SUMMARY
GB virus C (GBV-C) is frequently identified in patients co-infected with human immunodeficiency virus type 1 (HIV-1) due to the similar transmission routes. However, it remains unclear how these two viruses interact with each other and how one virus affects the replication of the other in the human body. In this study, we performed a case-control study to determine whether GBV-C viraemia could prevent the acquisition of HIV-1 infection, and a cohort study to determine the prevalence, genotypic characteristics and incidence of GBV-C infection in men who have sex with men (MSM) populations in Beijing, China. The prevalence of GBV-C infection in HIV-1-negative subjects was similar to that in HIV-1-positive subjects. Before HIV-1 acquisition, the prevalence of GBV-C was 17·7%, which increased to 27·2% at the acute stage and to 34% at the chronic stage. Genotype 3 was the major genotype of GBV-C in both groups. A significantly positive correlation was observed between the CD4+ T-cell counts and GBV-C viral loads at the acute stage of HIV infection. At the chronic stage (12 months later), this correlation was no longer significant, although it was still positive. Overall, this study demonstrated that pre-existing GBV-C viraemia could not prevent the acquisition of HIV-1 infection and transmission of HIV-1 significantly increased the prevalence of GBV-C viraemia.
Key words: Cohort study, GB virus C, human immunodeficiency virus type 1, men who have sex with men (MSM)
INTRODUCTION
GBV-C is a flavivirus that is closely related to hepatitis C virus (HCV) [1]. Transmission of GBV-C via parenteral, sexual, and vertical routes has been documented, and infection is common in many populations [2–7]. There are five major genotypes that are prevalent worldwide [8, 9]. Several studies have reported a strong association between the existence of GBV-C viraemia and the survival of HIV-infected patients [7, 10, 11]. However, other studies do not support this association [12–14]. In fact, two studies showed that patients co-infected with GBV-C and HIV-1 had an increased mortality rate [15, 16]. Mechanisms underlying the positive effects on the progression of HIV infection include decreased expression of CC chemokine receptor 5 (CCR5) on CD4+ T cells and direct inhibition of HIV replication by GBV-C [17, 18]. Recent findings suggest that HIV-1 patients with active GBV-C infection had a reduced percentage of T cells positive for CD38+CD4+, CD38+CD8+, CCR5+CD4+, and CCR5+CD8+, which might provide a key mechanism involved in the protection conferred by GBV-C against HIV-1 disease progression [19].
MSM are the second most vulnerable population for HIV-1 infections after intravenous drug users in China in recent years. In Beijing, 3·0–4·6% of MSM were found to be HIV-1 positive [20]. A cross-sectional study has shown a high prevalence of sexually transmitted infections and HCV to be present in HIV-1-positive MSM [21], but the infection rate of GBV-C in MSM has not been established. Here, we performed a cohort study to evaluate the prevalence of GBV-C viraemia in MSM for those with a clear date of HIV-1 infection and those without HIV-1 infection. We also analysed the association of GBV-C viraemia, syphilis, HCV and hepatitis B virus (HBV) with HIV-1 infection in this MSM cohort. Furthermore, we determined the potential effect of GBV-C viraemia on the progression of HIV-1 disease.
MATERIALS AND METHODS
Study sample
The subjects included in this study were selected from the participants in the AIDS High Risk Cohort Programme supported by the Beijing Science & Technology Committee and the study was approved by the Ethics Committee of the Beijing Youan Hospital. A total of 4236 MSM were enrolled in the study between 2007 and 2010. Written informed consent was obtained from all participants. At 2-month intervals, HIV-related clinical status was assessed, an interviewer-administered questionnaire was completed, and blood samples were obtained for detection of HIV-1 antibodies and HIV-1 RNA levels by immunoassay and RT–PCR (Roche, Sweden), respectively. Of 4236 MSM, 99 subjects who had a clear date of HIV-1 infection and 175 subjects without HIV-1 infection at the same visit as a control were selected from the cohort for the case-control study. The prevalence of GBV-C viraemia and antibodies against HCV, HBV surface antigen (HBsAg) and syphilis were compared between individuals with HIV-1 infection and controls without HIV-1 infection. In the cohort study, 99 HIV-1-positive subjects were detected for GBV-C viraemia at the acute stage of HIV infection and the chronic stage of HIV-1 infection. Acute stage is defined as negative HIV p24 antibody testing by enzyme-linked immunosorbent assay in the presence of detectable HIV-1 RNA or positive HIV p24 antibody testing and an evolving (⩽3 bands positive) HIV Western blot [22]. Chronic stage is defined as having been infected for at least 6 months and usually for several years [23]. In our study, we collected the samples at 12 months post-HIV infection. CD4+ T-cell counts and HIV-1 viral loads were also determined at each time point.
RT-PCR for detection of GBV-C RNA
GBV-C RNA was extracted from the samples by the Qiagen viral RNA Mini kit (Qiagen, Germany). A Superscript First-Strand Synthesis kit (Invitrogen, USA) was used to create GBV-C complementary DNAs (cDNAs). Both first- and second-round PCRs were performed using primers that hybridize to 5′ non-translated regions of GBV-C (GenBank accession no. NC_001710.1, 31–376 nt and 121–333 nt, respectively). Primers for the first-round RT–PCR were GBV-F1 (5′-ACT GGG TGC AAG CCC CAG AAA CC) and GBV-R1 (5′-CTG GTC CTT GTC AAC TCG CCG). Primers for the second-round PCR were GBV-F2 (5′-GTG ATG ACA GGG TTG GTA GGT CGT) and GBV-R2 (5′-GAC ATT GAA GGG CGA CGT GGA). PCR products were detected on 1·5% agarose gels containing 0·5 μg/ml of ethidium bromide. The expected band sizes were 366 and 233 bp for the first- and second-round PCRs, respectively.
