SUMMARY
We report an evaluation of the accuracy of ELISA for the detection of Leptospira-specific antibodies in humans. Eighty-eight studies published in 35 articles met all inclusion criteria and were submitted to meta-analysis. Pooled sensitivity and specificity were 0·779 (95% CI 0·770–0·789) and 0·913 (95% CI 0·908–0·917), respectively, and the area under the curve was 0·964. Heterogeneity across studies was statistically significant, but none of the sources of heterogeneity (disease stage, antigen used, antibody detected) could fully explain this finding. Although the convalescent stage of disease was significantly associated with higher diagnostic accuracy, IgM ELISA was the best choice, regardless of the stage of disease. Negative ELISAs (IgG or IgM) applied in the acute phase do not rule out leptospirosis due to the possibility of false-negative results. In this case it is advisable to request a second blood sample or to apply a direct method for leptospiral DNA.
Key words: Diagnostic accuracy, human leptospirosis, sensitivity, specificity
INTRODUCTION
Leptospirosis, a spirochaetal zoonotic disease of worldwide distribution, has been recognized as an important emerging infectious disease in the last decade [1, 2]. Humans are often exposed to leptospiral bacteria through contact with either animals or fresh water from rivers and lakes [3, 4]. Because of the wide variety of symptoms, leptospirosis can be easily confused with many other febrile illnesses including haemorrhagic fevers [5]. Timely diagnosis is essential since antibiotic therapy provides the greatest benefits when initiated early in the course of illness [1].
Humans react to leptospiral infection by producing specific anti-Leptospira antibodies. Seroconversion may occur as early as 5 days after the onset of disease, but may be delayed up to ⩾10 days [5]. During this acute phase, both IgM and IgG antibodies, can be detected in serum samples up to day 21. After 4 weeks, during the convalescent phase, a delayed immune response can arise in which both immunoglobulin classes can be detected, with a higher proportion of IgG than IgM [6].
The isolation of Leptospira spp. is inconvenient for clinical diagnosis because sensitivity of culture is usually low, is time-consuming and requires relatively elaborate laboratory facilities. Serological tests are therefore necessary to confirm clinically suspected cases. The microscopic agglutination test (MAT), which detects agglutinating antibodies produced against lipopolysaccharide outer antigens, is the reference ‘gold standard’ method for diagnosing human leptospirosis [3] but requires the maintenance of a large number of live Leptospira strains for use as antigens and technical expertise in reading and interpreting the results [2–7].
To overcome these problems, some potentially useful tests have been developed, particularly enzyme-linked immunosorbent assays (ELISA) to detect IgM and IgG antibodies [8]. However, the efficacy of ELISA for diagnosis of leptospirosis depends on the stage of the disease, the antigen used and the class of antibodies detected [9]. Many studies have reported the performance of ELISA applied to a variety of situations and patients. Systematic review, complemented with meta-analysis, is a recognized scientific technique of reviewing available literature using explicit methods to identify, select and critically evaluate studies which are relevant to a stated objective [10]. We have applied this approach to evaluate the accuracy of ELISA for the detection of Leptospira-specific antibodies in humans.
MATERIALS AND METHODS
Criteria for study selection
Papers evaluating the accuracy of ELISA to detect Leptospira-specific antibodies were selected. Published studies were considered eligible if they included patients (limited to human) with proven or suspected leptospirosis and compared an ELISA test compared with MAT as the reference standard. Eligible studies were required to provide absolute numbers of diagnostic accuracy test using 2 × 2 tables. Several scientific articles reported the evaluation of more than one ELISA test against a reference standard. In those cases each comparison (e.g. using different types of ELISA test or patients in different stages of the disease) was considered separately. Therefore, the number of studies subject to analysis exceeded the number of published papers. Studies not providing relevant data on diagnostic accuracy were excluded as were reviews or duplicate reports.
Outcomes and definitions
Sensitivity, specificity, likelihood ratio for a negative and a positive result and diagnostic odds ratio (DOR) values were calculated for the ELISA tests investigated in each study, along with their 95% confidence intervals (CIs), and displayed as forest plots. The likelihood ratio for a positive or a negative result is a measure of how much the odds of the disease increase or decrease when a test is positive or negative, respectively.
Data sources
PubMed, Scopus, LILACS, and Cochrane Library databases were searched from 1980 to 2010 for articles unrestricted by language. Search terms included leptospirosis*, human and ELISA*. We also searched references from reviews and key publications on the topic. Conference abstracts were included when sufficient data were reported. Abstracts were assessed and articles that met the a priori criteria for study selection were utilized. Initial selection was based on title and abstract contents. Further selection depended upon the analysis of the original publication and the selection of those deemed to be relevant according to the selection criteria.
Data extraction
Methodological and technical data, number of patients, criteria used to select control and case groups, number of true positives, false positives, true negatives and false negatives were extracted from each study [11].
Statistical analysis
Data analysis was performed using Meta-Disk 1.4® software (Unit of Clinical Biostatics, the Ramon y Cajal Hospital, Madrid, Spain). Pooled sensitivity, specificity, DOR, and likelihood ratios (considered as weighted average according to size of individual studies) were calculated. These parameters can be pooled by a fixed-effects model (Mantel–Haenszel method) or by a random-effects model (DerSimonian–Laird method) to incorporate variation among studies. The fixed-effects model assumes that all studies in the meta-analysis are drawn from a common population. The random-effects model assumes that the studies were drawn from populations that differ from each other in ways that could impact on the diagnostic accuracy. In this meta-analysis we used a random-effects model, assuming that diagnostic accuracy of the test varied between studies and the various degrees of accuracy were randomly distributed among a central value [11–13].
Each study in the meta-analysis contributed a pair of numbers: sensitivity and specificity. Since these measures tend to be correlated and vary with the thresholds used across individual studies, a summary receiver-operating characteristic (SROC) curve analysis was performed in order to explore the effect of thresholds on results. A shoulder-like curve indicates that heterogeneity between studies may be due to the threshold effect. Additionally, the Spearman rank correlation coefficient between the logit of sensitivity and logit of 1 – specificity was calculated.
