Abstract
Inflammation is associated with symptoms of anhedonia, a core feature of major depression (MD). We have shown that MD patients with high inflammation as measured by plasma C-reactive protein (CRP) and anhedonia display gene signatures of metabolic reprograming (e.g., shift to glycolysis) necessary to sustain cellular immune activation. To gain preliminary insight into the immune cell subsets and transcriptomic signatures that underlie increased inflammation and its relationship with behavior in MD at the single-cell (sc) level, herein we conducted scRNA-Seq on peripheral blood mononuclear cells from a subset of medically-stable, unmedicated MD outpatients. Three MD patients with high CRP (>3 mg/L) before and two weeks after anti-inflammatory challenge with the tumor necrosis factor antagonist infliximab and three patients with low CRP (≤3 mg/L) were studied. Cell clusters were identified using a Single Cell Wizard pipeline, followed by pathway analysis. CD14+ and CD16+ monocytes were more abundant in MD patients with high CRP and were reduced by 29% and 55% respectively after infliximab treatment. Within CD14+ and CD16+ monocytes, genes upregulated in high CRP patients were enriched for inflammatory (phagocytosis, complement, leukocyte migration) and immunometabolic (hypoxia-inducible factor [HIF]-1, aerobic glycolysis) pathways. Shifts in CD4+ T cell subsets included ∼30% and ∼10% lower abundance of CD4+ central memory (TCM) and naïve cells and ∼50% increase in effector memory-like (TEM-like) cells in high versus low CRP patients. TCM cells of high CRP patients displayed downregulation of the oxidative phosphorylation (OXPHOS) pathway, a main energy source in this cell type. Following infliximab, changes in the number of CD14+ monocytes and CD4+ TEM-like cells predicted improvements in anhedonia scores (r = 1.0, p < 0.001). In sum, monocytes and CD4+ T cells from MD patients with increased inflammation exhibited immunometabolic reprograming in association with symptoms of anhedonia. These findings are the first step toward determining the cellular and molecular immune pathways associated with inflammatory phenotypes in MD, which may lead to novel immunomodulatory treatments of psychiatric illnesses with increased inflammation.
Keywords: Depression, Inflammation, Single cell RNA-Sequencing, Anhedonia, Monocytes, T cells, Immunometabolism
1. Introduction (AHM)
My earliest interactions with Dr. Bruce McEwen were focused on the resistance of activated innate immune cells to the inhibitory effects of glucocorticoids in patients with major depression (MD) (Miller et al., 1987, 1991). Bruce was an amazing mentor and shared his insights and his laboratory as a platform for my work to further understand the cellular mechanisms of glucocorticoid resistance in the immune system. Many of the studies were carried out with my colleagues Dr. Robert Spencer in Bruce's laboratory at Rockefeller and Bob's graduate student Dr. Thaddeus Pace, who later joined me as a postdoctoral fellow at Emory University. Bob and Tad were two members of the vast McEwen legacy that span multiple generations of trainees, many of whom have made seminal contributions to the fields of stress neurobiology, neuroscience, endocrinology, and immunology. Together, Bob, Tad and I with help from Drs. Carmine Pariante, Xiaohong Wang and Charles Raison characterized the differential expression of glucocorticoid receptors (GRs) in various immune cell subtypes and ultimately delineated the impact of inflammatory cytokines on GR function (Pace et al., 2007; Pariante, 2004). In addition, we further instantiated the notion that chronic stress and MD were associated with glucocorticoid resistance in part as a result of the inhibitory effects of inflammatory cytokines on the translocation of the GR from cytoplasm to nucleus and GR binding to its response elements on DNA (Pace et al., 2007; Pariante, 2004). Glucocorticoid resistance in turn was seen as a primary contributor to unchecked innate immune responses and ultimately inflammation in MD (Raison and Miller, 2003). These studies could not have been possible without Bruce, who served as a major proponent of the work and contributed to many of the resulting scientific insights along the way. Bruce will always hold a special place in my heart for his guidance and support throughout my career, even in the last years of his life. Moreover, Bruce set me on a path that is reflected not only in my focus on the role of the immune system in psychiatric disorders but also in the cellular immune mechanisms involved, which is the topic of this brief review and preliminary data describing some of the first single cell RNA sequencing of peripheral blood mononuclear cells (PBMCs) from depressed patients.
1.1. Inflammation and depression
A rich literature has revealed that a significant proportion of patients with MD exhibit evidence of chronic inflammation as reflected by increased inflammatory markers in the peripheral blood and cerebrospinal fluid (CSF) with the peripheral blood cytokines tumor necrosis factor (TNF) and interleukin (IL)-6 and the acute phase reactant C-reactive protein (CRP) being some of the most reliably elevated in this regard (Dowlati et al., 2010; Howren et al., 2009; Miller and Raison, 2016). Reflecting the heterogeneity of depression, this increased inflammation occurs in ∼30% of otherwise medically stable MD patients, depending on multiple contributing factors including obesity, metabolic syndrome, aging, and childhood maltreatment, which are also known risk factors for depression (Miller and Raison, 2016; Miller et al., 2009; Osimo et al., 2019). In addition, increased inflammation is associated with treatment resistance to conventional antidepressants, and increased inflammatory markers predict response to advanced treatment strategies for depression including ketamine and electroconvulsive therapy in MD patients (Haroon et al., 2018a; Chamberlain et al., 2019; Yang et al., 2015; Kruse et al., 2018; Raison et al., 2013a). Supporting the notion that inflammation may play a cause and effect role in relevant depressive symptoms, peripheral administration of inflammatory stimuli including inflammatory cytokines such as interferon (IFN)-alpha as well as endotoxin or typhoid vaccination impact the brain to lead to symptoms of depression involving Research Domain Criteria positive and negative valence systems including anhedonia and psychomotor retardation as well as anxiety (Capuron et al., 2002, 2005; Harrison et al., 2009; Eisenberger et al., 2010). Moreover, blocking inflammation with anti-cytokine therapies has been shown to reverse these depressive symptoms in patients with autoimmune and inflammatory disorders and otherwise healthy depressed individuals with increased inflammation (Raison et al., 2013b; Kappelmann et al., 2018; Kohler-Forsberg et al., 2019; McIntyre et al., 2019).
Regarding the mechanisms by which inflammation affects the brain, much attention has been paid to the impact of inflammation on neurotransmitter systems and neurocircuits that regulate motivation and motor activity as well as anxiety, arousal, and alarm. Neuroimaging studies examining the impact of inflammation on neurocircuits have largely focused on subjects administered inflammatory stimuli (Harrison et al., 2009, 2015, 2016; Eisenberger et al., 2010; Capuron et al., 2005, 2012), but increasing data from our group and others have demonstrated that endogenous inflammation in MD (often indexed by blood concentrations of CRP) is also associated with alterations in activity of and functional connectivity (FC) within reward and motor circuits as well as circuits involving threat sensitivity that are in turn related to symptoms of anhedonia, psychomotor retardation and anxiety (Burrows et al., 2021; Felger et al., 2016; Mehta et al., 2018; Rengasamy et al., 2021; Savitz et al., 2013; Yin et al., 2019; Costi et al., 2021). A wealth of evidence from clinical and laboratory animal studies supports the idea that relationships between elevated inflammatory markers and deficits in these neurocircuits are driven by the impact of circulating inflammatory cytokines and peripheral blood immune cells on the brain to reduce availability and release of monoamines, notably dopamine, and increase synaptic and extrasynaptic glutamate in the anterior cingulate cortex and basal ganglia nuclei (Felger and Treadway, 2017; Felger et al., 2013a, 2013b, 2015; Yohn et al., 2016; Kitagami et al., 2003; Haroon et al., 2014, 2016, 2018b; Walker et al., 2013; Dantzer and Walker, 2014). Nevertheless, despite the vast database on the clinical and neurobiological effects of inflammation on the brain and behavior, there remain substantial gaps in our knowledge regarding the specific immune cell subtypes and their intracellular pathways that sustain inflammatory responses and/or support peripheral blood immune cell access to the brain that modulate CNS function and contribute to depressive symptoms.
1.2. Cellular immune mechanisms of inflammation in depression
Most studies examining the effects of endogenous inflammation on the brain in depression and other psychiatric disorders have utilized relatively non-specific inflammatory markers such as CRP to index inflammation. Though markers like CRP or circulating cytokines may be excellent proxies for inflammation, they give little information about the cellular and molecular pathways that comprise the inflammatory response and can ultimately serve as specific targets for biomarkers and therapeutic intervention. Of relevance in this regard, a recent study demonstrated that MD patients with increased CRP included two subgroups, one with an overrepresentation of peripheral blood myeloid cells and another with an overrepresentation of lymphoid cells (Lynall et al., 2020). These data suggest that increased inflammation in MD and ostensibly other psychiatric disorders is not a monolithic process and likely involves multiple cell subtypes and molecular pathways, each of which may have specific and potentially interacting effects and longitudinal trajectories that influence the brain and behavior (Felger and Miller, 2020). Given that nuanced immunologic changes and mediators are associated with various inflammatory states, it is not surprising that current anti-inflammatory therapies that have been tested in MD are largely non-specific and poorly targeted with modest effects at best (Kohler-Forsberg et al., 2019; Husain et al., 2020).
Recent data instantiating the relevance of peripheral blood immune cells to the CNS effects of inflammation reinforce the paramount need to identify cellular and molecular mechanisms of increased peripheral inflammation in depression. For example, studies in laboratory animals demonstrate that CD14+ peripheral blood monocytes expressing C–C Motif Chemokine Receptor 2 (CCR2) traffic to perivascular spaces in the brain and lead to the production of inflammatory cytokines that are essential for the development of stress-induced behavioral changes (McKim et al., 2018; Weber et al., 2017). Increased perivascular monocytes/macrophages have also been identified in postmortem brain samples of depressed patients who committed suicide (Torres-Platas et al., 2014). In addition, vulnerability to stress-induced depressive-like behavior in laboratory animals can be transferred from stress-vulnerable to stress-resilient animals through the transplant of bone marrow-derived immune cells, which reside in the peripheral blood and are distinct from yolk-sac derived microglia, the resident immune cells of the brain (Hodes et al., 2014). Moreover, in the context of stress and inflammation, cytokines produced by peripheral blood immune cells can access specific regions of the brain via disruption in the blood brain barrier (BBB) including notably the nucleus accumbens, a brain region that is a reliable target of inflammation in neuroimaging studies and plays a pivotal role in reward processing (Menard et al., 2017). Furthermore, inhibition of peripheral blood cytokines using anti-cytokine treatments that do not cross the BBB have been shown to block depressive-like behavior in rodents (Bayramgurler et al., 2013; O'Connor et al., 2009), while also reducing depressive symptoms including reward deficits, psychomotor slowing and anxiety in patients with autoimmune and inflammatory disorders as well as otherwise healthy depressed patients with increased inflammation (Raison et al., 2013b; Kappelmann et al., 2018; McIntyre et al., 2019; Salvadore et al., 2018; Davies et al., 2021). Finally, increasing data has characterized the circulation of peripheral blood immune cells, including both lymphocytes and monocytes, through perivascular spaces and the meninges, where they play a pivotal role in immunosurveillance of the brain under steady-state conditions and can modulate CNS function through release of inflammatory mediators during stress and disease (Rustenhoven and Kipnis, 2019; Louveau et al., 2015). Taken together, these data indicate that peripheral blood immune cells and their molecular pathways that drive inflammation and its effects on the brain represent a rich array of therapeutic targets and biomarkers for developing novel immunomodulatory approaches to treating psychiatric disorders.
1.3. Molecular pathways activated in peripheral blood immune cells from depressed patients
Previous studies have evaluated gene expression profiles in peripheral blood immune cells of MD patients versus healthy controls. Studies focusing on candidate gene expression have revealed activation of canonical inflammatory signaling pathways including toll-like receptors (TLRs) (Hung et al., 2014, 2017; Chen et al., 2017; Hajebrahimi et al., 2014; Keri et al., 2014), nuclear factor kappa B (NF-kB) (Weber et al., 2017; Keri et al., 2014; Guardado et al., 2016; Miklowitz et al., 2016), the NOD- LRR- and pyrin domain-containing protein (NLRP3) inflammasome complex, which via caspase-1 cleaves pro-interleukin (IL)-1beta into its mature form (Hasegawa et al., 2009; Momeni et al., 2016; Alcocer-Gomez et al., 2014), markers of oxidative stress (including reactive oxygen and nitrogen species [ROS, RNS]) (Galecki et al., 2012; Lukic et al., 2014) and the inflammatory cytokines themselves including TNF, IL-1beta, IL-6 and IFN-gamma (Tsao et al., 2006; Bekhbat et al., 2018). Of note, increased mRNA expression of TNF and IL-1beta have both been associated with antidepressant treatment non-response (Cattaneo et al., 2013, 2016; Belzeaux et al., 2012). Finally, studies in peripheral blood T cells have indicated that depressed patients exhibit an increased frequency of T regulatory cells expressing FOXP3 (Patas et al., 2018; Suzuki et al., 2017), although there are contradictory results, with at least one study indicating a decrease in T regulatory cells and an increased frequency of Th17 cells in depressed patients accompanied by higher mRNA levels of retinoic acid-related orphan receptor-γt, the specific transcription factor of Th17 cells (Chen et al., 2011).