Quantification of GBV-C viral load
Absolute quantification requires a standard curve of known amounts of the amplicon per reaction. The substrates for standard curve were generated by subcloning the amplicon in a plasmid (pGEM-T easy, Promega). A standard curve of the GBV-C NCR plasmid was quantified using the universal QPCR method in a 25-μl reaction containing 1× Taqman Universal PCR Master Mix (Applied Biosystems, USA), 400 nm of primer gbv-p1 (5′-AGC GCA CGG TCC ACA GGT GTT-3′), 400 nm of primer gbv-p2 (5′-GAC ATT GAA GGG CGA CGT GGA-3′), and 200 nm of fluorescent probe gbv-probe (5′-FAM-CCC TAC CGG TGG GAA TAA GGG CCC GAC TAMRA-3′) (Invitrogen). Serial tenfold dilutions of GBV-C NCR plasmid were used to cover a range of 104–107 molecules per reaction. Triplicate reactions of each dilution were pipetted into a 96-well plate and amplified in an ABI Prism 7900HT real-time PCR system (Applied Biosystems) at 50 °C for 30 min and then 94 °C for 2 min for the inactivation of the AMV reverse transcription enzyme, followed by 40 cycles of 94 °C for 10 s and 60 °C for 1 min. The data was analysed with Sequence Detection Systems (SDS) software (Applied Biosystems).
Determination of nucleotide sequences and GBV-C genotypes
The 627-bp PCR outer fragments of 5′-UTR were obtained by the primer pairs of GBV-OF1 (5′-GTG CAA GCC CCA GAA ACC GAC-3′, 36 nt, NC_001710.1) and GBV-OR1 (5′-GGG GCG CAA CAG TTT GTG AGG-3′, 641 nt, NC_001710.1). Subsequently, the 366-bp PCR products were obtained by primer pairs GBV-F1 and GBV-R1. The nucleotide sequences were determined using an ABI PRISM3730 XL Genetic Analyzer (ABI Biosystems, USA). The sequences were aligned with the sequences from reference isolates of GBV-C genotype 1 (U36380), genotype 2 (U44402 and AF081782), genotype 3 (D90601) and genotype 5 (AY949711) by Mega software (version 5.0). The phylogenetic trees were constructed by neighbour-joining as implemented in the software and the evolutionary distances were computed using the maximum composite likelihood method.
Statistical analyses
The odds ratios (OR) and 95% confidence intervals (CI) for the association between the occurrence of GBV-C viraemia and HIV acquisition was calculated. The occurrence of GBV-C viraemia between subjects prior to HIV-1 infection and those after acquisition of HIV-1 infection were compared by Pearson's χ2 test. The GBV-C viral loads at two time points were compared by paired Student's t test. The median of HIV viral loads and CD4+ T-cell counts between the GBV-C viraemia group and the GBV-C RNA-negative group in HIV-1-positive subjects were compared by Mann–Whitney U test. The relationship between HIV-1 viral loads and GBV-C viral loads, and CD4+ T-cell counts were evaluated by linear regression analysis.
RESULTS
HIV-1 acquisition was independent of GBV-C viraemia
To determine the impact of GBV-C viraemia on HIV-1 transmission, we followed up 99 MSM subjects before and after they acquired HIV-1. Based on a cross-sectional analysis, we found that the prevalence of GBV-C viraemia in these 99 study subjects was 17·7% before HIV-1 infection, which was not significantly different from that (12·6%) of the 175 HIV-1-negative subjects (OR 1·36, P = 0·29) (Table 1), indicating that a pre-existing GBV-C infection probably did not play a significant role in preventing the acquisition of HIV-1. Similarly, the prevalence of HBV and HCV in the 99 HIV-1-positive subjects was not significantly different from that in the 175 HIV-1-negative subjects (Table 1). In contrast, the prevalence of syphilis in the 99 HIV-1-positive subjects (20·20%) was significantly higher than that in the HIV-1-negative subjects (3·42%, OR 5·89). These results indicated that HIV-1 acquisition was independent of the presence of HBV, HCV and GBV-C viraemia, but was highly associated with the presence of syphilis in the MSM population in Beijing.
Table 1.
Association of HIV-1 co-infection with other pathogens including HBV, HCV, syphilis and GBV-C
| Characteristic | Cases (n = 99) | Controls (n = 175) | OR (95% CI) |
|---|---|---|---|
| HBV, n (%) | 2 (2·02) | 3 (1·71) | 1·2 (0·20–7·08) |
| HCV, n (%) | 1 (1·01) | 2 (1·14) | 0·88 (0·08–9·62) |
| Syphilis, n (%) | 20 (20·20) | 6 (3·42) | 5·89 (2·45–14·18) |
| GBV-C viraemia, n (%) | 17 (17·7) | 22 (12·6) | 1·37 (0·76–2·44) |
OR, Odds ratio, CI, confidence interval.