Apart from the variations due to thresholds, there are several other factors that can result in variations in accuracy estimates in different test accuracy studies. Heterogeneity among studies was evaluated using the DerSimonian–Laird test (Q statistic) and inconsistency index (I2 statistic [12]. A classification of I2 values was used to interpret its magnitude. Values of 25%, 50% and 75% were considered as low, medium, and high heterogeneity, respectively [12]. Further reasons for heterogeneity were investigated by pre-specified subgroup (stratified) analysis. In the subgroup analysis, data was stratified according to the stage of the disease (acute or convalescent phase), class of antibodies detected by the ELISA (IgM, IgG, IgA) and type of antigen used in the development of each ELISA (crude extract or specific antigen).
Additionally, meta-regression was conducted to investigate heterogeneity. In this unweighted linear regression model, studies were the units of analysis and the DOR was the outcome variable. DOR is a unitary measure of diagnostic performance that encompasses both sensitivity and specificity and it is a suitable method to compare the overall diagnostic accuracy of different tests [13]. The independent variables (stage of the disease, type of immunoglobulin, type of antigen) were the covariates that might be associated with the variability in DORs. The result of the meta-regression model was reported as relative diagnostic odds ratio (RDOR) [14].
RESULTS
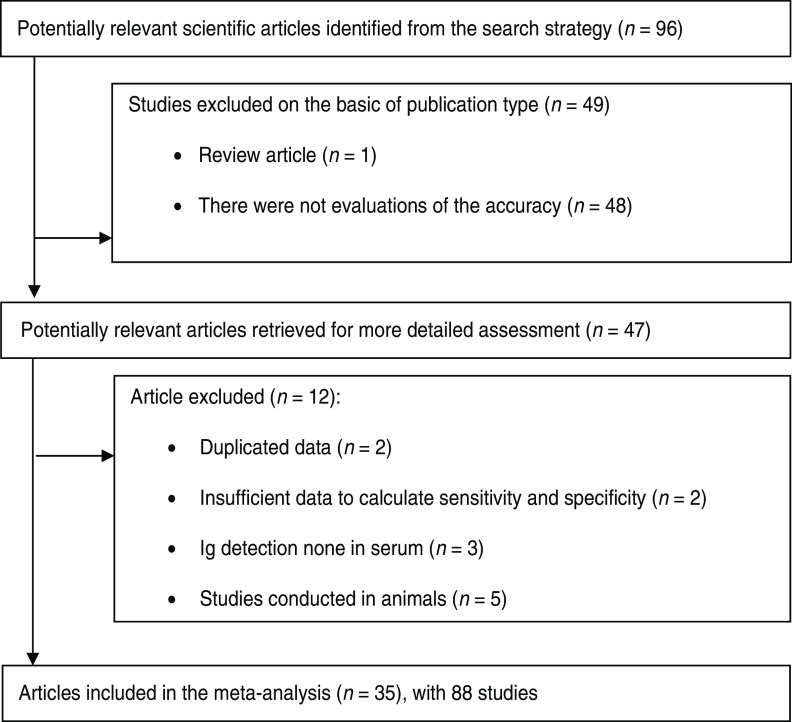
The literature search yielded 96 scientific papers on ELISA used to detect Leptospira antibodies in humans; of these 49 failed to meet one or more of the inclusion criteria (Fig. 1). A further 12 articles were excluded because they either had other objectives, were conducted in animals, showed duplicate data, or lacked sufficient statistical information to conduct a meta-analysis. Thirty-five articles describing 88 studies involving 21 494 patients were therefore available for further analysis. Two studies were conducted before 1990, eight between 1991 and 2000 and the remaining 25 after 2001. IgM ELISAs were evaluated in 21 instances whereas only three studies evaluated IgG ELISAs. Nine papers reported evaluations of IgM and IgG ELISAs and one IgG, IgM and IgA ELISA; one study only used an IgA ELISA.
Fig. 1.
Study selection flow chart.
In 17 studies the stage of disease was unknown or not specified. Three and two studies were reported on patients who had leptospirosis in acute and convalescent phases, respectively and 13 were conducted in patients with both stages of the disease (Table 1). Thirty studies utilized ELISA using whole-cell Leptospira-based antigens and five using recombinant/synthetic antigens.
Table 1.
Description of studies included in the meta-analysis
| Year | Sensitivity | Specificity | Type of antibody | Stage of the disease | Type of antigen | Reference |
|---|---|---|---|---|---|---|
| 1980 | 0·948 | 0·995 | IgM | Unspecified | Crude extract | [15] |
| 1988 | 0·820 | 0·772 | IgM | Unspecified | Crude extract | [16] |
| 1992 | 1·000 | 1·000 | IgM | Unspecified | Crude extract | [17] |
| 0·500 | 1·000 | IgA | Unspecified | Crude extract | ||
| 1995 | 0·921 | 0·975 | IgM | Unspecified | Crude extract | [18] |
| 1996 | 0·899 | 0·974 | IgM | Unspecified | Crude extract | [19] |
| 1997 | 0·959 | 0·462 | IgM + IgG | Unspecified | Crude extract | [20] |
| 0·984 | 0·979 | IgM | Acute | Crude extract | [21] | |
| 1997 | 0·698 | 1·000 | IgG | Acute | Crude extract | |
| 0·762 | 0·979 | IgA | Acute | Crude extract | ||
| 1998 | 0·991 | 0·988 | IgM | Unspecified | Crude extract | [22] |
| 1999 | 0·360 | 0·968 | IgM | Acute | Crude extract | [23] |
| 0·760 | 0·968 | IgM | Convalescent | Crude extract | ||
| 0·522 | 0·952 | IgM | Acute | Crude extract | [8] | |
| 1999 | 0·893 | 0·977 | IgM | Acute | Crude extract | |
| 0·969 | 0·939 | IgM | Convalescent | Crude extract | ||
| 2000 | 0·548 | 0·969 | IgM | Unspecified | Crude extract | [24] |
| 0·837 | 0·938 | IgM + IgG | Unspecified | Crude extract | ||
| 2001 | 0·965 | 0·986 | IgM | Unspecified | Crude extract | [25] |
| 0·930 | 0·824 | IgM | Unspecified | Crude extract | ||
| 2001 | 0·560 | 0·911 | IgG | Acute | Sepecific antigen | [26] |
| 0·940 | 0·911 | IgG | Convalescent | Sepecific antigen | ||
| 2001 | 0·596 | 0·958 | IgM | Acute | Crude extract | [5] |
