SUMMARY
Phylogenetic analyses suggest lyssaviruses, including Rabies virus, originated from bats. However, the role of bats in the maintenance, transmission and evolution of lyssaviruses is poorly understood. A number of genetically diverse lyssaviruses are present in Africa, including Lagos bat virus (LBV). A high seroprevalence of antibodies against LBV was detected in Eidolon helvum bats. Longitudinal seroprevalence and age-specific seroprevalence data were analysed and capture–mark–recapture (CMR) analysis used to follow 98 bats over 18 months. These data demonstrate endemic infection, with evidence of horizontal transmission, and force of infection was estimated for differing age categories. The CMR analysis found survival probabilities of seronegative and seropositive bats were not significantly different. The lack of increased mortality in seropositive animals suggests infection is not causing disease after extended incubation. These key findings point towards acute transmission of bat lyssaviruses in adapted bat hosts that occurs at a far higher rate than the occurrence of disease.
Key words: Capture–mark–recapture, Lyssavirus, multi-state model, rabies, straw-coloured fruit bat, time-series data
INTRODUCTION
Phylogenetic analyses and virus–host relationships suggest that all lyssaviruses (family Rhabdoviridae), including Rabies virus (RABV) which causes classical carnivore rabies, originated from bats [1]. Eleven of the 12 currently known viruses of the Lyssavirus genus have been isolated from bats [1–4]. Lyssaviruses can be grouped into phylogroups, which are separately evolving lineages below the genus level [5]. Phylogroup I includes all the species except Lagos bat virus (LBV), Shimoni bat virus (SHIV), and Mokola virus (MOKV) (phylogroup II); West Caucasian bat virus (WCBV) probably forms a new phylogroup (III) [2–5]. All anti-rabies vaccines are derived from RABV strains, and confer little or no protection against members of other phylogroups in experimental studies [5–7].
The majority of Lyssavirus spill-over events are dead-end infections, but host switching of lyssaviruses from bats to other mammals has occurred repeatedly in history [1]. Two recent examples occurred in North America with bat-derived RABV infecting foxes in Canada [8] and skunks in the USA [9]. Currently all three Lyssavirus phylogroups are probably present in African bats and LBV apparently circulates among bats in sub-Saharan Africa [10–13]. LBV spill-over from bats into other mammals has been reported infrequently [14–16], with infection of humans never having been reported [15]. However, while LBV has not been implicated in human fatalities, rabies cases are grossly under-diagnosed in Africa and virus characterization rarely performed [16, 17]. Despite vaccination against RABV being available for over a century, control is lacking across Africa [17, 18], and the impact of the emergence of a lyssavirus against which the vaccines are ineffective in Africa could be substantial.
Phylogenetic analyses and isolation histories of lyssaviruses provide substantial evidence for the importance of bats in lyssavirus evolution [1], but the role of bats in the maintenance and evolution of lyssaviruses is complex and poorly understood. Both experimental and field studies in putative reservoir hosts are difficult, partly because many species are protected in the developed world, but mainly because bats are often small, are always nocturnal, fly, and frequently migrate or hibernate. A recent study of a bat–RABV system in North America found that hibernation was a crucial factor in RABV persistence in the North American species, Eptesicus fuscus [19]. However, the extended incubation periods due to hibernation that allow RABV maintenance within Ep. fuscus colonies [19] are absent in tropical fruit bats and tropical species remain largely unstudied. Cross-sectional studies have revealed high seroprevalence of antibodies against LBV in two colonial fruit-bat species, Eidolon helvum and Rousettus aegyptiacus [10–12, 20]. Therefore, if the serological findings are evidence of endemic transmission, mechanisms other than the use of extended torpor must exist that allow infection persistence within these populations.
The objectives of this study were to understand the ecology and dynamics of LBV infection in E. helvum, the straw-coloured fruit bat, one of the virus's potential reservoir hosts. This Old World fruit-bat species was chosen because of the high anti-LBV seroprevalence, the history of LBV isolation from the species, its wide sub-Saharan distribution, propensity to form roosts in urban areas and its use as a source of bushmeat [10–12, 20–22]. The LBV–E. helvum system provides an interesting comparative study to Ep. fuscus due to the lack of seasonal torpor [19], but it exhibits additional long-distance migratory behaviour which may alter seasonal contact rates and affect transmission dynamics.