Occurrence of GBV-C viraemia was increased after HIV-1 acquisition
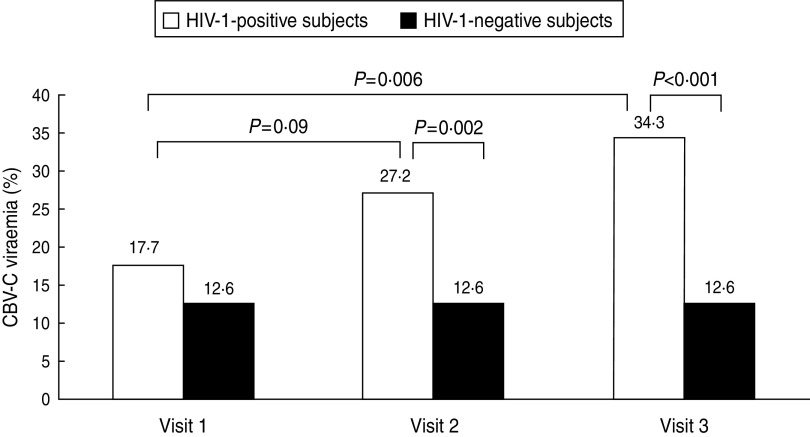
During follow-up studies, the occurrence of GBV-C viraemia in the 99 HIV-1-positive subjects increased from 17·7% prior to HIV-1 acquisition (visit 1) to 27·2% at the time of HIV-1 acquisition (visit 2) (P = 0·09). After 12 months (visit 3), GBV-C RNA was still positive in the 27 subjects that were positive at visit 2. Furthermore, an additional seven subjects had acquired GBV-C viraemia at visit 3, leading to a significant increase of GBV-C viraemia occurrence compared to visit 1 (34·3%, P < 0·01). However, for the HIV-negative subjects, newly acquired GBV-C viraemia was not observed after 12 months. The occurrence of GBV-C viraemia was significantly higher in the HIV-1-positive subjects than in the HIV-1-negative subjects at visits 2 and 3, respectively (Fig. 1). For HIV-1-positive subjects at visit 2, the distribution of GBV-C RNA (27·2%) was similar with respect to age, HBV, HCV, syphilis co-infection and HIV-1 subtypes. The baseline clinical characteristics of 99 MSM with HIV-1 infections are given in Table 2.
Fig. 1.
The occurrence of GBV-C viraemia in subjects at different stages of HIV infection (n = 99) and HIV-negative subjects (n = 175). The x-axis represents the time during which the samples were collected. The y-axis represents the percentage of GBV-C viraemia. P values represent the statistical analysis between each comparison as shown in the figure. Compared by Pearson's χ2 test, the significance level was set at 0·05. Visit 1: before HIV infection; visit 2: near HIV infection; visit 3: 12 months post-HIV infection.
Table 2.
Baseline clinical characteristics of 99 HIV-positive MSM with or without GBV-C infection
| GBV-C viraemia status | GBV-C positive (n = 27) | GBV-C negative (n = 72) | P |
|---|---|---|---|
| Median age (years) | 25 (24–31) | 30 (24–39) | 0·064* |
| Median days of HIV infection | 30 (17–30) | 30 (28–46) | 0·11* |
| HIV subtype | |||
| CRF01-AE | 15/27 (55·6%) | 36/72 (50%) | 0·62† |
| B | 9/27 (33·3%) | 21/72 (29·2%) | 0·69† |
| B/C | 3/27 (11·1%) | 7/72 (9·7%) | 1† |
| C | 0 | 2/72 (1·4%) | 1† |
| Not detected | 0 | 6/72 (8·3%) | |
| HBsAg positive | 1/27 (3·7%) | 1/72 (1·39%) | 0·47† |
| Anti-HCV positive | 0 | 1/72 (1·39%) | 1† |
| Syphilis | 7/27 (25·9%) | 13/72 (18·1%) | 0·385† |
| Median (IQR 25–75%) (log10 copies/ml) | 4·29 (3·69–5·51) | 4·83 (3·97–5·58) | 0·31* |
| Median (IQR 25–75%) CD4 counts (cells/mm3) | 423 (361–587) | 506 (334–602) | 0·46* |
MSM, Men who have sex with men; IQR, interquartile range.
Mann–Whitney test.
Two-sided Pearson's χ2 test.
GBV-C genotypes
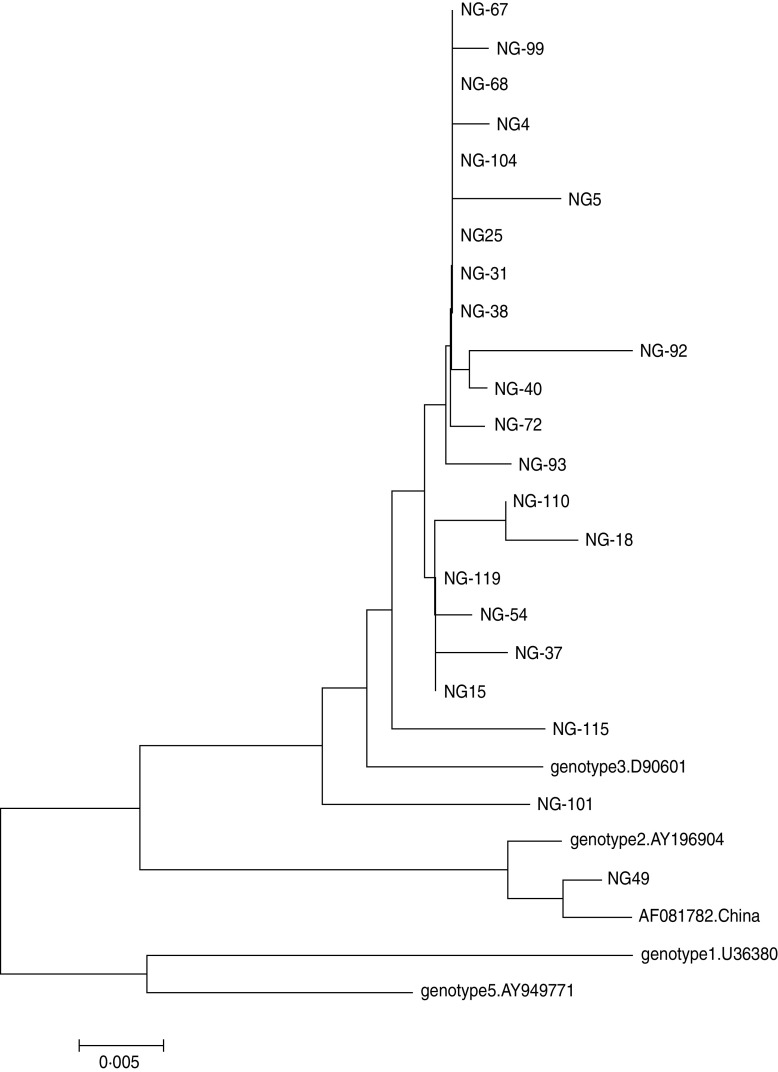
Following the determination of GBV-C prevalence in 99 HIV-1-positive and 175 HIV-1-negative subjects, the nucleotide sequences of PCR products were analysed to determine the GBV-C genotypes. Reference sequences of four genotypes (1, 2, 3, 5) were included in this analysis. Of the 34 samples from HIV-1-positive subjects, 30 (88·2%) were GBV-C genotype 3, while the remaining four (11·8%) were genotype 2 according to comparison with the reference strains (Fig. 2). Of the 22 samples from HIV-1-negative subjects, one (4·5%) sample was genotype 2, and 21 (95·5%) samples belonged to genotype 3. The genetic distances were small between the viruses from HIV-1-positive subjects and those from HIV-1-negative subjects (Figs 2, 3). These results indicated that the majority of GBV-C in MSM in Beijing belonged to genotype 3. During HIV-1 infection, both genotypes 2 and 3 were transmissible.