Studies have also examined whole genome expression including bulk RNA sequencing (RNAseq), which like whole genome expression in PBMCs or whole blood provides transcriptome profiling across immune cell types without sufficient power to detect cell-type specific expression differences. Results indicate that peripheral blood immune cells from MD patients exhibit increased activation of signaling pathways related to inflammation and innate immune responses including pathways enriched for IL-6, Type I IFN, and the TNF receptor gene as well as accelerated aging (Spijker et al., 2010; Yi et al., 2012; Mostafavi et al., 2014; Guilloux et al., 2015; Jansen et al., 2016; Hori et al., 2016; Leday et al., 2018; Le et al., 2018; Cole et al., 2021). Moreover, consistent with the impact of inflammation on neurocircuits involving subcortical nuclei, gene networks interconnecting TNF and NF-kB have been linked with the morphology of the caudate (Savitz et al., 2013). Finally, a study in women with postpartum depression identified gene pathways in depressed subjects associated with bioenergetics including glycolysis/gluconeogenesis and lipid metabolism in conjunction with pathways related to TLR signaling and cytokine/cytokine receptor interactions (Pan et al., 2018). These latter two findings are consistent with our previous results in bulk PBMCs in MD patients indicating that canonical inflammatory signaling pathways including TNF, TLRs and NF-kB as well as metabolic pathways involving insulin signaling, glycolysis and lipid metabolism are associated with anhedonia and psychomotor slowing, as well as both fMRI FC between both ventral and dorsal striatum (respectively) and ventromedial prefrontal cortex (vmPFC) and the behavioral response to the TNF antagonist infliximab (Raison et al., 2013b; Mehta et al., 2013; Bekhbat et al., 2020, 2021; Goldsmith et al., 2020). For example, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways related to insulin signaling, insulin resistance, hypoxia-inducible factor 1-alpha (HIF-1 alpha) signaling and the phosphoinositide 3-kinase (PI3K)/protein kinase B (also referred to as Akt) signaling pathway were significantly associated with anhedonia as well as psychomotor slowing in MD patients with high inflammation (Bekhbat et al., 2020, 2021). Taken together, these data indicate that, in addition to canonical activation of inflammatory signaling pathways, there is molecular activation of pathways associated with metabolism involving insulin signaling, glycolysis and lipid metabolism under aerobic conditions that reflect the immunometabolic shifts that have been shown to occur in activated immune cells and represent a “Warburg” effect (O'Neill et al., 2016). Although evolutionarily designed to address energy demands of a robust protective response against perceived or actual external or internal threats, this shift in metabolism is highly energy inefficient and may contribute to a shift in behavioral priorities away from energy expenditure and towards energy conservation as reflected by reduced motivation and motor activity (Miller and Raison, 2016; Wang et al., 2019; Treadway et al., 2019). However, the relationship between immunometabolic shifts in specific immune cell subtypes and behavioral measures of motivation has not been examined. Finally, despite many studies using flow cytometric analyses to enumerate peripheral blood immune cells in MD, there is a dearth of studies characterizing immune cells subsets using gene expression analyses as represented by single cell RNA sequencing (scRNA-Seq).
Thus, significant knowledge gaps in the literature include: 1) few studies have characterized immune pathways and mechanisms in individual cell subsets in MD patients with increased inflammation; 2) no study to our knowledge has taken advantage of the technological advances in immunology including scRNA-Seq to allow identification of relevant signaling pathways within specific immune cell subtypes; 3) no studies have mapped gene expression pathways in specific immune cell subtypes to behavior; and 4) virtually all studies have compared MD patients to HC, which presupposes that MD is a solitary disorder, with limited appreciation for the heterogeneity of the disease as it relates to inflammation or otherwise (wherein lies the opportunity for subgrouping and precision medicine).
To gain preliminary insight into these knowledge gaps and reveal the immune cell subsets as reflected by their transcriptomic signatures that underlie increased inflammation and its relationship with behavior in MD at the single-cell level, herein we conducted scRNA-Seq on a subset of previously collected, cryopreserved PBMC samples from a small but well-characterized sample of medically-stable, unmedicated MD outpatients.
2. Methods
2.1. Participants
Samples were analyzed from MD patients with low (≤3 mg/L; n = 3) and high plasma CRP (>3 mg/L; n = 3). Samples from patients with high CRP were obtained both before and after anti-inflammatory challenge with infliximab (two weeks after a single infusion of 5 mg/kg) (Raison et al., 2013b; Mehta et al., 2013), for a total of 9 samples. To limit demographic differences, patients in the low and high CRP groups were selected to be female and from one racial and ethnic background (African American, non-Hispanic). Age and BMI also did not differ between groups (p = 0.2–0.5) and were within 1 standard deviation of the means of MD patients in our previous studies (Felger et al., 2016, 2020; Mehta et al., 2013; Bekhbat et al., 2020).
Subjects were recruited from a community sample of MD patients through social media campaigns, and subjects signed written informed consent before study participation. All studies were a priori approved by the Emory Institutional Review Board. Screening included: 1) Past psychiatric history and current symptom severity using Structured Clinical Interview for DSM-V (SCID-V) (First et al., 1997), 17-item HAM-D and the 30-item IDS-SR (Hamilton, 1960; Rush et al., 1996), 2) Medical history from patient interview and review of medical records, 3) Complete medical and neurological examination including Mini Mental State Exam (Folstein et al., 1975), 4) Screening laboratory evaluation, and 5) hsCRP [obtained twice over a 2-week period as per AHA/CDC guidelines to establish stability and rule out acute infection (Pearson et al., 2003)]. No patients were removed from medication for the purposes of the study. All subjects were off all psychotropic medications for at least 4 weeks (8 weeks for fluoxetine). Additionally, all subjects were free of any autoimmune or inflammatory disorders as well as acute or chronic infectious diseases or acute inflammation and must have been off all anti-inflammatory drugs and supplements for at least 2 weeks. Patients had a primary diagnosis of MD and did not have a co-morbid diagnosis of obsessive compulsive disorder, an eating disorder or antisocial personality disorder. Patients with an active suicidal plan or intent as determined by a score >3 on item #3 of the HAM-D were excluded, and suicide status was monitored throughout the study using the Columbia Suicide Severity Rating Scale (Posner et al., 2007).
2.2. Study procedures
Following screening, subjects underwent baseline fasting blood sampling and clinical assessment of anhedonia using the Snaith-Hamilton Pleasure Scale-Clinician rated (SHAPS-C) (Snaith et al., 1995; Ameli et al., 2014). For subjects who received infliximab or placebo, study drug was dispensed according to a computer-generated randomization list provided by the research pharmacist. A safety visit was conducted at week 1 to evaluate psychiatric and medical status, and at week 2, subjects underwent the same procedures as indicated for the baseline visit.
2.3. Collection of PBMCs and plasma
Fasting whole blood samples were obtained between 8 and 10 a.m. after 30 min of rest (to avoid circadian influences and effects of stress) into chilled EDTA tubes using standard sterile techniques. Plasma was obtained from whole blood by centrifugation at 4 °C, aliquoted and stored at −80 °C until batched assay of CRP. PBMCs were isolated from EDTA whole blood on Histopaque‐1077. After washing, cells were counted, and 3–5 million cells were resuspended in sterile freezing media (90% FBS and 10% DMSO). Cells were slowly frozen in an isopropanol-containing cryopreservation vessel at −80 °C for 24 h then stored in liquid nitrogen.
2.4. Inflammatory marker measurement
High sensitivity (hs) CRP was assayed using an immunoturbidometric assay with a Beckman AU480 chemistry analyzer and Ultra WR CRP kit (Sekisui Diagnostics) (Raison et al., 2013b; Felger et al., 2016).
2.5. scRNA sequencing and analyses
Carefully thawed samples exhibited >70% viability, and dead cells were removed to ensure consistency across samples. Viable cells were processed on a 10x Chromium Controller using the 3′ GEX v3.1 platform (10x Genomics). Single cell gene expression library was prepared per manufacturer's instructions and sequenced on Illumina HiSeq (Illumina, California, USA). scRNA-Seq reads for gene quantification were analyzed using Seurat 3.1.5 (Stuart et al., 2019) after quality control filtering, alignment to reference genome and normalization to perform unsupervised and supervised analysis (Tran et al., 2016; Raof et al., 2016) (see SI for details). Cells were partitioned into clusters based on transcriptome profiles, and batch correction was conducted using Harmony (Korsunsky et al., 2019). Nonlinear dimensional reduction of the sc-transcriptomes was performed with the uniform manifold approximation and projection (UMAP) technique (LJPvd Hinton, 2008). Data were visualized via two-dimensional UMAP plots. Cell type identities of each cluster were determined using automated annotation via SingleR (Aran et al., 2019), and refined using cell-type specific canonical markers (see SI, Figs. S1A–B). Differential expression analyses were conducted in Seurat followed by functional enrichment analysis using WikiPathways and KEGG (Kyoto Encyclopedia Gene and Genome) databases as implemented in clusterProfiler (Yu et al., 2012) as well as GeneGo MetaCore (St. Joseph, MI, USA) (see SI). An FDR significance threshold of q < 0.1 (Storey and Tibshirani, 2003) was used herein for pathway analysis to ensure biologically meaningful pathway results (Mostafavi et al., 2014; Jansen et al., 2016; Hulsegge et al., 2009; Yang et al., 2008, 2014; de Kluiver et al., 2019; Zhou et al., 2018).
2.6. Statistical analysis
Demographic and clinical variables were characterized using descriptive statistics. Associations between cell clusters and clinical variables were tested using Spearman correlations. Analyses were conducted in IBM SPSS Statistics 27.0 (New York, NY, USA).
3. Results
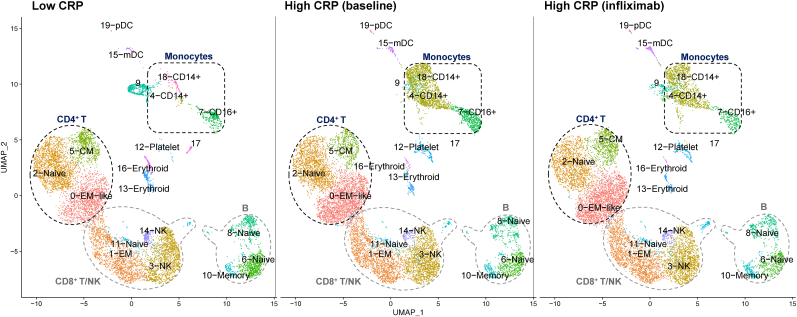
Demographic and clinical variables of the study sample are shown in Table 1. PBMC samples from MD patients with low and high CRP (before and after infliximab) comprised a total of 33,833 cells in 20 unique clusters (Fig. 1, Table S1A). Of these, 10,311 cells were from three patients with low CRP, 12,167 from three patients with high CRP at baseline, and 11,355 from the same three patients with high CRP two weeks after infliximab. In the UMAP plots, clusters were assigned unique colors according to the unsupervised clustering in Seurat and labeled according to their cell subtypes. Five clusters were identified as T cells (CD3D), two as NKs (KLRB1, NKG7) (Wang et al., 2021; Pizzolato et al., 2019), three as monocytes (CD68, LYZ, S100A9, CD14 or FCGR3A [CD16]) (Wang et al., 2021; Pizzolato et al., 2019; Zhu et al., 2020), two as dendritic cells (DCs)(LYZ, S100A9, FCER1A) (Pizzolato et al., 2019), and three as B cells (CD19, MS4A1 or CD20, CD79A, CD79B) (Wang et al., 2021). Five small clusters were identified as erythrocytes, platelets, or not classified as one of the major leukocyte subsets. Top-expressed genes in each cluster are shown in Table S1B. Based on differential cell counts between the low and high CRP groups, as well as subsequent modulation by infliximab (Table S1A), monocyte and CD4+ T cell clusters were of primary interest for subsequent analyses.
Table 1.
