitive to vancomycin, with MIC50/90 of 1/1 mg/l based on broth microdilution. There was no significant difference between the different clones with regard to vancomycin MIC.
The major limitations of this surveillance are that only a small number of isolates were typed, and the sampling was not systematic. There was also a lack of clinical and epidemiological detail that might have permitted better categorization. Nonetheless, the results were sufficient to highlight the trends of the major circulating MRSA clones, and perhaps to identify a potential emerging clone. Several ST30-IV isolates were identified, and on chart review, these represented community infections that necessitated hospitalization rather than in-hospital transmission. On the other hand, the majority of the ST45-IV and ST5-IV clones appeared to be isolated from healthcare-associated infections on chart review.
In conclusion, continued passive and molecular surveillance of MRSA will be useful in monitoring future trends of the major circulating clones. More effort is required in Singaporean hospitals in order to reduce the rates of MRSA infection significantly.
ACKNOWLEDGEMENTS
Other members of the Network for Antimicrobial Resistance Surveillance (Singapore) include Nancy Tee (KK Hospital), Roland Jureen (National University Health System), and Joey Chan (Khoo Teck Puat Hospital). We thank the many dedicated staff who helped with data collection and technical issues, in particular Ms. Mee-Lee Tan and Dr Andrea Kwa. L.Y.H. has received research funding and speakers' honoraria from Pfizer, AstraZeneca, Janssen & Cilag, and Merck, Sharpe & Dohme.
DECLARATION OF INTEREST
This work was funded by the following grants: SingHealth Foundation Grant 2006, Ministry of Health (Singapore) Healthcare Quality Improvement Fund 2006, and educational grants from Pfizer Singapore, Janssen-Cilag, Merck Sharpe & Dohme, and AstraZeneca.
REFERENCES
- 1.Gould IM, et al. Cost of healthcare-associated methicillin-resistant Staphylococcus aureus and its control. Clinical Microbiology and Infection 2010; 16: 1721–1728. [DOI] [PubMed] [Google Scholar]
- 2.Monecke S, et al. A field guide to pandemic, epidemic and sporadic clone of methicillin-resistant Staphylococcus aureus. PLoS One 2011; 6: e17936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hsu LY, et al. Evolving EMRSA-15 epidemic in Singapore hospitals. Journal of Medical Microbiology 2007; 56: 376–379. [DOI] [PubMed] [Google Scholar]
- 4.Clinical and Laboratory Standards Institute. Analysis and presentation of cumulative susceptibility test data. Wayne, PA, USA: CLSI, 2010. Approved guideline, 2nd edn, CLSI document M39-A2, vol. 25, no. 28. [Google Scholar]
- 5.Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. Wayne, PA, USA: CLSI, 2006. CLSI supplement M100-S14, vol. 24, no. 1. [Google Scholar]
- 6.François P, et al. Use of an automated multiple-locus, variable-number tandem repeat-based method for rapid and high-throughput genotyping of Staphylococcus aureus isolates. Journal of Clinical Microbiology 2005; 43: 3346–3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Enright MC, et al. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. Journal of Clinical Microbiology 2000; 38: 1008–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Milheiriço C, Oliveira DC, de Lencastre H. Update to the multiplex PCR strategy for assignment of mec element types in Staphylococcus aureus. Antimicrobial Agents and Chemotherapy 2007; 51: 3374–3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.United Kingdom Health Protection Agency. Summary points on methicillin resistant Staphylococcus aureus (MRSA) bacteremia. (http://www.hpa.org.uk/web/HPAwebFile/HPAweb_C/1278944283762). Accessed 4 September 2011.
- 10.Ho PL, et al. Changes in the epidemiology of methicillin-resistant Staphylococcus aureus associated with spread of the ST45 lineage in Hong Kong. Diagnostic Microbiology and Infectious Disease 2009; 64: 131–137. [DOI] [PubMed] [Google Scholar]