Fig. 2.
Phylogenetic tree of 34 GBV-C strains from 34 HIV-infected Chinese MSM subjects (indicated by pt. prefix) based on the sequences of the 5′-UTR gene (300 bp). The phylogenetic tree was constructed by the neighbour-joining method and the evolutionary distances were computed using the maximum composite likelihood method of Mega software (version 5.0). Reference strains of genotypes 1 (U36380), 2 (U44402 and AF081782 from China), 3 (D90601) and 5 (AY949711) were obtained from GenBank. The scale bar represents 0·5% genetic distance (0·005 substitution per site).
Fig. 3.
Phylogenetic tree of 22 GBV-C strains from 22 HIV-1-negative MSM subjects (indicated by NG prefix) based on the sequences of the 5′-UTR gene (300 bp). The phylogenetic tree was constructed by the neighbour-joining method and the evolutionary distances were computed using the maximum composite likelihood method of Mega software (version 5.0). Reference strains of genotypes 1 (U36380), 2 (U44402 and AF081782 from China), 3 (D90601) and 5 (AY949711) were obtained from GenBank. The scale bar represents 0·5% genetic distance (0·005 substitution per site).
GBV-C viral loads at the acute and chronic stages of HIV-1 infection
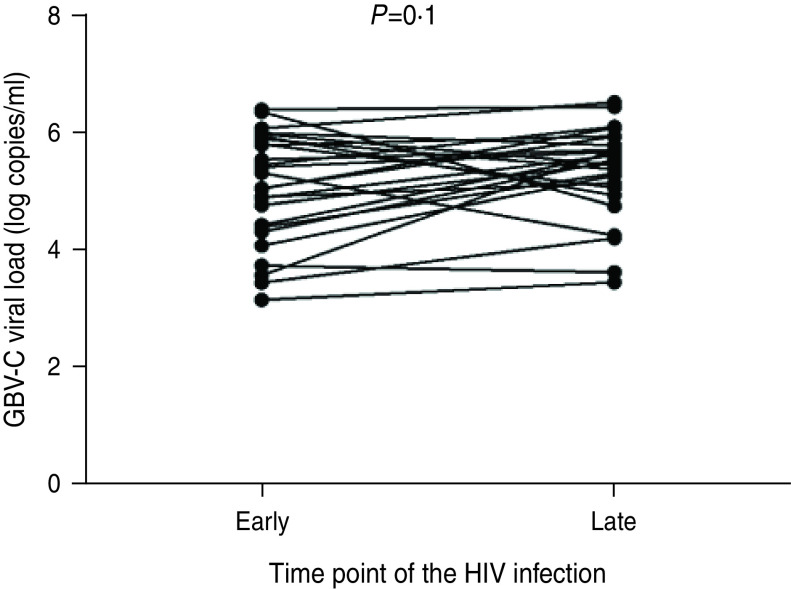
The GBV-C viral load in each of 26 HIV-1-positive subjects [one case started antiretroviral therapy (ART) 3 months after HIV-1 acquisition in the follow-up study and was excluded from the analysis] at two time points (acute and chronic stages) of HIV infection were quantified with a real-time PCR. The GBV-C viral loads at the acute stage of HIV-1 infection ranged from 1·62 × 103 to 2·82 × 106 copies/ml with a median of 5·24 log copies/ml. At 12 months post-HIV-1 acquisition (chronic stage), the viral loads ranged from 3·24 × 103 to 3·80 × 106 copies/ml with a median of 5·56 log copies/ml. No significant difference was found between the GBV-C viral loads at acute stage and those at the chronic stage (Fig. 4).
Fig. 4.
Comparison of the GBV-C viral loads at the acute and chronic stages of HIV infection in HIV-1-positive subjects (n = 26, one case started ART 3 months after acquiring HIV-1 and was excluded from the analysis.). The x-axis represents the stage of HIV infection, ‘Early’ indicates the acute stage of HIV infection and ‘Late’ indicates the chronic stage of HIV infection. The y-axis represents the GBV-C viral loads (log copies/ml). Evaluated by paired Student's t test; significance level was set at 0·05.