Demographic and clinical variables of the study sample.
| Low CRP (≤3 mg/L) n = 3 | High CRP (>3 mg/L) n = 3; Baseline | ||
|---|---|---|---|
| Age | Median (IQR) | 38 (0.5) | 43 (6.5) |
| BMI | Mean (SD) | 32.2 (5.1) | 35.5 (5.8) |
| hsCRP (mg/L) | Median (IQR) | 0.6 (0.7) | 4.8 (6.8) |
| HAM-D | Mean (SD) | 24 (2) | 22 (3) |
| IDS-SR | Mean (SD) | 33.3 (9.8) | 26 (8) |
| SHAPS-C | Mean (SD) | 32.3 (9.6) | 35.7 (7.8) |
IQR, interquartile range; SD, standard deviation; BMI, body mass index; hsCRP, high-sensitivity C-reactive protein; HAM-D, Hamilton Depression Rating Scale; IDS-SR, Inventory of Depressive Symptomology-Self Report; SHAPS-C, Clinician-rated Snaith-Hamilton Pleasure Scale.
Fig. 1.
Single-cell RNA-seq analysis of peripheral blood mononuclear cell (PBMC) samples from MD patients. Two-dimensional uniform manifold approximation and projection (UMAP) plots were derived from the single-cell transcriptomes of 10,311 cells from three patients with low CRP, 12,167 cells from three patients with High CRP before infliximab, and 11,355 cells from the same three patients with high CRP two weeks after infliximab. Each dot represents a single cell. Clusters were colored according to the unsupervised clustering in Seurat and annotated using canonical marker genes for cell types enriched in each cluster. CRP, C-reactive protein; EM, effector memory; CM, central memory; DC, dendritic cell; pDC, plasmacytoid DC; mDC, myeloid DC; NK, natural killer.
3.1. Activated monocytes/macrophages in MD with high CRP and response to anti-inflammatory challenge
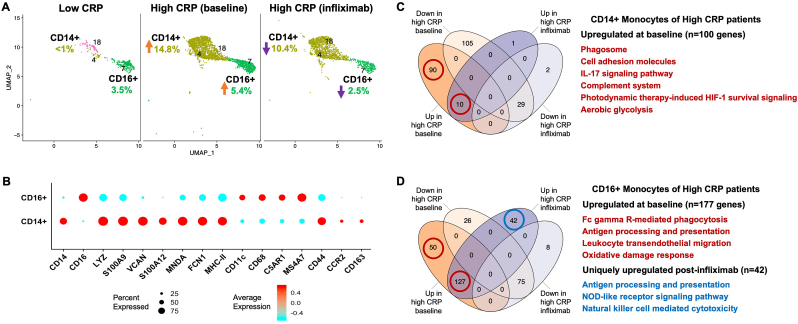
We identified three clusters of monocyte lineage cells (Fig. 2A). A large CD14+ cluster, which expressed markers consistent with classical CD14+CD16−monocytes (Fig. 2B), was dramatically increased in high CRP patients (from 0.5 to 14.8% of PBMCs, t(4) = 6.12, p = 0.004), and the number of cells was moderately reduced by 29% after infliximab (14.8 vs 10.4% of PBMCs, t(2) = 1.53, p = 0.27). Conversely, CD16+ monocytes, which expressed macrophage activation markers such as CD68, C5AR1 and MS4A7, were ∼74% more abundant in MD patients with high CRP (t(4) = 1.13, p = 0.32) and reduced by 55% after infliximab (5.4 vs 2.5% of PBMCs, t(2) = 3.04, p = 0.09). (Fig. 2A). Interestingly, both monocyte populations expressed molecules and pathways related to chemotaxis. For example, the CD14+CD16−cells (consistent with classical monocytes) were the only cluster to express CCR2, which was found in a subset of cells but at a high intensity (Fig. 2B). Genes upregulated in these CD14+ monocytes, which were unique to MD with high CRP, were significantly enriched for a number of pathways related to inflammation (Phagosome, Complement system), immune cell migration (Cell adhesion molecules, IL-17 signaling pathway (Garza-Reyes et al., 2020)), and immunometabolic shifts thought to play a role in myeloid-driven inflammation (HIF-1 signaling, aerobic glycolysis)(all p < 0.05 and q < 0.1; Fig. 2C, Figs. S2–3; see Table S2 for all pathway results). The CD16+ cells expressed C5AR1 and MS4A7, which are molecules expressed on myeloid cells that traffic to tissues including brain and are found to contribute to CNS inflammation in several disease states (Hernandez et al., 2017; Wang et al., 2017; Jordao et al., 2019; DePaula-Silva et al., 2019; Buckner et al., 2011). Genes upregulated in CD16+ monocytes of high CRP patients were enriched for pathways including Fc gamma R-mediated phagocytosis, Antigen processing and presentation, Leukocyte transendothelial migration, and the Oxidative damage response (all p < 0.05 and q < 0.1; Fig. 2D). While only 42 genes (and no pathways) in CD14+ cells were differentially expressed post-infliximab, the CD16+ cluster had evidence of both reduced cell percentage (from 5.4 to 2.5%) post-infliximab (Fig. 2A) and differential regulation of inflammatory genes (Tables S2–3). In addition to the 177 genes that were upregulated in CD16+ monocytes of high CRP patients at baseline, 42 genes were uniquely up-regulated post-infliximab, and enriched pathways including Antigen processing and presentation, NOD-like receptor signaling pathway, and Natural killer cell mediated cytotoxicity (all p < 0.05 and q < 0.1; Fig. 2D; see Table S3 for full pathway results). Additionally, exploratory analyses suggested that CD16+ monocytes, and to a lesser extent CD14+ monocytes, in MD patients with high versus low CRP showed indices of glucocorticoid resistance such as lower expression of the glucocorticoid-inducible anti-inflammatory genes TSC22D3, DUSP1 and KLF2 in CD16+ monocytes and KLF2 in CD14+ monocytes (Fig. S4 and Table S5). (Hasselmann et al., 2018; Menke et al., 2012; Das et al., 2006).
Fig. 2.
CD14+ and CD16+ monocyte subsets were increased in MD with high vs. low inflammation and reduced by infliximab. Whereas CD14+ monocytes were only present in high vs. low CRP (>3 versus ≤3 mg/L) and were moderately reduced by infliximab (29%), CD16+ monocytes were both increased by 74% in high CRP and reduced by 55% post-infliximab (N = 3 per group). Orange and purple arrows indicate direction of changes in cell abundance as % of PBMCs in high vs low CRP and high CRP baseline vs post-infliximab comparisons (A). Dot plot depicting the average expression level and % of cells expressing activation- and chemotaxis-related markers across CD14+ and CD16+ monocytes (B). The number of differentially expressed genes in high CRP baseline and post-infliximab conditions, and representative enriched pathways for both CD14+ (C) and CD16+ (D) monocyte subsets. Red and blue circles indicate genes upregulated in high CRP patients at baseline or uniquely upregulated post-infliximab, respectively, and enrich pathways corresponding to their colors. CRP, C-reactive protein; UMAP, uniform manifold approximation and projection; HIF-1, hypoxia-inducible factor-1; NOD-like, nucleotide-binding oligomerization domain-like. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.2. CD4+ T cells in MD with high CRP and response to anti-inflammatory challenge
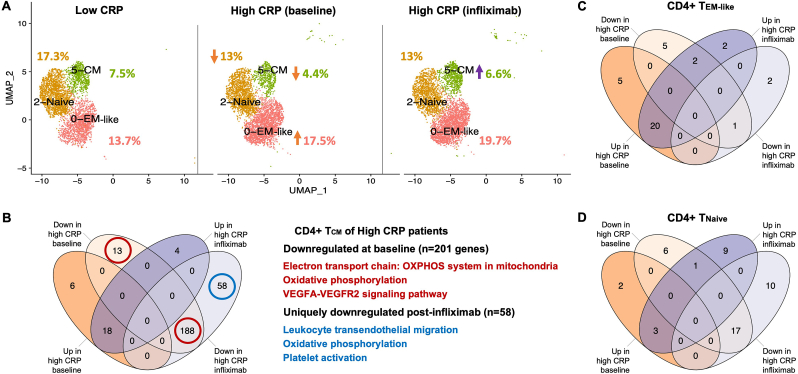
Differences in CD4+ T cell subpopulations were revealed between MD patients with low vs. high CRP, which also differentially responded to infliximab (Fig. 3A). Shifts in the CD4+ T cell compartment included ∼30% and ∼10% lower abundance of CD4+ central memory T cells (TCM), and Tnaive cells respectively (t(4) = 3.87, p = 0.018; t(4) = 0.73, p = 0.51), along with a ∼50% increase in a cluster consistent with effector memory-like (TEM-like) cells (t(4) = 2.63, p = 0.059) in high versus low CRP patients (Fig. 3A). While TCM cells are known to rely primarily upon oxidative phosphorylation (OXPHOS) as a main energy source (Bantug et al., 2018), CD4+ TCM cells of high CRP patients displayed downregulation of 201 genes involved in OXPHOS, electron transport chain, and VEGFA-VEGFR2 signaling pathways (all p < 0.05 and q < 0.1; Fig. 3B; see Table S4 for full pathway results). Although infliximab partially restored the percentage of CD4+ TCM cells, an additional 58 genes were uniquely downregulated in CD4+ TCM cells post-infliximab, and were enriched for Leukocyte transendothelial migration, OXPHOS, and Platelet activation pathways (all p < 0.05 and q < 0.1; Fig. 3B; see Table S4). TEM-like and Tnaive cells of high CRP patients displayed few differentially expressed genes at baseline or following infliximab (Fig. 3C–D).
Fig. 3.
Cellular and molecular shifts in CD4+ T cell clusters in MD with high inflammation and response to infliximab. Shifts in the CD4+ T cell compartment included ∼30% and ∼10% lower abundance of CD4+ central memory T cells (TCM) and Tnaive cells respectively as well as ∼50% higher abundance in a cluster consistent with effector memory-like (TEM-like) cells in high CRP patients (N = 3 per group). Orange and purple arrows indicate direction of changes in cell abundance as % of PBMCs in high vs low CRP and high CRP baseline vs post-infliximab comparisons (A). The numbers of differentially expressed genes in TCM cells of high CRP patients at baseline and post-infliximab are shown along with representative pathways enriched by genes downregulated at baseline (red circles) or uniquely downregulated post-infliximab (blue circle)(B). The numbers of differentially expressed in TEM-like (C) and Tnaive cells (D) of high CRP patients at baseline and post-infliximab. CRP, C-reactive protein; UMAP, uniform manifold approximation and projection; HIF-1, hypoxia-inducible factor-1; EM, effector memory; CM, central memory; OXPHOS, oxidative phosphorylation; VEGF, vascular endothelial growth factor A; VEGFR, vascular endothelial growth factor receptor. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.3. Relationships between monocytes and CD4+ T cells and severity of anhedonia and the response to anti-inflammatory challenge
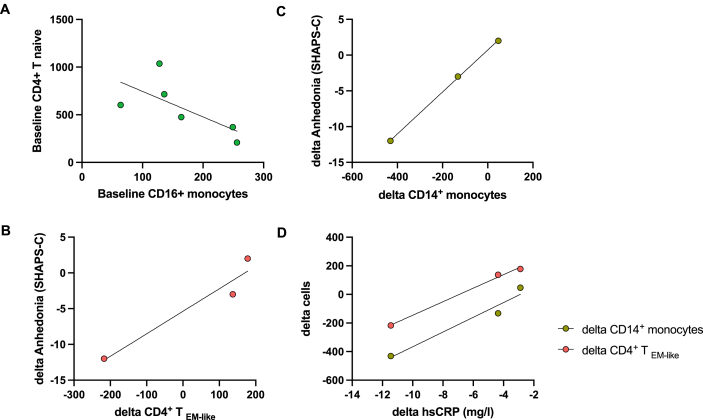
Baseline numbers of CD16+ monocytes negatively correlated with CD4+ Tnaive cells in all patients (rs = 1.0, p < 0.001)(Fig. 4A). MD patients for the current study were selected to minimize variability in sex, age, race, ethnicity, and BMI (see 2.1 above). Interestingly, the subjects with high CRP varied with respect to their anhedonia response to infliximab, which allowed for a preliminary assessment of relationships between immune cell subsets and response to infliximab. Change in the number of CD14+ monocytes (rs = 1.0, p < 0.001), but not the CD16+ cluster (rs = −0.5, p > 0.05), predicted change in anhedonia scores as measured by SHAPS-C (Fig. 4B). Similarly, change in the number of CD4+ TEM-like cells predicted change in anhedonia scores (Fig. 4C). Interestingly, infliximab-induced change in hsCRP was significantly associated with change in the number of both CD14+ monocytes and CD4+ TEM-like cells (rs = 1.0, p < 0.001 each)(Fig. 4D).
Fig. 4.
Monocyte and CD4+ T cell clusters at baseline and post-infliximab: relationships with changes in CRP and anhedonia. At baseline, the number of CD16+ monocytes negatively correlated with CD4+ Tnaive cells (rs = 1.0, p < 0.001)(A). Change in the number of CD14+ monocytes (B) and CD4+ TEM-like cells (C) predicted change in anhedonia scores as measured by SHAPS-C (rs = 1.0, p < 0.001 each). Infliximab-induced decrease in hsCRP was also associated with change in the number of both CD14+ monocytes and CD4+ TEM-like cells (rs = 1.0, p < 0.001 each)(D). hsCRP, high sensitivity C-reactive protein; EM, effector memory; SHAPS-C, Clinician-rated Snaith-Hamilton Pleasure Scale.