We performed studies using similar model structures to George et al. [19] to estimate key epidemiological parameters that would enable us to determine if LBV was endemic and if infection varied seasonally. Due to the enormous colony sizes formed by E. helvum and the species' migratory habits [23–27], physical recapture for capture–mark–recapture (CMR) studies was impractical, therefore radiotelemetry was used to collect CMR data providing evidence of long-term survival of seropositive bats in the wild. We also present the results of a cross-sectional survey of age-specific seroprevalence in a wild colony of E. helvum, allowing force of infection to be estimated.
MATERIALS AND METHODS
Ethical approval for this project (WLE/0467) was received from the Zoological Society of London Ethics Committee and locally from the Wildlife Division of the Forestry Commission, Ghana.
Locations and sampling
All animal studies were conducted in the southern half of the Republic of Ghana, in West Africa, as described previously [10, 12]. Animals were classified by age, with neonates (pups) weighing ∼45 g at birth, juvenile bats ranging from 120 to 150 g and sexually immature and sexually mature adult bats (240–250 g body mass for males, and 200 g for females) being differentiated on the basis of testicular and nipple development. All samples and data were collected from the E. helvum roost in central Accra, except where stated from the Kumasi (∼200 km from Accra) and Tanoboase (∼300 km from Accra) E. helvum colonies [12]. Because E. helvum is migratory, large numbers of E. helvum bats are present at each colony during the dry season only [26, 27].
Between 2007 and March 2010, 1306 E. helvum bats were caught (Supplementary Table S1). Blood was taken by phlebotomy from the propatagial vein as described elsewhere [10, 12]. Oropharyngeal swabs were taken and placed immediately in RNALater® (Ambion, USA), before being frozen at −70 °C until analysed using polymerase chain reaction (PCR). Initially only serological surveys were performed, and therefore more serological samples were available for testing than swabs. During 2008 and 2009, 122 (68 and 54, respectively) bats were killed, including one sick animal, from the Accra (n = 116), Kumasi (n = 3), and Tanoboase colonies (n = 3). The sick E. helvum bat appeared to have muscle loss, and possibly signs of pyrexia, with reduced mental awareness and constant panting. Killed bats were examined post-mortem to estimate virus prevalence. Bats were anaesthetized with an excess of ketamine and medetomidine and euthanased by exsanguination via cardiac puncture. Serum and oropharyngeal swabs were taken. Samples of brain, kidney, salivary gland and spleen were collected immediately in duplicate and preserved frozen (−70 °C), in RNALater (Ambion) or in neutral-buffered 10% formalin. Canine teeth were removed post-mortem for age determination by cementum annuli analysis (conducted at Matson's Laboratory, USA) [28].
Serology
Serological testing for anti-LBV antibodies was conducted using a modified fluorescent antibody virus neutralization test (mFAVN), as described previously [10, 12], using the LBVNig56 isolate [22]. Negative control sera [a pool of naive (unvaccinated) dog sera] and two positive anti-LBVNig56 rabbit sera were included and control titres kept within ±2 standard deviations. The bat sera were heat-inactivated for 30 min at 56 °C and tested in duplicate using threefold serial dilutions. Reciprocal titres were calculated using the Spearman–Karber equation [29]. Animals were considered seropositive if the serum sample neutralized LBV at a reciprocal titre of >9.
Molecular studies
The presence of infection was determined from oropharyngeal swabs using a sensitive pan-Lyssavirus species SYBR® Green (Applied Biosystems, USA) real-time reverse transcription PCR (RT–qPCR), which is able to detect 25–195 LBV copies, as described previously [30]. High numbers of oropharyngeal swabs led to the pooling of aliquots of ten samples at a time prior to RNA extraction using High Pure™ (Roche, Germany), according to the manufacturer's instructions. This gives a tenfold lower sensitivity than individual extractions, although each sample was individually identifiable and could be traced back to the original swab. To allow us to estimate and incorporate the sensitivity of the oropharyngeal sampling into our prevalence studies, we determined what proportion of oropharyngeal swabs was positive for host RNA by testing a subset of 25 individual oropharyngeal swabs from bats stored without RNALater (Ambion) using the hemi-nested (hn) pan-Lyssavirus RT–PCR and 18S controls as described previously [31].
Total RNA was extracted from the frozen bat tissues sampled post-mortem (n = 122) using MELT™ (Ambion) and KingFisher 96® (Thermo Electron Corporation, USA) technology according to the manufacturer's instructions. RNA was reverse-transcribed and analysed using RT–qPCR as described previously [30]. All 68 brain, kidney, salivary gland and spleen samples from the 2008 sample of euthanized bats were also tested using hnRT–PCR [31].