Comparison of HIV-1 viral loads and CD4+ T-cell counts between subjects with and without GBV-C infection
To evaluate the potential effect of GBV-C co-infection on the replication of HIV-1, we analysed the HIV viral loads and CD4+ T-cell counts in the GBV-C-positive and -negative subjects at the acute and chronic stages of HIV-1 infection. In the GBV-C RNA-positive subjects, the median HIV viral load was 4·29 log copies/ml [25–75%, interquartile range (IQR) 3·69–5·51], and the median CD4+ T-cell count was 423 cells/mm3 (IQR 361–587) at the acute stage. In the GBV-C RNA-negative subjects, the median viral load was 4·81 log copies/ml (IQR 4·00–5·57), and the median CD4+ T-cell count was 502 cells/mm3 (IQR 327–622). Statistical analysis showed that both CD4+ T-cell counts and HIV-1 viral load were not significantly different between the two groups at the acute stage of HIV-1 infection (P = 0·34, P = 0·5, respectively) (Table 2).
In the follow-up study, one case in the GBV-C-positive group and two cases in the GBV-C-negative group started ART 3 months after acquiring HIV-1. Seven cases in the GBV-C-negative group acquired GBV-C viraemia 12 months post-HIV-1 acquisition. Thus, the case numbers in the GBV-C-positive and -negative groups became 26 and 63, respectively. The median of HIV-1 viral load for 26 GBV-C-positive subjects and 63 GBV-C-negative subjects was 4·65 log10 copies/ml (IQR 3·86–5·03) and 4·41 log10 copies/ml (IQR 3·99–4·97), respectively, at 12 months post-HIV-1 acquisition. Statistical analysis showed that HIV-1 viral load was not significantly different between these two groups (P = 0·47). The median CD4+ T-cell counts were 431 cells/mm3 (IQR 281–542) and 435 cells/mm3 (IQR 313–613) in the GBV-C positive and negative groups, respectively. Statistical analysis showed that there was no significant difference between CD4+ T-cell counts in the GBV-C-positive and GBV-C-negative subjects (P = 0·31).
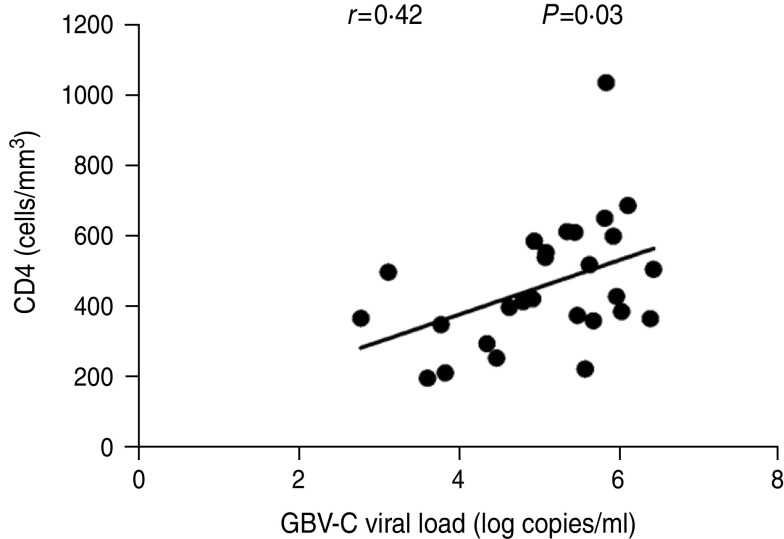
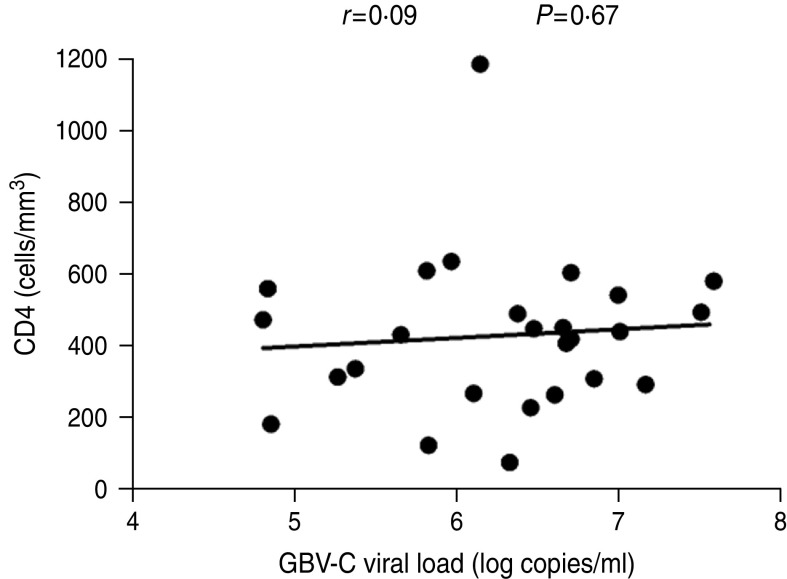
We then conducted a correlation analysis between GBV-C viral load and HIV-1 viral load or CD4+ T-cell count at the acute or chronic stages of HIV-1 infection. At the acute stage, a significant positive correlation was observed between the GBV-C viral load and CD4+ T-cell count (r = 0·42, P = 0·03) (Fig. 5). HIV-1 viral load was inversely and not significantly correlated with the GBV-C viral load (r = − 0·17, P = 0·39). At the chronic stage (12 months post-HIV-1 acquisition), we performed the same analyses excluding the ART cases, and found a persistent positive correlation between GBV-C viral loads and CD4+ T-cell counts at this time point, but the correlation was not significant (r = 0·09, P = 0·67) (Fig. 6). There was a persistent negative, but not significant, correlation between GBV-C viral load and HIV-1 viral load (r = − 0·2, P = 0·34) (data not shown) at the chronic stage.
Fig. 5.
Correlation between GBV-C viral loads and CD4+ T-cell counts at the acute stage of HIV-1 infection in ART-naive Chinese MSM subjects (n = 27). The x-axis represents the GBV-C viral loads (log copies/ml). The y-axis represents the CD4 cell counts (cells/mm3). The GBV-C viral loads showed a significant positive correlation (r = 0·42, P = 0·03) with the CD4+ T-cell counts. The significance level was set at 0·05 in the linear regression analysis.