| 0·895 | 0·992 | IgM | Convalescent | Crude extract | ||
| 2001 | 0·570 | 0·958 | IgM | Acute | Crude extract | [27] |
| 0·844 | 0·989 | IgM | Convalescent | Crude extract | ||
| 2002 | 0·519 | 0·951 | IgM | Unspecified | Crude extract | [28] |
| 0·500 | 0·902 | IgM | Unspecified | Crude extract | ||
| 0·346 | 0·978 | IgM | Unspecified | Crude extract | ||
| 0·423 | 0·978 | IgM | Unspecified | Crude extract | ||
| 2002 | 0·975 | 0·988 | IgM | Unspecified | Crude extract | [29] |
| 2002 | 0·896 | 0·927 | IgG | Unspecified | Crude extract | [30] |
| 0·875 | 0·964 | IgM | Unspecified | Crude extract | ||
| 2003 | 0·486 | 0·969 | IgM | Acute | Crude extract | [31] |
| 0·750 | 0·969 | IgM | Convalescent | Crude extract | ||
| 2003 | 0·500 | 0·787 | IgM | Acute | Crude extract | [2] |
| 0·877 | 0·872 | IgM | Convalescent | Crude extract | ||
| 2004 | 0·835 | 0·802 | IgM | Unspecified | Crude extract | [9] |
| 0·541 | 0·634 | IgG | Unspecified | Crude extract | ||
| 0·976 | 0·965 | IgM | Unspecified | Crude extract | ||
| 0·788 | 0·713 | IgG | Unspecified | Crude extract | ||
| 0·965 | 0·941 | IgM | Unspecified | Crude extract | ||
| 0·847 | 0·792 | IgG | Unspecified | Crude extract | ||
| 2004 | 1·000 | 0·962 | IgM | Unspecified | Crude extract | [32] |
| 2005 | 0·875 | 0·976 | IgM | Acute | Crude extract | [33] |
| 0·990 | 0·939 | IgM | Acute | Crude extract | ||
| 2006 | 0·936 | 0·933 | IgM | Unspecified | Crude extract | [7] |
| 2006 | 0·883 | 0·891 | IgM | Unspecified | Crude extract | [34] |
| 2006 | 0·609 | 0·656 | IgM | Acute | Crude extract | [35] |
| 0·652 | 0·454 | IgM | Convalescent | Crude extract | ||
| 2007 | 0·792 | 0·950 | IgM | Acute | Crude extract | [36] |
| 0·875 | 0·875 | IgM | Acute | Crude extract | ||
| 0·960 | 0·950 | IgM | Convalescent | Crude extract | ||
| 0·920 | 0·875 | IgM | Convalescent | Crude extract | ||
| 0·755 | 0·977 | IgM | Acute | Crude extract | ||
| 2007 | 0·681 | 0·963 | IgG | Acute | Crude extract | [37] |
| 0·932 | 0·993 | IgG | Convalescent | Crude extract | ||
| 0·788 | 1·000 | IgG | Convalescent | Crude extract | ||
| 2008 | 1·000 | 0·977 | IgM | Convalescent | Crude extract | [36] |
| 2008 | 0·696 | 0·968 | IgM | Acute | Sepecific antigen | [38] |
| 0·667 | 0·968 | IgM | Convalescent | Sepecific antigen | ||
| 0·630 | 1·000 | IgG | Acute | Sepecific antigen | ||
| 0·861 | 1·000 | IgG | Convalescent | Sepecific antigen | ||
| 2008 | 0·898 | 0·981 | IgM | Unspecified | Crude extract | [39] |
| 2008 | 0·963 | 0·911 | IgM | Unspecified | Sepecific antigen | [40] |
| 2009 | 0·856 | 0·993 | IgM | Acute | Sepecific antigen | [41] |
| 1·000 | 0·993 | IgM | Convalescent | Sepecific antigen | ||
| 0·833 | 0·993 | IgM | Acute | Sepecific antigen | ||
| 0·922 | 0·993 | IgM | Convalescent | Sepecific antigen | ||
| 2009 | 0·850 | 0·933 | IgM | Unspecified | Crude extract | [42] |
| 2010 | 0·434 | 0·882 | IgM | Unspecified | Sepecific antigen | [3] |
| 0·943 | 1·000 | IgG | Unspecified | Sepecific antigen | ||
| 2011 | 0·777 | 0·820 | IgG | Unspecified | Crude extract | [43] |
| 0·777 | 0·680 | IgG | Unspecified | Sepecific antigen | ||
| 0·690 | 0·831 | IgG | Unspecified | Sepecific antigen | ||
| 0·642 | 0·895 | IgG | Unspecified | Sepecific antigen | ||
| 0·624 | 0·930 | IgG | Acute | Crude extract | ||
| 0·624 | 0·775 | IgG | Acute | Sepecific antigen | ||
| 0·661 | 0·873 | IgG | Acute | Sepecific antigen | ||
| 0·514 | 0·944 | IgG | Acute | Sepecific antigen | ||
| 0·956 | 0·977 | IgG | Convalescent | Crude extract | ||
| 0·824 | 0·724 | IgG | Convalescent | Sepecific antigen | ||
| 0·758 | 0·782 | IgG | Convalescent | Sepecific antigen | ||
| 0·725 | 0·872 | IgG | Convalescent | Sepecific antigen | ||
| 0·828 | 1·000 | IgG | Convalescent | Crude extract | ||
| 0·793 | 0·864 | IgG | Convalescent | Sepecific antigen | ||
| 0·724 | 0·773 | IgG | Convalescent | Sepecific antigen | ||
| 0·862 | 0·773 | IgG | Convalescent | Sepecific antigen |
Most of the studies were conducted in Brazil (n = 11), and others were conducted in Thailand (n = 3), India (n = 3), Argentina (n = 2), USA (n = 2), and Barbados (n = 2). Two studies used samples from different regions (Hawaii, Indonesia, Seychelles, The Netherlands). Single studies were conducted in each of the following countries: Venezuela, Seychelles, The Netherlands, Singapore, Laos, UK, Peru, Italy, France, and Iran.