4. Discussion
The preliminary data reported herein indicate that both myeloid and lymphoid cells including CD14+ and CD16+ monocyte as well as CD4+ T effector memory-like (TEM-like) populations were enriched in MD patients with increased inflammation (as reflected by a CRP >3 mg/L). Moreover, the data indicated that enriched monocyte subsets expressed signaling pathways involving inflammatory networks and/or metabolic reprograming (e.g., HIF-1 signaling, aerobic glycolysis). Improvement in anhedonia in response to infliximab in patients with high inflammation was associated with reduction in the CD14+ monocyte cell population as well as the TEM-like cell population. These findings are the first step toward determining the cellular and molecular immune pathways and networks associated with high inflammation in MD, as well as those that are impacted by inhibition of TNF by infliximab in relation to brain and behavioral responses. Future analyses in a larger sample may lead to identification of new immunomodulatory targets and biomarkers for treatments that may have greater specificity and efficacy (and therefore safety) than available anti-inflammatory therapies including cytokine antagonists that carry a high risk of infection, exacerbation of demyelinating conditions and the possibility of cancer.
Relevant to the extant literature, these data are the first evidence from humans that cells with a similar phenotype as the Ly6ChiCCR2+ monocytes frequently reported in the rodent literature to traffic to the brain and mediate the inflammatory effects of stress on anxiety and depressive-like behavior (consistent with CD14+CD16−negative classical monocytes that express high CCR2) (Weber et al., 2017; Wohleb et al., 2014a, 2014b) are selectively increased in MD patients that have high inflammation. Of note, consistent with these data, an increase in classical monocytes in the peripheral blood of depressed patients with increased inflammation has been previously described, however CCR2 was not measured in this study (Lynall et al., 2020). Additionally, a second population of CD16+ monocytes with increased expression of markers characteristic of activated macrophages, was both higher in MD with high CRP and transcriptionally impacted by anti-TNF challenge with infliximab in association with a 55% reduction in cell percentage. Interestingly, these cells expressed markers found on monocytes that infiltrate the brain or perivascular associated-monocytes (Hernandez et al., 2017; Wang et al., 2017; Jordao et al., 2019; DePaula-Silva et al., 2019; Buckner et al., 2011). Of note, the CD14+CD16−cluster was only moderately impacted by infliximab in terms of cell percentage and transcriptomics, suggesting that TNF inhibition resolved only a part of the immunologic and transcriptomic landscape associated with high CRP in MD, possibly contributing to the limited overall responsiveness of depressive symptoms to infliximab in treatment resistant MD subjects.
The evidence of shifts in CD4+ T cell subsets in favor of the more activated TEM-like (e.g. Th1 and Th17 cell populations) and a decline in naïve and central memory T subsets in MD patients with high inflammation as revealed by scRNA-Seq is important in that it highlights the strength of sc-transcriptomic approaches to identify subsets of larger cell populations and their metabolic profiles that might otherwise be missed by techniques such as flow cytometry. These preliminary data also suggest that the mechanism of inflammation in MD goes beyond myeloid lineage cells to include the T cell compartment, and that inflammatory shifts toward TEM-like and away from TCM may be modified by TNF blockade. Interestingly, myeloid and lymphoid cell lineages may interact as higher numbers of CD14−CD16+ monocytes were associated with fewer CD4+ naïve T cells, consistent with reports showing that CD16+ monocytes promote pro-inflammatory responses in co-cultured CD4+ naïve T cells such as increased IFN-γ and IL-17 expression, while suppressing T regulatory cell proliferation (Roberts et al., 2015). Our findings of bioenergetic transcriptomic shifts in relation to anhedonia are consistent with the role of immune-metabolic interactions in reward deficits (Bekhbat et al., 2020; Goldsmith et al., 2020, 2021; Nettis et al., 2019), and suggest that MD with high inflammation may involve a sustained activation of immune cells (trained immunity) that impact the brain to drive symptoms. While the specific immune cells that are driving these effects remain unclear, our preliminary data herein indicate that they predominantly occur in CD14+CD16−monocytes as well as CD4+ TEM-like cells. Of note, the data presented also revealed that the patient with the largest decrease in both CD14+CD16−monocytes and CD4+ TEM-like cells had the greatest improvement in anhedonia in response to infliximab, and that the patient who had no change in CD14+CD16−monocytes had no change in anhedonia. Despite these differential responses, all patients exhibited infliximab-induced decreases in CRP. Although it remains possible that infliximab-related changes in anhedonia mediated the changes in immune cell subsets, these data support the notion that variability in the cellular response to anti-inflammatory treatments within subpopulations of immune cells may serve as a more accurate biomarker of whether the immune system has been sufficiently engaged to effect a behavioral change, compared to more non-specific markers of inflammation such as CRP.
There are several strengths and limitations of the data presented. The sample represents a homogenous group of subjects who were medically stable and without inflammatory conditions or treatment with psychotropic drugs. Moreover, subjects were studied under controlled conditions, and patients with high inflammation were assessed both before and after administration of a highly specific biologic antagonist of TNF. Nevertheless, the sample size was very small and was limited to a discrete population of Black females. Thus, despite the relatively large effect sizes and correlations with behavior, there may be variability as a function of sex and race. In addition, no protein markers were obtained so that the identification of specific cell subsets was based on bioinformatic analysis of gene expression data only. Studies combining proteomics and transcriptomics (e.g., by using Cellular Indexing of Transcriptomes and Epitopes by Sequencing [CITE-Seq]) would be an important complement to future work. Nevertheless, given the limited repertoire of protein markers currently available for flow cytometry and barcodes for CITE-Seq, advanced analytic strategies combining both bulk and single cell sequencing data (Sun et al., 2021) represent a powerful alternative to identify cell subpopulations driving a behavioral phenotype at the single cell level while leveraging greater statistical power afforded by bulk sequencing and phenotypic data.
Taken together, our data and the review of the literature indicate that multiple immune cell types and pathways appear to participate in the chronic inflammatory response as well as behavioral changes found in MD patients. Many of these pathways and cells can be uniquely targeted by existing drugs (see Table 2). (Kubo et al., 2018; Palsson-McDermott and O'Neill, 2020) Moreover, evolving immunotherapies are becoming increasingly specific, including for example drugs that target cytokines that activate specific immune cell subpopulations (e.g. IL-12 and IL-23 activation of Th1 and Th17 cells, respectively)(Iwakura and Ishigame, 2006) as well as drugs that target the specific cells (e.g. nanotechnology to target macrophages)(Hu et al., 2019) and molecular pathways involved in inflammation. Finally, treatments targeting immunometabolism are also rapidly evolving, and many are available including for example rapamycin, which blocks mammalian target of rapamycin (mTOR) pathways and has been shown to augment the antidepressant response to ketamine (Abdallah et al., 2020), as well as dimethyl fumarate, which blocks glycolysis (Kornberg et al., 2018). Nevertheless, to take full advantage of these advances in anti-inflammatory therapeutics, a more nuanced understanding of the cellular and molecular immunologic processes that drive inflammation in depression and other psychiatric disorders is required. Such an understanding will allow the development of better, more precise biomarkers and treatments, while also providing informed target engagement strategies to monitor treatment efficacy of novel agents at the immune level.
Table 2.
Relevant pathways and cells that can be targeted by existing drugs.
| Drug | Target | Immune Cells Affected |
|---|---|---|
| Dimethyl Fumarate | Keap1/Nrf2, NF-kB, IRAK4 | T and B cells, NK cells, DC, macrophages, neutrophils |
| Metformin | AMP kinase | T and B cells, macrophages |
| Methotrexate | Dihydrofolate reductase | T and B cells, neutrophils, macrophages |
| Rapamycin | mTORC1 | T cells, macrophages, NK cells |
| TEPP-46 | Pyruvate kinase M2 | CD4+ T cells, B cells, NK cells, macrophages |
| Baricitinib | JAK1/JAK2 | T and B cells, DC |
Abbreviations: Keap1, Kelch Like ECH Associated Protein 1; Nrf2, Nuclear factor-erythroid factor 2-related factor 2; NF-kB, Nuclear Factor kappa-light-chain-enhancer of activated B cells; IRAK4, interleukin-1 receptor-associated kinase 4; AMP kinase, AMP-activated protein kinase; mTORC1, mammalian target of rapamycin complex 1; TEPP-46, a small molecular agonist of pyruvate kinase M2; JAK, Janus Kinase; NK, natural killer; DC, dendritic cells.
Funding and disclosure
This work was supported by grants R61MH121625 (JCF); R21MH121891 and R01MH128872 (JCF/AHM); R01MH112076 (AHM); and F32MH119750 (MB) from the National Institute of Mental Health, Shariatzadeh and Sanei Award for Innovative Research in Mood Disorders from Emory University Department of Psychiatry and Behavioral Sciences, CADF49143 from the Dana Foundation and grants BBRF22296 and BBRF26983 (a NARSAD Distinguished Investigator Grant) from the Brain & Behavior Research Foundation. In addition, the study was supported in part by PHS Grants UL1TR000454, UL1TR002378, KL2TR000455, and TL1TR002382 from the National Center for Advancing Translational Sciences, and P30CA138292 from the National Cancer Institute. All funding sources had no role in study design; in the collection, analysis and interpretation of data; in the writing of the report; and in the decision to submit the article for publication. In the past 3 years, JCF has consulted with Otsuka; MKB has consulted with Canomiks Inc, Anxomics LLC, Sanofi-Genzyme Inc; and AHM has consulted with Cerevel; all on topics unrelated to this work. The remaining authors have nothing to disclose.