Fig. 6.
Correlation between GBV-C viral loads and the CD4+ T-cell counts at the chronic stage (12 months post-HIV-1 acquisition) of HIV-1 infection in Chinese MSM subjects (n = 26, one case started ART 3 months after acquiring HIV-1 and was excluded from the analysis). The correlation was still positive but no longer significant (r = 0·09, P = 0·67). The x-axis represents the GBV-C viral loads (log copies/ml). The y-axis represents the CD4 cell counts (cells/mm3). The significance level was set at 0·05 in the linear regression analysis.
DISCUSSION
Previous studies have shown that GBV-C is prevalent in haemodialysis patients, commercial blood donors [24], intravenous drug users [25] and female commercial sex workers [26] in China. To our knowledge, the current study is the first to describe the prevalence of GBV-C viraemia in the MSM population with or without HIV-1 infection in China. The positive rate of GBV-C viraemia in subjects with HIV-1 infections was 27·2% (27/99) at the time of HIV-1 acquisition, which was much higher than that in the HIV-1-negative subjects (P = 0·002). These results are consistent with previous reports showing that the prevalence of GBV-C viraemia in HIV-1-positive Americans and Europeans was higher than in HIV-1-negative subjects [14, 27–29]. Our data also indicate that GBV-C infection is common in recently infected HIV-1 MSM populations in Beijing, China.
The purpose of this study was to determine if pre-existence of GBV-C viraemia confers beneficial effects for resistance to HIV-1 infection. Our case-control study showed that the positive rate of GBV-C in the 99 HIV-1-positive subjects prior to acquisition of HIV-1 infection was not significantly different from that in HIV-1-negative subjects (P = 0·29), suggesting that the pre-existence of GBV-C viraemia does not play a significant role in resistance to HIV-1 infection. These results are consistent with a previous study showing that the prevalence of GBV-C viraemia in HIV-uninfected individuals who did and did not acquire HIV was similar in American populations [30]. Our results also showed that after HIV-1 acquisition, the positive rate of GBV-C significantly increased (Fig. 1), indicating GBV-C infection may occur concurrently with HIV-1 acquisition. In the follow-up study, seven cases acquired GBV-C viraemia among HIV-1-infected individuals at 12 months post-HIV-1 infection, resulting in an incidence density of 9·7/100 person-years. None of the subjects acquired new GBV-C viraemia in the HIV-negative group during this period, indicating the high incidence density of GBV-C viraemia in the MSM populations with HIV infection. In our study, the occurrence of syphilis was high in HIV-1-positive subjects. This result was in accord with a previous cross-sectional study on MSM in Beijing [21]. In addition, we did not find a high prevalence of HCV and HBV in MSM with or without HIV-1 infections.
According to some reports of the past two decades, the major genotype of GBV-C is genotype 3. Few virus strains of genotypes 2 and 1 have been reported in different populations in China [31, 32]. Genotypic analysis in this study showed that MSM subjects were mainly infected with the genotype 3 strain of GBV-C (88·2%). Only four subjects were infected with genotype 2 strain and no subjects were infected with genotype 1 strain. Therefore, we concluded that the major GBV-C genotype prevalent in the MSM population was similar to that in other populations in China. Our results also agree with the viral genotype prevalent in other Asian countries [32, 34]. We attempted to identify if the genotypes had any effect on the viral loads of GBV-C. Our results showed that there was no significant relationship between the genotype and viral loads of GBV-C (data not shown). This differs from a previous report [34] showing that GBV-C viral loads were associated with genotype. This difference may be due to the small numbers of genotype 2 in our study or differences in the progress of HIV-1 within the study population.
Previous studies have shown that GBV-C replication in peripheral blood mononuclear cells decreases the expression of HIV-1 co-receptors (chemokine receptors CCR5 and CXCR4) on the surface of CD4+ T cells and increases the production of various chemokines that serve as competitive inhibitors of the HIV-1 co-receptors [18, 35, 36]. It is known that there is vigorous viral replication at the acute stage during the natural history of HIV-1 infection. Thus, we speculate that there are some complicated interactions between the two viruses at the acute stage of HIV-1 infection. Our results showed that the GBV-C viral loads had an ascending trend from the acute (5·24 log copies/ml) to chronic (5·56 log copies/ml) stage (Fig. 4). Statistical analysis showed a sustained negative correlation between the GBV-C viral loads and HIV-1 viral loads in the acute and chronic stages of HIV-1 infection. Although, we were unable to conclude that the lower GBV-C viral loads at the acute stage of HIV-1 infection was directly caused by higher HIV-1 viral loads, our results provide some evidence that an inhibitory interaction between these two viruses might exist during the period of co-infection.
Previous studies have suggested that HIV RNA levels at later time points are better indicators of long-term disease progression compared to the levels when HIV-1 was acquired because the viral load reaches a stable mean or ‘set point’ around 1 year post-infection [37, 38]. In this study, both plasma HIV RNA and the CD4+ T-cell counts did not differ between GBV-C-positive and GBV-C-negative subjects at 1 year post-HIV infection. These results indicate that GBV-C viraemia could not increase CD4+ T-cell counts or reduce HIV-1 viral load during the 1-year period following HIV-1 infection. These results also suggest that there was no significant suppressive effect by GBV-C on HIV-1 replication in the year following HIV-1 infection. Williams et al. reported that GBV-C viraemia was significantly associated with the prolonged survival of HIV-positive men 5–6 years post-HIV-1 infection. However, the survival rate was not increased 12–18 months post-HIV infection [39]. Thus, a long-term follow-up study should be conducted on this cohort.