Accuracy of assays
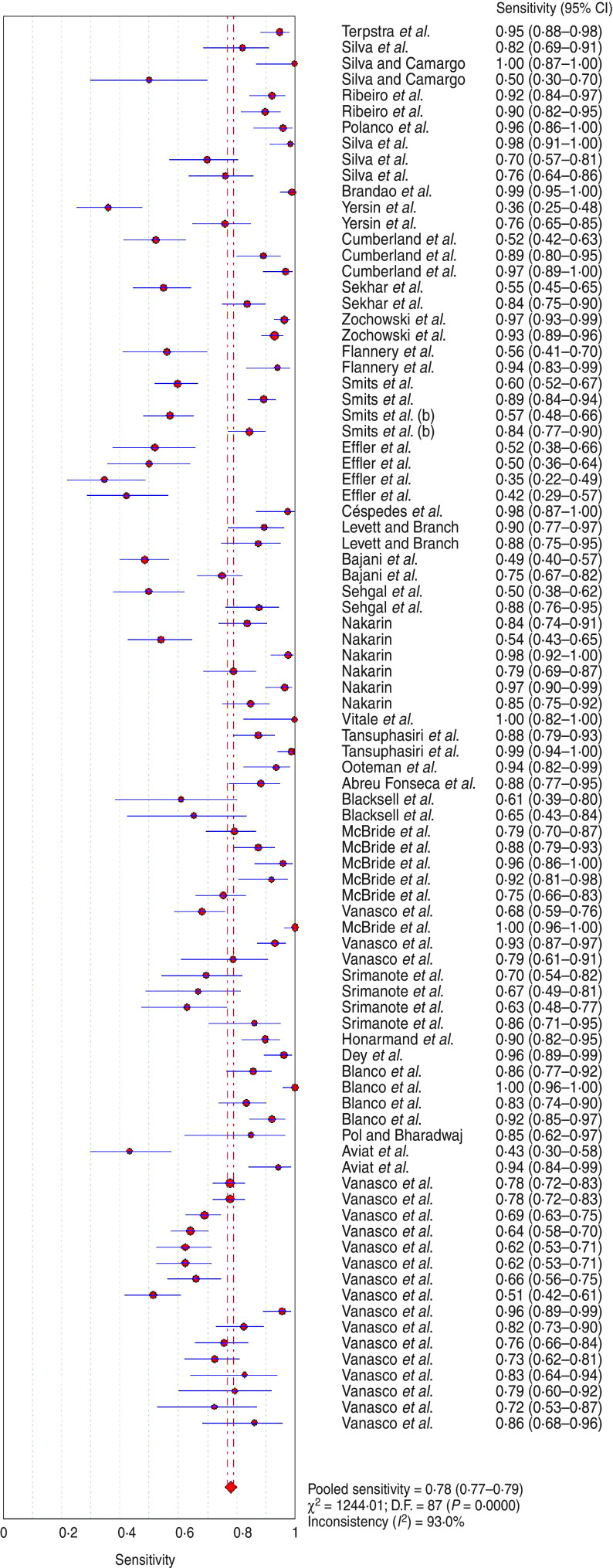
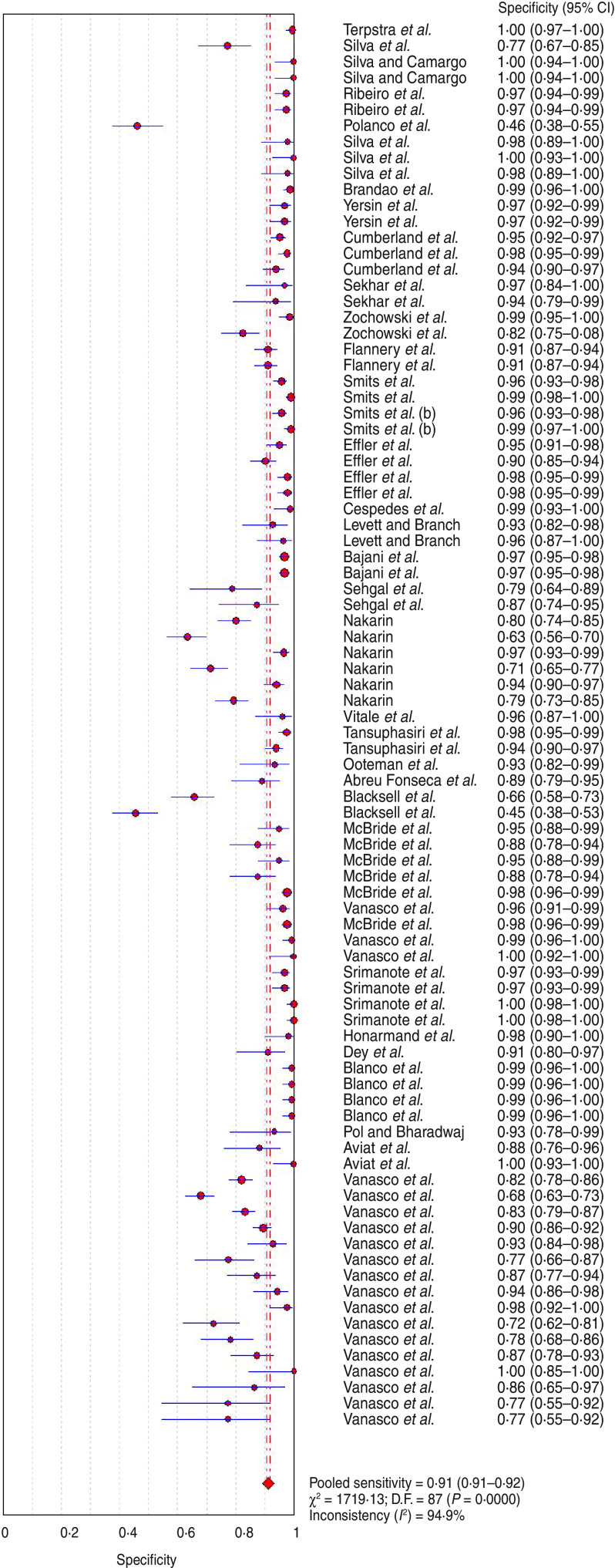
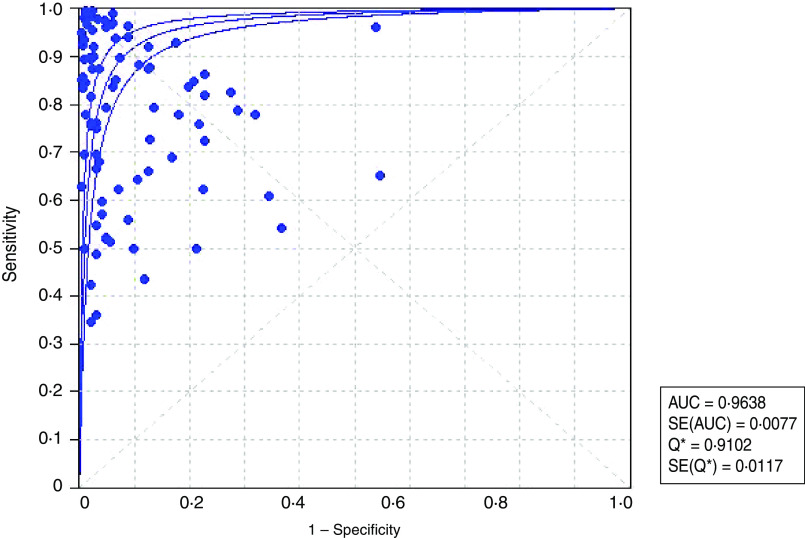
The analysis of the 88 assays yielded a pooled sensitivity of 0·779 (95% CI 0·770–0·789) (Fig. 2) and specificity of 0·913 (95% CI 0·908–0·917) (Fig. 3) for the end-point prediction. Positive and negative likelihood ratios were 12·9 (95% CI 9·935–16·750) and 0·206 (95% CI 0·175–0·243), respectively. The DOR was 75·057 (95% CI 53·181–105·930) indicating high accuracy. SROC analysis showed that the area under the curve (AUC) was 0·964, Q* value was 0·910 (SE(Q*) = 0·0117). Asymmetric SROC analysis yielded no difference in the result (Fig. 4). The significant heterogeneity in sensitivity and specificity estimates precluded the determination of clinically useful summary measures.
Fig. 2.
[color online]. Forest plot of sensitivity estimate for ELISA diagnosis of human leptospirosis. •, Point estimates of sensitivity from each study (proportional to size of the study); ——, 95% confidence intervals;◆, pooled sensitivity estimated.
Fig. 3.
[color online]. Forest plot of specificity estimate for ELISA diagnosis of human leptospirosis. •, Point estimates of specificity from each study (proportional to size of the study); ——, 95% confidence intervals;◆, pooled specificity estimated.
Fig. 4.
[color online]. Summary receiver-operating characteristic (SROC) plot for ELISA diagnosis of human leptospirosis. •, Each study in the meta-analysis, size proportional to size of study; ——, regression line that summarizes the overall diagnostic accuracy.
Heterogeneity and stratified analyses
One of the primary causes of heterogeneity in test accuracy studies is the threshold effect. The latter arises when differences in sensitivities and specificities are due to the selection of different cut-off values to define a positive (or negative) test result [13]. The Spearman correlation coefficient between the logit of sensitivity and log of 1 – specificity was not significant (P = 0·062). Studies clearly showed a high degree of variability in sensitivity (P < 0·001, I2 = 93·72%) and specificity (P < 0·001, I2 = 95·02%) estimates. This heterogeneity may result from differences in test methods (type of immunoglobulin identified by the ELISA test, and type of antigen used in the development of each test) or population characteristics (patients sampled in the acute or convalescent stage). In order to identify factors associated with heterogeneity, stratified (subgroup) analyses were performed (Table 2).
Table 2.
Summary of measures for sub-analysis comparison
| Sub-analysis | Global effect and subgroup analysis restricted | Studies (n) | Sensitivity | Specificity | Diagnostic odds ratio | ||
|---|---|---|---|---|---|---|---|
| Pooled estimate (95% CI) | Q (P) | Pooled estimate (95% CI) | Q (P) | ||||
| Global effect | 88 | 0·779 (0·770–0·789) | <0·001 | 0·913 (0·908–0·917) | <0·001 | 75·05 (53·18–105·93) | |
| Stage of the disease | |||||||
| Stages not identified | 37 | 0·802 (0·788–0·816) | <0·001 | 0·869 (0·860–0·878) | <0·001 | 70·94 (42·33–118–89) | |
| Acute | 26 | 0·683 (0·663–0·701) | <0·001 | 0·946 (0·939–0·952) | <0·001 | 50·20 (29·11–86·56) | |
| Convalescent | 25 | 0·864 (0·847–0·879) | <0·001 | 0·934 (0·925–0·941) | <0·001 | 128·40 (58·82–280·30) | |
| Type of immunoglobulin identified | |||||||
| IgM + IgG | 2 | 0·876 (0·860–0·995) | 0·019 | 0·555 (0·475–0·632) | <0·001 | 38·63 (10·22–146·01) | |
| IgM | 55 | 0·804 (0·792–0·815) | <0·001 | 0·944 (0·939–0·949) | <0·001 | 123·45 (77·72–196·09) | |
| IgG | 29 | 0·736 (0·719–0·752) | <0·001 | 0·848 (0·837–0·859) | 0·001 | 25·85 (16·99–39·33) | |
| IgA | 2 | 0·685 (0·578–0·780) | 0·018 | 0·990 (0·948–1·000) | 0·209 | 137·33 (25·63–735·70) | |
| Type of antigen | |||||||
| Crude extract | 63 | 0·794 (0·783–0·805) | <0·001 | 0·920 (0·914–0·925) | 0·001 | 95·44 (61·75–147·51) | |
| Specific | 25 | 0·744 (0·725–0·762) | <0·001 | 0·890 (0·880–0·901) | <0·001 | 37·27 (22·55–61·61) | |
CI, Confidence interval.