CRediT authorship contribution statement
Mandakh Bekhbat: Conceptualization, Methodology, Software, Validation, Formal analysis, Data curation, Writing – original draft, Writing – review & editing, Visualization, Project administration, Funding acquisition. G. Bengü Ulukaya: Methodology, Software, Validation, Formal analysis, Data curation, Visualization. Manoj K. Bhasin: Conceptualization, Methodology, Software, Validation, Formal analysis, Resources, Data curation, Visualization, Supervision. Jennifer C. Felger: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Resources, Data curation, Writing – review & editing, Visualization, Supervision, Project administration, Funding acquisition. Andrew H. Miller: Conceptualization, Investigation, Resources, Writing – original draft, Writing – review & editing, Supervision, Project administration, Funding acquisition.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ynstr.2022.100462.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Abdallah C.G., Averill L.A., Gueorguieva R., Goktas S., Purohit P., Ranganathan M., Sherif M., Ahn K.H., D'Souza D.C., Formica R., Southwick S.M., Duman R.S., Sanacora G., Krystal J.H. Modulation of the antidepressant effects of ketamine by the mTORC1 inhibitor rapamycin. Neuropsychopharmacology: Off. Publ. Am. Coll. Neuropsychopharmacol. 2020;45(6):990–997. doi: 10.1038/s41386-020-0644-9. PMCID: PMC7162891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcocer-Gomez E., de Miguel M., Casas-Barquero N., Nunez-Vasco J., Sanchez-Alcazar J.A., Fernandez-Rodriguez A., Cordero M.D. NLRP3 inflammasome is activated in mononuclear blood cells from patients with major depressive disorder. Brain Behav. Immun. 2014;36:111–117. doi: 10.1016/j.bbi.2013.10.017. [DOI] [PubMed] [Google Scholar]
- Ameli R., Luckenbaugh D.A., Gould N.F., Holmes M.K., Lally N., Ballard E.D., Zarate C.A., Jr. SHAPS-C: the Snaith-Hamilton pleasure scale modified for clinician administration. PeerJ. 2014;2 doi: 10.7717/peerj.429. PMCID: PMC4081294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aran D., Looney A.P., Liu L., Wu E., Fong V., Hsu A., Chak S., Naikawadi R.P., Wolters P.J., Abate A.R., Butte A.J., Bhattacharya M. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 2019;20(2):163–172. doi: 10.1038/s41590-018-0276-y. PMCID: PMC6340744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bantug G.R., Galluzzi L., Kroemer G., Hess C. The spectrum of T cell metabolism in health and disease. Nat. Rev. Immunol. 2018;18(1):19–34. doi: 10.1038/nri.2017.99. [DOI] [PubMed] [Google Scholar]
- Bayramgurler D., Karson A., Ozer C., Utkan T. Effects of long-term etanercept treatment on anxiety- and depression-like neurobehaviors in rats. Physiol. Behav. 2013;119:145–148. doi: 10.1016/j.physbeh.2013.06.010. [DOI] [PubMed] [Google Scholar]
- Bekhbat M., Chu K., Le N.A., Woolwine B.J., Haroon E., Miller A.H., Felger J.C. Glucose and lipid-related biomarkers and the antidepressant response to infliximab in patients with treatment-resistant depression. Psychoneuroendocrinology. 2018 doi: 10.1016/j.psyneuen.2018.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekhbat M., Treadway M.T., Goldsmith D.R., Woolwine B.J., Haroon E., Miller A.H., Felger J.C. Gene signatures in peripheral blood immune cells related to insulin resistance and low tyrosine metabolism define a sub-type of depression with high CRP and anhedonia. Brain Behav. Immun. 2020;88:161–165. doi: 10.1016/j.bbi.2020.03.015. PMCID: PMC7415632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekhbat M., Goldsmith D.R., Woolwine B.J., Haroon E., Miller A.H., Felger J.C. Transcriptomic signatures of psychomotor slowing in peripheral blood of depressed patients: evidence for immunometabolic reprogramming. Mol. Psychiatr. 2021;26(12):7384–7392. doi: 10.1038/s41380-021-01258-z. PMCID: PMC8881295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belzeaux R., Bergon A., Jeanjean V., Loriod B., Formisano-Treziny C., Verrier L., Loundou A., Baumstarck-Barrau K., Boyer L., Gall V., Gabert J., Nguyen C., Azorin J.M., Naudin J., Ibrahim E.C. Responder and nonresponder patients exhibit different peripheral transcriptional signatures during major depressive episode. Transl. Psychiatry. 2012;2 doi: 10.1038/tp.2012.112. PMCID: PMC3565773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckner C.M., Calderon T.M., Willams D.W., Belbin T.J., Berman J.W. Characterization of monocyte maturation/differentiation that facilitates their transmigration across the blood-brain barrier and infection by HIV: implications for NeuroAIDS. Cell. Immunol. 2011;267(2):109–123. doi: 10.1016/j.cellimm.2010.12.004. PMCID: PMC4335637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrows K., Stewart J.L., Kuplicki R., Figueroa-Hall L., Spechler P.A., Zheng H., Guinjoan S.M., Tulsa I., Savitz J.B., Kent Teague T., Paulus M.P. Elevated peripheral inflammation is associated with attenuated striatal reward anticipation in major depressive disorder. Brain Behav. Immun. 2021;93:214–225. doi: 10.1016/j.bbi.2021.01.016. PMCID: PMC7979507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capuron L., Gumnick J.F., Musselman D.L., Lawson D.H., Reemsnyder A., Nemeroff C.B., Miller A.H. Neurobehavioral effects of interferon-alpha in cancer patients: phenomenology and paroxetine responsiveness of symptom dimensions. Neuropsychopharmacology: Off. Publ. Am. Coll. Neuropsychopharmacol. 2002;26(5):643–652. doi: 10.1016/S0893-133X(01)00407-9. [DOI] [PubMed] [Google Scholar]
- Capuron L., Pagnoni G., Demetrashvili M., Woolwine B.J., Nemeroff C.B., Berns G.S., Miller A.H. Anterior cingulate activation and error processing during interferon-alpha treatment. Biol. Psychiatr. 2005;58(3):190–196. doi: 10.1016/j.biopsych.2005.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capuron L., Pagnoni G., Drake D.F., Woolwine B.J., Spivey J.R., Crowe R.J., Votaw J.R., Goodman M.M., Miller A.H. Dopaminergic mechanisms of reduced basal ganglia responses to hedonic reward during interferon alfa administration. Arch. Gen. Psychiatr. 2012;69(10):1044–1053. doi: 10.1001/archgenpsychiatry.2011.2094. PMCID: PMC3640298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattaneo A., Gennarelli M., Uher R., Breen G., Farmer A., Aitchison K.J., Craig I.W., Anacker C., Zunsztain P.A., McGuffin P., Pariante C.M. Candidate genes expression profile associated with antidepressants response in the GENDEP study: differentiating between baseline 'predictors' and longitudinal 'targets. Neuropsychopharmacology: Off. Publ. Am. Coll. Neuropsychopharmacol. 2013;38(3):377–385. doi: 10.1038/npp.2012.191. PMCID: 3547188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattaneo A., Ferrari C., Uher R., Bocchio-Chiavetto L., Riva M.A., Consortium M.R.C.I., Pariante C.M. Absolute measurements of macrophage migration inhibitory factor and interleukin-1-beta mRNA levels accurately predict treatment response in depressed patients. Int. J. Neuropsychopharmacol. 2016;19(10) doi: 10.1093/ijnp/pyw045. PMCID: PMC5091822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain S.R., Cavanagh J., de Boer P., Mondelli V., Jones D.N.C., Drevets W.C., Cowen P.J., Harrison N.A., Pointon L., Pariante C.M., Bullmore E.T. Treatment-resistant depression and peripheral C-reactive protein. Br. J. Psychiatr. : J. Ment. Sci. 2019;214(1):11–19. doi: 10.1192/bjp.2018.66. PMCID: PMC6124647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Jiang T., Chen P., Ouyang J., Xu G., Zeng Z., Sun Y. Emerging tendency towards autoimmune process in major depressive patients: a novel insight from Th17 cells. Psychiatr. Res. 2011;188(2):224–230. doi: 10.1016/j.psychres.2010.10.029. [DOI] [PubMed] [Google Scholar]
- Chen R.A., Huang T.L., Huang K.W., Hung Y.Y. TNFAIP3 mRNA level is associated with psychological anxiety in major depressive disorder. Neuroimmunomodulation. 2017;24(4–5):271–275. doi: 10.1159/000486860. [DOI] [PubMed] [Google Scholar]
- Cole J.J., McColl A., Shaw R., Lynall M.E., Cowen P.J., de Boer P., Drevets W.C., Harrison N., Pariante C., Pointon L., Goodyear C., Bullmore E., Cavanagh J. No evidence for differential gene expression in major depressive disorder PBMCs, but robust evidence of elevated biological ageing. Transl. Psychiatry. 2021;11(1):404. doi: 10.1038/s41398-021-01506-4. PMCID: PMC8298604 Mood Disorders and Alzheimer’s Disease (NIMA) Consortium, which is also funded by Janssen, GlaxoSmithKline, Lundbeck and Pfizer. Dr. Drevets and Dr. De Boer are employees of Janssen Research & Development, LLC, of Johnson & Johnson and hold equity in Johnson & Johnson. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costi S., Morris L.S., Collins A., Fernandez N.F., Patel M., Xie H., Kim-Schulze S., Stern E.R., Collins K.A., Cathomas F., Parides M.K., Whitton A.E., Pizzagalli D.A., Russo S.J., Murrough J.W. Peripheral immune cell reactivity and neural response to reward in patients with depression and anhedonia. Transl. Psychiatry. 2021;11(1):565. doi: 10.1038/s41398-021-01668-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantzer R., Walker A.K. Is there a role for glutamate-mediated excitotoxicity in inflammation-induced depression? J. Neural. Transm. (Vienna) 2014;121(8):925–932. doi: 10.1007/s00702-014-1187-1. PMCID: PMC4134384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das H., Kumar A., Lin Z., Patino W.D., Hwang P.M., Feinberg M.W., Majumder P.K., Jain M.K. Kruppel-like factor 2 (KLF2) regulates proinflammatory activation of monocytes. Proc. Natl. Acad. Sci. U. S. A. 2006;103(17):6653–6658. doi: 10.1073/pnas.0508235103. PMCID: PMC1458936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies K.A., Cooper E., Voon V., Tibble J., Cercignani M., Harrison N.A. Interferon and anti-TNF therapies differentially modulate amygdala reactivity which predicts associated bidirectional changes in depressive symptoms. Mol. Psychiatr. 2021;26(9):5150–5160. doi: 10.1038/s41380-020-0790-9. PMCID: PMC8589643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kluiver H., Jansen R., Milaneschi Y., Penninx B. Involvement of inflammatory gene expression pathways in depressed patients with hyperphagia. Transl. Psychiatry. 2019;9(1):193. doi: 10.1038/s41398-019-0528-0. PMCID: PMC6702221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePaula-Silva A.B., Gorbea C., Doty D.J., Libbey J.E., Sanchez J.M.S., Hanak T.J., Cazalla D., Fujinami R.S. Differential transcriptional profiles identify microglial- and macrophage-specific gene markers expressed during virus-induced neuroinflammation. J. Neuroinflammation. 2019;16(1):152. doi: 10.1186/s12974-019-1545-x. PMCID: PMC6642742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowlati Y., Herrmann N., Swardfager W., Liu H., Sham L., Reim E.K., Lanctot K.L. A meta-analysis of cytokines in major depression. Biol. Psychiatr. 2010;67(5):446–457. doi: 10.1016/j.biopsych.2009.09.033. [DOI] [PubMed] [Google Scholar]
- Eisenberger N.I., Berkman E.T., Inagaki T.K., Rameson L.T., Mashal N.M., Irwin M.R. Inflammation-induced anhedonia: endotoxin reduces ventral striatum responses to reward. Biol. Psychiatr. 2010;68(8):748–754. doi: 10.1016/j.biopsych.2010.06.010. PMCID: 3025604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felger J.C., Miller A.H. Identifying immunophenotypes of inflammation in depression: dismantling the monolith. Biol. Psychiatr. 2020;88(2):136–138. doi: 10.1016/j.biopsych.2020.04.024. [DOI] [PubMed] [Google Scholar]