The molecular mechanisms by which GBV-C suppresses the replication of HIV-1 are poorly understood and the effect of GBV-C viraemia on the progress of HIV disease is currently controversial. Several studies have shown that GBV-C viraemia could assist the elevation of CD4+ T-cell counts because a positive correlation between GBV-C viral load and CD4+ T-cell counts was observed [40, 41]. Interestingly, in our study, a significant positive correlation between CD4+ T-cell counts and GBV-C viral loads (r = 0·42, P = 0·03) (Fig. 5) was observed at the acute stage of HIV-1 infection after 1 year, and the correlation was still positive but no longer significant (r = 0·09, P = 0·67) (Fig. 6). Because GBV-C RNA can replicate in CD4+ T cells, the decrease of CD4+ T cells during the course of HIV-1 infection implies a loss of target cells for GBV-C RNA. This might explain why the correlation between CD4+ T cells and GBV-C viral loads was weakened 1 year post-HIV infection as a consequence of a decrease in CD4+ T-cell counts. Our study can only reflect on the instance of male homosexuals who sexually transmit GBV-C, and does not include bloodborne transmission of HIV-1. In addition, the period of the current study is not long enough to observe the loss of GBV-C viraemia and the effect of sustained GBV-C viraemia on the progress of HIV/AIDS. In conclusion, this is the first cohort study, to the best of our knowledge, on GBV-C co-infection with HIV-1 in the MSM population of Beijing, China. Long-term observation is needed on this cohort to further reveal the correlation between GBV-C and HIV infection.
ACKNOWLEDGEMENTS
This study was supported by Beijing Science and Technology Programme (D09050703590901 to H. Wu); Natural Science Foundation of China (30870853 to D. Chen); HKU-UDF and LSK Faculty of Medicine Matching Fund for financial supports to HKU AIDS Institute (to Z. Chen).
DECLARATION OF INTEREST
None.
REFERENCES
- 1.Stapleton JT. GB virus type C/Hepatitis G virus. Seminars in Liver Disease 2003; 23: 137–148. [DOI] [PubMed] [Google Scholar]
- 2.Dawson GJ, et al. Prevalence studies of GB virus-C infection using reverse transcriptase-polymerase chain reaction. Journal of Medical Virology 1996; 50: 97–103. [DOI] [PubMed] [Google Scholar]
- 3.Linnen J, et al. Molecular cloning and disease association of hepatitis G virus: a transfusion-transmissible agent. Science 1996; 271: 505–508. [DOI] [PubMed] [Google Scholar]
- 4.Fischler B, et al. Genetic evidence for mother-to-infant transmission of hepatitis G virus. Journal of Infectious Diseases 1997; 176: 281–285. [DOI] [PubMed] [Google Scholar]
- 5.Rubio A, et al. Is hepatitis G virus transmitted sexually? Japan Automobile Manufacturers Association 1997; 277: 532–533. [DOI] [PubMed] [Google Scholar]
- 6.Semprini AE, et al. Absence of hepatitis C virus and detection of hepatitis G virus/GB virus C RNA sequences in the semen of infected men. Journal of Infectious Diseases 1998; 177: 848–854. [DOI] [PubMed] [Google Scholar]
- 7.Xiang J, et al. Effect of coinfection with GB virus C on survival among patients with HIV infection. New England Journal of Medicine 2001; 345: 707–714. [DOI] [PubMed] [Google Scholar]
- 8.Muerhoff AS, et al. Sequence heterogeneity within the 5′-terminal region of the hepatitis GB virus C genome and evidence for genotypes. Journal of Hepatology 1996; 25: 379–384. [DOI] [PubMed] [Google Scholar]
- 9.Naito H, Hayashi S, Abe K. The entire nucleotide sequence of two hepatitis G virus isolates belonging to a novel genotype: isolation in Myanmar and Vietnam. Journal of General Virology 2000; 81: 189–194. [DOI] [PubMed] [Google Scholar]
- 10.Lefrère JJ, et al. Carriage of GB virus C/hepatitis G virus RNA is associated with a slower immunologic, virologic, and clinical progression of human immunodeficiency virus disease in coinfected persons. Journal of Infectious Diseases 1999; 179: 783–789. [DOI] [PubMed] [Google Scholar]
- 11.Tillmann HL, et al. Infection with GB virus C and reduced mortality among HIV-infected patients. New England Journal of Medicine 2001; 345: 715–724. [DOI] [PubMed] [Google Scholar]
- 12.Birk M, Lindbäck S, Lidman C. No influence of GB virus C replication on the prognosis in a cohort of HIV-1-infected patients. Acquired Immune Deficiency Syndrome 2002; 16: 2482–2485. [DOI] [PubMed] [Google Scholar]
- 13.Björkman P, et al. GB virus C during the natural course of HIV-1 infection: viremia at diagnosis does not predict mortality. Acquired Immune Deficiency Syndrome 2004; 18: 877–886. [DOI] [PubMed] [Google Scholar]
- 14.Van der Bij AK, et al. GB virus C coinfection and HIV-1 disease progression: The Amsterdam Cohort Study. Journal of Infectious Diseases 2005; 191: 678–685. [DOI] [PubMed] [Google Scholar]
- 15.Quiros-Roldan E, et al. No evidence of benefical effect of GB virus type C infection on the course of HIV infection. Acquired Immune Deficiency Syndrome 2002; 16: 1430–1431. [DOI] [PubMed] [Google Scholar]
- 16.Ryt-Hansen R, et al. No influence of GB virus C on disease progression in a Danish cohort of HIV-infected men. AIDS Research and Human Retroviruses 2006; 22: 496–498. [DOI] [PubMed] [Google Scholar]
- 17.Nattermann J, et al. Regulation of CC chemokine receptor 5 in hepatitis G virus infection. Acquired Immune Deficiency Syndrome 2003; 17: 1457–1462. [DOI] [PubMed] [Google Scholar]