Three subgroup analyses restricted to the stage of the disease revealed 37 studies where disease stage was not identified or included patients in different stages. The sensitivity and specificity values in this subgroup were 0·802 (95% CI 0·788–0·816) and 0·869 (95% CI 0·860–0·878), respectively. When patients in the acute stage were considered (n = 26), the pooled sensitivity was 0·683 (95% CI 0·663–0·701) and the pooled specificity 0·946 (95% CI 0·939–0·952). However, when the ELISA was applied in convalescent patients (n = 25) sensitivity was higher (0·863, 95% CI 0·847–0·879) than in the acute phase, while specificity remained at similar levels (0·934, 95% CI 0·925–0·941). However, all the stratified analyses showed significant heterogeneity (Table 2).
Four subgroup analyses restricted to the type of immunoglobulin identified by the ELISA test (IgM, IgG, IgA, or IgM + IgG), were conducted. Two assays using IgM and IgG in the same ELISA showed a pooled sensitivity and specificity of 0·876 (95% CI 0·860–0·995) and 0·555 (95% CI 0·475–0·632), respectively. The pooled sensitivity (0·804, 95% CI 0·792–0·815) and specificity (0·944, 95% CI 0·939–0·949) were higher in the assays which used an IgM rather than IgG ELISA (sensitivity = 0·736 and specificity = 0·848). Another subgroup analysis restricted to only two assays based on IgA ELISA showed the lowest sensitivity (0·685, 95% CI 0·578–0·780) and the highest specificity (0·990, 95% CI 0·948–1·000). However, the stratified analysis did not reduce the heterogeneity in the studies (Table 2).
Taking into account the type of antigen used in the development of each ELISA test, the subgroup analysis indicated no effect on the sensitivity when crude extract antigen (0·794, 95% CI 0·783–0·805) or specific antigens (0·744, 95% CI 0·725–0·762) were used. However, those assays which used crude extract antigen had increased specificity (0·920, 95% CI 0·914–0·925) over ELISA based on specific antigens (0·890, 95% CI 0·880–0·901). Nevertheless, the type of antigen used had no significant influence on heterogeneity (P < 0·001) (Table 2).
None of the stratified analyses results fully explained the significant heterogeneity across studies in this review. The statistical tests for heterogeneity were significant even within the different strata (Table 2). Therefore, a meta-regression analysis was performed to evaluate multiple factors in the same analysis. The outcome of the regression analysis was the RDOR (Table 3). Studies that included patients in the convalescent stage of the disease showed a RDOR significantly higher (1·84 times) than those that included patients in the acute stage or where disease stage was not identified. Studies that utilized an IgM ELISA produced RDOR values significantly higher (7·14 times) than assays that detected IgG. The type of antigen used in the ELISA did not produce a significant RDOR, indicating that the use of crude extract or specific antigen did not substantially affect diagnostic accuracy.
Table 3.
Meta-regression analysis to determine sources of heterogeneity
| Covariate | Coefficient | P value | RDOR | 95% CI |
|---|---|---|---|---|
| Intercept | 7·047 | <0·0001 | – | – |
| Threshold (S) | 0·221 | 0·1234 | – | – |
| General + acute vs. convalescence | 0·611 | 0·0385 | 1·84 | (1·03–3·28) |
| IgM vs. IgG | −1·983 | 0·0008 | 0·14 | (0·04–0·43) |
| Crude extract vs. specific antigen | 0·115 | 0·847 | 1·12 | (0·34–3·66) |
RDOR, Relative diagnostic odds ratio; CI, confidence interval.
Intercept = constant in the model.
S, Indicator of threshold (logit true positive rate + logit false positive rate).
DISCUSSION
This meta-analysis was conducted taking into account the standard protocol for systematic reviews, considering studies published in different languages. Two reviewers independently performed the article selection and data extraction. Several methodologies were applied, including SROC analyses, methods for exploring heterogeneity and meta-regression.
Our meta-analysis, based on 88 published ELISA evaluations, showed a SROC curve with an AUC of 91%, indicating a high efficiency for the detection of leptospirosis in human patients. However, the high heterogeneity in the overall sensitivity and specificity estimates may hamper their usefulness as summary measures for clinical detection. In an attempt to explain the observed variability, we performed a stratified analysis with three major sources of variability: (1) phase of the disease (not identified, acute and convalescent), (2) type of immunoglobulin detected by the ELISA (IgM, IgG, IgA, or IgM + IgG) and (3) type of antigen used in the development of each assay (whole cell or recombinant/synthetic). Heterogeneity was evident in the results and could not be completely explained by the different sources of variability analysed.
The variability and quality in study designs can be a source of heterogeneity of results. However, no categorization taking into account the quality of studies was included in our analysis due to the lack of sufficient elements for such an assessment. Several other variables that could generate heterogeneity among the diagnostic methods include demographics (age, sex, occupation), clinical severity and morbidity, and regional characteristics such as climate and flooding. These sources could not be quantified or measured in this study owing to the lack of information available and merit further investigation.
Due to the heterogeneity in sensitivity and specificity, it is difficult to determine clinically useful estimates of accuracy. Nevertheless, meta-regression analysis allowed us to highlight some variables that apparently affect the estimates of efficiency of ELISAs in diagnosing human leptospirosis. This technique showed a significantly higher efficiency when applied to patients in convalescent rather than acute or unidentified phases. The latter is expected for any serological test because of the time lapse for the appearance of serum antibodies after an infection and increased sensitivity does not necessarily correlate with time [43].
Another factor associated with the effectiveness of an ELISA is the type of antibody detected. According to their DOR, assays which identified IgM were seven times more efficient than those that detected IgG. This marked difference indicates that, in most studies, sensitivity and specificity of IgM ELISA were significantly greater than for IgG assays. Anti-Leptospira IgM antibodies appear earlier than IgG and remain detectable for months or years at low titres. On the other hand, IgG levels may vary from non-detectable, or positive for short time periods or several years [44].