- Felger J.C., Treadway M.T. Inflammation effects on motivation and motor activity: role of dopamine. Neuropsychopharmacology: Off. Publ. Am. Coll. Neuropsychopharmacol. 2017;42(1):216–241. doi: 10.1038/npp.2016.143. PMCID: PMC5143486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felger J.C., Mun J., Kimmel H.L., Nye J.A., Drake D.F., Hernandez C.R., Freeman A.A., Rye D.B., Goodman M.M., Howell L.L., Miller A.H. Chronic interferon-alpha decreases dopamine 2 receptor binding and striatal dopamine release in association with anhedonia-like behavior in nonhuman primates. Neuropsychopharmacology: Off. Publ. Am. Coll. Neuropsychopharmacol. 2013;38(11):2179–2187. doi: 10.1038/npp.2013.115. PMCID: 3773667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felger J.C., Li L., Marvar P.J., Woolwine B.J., Harrison D.G., Raison C.L., Miller A.H. Tyrosine metabolism during interferon-alpha administration: association with fatigue and CSF dopamine concentrations. Brain Behav. Immun. 2013;31:153–160. doi: 10.1016/j.bbi.2012.10.010. PMCID: 3578984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felger J.C., Hernandez C.R., Miller A.H. Levodopa reverses cytokine-induced reductions in striatal dopamine release. Int. J. Neuropsychopharmacol./Off. Sci. J. Collegium Internationale Neuropsychopharmacol. 2015;18(4):1–5. doi: 10.1093/ijnp/pyu084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felger J.C., Li Z., Haroon E., Woolwine B.J., Jung M.Y., Hu X., Miller A.H. Inflammation is associated with decreased functional connectivity within corticostriatal reward circuitry in depression. Mol. Psychiatr. 2016;21(10):1358–1365. doi: 10.1038/mp.2015.168. PMCID: PMC4862934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felger J.C., Haroon E., Patel T.A., Goldsmith D.R., Wommack E.C., Woolwine B.J., Le N.A., Feinberg R., Tansey M.G., Miller A.H. What does plasma CRP tell us about peripheral and central inflammation in depression? Mol. Psychiatr. 2020;25(6):1301–1311. doi: 10.1038/s41380-018-0096-3. PMCID: PMC6291384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- First M.B., Spitzer R.L., Gibbon M., Williams J.B. American Psychiatric Press; Washington DC: 1997. Structured Clinical Interview for DSM-IV. [Google Scholar]
- Folstein M.F., Folstein S.E., McHugh P.R. Mini-mental state". A practical method for grading the cognitive state of patients for the clinician. J. Psychiatr. Res. 1975;12(3):189–198. doi: 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
- Galecki P., Galecka E., Maes M., Chamielec M., Orzechowska A., Bobinska K., Lewinski A., Szemraj J. The expression of genes encoding for COX-2, MPO, iNOS, and sPLA2-IIA in patients with recurrent depressive disorder. J. Affect. Disord. 2012;138(3):360–366. doi: 10.1016/j.jad.2012.01.016. [DOI] [PubMed] [Google Scholar]
- Garza-Reyes M.G., Mora-Ruiz M.D., Chavez-Sanchez L., Madrid-Miller A., Cabrera-Quintero A.J., Maravillas-Montero J.L., Zentella-Dehesa A., Moreno-Ruiz L., Pastor-Salgado S., Ramirez-Arias E., Perez-Velazquez N., Chavez-Rueda A.K., Blanco-Favela F., Vazquez-Gonzalez W.G., Contreras-Rodriguez A. Effect of interleukin-17 in the activation of monocyte subsets in patients with ST-segment elevation myocardial infarction. J. Immunol. Res. 2020 doi: 10.1155/2020/5692829. 2020. PMCID: PMC7336211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsmith D.R., Bekhbat M., Le N.A., Chen X., Woolwine B.J., Li Z., Haroon E., Felger J.C. Protein and gene markers of metabolic dysfunction and inflammation together associate with functional connectivity in reward and motor circuits in depression. Brain Behav. Immun. 2020;88:193–202. doi: 10.1016/j.bbi.2020.05.013. PMCID: PMC7415617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsmith D.R., Massa N., Miller B.J., Miller A.H., Duncan E. The interaction of lipids and inflammatory markers predict negative symptom severity in patients with schizophrenia. NPJ schizophrenia. 2021;7(1):50. doi: 10.1038/s41537-021-00179-8. PMCID: PMC8528914 Pharmaceuticals, Inc. and Teva Pharmaceuticals, Inc. E.D. is a full-time attending psychiatrist and Nicholas Massa is a full-time research associate at the Atlanta Veterans Affairs Health Care System, Decatur, GA. The content is solely the responsibility of the authors and does not necessarily represent the official views of the Department of Veterans Affairs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guardado P., Olivera A., Rusch H.L., Roy M., Martin C., Lejbman N., Lee H., Gill J.M. Altered gene expression of the innate immune, neuroendocrine, and nuclear factor-kappa B (NF-kappaB) systems is associated with posttraumatic stress disorder in military personnel. J. Anxiety Disord. 2016;38:9–20. doi: 10.1016/j.janxdis.2015.12.004. [DOI] [PubMed] [Google Scholar]
- Guilloux J.P., Bassi S., Ding Y., Walsh C., Turecki G., Tseng G., Cyranowski J.M., Sibille E. Testing the predictive value of peripheral gene expression for nonremission following citalopram treatment for major depression. Neuropsychopharmacology: Off. Publ. Am. Coll. Neuropsychopharmacol. 2015;40(3):701–710. doi: 10.1038/npp.2014.226. PMCID: PMC4289958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajebrahimi B., Bagheri M., Hassanshahi G., Nazari M., Bidaki R., Khodadadi H., Arababadi M.K., Kennedy D. The adapter proteins of TLRs, TRIF and MYD88, are upregulated in depressed individuals. Int. J. Psychiatr. Clin. Pract. 2014;18(1):41–44. doi: 10.3109/13651501.2013.859708. [DOI] [PubMed] [Google Scholar]
- Hamilton M. A rating scale for depression. J. Neurol. Neurosurg. Psychiatry. 1960;23:56–62. doi: 10.1136/jnnp.23.1.56. PMCID: PMC495331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haroon E., Woolwine B.J., Chen X., Pace T.W., Parekh S., Spivey J.R., Hu X.P., Miller A.H. IFN-alpha-induced cortical and subcortical glutamate changes assessed by magnetic resonance spectroscopy. Neuropsychopharmacology: Off. Publ. Am. Coll. Neuropsychopharmacol. 2014;39(7):1777–1785. doi: 10.1038/npp.2014.25. PMCID: PMC4023151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haroon E., Fleischer C.C., Felger J.C., Chen X., Woolwine B.J., Patel T., Hu X.P., Miller A.H. Conceptual convergence: increased inflammation is associated with increased basal ganglia glutamate in patients with major depression. Mol. Psychiatr. 2016;21(10):1351–1357. doi: 10.1038/mp.2015.206. PMCID: PMC4940313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haroon E., Daguanno A.W., Woolwine B.J., Goldsmith D.R., Baer W.M., Wommack E.C., Felger J.C., Miller A.H. Antidepressant treatment resistance is associated with increased inflammatory markers in patients with major depressive disorder. Psychoneuroendocrinology. 2018;95:43–49. doi: 10.1016/j.psyneuen.2018.05.026. PMCID: PMC6427066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haroon E., Chen X., Li Z., Patel T., Woolwine B.J., Hu X.P., Felger J.C., Miller A.H. Increased inflammation and brain glutamate define a subtype of depression with decreased regional homogeneity, impaired network integrity, and anhedonia. Transl. Psychiatry. 2018;8(1):189. doi: 10.1038/s41398-018-0241-4. PMCID: PMC6131242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison N.A., Brydon L., Walker C., Gray M.A., Steptoe A., Critchley H.D. Inflammation causes mood changes through alterations in subgenual cingulate activity and mesolimbic connectivity. Biol. Psychiatr. 2009;66(5):407–414. doi: 10.1016/j.biopsych.2009.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison N.A., Cercignani M., Voon V., Critchley H.D. Effects of inflammation on hippocampus and substantia nigra responses to novelty in healthy human participants. Neuropsychopharmacology : Off. Publ. Am. Coll. Neuropsychopharmacol. 2015;40(4):831–838. doi: 10.1038/npp.2014.222. PMCID: Pmc4264953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison N.A., Voon V., Cercignani M., Cooper E.A., Pessiglione M., Critchley H.D. A neurocomputational account of how inflammation enhances sensitivity to punishments versus rewards. Biol. Psychiatr. 2016;80(1):73–81. doi: 10.1016/j.biopsych.2015.07.018. PMCID: PMC4918729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa M., Imamura R., Motani K., Nishiuchi T., Matsumoto N., Kinoshita T., Suda T. Mechanism and repertoire of ASC-mediated gene expression. J. Immunol. 2009;182(12):7655–7662. doi: 10.4049/jimmunol.0800448. [DOI] [PubMed] [Google Scholar]
- Hasselmann H., Gamradt S., Taenzer A., Nowacki J., Zain R., Patas K., Ramien C., Paul F., Wingenfeld K., Piber D., Gold S.M., Otte C. Pro-inflammatory monocyte phenotype and cell-specific steroid signaling alterations in unmedicated patients with major depressive disorder. Front. Immunol. 2018;9:2693. doi: 10.3389/fimmu.2018.02693. PMCID: PMC6265986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez M.X., Jiang S., Cole T.A., Chu S.H., Fonseca M.I., Fang M.J., Hohsfield L.A., Torres M.D., Green K.N., Wetsel R.A., Mortazavi A., Tenner A.J. Prevention of C5aR1 signaling delays microglial inflammatory polarization, favors clearance pathways and suppresses cognitive loss. Mol. Neurodegener. 2017;12(1):66. doi: 10.1186/s13024-017-0210-z. PMCID: PMC5604420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodes G.E., Pfau M.L., Leboeuf M., Golden S.A., Christoffel D.J., Bregman D., Rebusi N., Heshmati M., Aleyasin H., Warren B.L., Lebonte B., Horn S., Lapidus K.A., Stelzhammer V., Wong E.H., Bahn S., Krishnan V., Bolanos-Guzman C.A., Murrough J.W., Merad M., Russo S.J. Individual differences in the peripheral immune system promote resilience versus susceptibility to social stress. Proc. Natl. Acad. Sci. U. S. A. 2014;111(45):16136–16141. doi: 10.1073/pnas.1415191111. PMCID: Pmc4234602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori H., Sasayama D., Teraishi T., Yamamoto N., Nakamura S., Ota M., Hattori K., Kim Y., Higuchi T., Kunugi H. Blood-based gene expression signatures of medication-free outpatients with major depressive disorder: integrative genome-wide and candidate gene analyses. Sci. Rep. 2016;6:18776. doi: 10.1038/srep18776. PMCID: PMC4700430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howren M.B., Lamkin D.M., Suls J. Associations of depression with C-reactive protein, IL-1, and IL-6: a meta-analysis. Psychosom. Med. 2009;71(2):171–186. doi: 10.1097/PSY.0b013e3181907c1b. [DOI] [PubMed] [Google Scholar]
- Hu G., Guo M., Xu J., Wu F., Fan J., Huang Q., Yang G., Lv Z., Wang X., Jin Y. Nanoparticles targeting macrophages as potential clinical therapeutic agents against cancer and inflammation. Front. Immunol. 2019;10:1998. doi: 10.3389/fimmu.2019.01998. PMCID: PMC6712945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulsegge I., Kommadath A., Smits M.A. Globaltest and GOEAST: two different approaches for Gene Ontology analysis. BMC Proc. 2009;3(Suppl. 4:S10) doi: 10.1186/1753-6561-3-S4-S10. PMCID: PMC2712740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung Y.Y., Kang H.Y., Huang K.W., Huang T.L. Association between toll-like receptors expression and major depressive disorder. Psychiatr. Res. 2014;220(1–2):283–286. doi: 10.1016/j.psychres.2014.07.074. [DOI] [PubMed] [Google Scholar]
- Hung Y.Y., Lin C.C., Kang H.Y., Huang T.L. TNFAIP3, a negative regulator of the TLR signaling pathway, is a potential predictive biomarker of response to antidepressant treatment in major depressive disorder. Brain Behav. Immun. 2017;59:265–272. doi: 10.1016/j.bbi.2016.09.014. [DOI] [PubMed] [Google Scholar]
- Husain M.I., Chaudhry I.B., Khoso A.B., Husain M.O., Hodsoll J., Ansari M.A., Naqvi H.A., Minhas F.A., Carvalho A.F., Meyer J.H., Deakin B., Mulsant B.H., Husain N., Young A.H. Minocycline and celecoxib as adjunctive treatments for bipolar depression: a multicentre, factorial design randomised controlled trial. Lancet Psychiatr. 2020;7(6):515–527. doi: 10.1016/S2215-0366(20)30138-3. [DOI] [PubMed] [Google Scholar]
- Iwakura Y., Ishigame H. The IL-23/IL-17 axis in inflammation. J. Clin. Invest. 2006;116(5):1218–1222. doi: 10.1172/JCI28508. PMCID: PMC1451213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R., Penninx B.W., Madar V., Xia K., Milaneschi Y., Hottenga J.J., Hammerschlag A.R., Beekman A., van der Wee N., Smit J.H., Brooks A.I., Tischfield J., Posthuma D., Schoevers R., van Grootheest G., Willemsen G., de Geus E.J., Boomsma D.I., Wright F.A., Zou F., Sun W., Sullivan P.F. Gene expression in major depressive disorder. Mol. Psychiatr. 2016;21(3):339–347. doi: 10.1038/mp.2015.57. [DOI] [PubMed] [Google Scholar]
- Jordao M.J.C., Sankowski R., Brendecke S.M., Sagar, Locatelli G., Tai Y.H., Tay T.L., Schramm E., Armbruster S., Hagemeyer N., Gross O., Mai D., Cicek O., Falk T., Kerschensteiner M., Grun D., Prinz M. Single-cell profiling identifies myeloid cell subsets with distinct fates during neuroinflammation. Science. 2019;363(6425) doi: 10.1126/science.aat7554. [DOI] [PubMed] [Google Scholar]
- Kappelmann N., Lewis G., Dantzer R., Jones P.B., Khandaker G.M. Antidepressant activity of anti-cytokine treatment: a systematic review and meta-analysis of clinical trials of chronic inflammatory conditions. Mol. Psychiatr. 2018;23(2):335–343. doi: 10.1038/mp.2016.167. PMCID: PMC5794896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keri S., Szabo C., Kelemen O. Expression of Toll-Like Receptors in peripheral blood mononuclear cells and response to cognitive-behavioral therapy in major depressive disorder. Brain Behav. Immun. 2014;40:235–243. doi: 10.1016/j.bbi.2014.03.020. [DOI] [PubMed] [Google Scholar]