- 18.Xiang J, et al. Inhibition of HIV-1 replication by GB virus C infection through increases in RANTES, MIP-1alpha, MIP-1beta, and SDF-1. Lancet 2004; 363: 2040–2046. [DOI] [PubMed] [Google Scholar]
- 19.Maidana-Giret MT, et al. GB virus type C infection modulates T-cell activation independently of HIV-1 viral load. Acquired Immune Deficiency Syndrome 2009; 23: 2277–2287. [DOI] [PubMed] [Google Scholar]
- 20.Zhang X, et al. Characterization of HIV-1 subtypes and viral antiretroviral drug resistance in men who have sex with men in Beijing, China. Acquired Immune Deficiency Syndrome 2007; 21 (Suppl. 8): S59–65. [DOI] [PubMed] [Google Scholar]
- 21.Zhang X, et al. Risk factors of HIV infection and prevalence of co-infections among men who have sex with men in Beijing, China. Acquired Immune Deficiency Syndrome 2007; 21 (Suppl. 8): S53–57. [DOI] [PubMed] [Google Scholar]
- 22.Streeck H, et al. Human immunodeficiency virus type 1-specific CD8+ T-cell responses during primary infection are major determinants of the viral set point and loss of CD4+ T cells. Journal of Virology 2009; 83: 7641–7648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moir S, et al. B cells in early and chronic HIV infection: evidence for preservation of immune function associated with early initiation of antiretroviral therapy. Blood 2010; 116: 5571–5579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y, et al. Infection with GB virus C and hepatitis C virus in hemodialysis patients and blood donors in Beijing. Journal of Medical Virology 1997; 52: 26–30. [DOI] [PubMed] [Google Scholar]
- 25.Wu RR, et al. GB virus C/hepatitis G virus infection in southern China. Journal of Infectious Diseases 1997; 175: 168–171. [DOI] [PubMed] [Google Scholar]
- 26.Wu JC, et al. Prevalence and risk factor analysis of GBV-C/HGV infection in prostitutes. Journal of Medical Virology 1997; 52: 83–85. [PubMed] [Google Scholar]
- 27.Ibáñez A, et al. Prevalence and genotypes of GB virus C/hepatitis G virus (GBV-C/HGV) and hepatitis C virus among patients infected with human immunodeficiency virus: evidence of GBV-C/HGV sexual transmission. Journal of Medical Virology 1998; 55: 293–299. [DOI] [PubMed] [Google Scholar]
- 28.Wächtler M, et al. Prevalence of GB virus C/hepatitis G virus RNA and anti-E2 glycoprotein antibodies in homosexual men with HIV coinfection. Infection 2000; 28: 297–300. [DOI] [PubMed] [Google Scholar]
- 29.Berzsenyi MD, et al. Male to male sex is associated with a high prevalence of exposure to GB virus C. Journal of Clinical Virology 2005; 33: 243–246. [DOI] [PubMed] [Google Scholar]
- 30.Bisson GP, et al. Effect of GB virus C viremia on HIV acquisition and HIV set-point. Acquired Immune Deficiency Syndrome 2005; 19: 1910–1912. [DOI] [PubMed] [Google Scholar]
- 31.Cao K, et al. GB virus C/hepatitis G virus infection among patients with hepatocellular carcinoma in the inshore area of the Yangtze River, China. Journal of Gastroenterology and Hepatology 1998; 13: 1241–1248. [PubMed] [Google Scholar]
- 32.Yu ML, et al. The serological and molecular epidemiology of GB virus C/hepatitis G virus infection in a hepatitis C and B endemic area. Journal of Infection 2001; 42: 61–66. [DOI] [PubMed] [Google Scholar]
- 33.Hattori J, et al. Prevalence of infection and genotypes of GBV-C/HGV among homosexual men. Microbiology and Immunology 2003; 47: 759–763. [DOI] [PubMed] [Google Scholar]
- 34.Hattori J, et al. Beneficial effect of GB virus C co-infection in human immunodeficiency virus type 1-infected individuals. Microbiology and Immunology 2007; 51: 193–200. [DOI] [PubMed] [Google Scholar]
- 35.Jung S, et al. Inhibition of HIV strains by GB virus C in cell culture can be mediated by CD4 and CD8 T-lymphocyte derived soluble factors. Acquired Immune Deficiency Syndrome 2005; 19: 1267–1272. [DOI] [PubMed] [Google Scholar]
- 36.Schwarze-Zander C, et al. GB virus C coinfection in advanced HIV type-1 disease is associated with low CCR5 and CXCR4 surface expression on CD4(+) T-cells. Antiviral Therapy 2010; 15: 745–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Wolf F, et al. AIDS prognosis based on HIV-1 RNA, CD4+ T-cell count and function: markers with reciprocal predictive value over time after seroconversion. Acquired Immune Deficiency Syndrome 1997; 11: 1799–1806. [DOI] [PubMed] [Google Scholar]
- 38.Hubert JB, et al. Natural history of serum HIV-1 RNA levels in 330 patients with a known date of infection. The SEROCO Study Group. Acquired Immune Deficiency Syndrome 2000; 14: 123–131. [DOI] [PubMed] [Google Scholar]
- 39.Williams CF, et al. Persistent GB virus C infection and survival in HIV-infected men. New England Journal of Medicine 2004; 350: 981–990. [DOI] [PubMed] [Google Scholar]
- 40.Smith SM, et al. Prevalence of GB virus type C in urban Americans infected with human immunodeficiency virus type 1. Retrovirology 2005; 2: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Giret MT, et al. Prevalence, Incidence density, and genotype distribution of GB virus C infection in a cohort of recently HIV-1-infected subjects in Sao Paulo, Brazil. PLoS One 2011; 6: e18407. [DOI] [PMC free article] [PubMed] [Google Scholar]