However, the higher specificity of IgM over IgG ELISA is difficult to explain because IgM in general is characterized by a lower affinity than IgG for antigens [44]. In this system patient IgM and IgG responses compete for binding to antigen and the higher affinity IgG molecules are more readily captured. Non-specific IgM is unable to bind and is therefore not detected. In this case, non-specific IgG is reflected as a false-positive test result for IgG which is not shown for IgM. Thus, what constitutes a classical advantage for IgG detection could become a disadvantage that indirectly favours IgM detection. A possible explanation for this is that patients with febrile diseases other than leptospirosis may have non-specific IgG antibodies due to a false-positive test result.
Finally, the type of antigen (whole cell or recombinant/synthetic) used in the ELISA test did not produce a significant change in the RDOR. These results are consistent with recent studies suggesting that recombinant and synthetic antigens would fail to overcome the diagnostic efficiency of crude extract antigen, which is a whole-cell antigen of Leptospira [45]. Faced with a suspected case of leptospirosis, an IgM ELISA will show good efficacy and be the best choice of test; the type of antigen would not be of great importance nor the stage of disease. However, if the patient is in the acute phase and both IgG and IgM are negative, leptospirosis can not be ruled out due to the possibility of false-negative results in the early stages of the disease. In this case it is advisable to request a second blood sample or apply a direct method such as real-time PCR for leptospiral DNA [46].
In conclusion the meta-analysis identified the type of detected immunoglobulin and the phase of the disease in the patient to be two important sources of variability for the efficiency of ELISA for the diagnosis of human leptospirosis. Our results suggest that an IgM ELISA would be the best choice of method for early detection of cases during the acute phase of infection and serve as a good screening tool at all stages of the disease. Additional studies should be performed to analyse the effect of other potential sources of variability, especially those related to regional aspects.
ACKNOWLEDGMENTS
Marcelo L. Signorini is a Research Career Member from the Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET, Argentina).
DECLARATION OF INTEREST
None.
REFERENCES
- 1.McBride AJA, et al. Leptospirosis. Current Opinion on Infectious Diseases 2005; 18: 376–386. [DOI] [PubMed] [Google Scholar]
- 2.Sehgal SC, et al. Field application of Lepto lateral flow for rapid diagnosis of leptospirosis. Journal of Medicine and Microbiology 2003; 52: 897–901. [DOI] [PubMed] [Google Scholar]
- 3.Aviat F, et al. Synthetic peptide issued from Hap1/LipL32 for new early serodiagnosis of human leptospirosis. Comparative Immunology, Microbiology and Infectious Diseases 2010; 33: 375–387. [DOI] [PubMed] [Google Scholar]
- 4.Vanasco NB, et al. Clinical characteristics and risk factors of human leptospirosis in Argentina (1999–2005). Acta Tropica 2008; 107: 255–258. [DOI] [PubMed] [Google Scholar]
- 5.Smits HL, et al. Latex based, rapid and easy assay for human leptospirosis in a single test format. Tropical Medicine and International Health 2001; 6: 114–118. [DOI] [PubMed] [Google Scholar]
- 6.Levett PN. Leptospirosis. Clinical Microbiology Reviews 2001; 14: 296–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ooteman MC, Ravara Vago A, Koury MC. Evaluation of MAT, IgM ELISA and PCR methods for the diagnosis of human leptospirosis. Journal of Microbiological Methods 2006; 65: 247–257. [DOI] [PubMed] [Google Scholar]
- 8.Cumberland P, Everard COR, Levett PN. Assessment of the efficacy of an IgM-ELISA and microscopic agglutination test (MAT) in the diagnosis of acute leptospirosis. American Journal of Tropical Medicine and Hygiene 1999; 61: 731–734. [DOI] [PubMed] [Google Scholar]
- 9.Nakarin J, Pradutkanchana S. Evaluation of enzyme-linked immunosorbent assay and indirect hemagglutination assay for detection of leptospiral antibody by using three different antigens. Journal of the Medical Association of Thailand 2004; 87: 1218–1223. [PubMed] [Google Scholar]
- 10.Faria Filho DE, et al. Probiotics for broiler chickens in Brazil: systematic review and meta-analysis. Brazilian Journal of Poultry Science 2006; 8: 89–98. [Google Scholar]
- 11.Lean IJ, et al. Invited review: Use of meta-analysis in animal health and reproduction: Methods and applications. Journal of Dairy Science 2009; 92: 3545–3565. [DOI] [PubMed] [Google Scholar]
- 12.Higgins JPT, Thompson SG. Quantifying heterogeneity in a meta-analysis. Statistics in Medicine 2002; 21: 1539–1558. [DOI] [PubMed] [Google Scholar]
- 13.Zamora J, et al. Meta-DiSc: a software for meta-analysis of test accuracy data. BMC Medical Research Methodology 2006; 6: 31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flores LL, et al. In-house nucleic acid amplification tests for the detection of Mycobacterium tuberculosis in sputum specimens: meta-analysis and meta-regression. BMC Microbiology 2005; 5: 55–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Terpstra WJ, Ligthart GS, Schoone GJ. Serodiagnosis of human leptospirosis by enzyme-linked-immunosorbent-assay (ELISA). Zentralblatt fur Bakteriologie, Mikrobiologie, und Hygiene. Series A, Medical Microbiology, Infectious Diseases, Virology, Parasitology 1980; 247: 400–405. [PubMed] [Google Scholar]
- 16.Silva MV, et al. Enzyme-linked immunosorbent assay for the detection of IgM antibodies in human leptospirosis. Revista Instituto de Medicina Tropical de Sao Paulo 1988; 30: 95–100. [Google Scholar]
- 17.Silva MV, Camargo ED. Enzyme-linked immunosorbent assay ELISA for the detection of IM antibodies in human leptospirosis. Revista Instituto de Medicina Tropical de Sao Paulo 1992; 34: 239–242. [DOI] [PubMed] [Google Scholar]