- Kitagami T., Yamada K., Miura H., Hashimoto R., Nabeshima T., Ohta T. Mechanism of systemically injected interferon-alpha impeding monoamine biosynthesis in rats: role of nitric oxide as a signal crossing the blood-brain barrier. Brain Res. 2003;978(1–2):104–114. doi: 10.1016/s0006-8993(03)02776-8. [DOI] [PubMed] [Google Scholar]
- Kohler-Forsberg O., Nicolaisen Lydholm C., Hjorthoj C., Nordentoft M., Mors O., Benros M.E. Efficacy of anti-inflammatory treatment on major depressive disorder or depressive symptoms: meta-analysis of clinical trials. Acta Psychiatr. Scand. 2019 doi: 10.1111/acps.13016. [DOI] [PubMed] [Google Scholar]
- Kornberg M.D., Bhargava P., Kim P.M., Putluri V., Snowman A.M., Putluri N., Calabresi P.A., Snyder S.H. Dimethyl fumarate targets GAPDH and aerobic glycolysis to modulate immunity. Science. 2018;360(6387):449–453. doi: 10.1126/science.aan4665. PMCID: PMC5924419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korsunsky I., Millard N., Fan J., Slowikowski K., Zhang F., Wei K., Baglaenko Y., Brenner M., Loh P.R., Raychaudhuri S. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods. 2019;16(12):1289–1296. doi: 10.1038/s41592-019-0619-0. PMCID: PMC6884693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse J.L., Congdon E., Olmstead R., Njau S., Breen E.C., Narr K.L., Espinoza R., Irwin M.R. Inflammation and improvement of depression following electroconvulsive therapy in treatment-resistant depression. J. Clin. Psychiatr. 2018;79(2) doi: 10.4088/JCP.17m11597. PMCID: PMC6013272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo S., Nakayamada S., Sakata K., Kitanaga Y., Ma X., Lee S., Ishii A., Yamagata K., Nakano K., Tanaka Y. Janus kinase inhibitor baricitinib modulates human innate and adaptive immune system. Front. Immunol. 2018;9:1510. doi: 10.3389/fimmu.2018.01510. PMCID: PMC6031708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le T.T., Savitz J., Suzuki H., Misaki M., Teague T.K., White B.C., Marino J.H., Wiley G., Gaffney P.M., Drevets W.C., McKinney B.A., Bodurka J. Identification and replication of RNA-Seq gene network modules associated with depression severity. Transl. Psychiatry. 2018;8(1):180. doi: 10.1038/s41398-018-0234-3. PMCID: PMC6125582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leday G.G.R., Vertes P.E., Richardson S., Greene J.R., Regan T., Khan S., Henderson R., Freeman T.C., Pariante C.M., Harrison N.A., Perry V.H., Drevets W.C., Wittenberg G.M., Bullmore E.T. Replicable and coupled changes in innate and adaptive immune gene expression in two case-control studies of blood microarrays in major depressive disorder. Biol. Psychiatr. 2018;83(1):70–80. doi: 10.1016/j.biopsych.2017.01.021. PMCID: PMC5720346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LJPvd Maaten, Hinton G.E. Visualizing high-dimensional data using t-SNE. J. Mach. Learn. Res. 2008:2579–2605. [Google Scholar]
- Louveau A., Smirnov I., Keyes T.J., Eccles J.D., Rouhani S.J., Peske J.D., Derecki N.C., Castle D., Mandell J.W., Lee K.S., Harris T.H., Kipnis J. Structural and functional features of central nervous system lymphatic vessels. Nature. 2015;523(7560):337–341. doi: 10.1038/nature14432. PMCID: PMC4506234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukic I., Mitic M., Djordjevic J., Tatalovic N., Bozovic N., Soldatovic I., Mihaljevic M., Pavlovic Z., Radojcic M.B., Maric N.P., Adzic M. Lymphocyte levels of redox-sensitive transcription factors and antioxidative enzymes as indicators of pro-oxidative state in depressive patients. Neuropsychobiology. 2014;70(1):1–9. doi: 10.1159/000362841. [DOI] [PubMed] [Google Scholar]
- Lynall M.E., Turner L., Bhatti J., Cavanagh J., de Boer P., Mondelli V., Jones D., Drevets W.C., Cowen P., Harrison N.A., Pariante C.M., Pointon L., Clatworthy M.R., Bullmore E., Neuroimmunology of Mood D., Alzheimer's Disease C. Peripheral blood cell-stratified subgroups of inflamed depression. Biol. Psychiatr. 2020;88(2):185–196. doi: 10.1016/j.biopsych.2019.11.017. [DOI] [PubMed] [Google Scholar]
- McIntyre R.S., Subramaniapillai M., Lee Y., Pan Z., Carmona N.E., Shekotikhina M., Rosenblat J.D., Brietzke E., Soczynska J.K., Cosgrove V.E., Miller S., Fischer E.G., Kramer N.E., Dunlap K., Suppes T., Mansur R.B. Efficacy of adjunctive infliximab vs placebo in the treatment of adults with bipolar I/II depression: a randomized clinical trial. JAMA Psychiatr. 2019;76(8):783–790. doi: 10.1001/jamapsychiatry.2019.0779. PMCID: PMC6506894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim D.B., Weber M.D., Niraula A., Sawicki C.M., Liu X., Jarrett B.L., Ramirez-Chan K., Wang Y., Roeth R.M., Sucaldito A.D., Sobol C.G., Quan N., Sheridan J.F., Godbout J.P. Microglial recruitment of IL-1beta-producing monocytes to brain endothelium causes stress-induced anxiety. Mol. Psychiatr. 2018;23(6):1421–1431. doi: 10.1038/mp.2017.64. PMCID: PMC5628107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta D., Raison C.L., Woolwine B.J., Haroon E., Binder E.B., Miller A.H., Felger J.C. Transcriptional signatures related to glucose and lipid metabolism predict treatment response to the tumor necrosis factor antagonist infliximab in patients with treatment-resistant depression. Brain Behav. Immun. 2013;31:205–215. doi: 10.1016/j.bbi.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta N.D., Haroon E., Xu X., Woolwine B.J., Li Z., Felger J.C. Inflammation negatively correlates with amygdala-ventromedial prefrontal functional connectivity in association with anxiety in patients with depression: preliminary results. Brain Behav. Immun. 2018;73:725–730. doi: 10.1016/j.bbi.2018.07.026. PMCID: PMC6129411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menard C., Pfau M.L., Hodes G.E., Kana V., Wang V.X., Bouchard S., Takahashi A., Flanigan M.E., Aleyasin H., LeClair K.B., Janssen W.G., Labonte B., Parise E.M., Lorsch Z.S., Golden S.A., Heshmati M., Tamminga C., Turecki G., Campbell M., Fayad Z.A., Tang C.Y., Merad M., Russo S.J. Social stress induces neurovascular pathology promoting depression. Nat. Neurosci. 2017;20(12):1752–1760. doi: 10.1038/s41593-017-0010-3. PMCID: PMC5726568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menke A., Arloth J., Putz B., Weber P., Klengel T., Mehta D., Gonik M., Rex-Haffner M., Rubel J., Uhr M., Lucae S., Deussing J.M., Muller-Myhsok B., Holsboer F., Binder E.B. Dexamethasone stimulated gene expression in peripheral blood is a sensitive marker for glucocorticoid receptor resistance in depressed patients. Neuropsychopharmacology : Off. Publ. Am. Coll. Neuropsychopharmacol. 2012;37(6):1455–1464. doi: 10.1038/npp.2011.331. PMCID: PMC3327850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miklowitz D.J., Portnoff L.C., Armstrong C.C., Keenan-Miller D., Breen E.C., Muscatell K.A., Eisenberger N.I., Irwin M.R. Inflammatory cytokines and nuclear factor-kappa B activation in adolescents with bipolar and major depressive disorders. Psychiatr. Res. 2016;241:315–322. doi: 10.1016/j.psychres.2016.04.120. PMCID: PMC4912920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller A.H., Raison C.L. The role of inflammation in depression: from evolutionary imperative to modern treatment target. Nat. Rev. Immunol. 2016;16(1):22–34. doi: 10.1038/nri.2015.5. PMCID: PMC5542678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller A.H., Asnis G.M., Lackner C., Norin A.J. The in vitro effect of cortisol on natural killer cell activity in patients with major depressive disorder. Psychopharmacol. Bull. 1987;23:502–504. [Google Scholar]
- Miller A.H., Asnis G.M., Lackner C., Halbreich U., Norin A.J. Depression, natural killer cell activity, and cortisol secretion. Biol. Psychiatr. 1991;29(9):878–886. doi: 10.1016/0006-3223(91)90054-p. [DOI] [PubMed] [Google Scholar]
- Miller A.H., Maletic V., Raison C.L. Inflammation and its discontents: the role of cytokines in the pathophysiology of major depression. Biol. Psychiatr. 2009;65(9):732–741. doi: 10.1016/j.biopsych.2008.11.029. PMCID: Pmc2680424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momeni M., Ghorban K., Dadmanesh M., Khodadadi H., Bidaki R., Kazemi Arababadi M., Kennedy D. ASC provides a potential link between depression and inflammatory disorders: a clinical study of depressed Iranian medical students. Nord. J. Psychiatr. 2016;70(4):280–284. doi: 10.3109/08039488.2015.1100328. [DOI] [PubMed] [Google Scholar]
- Mostafavi S., Battle A., Zhu X., Potash J.B., Weissman M.M., Shi J., Beckman K., Haudenschild C., McCormick C., Mei R., Gameroff M.J., Gindes H., Adams P., Goes F.S., Mondimore F.M., MacKinnon D.F., Notes L., Schweizer B., Furman D., Montgomery S.B., Urban A.E., Koller D., Levinson D.F. Type I interferon signaling genes in recurrent major depression: increased expression detected by whole-blood RNA sequencing. Mol. Psychiatr. 2014;19(12):1267–1274. doi: 10.1038/mp.2013.161. PMCID: PMC5404932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nettis M.A., Pergola G., Kolliakou A., O'Connor J., Bonaccorso S., David A., Gaughran F., Di Forti M., Murray R.M., Marques T.R., Blasi G., Bertolino A., Pariante C.M., Dazzan P., Mondelli V. Metabolic-inflammatory status as predictor of clinical outcome at 1-year follow-up in patients with first episode psychosis. Psychoneuroendocrinology. 2019;99:145–153. doi: 10.1016/j.psyneuen.2018.09.005. [DOI] [PubMed] [Google Scholar]
- O'Connor J.C., Andre C., Wang Y., Lawson M.A., Szegedi S.S., Lestage J., Castanon N., Kelley K.W., Dantzer R. Interferon-gamma and tumor necrosis factor-alpha mediate the upregulation of indoleamine 2,3-dioxygenase and the induction of depressive-like behavior in mice in response to bacillus Calmette-Guerin. J. Neurosci. : Off. J. Soc. Neurosci. 2009;29(13):4200–4209. doi: 10.1523/JNEUROSCI.5032-08.2009. PMCID: 2835569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill L.A., Kishton R.J., Rathmell J. A guide to immunometabolism for immunologists. Nat. Rev. Immunol. 2016;16(9):553–565. doi: 10.1038/nri.2016.70. PMCID: PMC5001910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osimo E.F., Baxter L.J., Lewis G., Jones P.B., Khandaker G.M. Prevalence of low-grade inflammation in depression: a systematic review and meta-analysis of CRP levels. Psychol. Med. 2019;49(12):1958–1970. doi: 10.1017/S0033291719001454. PMCID: PMC6712955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace T.W., Hu F., Miller A.H. Cytokine-effects on glucocorticoid receptor function: relevance to glucocorticoid resistance and the pathophysiology and treatment of major depression. Brain Behav. Immun. 2007;21(1):9–19. doi: 10.1016/j.bbi.2006.08.009. PMCID: 1820632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palsson-McDermott E.M., O'Neill L.A.J. Targeting immunometabolism as an anti-inflammatory strategy. Cell Res. 2020;30(4):300–314. doi: 10.1038/s41422-020-0291-z. PMCID: PMC7118080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D., Xu Y., Zhang L., Su Q., Chen M., Li B., Xiao Q., Gao Q., Peng X., Jiang B., Gu Y., Du Y., Gao P. Gene expression profile in peripheral blood mononuclear cells of postpartum depression patients. Sci. Rep. 2018;8(1):10139. doi: 10.1038/s41598-018-28509-4. PMCID: PMC6031634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pariante C.M. Glucocorticoid receptor function in vitro in patients with major depression. Stress. 2004;7(4):209–219. doi: 10.1080/10253890500069650. [DOI] [PubMed] [Google Scholar]
- Patas K., Willing A., Demiralay C., Engler J.B., Lupu A., Ramien C., Schafer T., Gach C., Stumm L., Chan K., Vignali M., Arck P.C., Friese M.A., Pless O., Wiedemann K., Agorastos A., Gold S.M. T cell phenotype and T cell receptor repertoire in patients with major depressive disorder. Front. Immunol. 2018;9:291. doi: 10.3389/fimmu.2018.00291. PMCID: PMC5826233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson T.A., Mensah G.A., Alexander R.W., Anderson J.L., Cannon R.O., 3rd, Criqui M., Fadl Y.Y., Fortmann S.P., Hong Y., Myers G.L., Rifai N., Smith S.C., Jr., Taubert K., Tracy R.P., Vinicor F. Markers of inflammation and cardiovascular disease: application to clinical and public health practice: a statement for healthcare professionals from the Centers for Disease Control and Prevention and the American Heart Association. Circulation. 2003;107(3):499–511. doi: 10.1161/01.cir.0000052939.59093.45. [DOI] [PubMed] [Google Scholar]