- 18.Ribeiro MA, Souza CC, Almeida SHP. Dot-ELISA for human leptospirosis employing immunodominant antigen. Journal of Tropical Medicine and Hygiene 1995; 98: 452–456. [PubMed] [Google Scholar]
- 19.Ribeiro MA, Brandao AP, Romero EC. Evaluation of diagnostic test for human leptospirosis. Brazilian Journal of Medicine and Biological Research 1996; 29: 773–777. [PubMed] [Google Scholar]
- 20.Polanco J, et al. Diagnosis of human leptospirosis using dot enzyme-linked immunosorbent assay. Veterinaria Tropical 1997; 22: 65–75. [Google Scholar]
- 21.Da Silva MV, et al. Immunodiagnosis of human leptospirosis by dot-ELISA for the detection of IgM, IgG and IgA antibodies. American Journal of Tropical Medicine and Hygiene 1997; 56: 650–655. [DOI] [PubMed] [Google Scholar]
- 22.Brandao AP, et al. Macroscopic agglutination test for rapid diagnosis of human leptospirosis. Journal of Clinical Microbiology 1998; 36: 3138–3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yersin C, et al. Field evaluation of a one-step dipstick assay for the diagnosis of human leptospirosis in the Seychelles. Tropical Medicine and International Health 1999; 4: 38–45. [DOI] [PubMed] [Google Scholar]
- 24.Sekhar WY, et al. Leptospirosis in Kuala Lumpur and the comparative evaluation of two rapid commercial diagnostic kits against the MAT test for the detection of antibodies to Leptospira interrogans. Singapore Medical Journal 2000; 41: 370–375. [PubMed] [Google Scholar]
- 25.Zochowski WJ, Palmer MF, Coleman TJ. An evaluation of three comercial kits for use as screening methods for the detection of leptospiral antibodies in the UK. Journal of Clinical Pathology 2001; 54: 25–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Flannery B, et al. Evaluation of recombinant Leptospira antigen based enzyme-linked immunosorbent assay for the serodiagnosis of leptospirosis. Journal of Clinical Microbiology 2001; 39: 3303–3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smits HL, et al. Lateral-flow assay for rapid serodiagnosis of human leptospirosis. Clinical and Diagnostic Laboratory Immunology 2001; 8: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Effler PV, et al. Evaluation of eight rapid screening test for acute leptospirosis in Hawaii. Journal of Clinical Microbiology 2002; 40: 1464–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Céspedes M, et al. Enzyme-linked immunosorbent assay for the detection of IgM in the diagnostic of human leptospirosis. Revista Peruana de Medicina Experimental y Salud Publica 2002; 19: 1–6. [Google Scholar]
- 30.Levett PN, Branch SL. Evaluation of two enzyme-linked immunosorbent assay methods for detection of immunoglobulin M antibodies in acute leptospirosis. American Journal of Tropical Medicine and Hygiene 2002; 66: 745–748. [DOI] [PubMed] [Google Scholar]
- 31.Bajani MD, et al. Evaluation of four commercially available rapid serologic tests for diagnosis of leptospirosis. Journal of Clinical Microbiology 2003; 41: 803–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vitale G, et al. Evaluation of an IgM-ELISA test for the diagnosis of human leptospirosis. New Microbiologica 2004; 27: 149–154. [PubMed] [Google Scholar]
- 33.Tansuphasiri U, et al. Two simple immunoassays using endemic leptospiral antigens for serodiagnosis of human leptospirosis. Southeast Asian Journal of Tropical Medicine and Public Health 2005; 36: 302–311. [PubMed] [Google Scholar]
- 34.Abreu Fonseca C, et al. Polymerase chain reaction in comparison with serological tests for early diagnosis of human leptospriosis. Tropical Medicine & International Health 2006; 11: 1699–1707. [DOI] [PubMed] [Google Scholar]
- 35.Blacksell SD, et al. Limited diagnostic capacities of two commercial assays for the detection of Leptospira immunoglobulin M antibodies in Laos. Clinical Vaccine Immunology 2006; 13: 1166–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McBride AJA, et al. Evaluation of four whole-cell Leptospira-based serological tests for diagnosis of urban leptospirosis. Clinical Vaccine and Immunology 2007; 14: 1245–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vanasco NB, et al. Diagnosis of leptospirosis evaluation of a solid-phase enzyme immunoassay in different stages of the disease. Revista Panamericana de Salud Publica 2007; 21: 388–395. [DOI] [PubMed] [Google Scholar]
- 38.Srimanote P, et al. Recominant ligA for leptospirosis diagnosis and ligA among the Leptospira spp. clinical isolates. Journal of Microbiological Methods 2008; 72: 73–81. [DOI] [PubMed] [Google Scholar]
- 39.Honarmand H, et al. Evaluation an in-house IgM-ELISA for the diagnosis of human leptospirosis. Journal of Semnan University of Medical Sciences 2008; 9: 309–314. [Google Scholar]
- 40.Dey S, et al. Diagnosis of leptospirosis by recombinant antigen based single serum dilution ELISA. Indian Journal of Medical Research 2008; 128: 172–177. [PubMed] [Google Scholar]
- 41.Blanco RM, Takei K, Romero EC. Leptospiral glycolipoprotein as a candidate antigen for serodiagnosis of human leptospirosis. Letters in Applied Microbiology 2009; 49: 267–273. [DOI] [PubMed] [Google Scholar]
- 42.Pol S, Bharadwaj R. Evaluation of high performance liquid chromatography purified leptospiral antigen for the diagnosis of leptospirosis. Japanese Journal of Infectious Diseases 2009; 62: 428–431. [PubMed] [Google Scholar]
- 43.Fainboim L, Geffner J. Introducción a la inmunología humana, 5th edn. Buenos Aires, Argentina. Panamericana, 2005, pp. 504. [Google Scholar]
- 44.ILS-WHO. Human Leptospirosis: Guidance for Diagnosis, Surveillance and Control. Malta. WHO, 2003, pp. 109. [Google Scholar]
- 45.Vanasco NB, et al. Evaluation of ELISA antigens for leptospiral serodiagnosis in different stages of the disease. VII Reunión de la Sociedad Internacional de Leptospirosis, Merida, Yucatan, Mexico, 19–22 September 2011. Oral presentation.
- 46.Vanasco NB, et al. Usefulness of direct and indirect methods for early diagnosis of leptospirosis. VII ILS Meeting, Merida, Mexico, 19–22 September, 2011.