- Pizzolato G., Kaminski H., Tosolini M., Franchini D.M., Pont F., Martins F., Valle C., Labourdette D., Cadot S., Quillet-Mary A., Poupot M., Laurent C., Ysebaert L., Meraviglia S., Dieli F., Merville P., Milpied P., Dechanet-Merville J., Fournie J.J. Single-cell RNA sequencing unveils the shared and the distinct cytotoxic hallmarks of human TCRVdelta1 and TCRVdelta2 gammadelta T lymphocytes. Proc. Natl. Acad. Sci. U. S. A. 2019;116(24):11906–11915. doi: 10.1073/pnas.1818488116. PMCID: PMC6576116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posner K., Oquendo M.A., Gould M., Stanley B., Davies M. Columbia Classification Algorithm of Suicide Assessment (C-CASA): classification of suicidal events in the FDA's pediatric suicidal risk analysis of antidepressants. Am. J. Psychiatr. 2007;164(7):1035–1043. doi: 10.1176/appi.ajp.164.7.1035. PMCID: PMC3804920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raison C.L., Miller A.H. When not enough is too much: the role of insufficient glucocorticoid signaling in the pathophysiology of stress-related disorders. Am. J. Psychiatr. 2003;160(9):1554–1565. doi: 10.1176/appi.ajp.160.9.1554. [DOI] [PubMed] [Google Scholar]
- Raison C.L., Felger J.C., Miller A.H. 2013. (Inflammation and Treatment Resistance in Major Depression: A Perfect Storm Psychiatric Times). september 12. [Google Scholar]
- Raison C.L., Rutherford R.E., Woolwine B.J., Shuo C., Schettler P., Drake D.F., Haroon E., Miller A.H. A randomized controlled trial of the tumor necrosis factor antagonist infliximab for treatment-resistant depression: the role of baseline inflammatory biomarkers. JAMA Psychiatr. 2013;70(1):31–41. doi: 10.1001/2013.jamapsychiatry.4. PMCID: PMC4015348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raof N.A., Rajamani D., Chu H.C., Gurav A., Johnson J.M., LoGerfo F.W., Pradhan-Nabzdyk L., Bhasin M. The effects of transfection reagent polyethyleneimine (PEI) and non-targeting control siRNAs on global gene expression in human aortic smooth muscle cells. BMC Genom. 2016;17(1):20. doi: 10.1186/s12864-015-2267-9. PMCID: 4700750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rengasamy M., Brundin L., Griffo A., Panny B., Capan C., Forton C., Price R.B. Cytokine and reward circuitry relationships in treatment-resistant depression. Biol. Psychiatr. Global Open Sci. 2021 doi: 10.1016/j.bpsgos.2021.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts C.A., Dickinson A.K., Taams L.S. The interplay between monocytes/macrophages and CD4(+) T cell subsets in rheumatoid arthritis. Front. Immunol. 2015;6:571. doi: 10.3389/fimmu.2015.00571. PMCID: PMC4652039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rush A.J., Gullion C.M., Basco M.R., Jarrett R.B., Trivedi M.H. The inventory of depressive symptomatology (IDS): psychometric properties. Psychol. Med. 1996;26(3):477–486. doi: 10.1017/s0033291700035558. [DOI] [PubMed] [Google Scholar]
- Rustenhoven J., Kipnis J. Bypassing the blood-brain barrier. Science. 2019;366(6472):1448–1449. doi: 10.1126/science.aay0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvadore G., Nash A., Bleys C., Hsu B., Saad Z., Gause A., Moyer J., Xi L., Manji H., Van Nueten L., Drevets W. American College of Neuropsychopharmacology; Hollywood, FL: 2018. A Double-Blind, Placebo-Controlled, Multicenter Study of Sirukumab as Adjunctive Treatment to a Monoaminergic Antidepressant in Adults with Major Depressive Disorder. [Google Scholar]
- Savitz J., Frank M.B., Victor T., Bebak M., Marino J.H., Bellgowan P.S., McKinney B.A., Bodurka J., Kent Teague T., Drevets W.C. Inflammation and neurological disease-related genes are differentially expressed in depressed patients with mood disorders and correlate with morphometric and functional imaging abnormalities. Brain Behav. Immun. 2013;31:161–171. doi: 10.1016/j.bbi.2012.10.007. PMCID: 3577998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snaith R.P., Hamilton M., Morley S., Humayan A., Hargreaves D., Trigwell P. A scale for the assessment of hedonic tone the Snaith-Hamilton Pleasure Scale. Br. J. Psychiatr. : J. Ment. Sci. 1995;167(1):99–103. doi: 10.1192/bjp.167.1.99. [DOI] [PubMed] [Google Scholar]
- Spijker S., Van Zanten J.S., De Jong S., Penninx B.W., van Dyck R., Zitman F.G., Smit J.H., Ylstra B., Smit A.B., Hoogendijk W.J. Stimulated gene expression profiles as a blood marker of major depressive disorder. Biol. Psychiatr. 2010;68(2):179–186. doi: 10.1016/j.biopsych.2010.03.017. [DOI] [PubMed] [Google Scholar]
- Storey J.D., Tibshirani R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. U. S. A. 2003;100(16):9440–9445. doi: 10.1073/pnas.1530509100. PMCID: 170937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., 3rd, Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive integration of single-cell data. Cell. 2019;177(7):1888–1902. doi: 10.1016/j.cell.2019.05.031. e21. PMCID: PMC6687398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D., Guan X., Moran A.E., Wu L.Y., Qian D.Z., Schedin P., Dai M.S., Danilov A.V., Alumkal J.J., Adey A.C., Spellman P.T., Xia Z. Identifying phenotype-associated subpopulations by integrating bulk and single-cell sequencing data. Nat. Biotechnol. 2021 doi: 10.1038/s41587-021-01091-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H., Savitz J., Kent Teague T., Gandhapudi S.K., Tan C., Misaki M., McKinney B.A., Irwin M.R., Drevets W.C., Bodurka J. Altered populations of natural killer cells, cytotoxic T lymphocytes, and regulatory T cells in major depressive disorder: association with sleep disturbance. Brain Behav. Immun. 2017;66:193–200. doi: 10.1016/j.bbi.2017.06.011. PMCID: PMC5650936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Platas S.G., Cruceanu C., Chen G.G., Turecki G., Mechawar N. Evidence for increased microglial priming and macrophage recruitment in the dorsal anterior cingulate white matter of depressed suicides. Brain Behav. Immun. 2014;42:50–59. doi: 10.1016/j.bbi.2014.05.007. [DOI] [PubMed] [Google Scholar]
- Tran M.T., Zsengeller Z.K., Berg A.H., Khankin E.V., Bhasin M.K., Kim W., Clish C.B., Stillman I.E., Karumanchi S.A., Rhee E.P., Parikh S.M. PGC1alpha drives NAD biosynthesis linking oxidative metabolism to renal protection. Nature. 2016;531(7595):528–532. doi: 10.1038/nature17184. PMCID: PMC4909121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treadway M.T., Cooper J.A., Miller A.H. Can't or won't? Immunometabolic constraints on dopaminergic drive. Trends Cognit. Sci. 2019;23(5):435–448. doi: 10.1016/j.tics.2019.03.003. PMCID: PMC6839942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsao C.W., Lin Y.S., Chen C.C., Bai C.H., Wu S.R. Cytokines and serotonin transporter in patients with major depression. Prog. Neuro-Psychopharmacol. Biol. Psychiatr. 2006;30(5):899–905. doi: 10.1016/j.pnpbp.2006.01.029. [DOI] [PubMed] [Google Scholar]
- Walker A.K., Budac D.P., Bisulco S., Lee A.W., Smith R.A., Beenders B., Kelley K.W., Dantzer R. NMDA receptor blockade by ketamine abrogates lipopolysaccharide-induced depressive-like behavior in C57BL/6J mice. Neuropsychopharmacology: Off. Publ. Am. Coll. Neuropsychopharmacol. 2013;38(9):1609–1616. doi: 10.1038/npp.2013.71. PMCID: 3717543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.A., Lee J.D., Lee K.M., Woodruff T.M., Noakes P.G. Complement C5a-C5aR1 signalling drives skeletal muscle macrophage recruitment in the hSOD1(G93A) mouse model of amyotrophic lateral sclerosis. Skeletal Muscle. 2017;7(1):10. doi: 10.1186/s13395-017-0128-8. PMCID: PMC5455206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A., Luan H.H., Medzhitov R. An evolutionary perspective on immunometabolism. Science. 2019;363(6423) doi: 10.1126/science.aar3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Xie L., Ding G., Song S., Chen L., Li G., Xia M., Han D., Zheng Y., Liu J., Xiao T., Zhang H., Huang Y., Li Y., Huang M. Single-cell RNA sequencing of peripheral blood mononuclear cells from acute Kawasaki disease patients. Nat. Commun. 2021;12(1):5444. doi: 10.1038/s41467-021-25771-5. PMCID: PMC8440575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M.D., Godbout J.P., Sheridan J.F. Repeated social defeat, neuroinflammation, and behavior: monocytes carry the signal. Neuropsychopharmacology : Off. Publ. Am. Coll. Neuropsychopharmacol. 2017;42(1):46–61. doi: 10.1038/npp.2016.102. PMCID: PMC5143478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohleb E.S., McKim D.B., Shea D.T., Powell N.D., Tarr A.J., Sheridan J.F., Godbout J.P. Re-establishment of anxiety in stress-sensitized mice is caused by monocyte trafficking from the spleen to the brain. Biol. Psychiatr. 2014;75(12):970–981. doi: 10.1016/j.biopsych.2013.11.029. PMCID: Pmc4084643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohleb E.S., McKim D.B., Sheridan J.F., Godbout J.P. Monocyte trafficking to the brain with stress and inflammation: a novel axis of immune-to-brain communication that influences mood and behavior. Front. Neurosci. 2014;8:447. doi: 10.3389/fnins.2014.00447. PMCID: Pmc4300916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D., Li Y., Xiao H., Liu Q., Zhang M., Zhu J., Ma W., Yao C., Wang J., Wang D., Guo Z., Yang B. Gaining confidence in biological interpretation of the microarray data: the functional consistence of the significant GO categories. Bioinformatics. 2008;24(2):265–271. doi: 10.1093/bioinformatics/btm558. [DOI] [PubMed] [Google Scholar]
- Yang Y., Han L., Yuan Y., Li J., Hei N., Liang H. Gene co-expression network analysis reveals common system-level properties of prognostic genes across cancer types. Nat. Commun. 2014;5:3231. doi: 10.1038/ncomms4231. PMCID: PMC3951205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J.J., Wang N., Yang C., Shi J.Y., Yu H.Y., Hashimoto K. Serum interleukin-6 is a predictive biomarker for ketamine's antidepressant effect in treatment-resistant patients with major depression. Biol. Psychiatr. 2015;77(3):e19–e20. doi: 10.1016/j.biopsych.2014.06.021. [DOI] [PubMed] [Google Scholar]
- Yi Z., Li Z., Yu S., Yuan C., Hong W., Wang Z., Cui J., Shi T., Fang Y. Blood-based gene expression profiles models for classification of subsyndromal symptomatic depression and major depressive disorder. PLoS One. 2012;7(2) doi: 10.1371/journal.pone.0031283. PMCID: PMC3278427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L., Xu X., Chen G., Mehta N.D., Haroon E., Miller A.H., Luo Y., Li Z., Felger J.C. Inflammation and decreased functional connectivity in a widely-distributed network in depression: centralized effects in the ventral medial prefrontal cortex. Brain Behav. Immun. 2019;80:657–666. doi: 10.1016/j.bbi.2019.05.011. PMCID: PMC6660411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yohn S.E., Arif Y., Haley A., Tripodi G., Baqi Y., Muller C.E., Miguel N.S., Correa M., Salamone J.D. Effort-related motivational effects of the pro-inflammatory cytokine interleukin-6: pharmacological and neurochemical characterization. Psychopharmacology (Berl) 2016;233(19–20):3575–3586. doi: 10.1007/s00213-016-4392-9. [DOI] [PubMed] [Google Scholar]
- Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. PMCID: PMC3339379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Lutz P.E., Wang Y.C., Ragoussis J., Turecki G. Global long non-coding RNA expression in the rostral anterior cingulate cortex of depressed suicides. Transl. Psychiatry. 2018;8(1):224. doi: 10.1038/s41398-018-0267-7. PMCID: PMC6193959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L., Yang P., Zhao Y., Zhuang Z., Wang Z., Song R., Zhang J., Liu C., Gao Q., Xu Q., Wei X., Sun H.X., Ye B., Wu Y., Zhang N., Lei G., Yu L., Yan J., Diao G., Meng F., Bai C., Mao P., Yu Y., Wang M., Yuan Y., Deng Q., Li Z., Huang Y., Hu G., Liu Y., Wang X., Xu Z., Liu P., Bi Y., Shi Y., Zhang S., Chen Z., Wang J., Xu X., Wu G., Wang F.S., Gao G.F., Liu L., Liu W.J. Single-cell sequencing of peripheral mononuclear cells reveals distinct immune response landscapes of COVID-19 and influenza patients. Immunity. 2020;53(3):685–696. doi: 10.1016/j.immuni.2020.07.009. e3. PMCID: PMC7368915. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.