All samples were tested using host 18S RNA RT–PCR to ensure RNA was extracted successfully. LBV-positive and -negative mouse brain or tissue culture supernatant-negative controls were used with each extraction and PCR run.
Capture–mark–recapture
To estimate relative rates of survival of LBV seropositive bats within the Accra colony, radiotelemetry was used as a method of marking individuals in a CMR study. Ninety-eight bats were each fitted with a radio transmitter [150/151 MHz range, 8·5–9 g weight (<4% body weight), with at least 420 days battery life, Wildlife Materials Inc., USA]. Sixty-three transmitters were fitted in January 2008 and a further 35 in March 2008. Detection was with a 138–174 MHz SIKA receiver (Biotrack Ltd, UK). Serum samples were available from 91 of 98 animals caught and used in the CMR study and these were tested for anti-LBV antibodies using mFAVN. ‘Recapture’ attempts using telemetry were made approximately weekly in the Accra colony from January 2008 to July 2009. These data were combined to monthly recaptures for CMR analysis (see below).
Age determination
Cementum annuli analysis of histological sections (Matson's Laboratory) was used to determine the age of 88 bats necropsied as part of this study. It was assumed annuli were deposited annually [27, 28]. Samples were taken during the pregnancy and birthing period of January–July in both 2008 and 2009. Six of the 88 individuals were not from Accra, but Kumasi (n = 3) and Tanoboase (n = 3); however, these were included in the study only once radiotelemetry data (see below) demonstrated animals moved between these colonies and the Accra colony.
Differential staining of cementum annuli was good and most individuals had a ±0–1 year age range where some annuli would split, merge or fade around the estimated number of cementum annuli. Three older animals, however, had a ±1·5 year age range around the estimated age (i.e. an estimated age of 13–15 years). Animals with no annuli were included in the 1-year-old class, as these were determined by morphometric observations to have been born the previous year [32]. An assumed birth date was taken as 1 March, due to fetuses from necropsied pregnant females being near term (∼45 g) in March.
Data analysis
All data obtained from bats from different calendar months, but the same sampling period (1–2 weeks) were combined for analysis. The probability of being seropositive in relation to sex, age, the location and date of sampling from the wild population was estimated using a generalized linear model (GLM) with binomial error distribution (glm function in R software). We started with the most complete model including two-way interaction between all explanatory variables and models were subsequently simplified using likelihood ratio tests to arrive at the minimal adequate model [33]. The strength of the relationship between each of the explanatory variables and serostatus was estimated using GLM analysis. The multiple comparisons for parametric models (including GLM) glht function in the multcomp package in R was then used to estimate the coefficients and their confidence limits, before conversion to odds ratios (OR). The χ2 test for trend in proportions (prop.trend.test function in R) was used to analyse the longitudinal seroprevalence data for systematic changes in seroprevalence.
The age-related risk of infection, or force of infection (λ), was inferred from seroprevalence data for each age group. Due to a relatively small sample size of known age animals (n = 88) we binned ages into four groups (1–3, 4–6, 7–9, ⩾10 years) for analysis to estimate λ for LBV in E. helvum. We also assumed that the overall proportion of bats that die from infection is very small (see prevalence and CMR results below) and therefore that λ is estimable from the age-specific seroprevalence data. Therefore, λ = [(Sx – Sx–1)/(1 – Sx)] [34], where Sx is the seroprevalence for the binned ages (e.g. x = 1–3 years). The binomial exact test was used to estimate 95% confidence limits.
Capture–mark–recapture analysis
We fitted a multi-state Arnason–Schwarz model [35, 36] to mark–recapture data in order to test the effects of serostatus on survival rate in free-living bats, while controlling for heterogeneity in capture rates arising from temporary emigration from the study site. Full details of the model structure are given elsewhere [27]. Data were collected weekly for 19 months. Weekly recapture data were combined to give the monthly recapture data and only the first 12 months of data used for analysis, due to low recapture rates during the second migratory period.
RESULTS
Serology
Between January 2007 and March 2010, 1169 E. helvum bats were tested using mFAVN for anti-LBV antibodies (Supplementary Table S4). The median reciprocal titre was 9 (seronegative) and first and third quartiles were 5·2 and 27, respectively. In March and April 2008, bats were sampled in three different geographical locations (Accra, Kumasi, Tanoboase) in an attempt to obtain blood samples from post-partum females, but none was caught. The overall seroprevalence between locations in March and April 2008 differed substantially (21–60%), but this difference was not significant (P = 0·06), possibly due to low capture numbers from Tanoboase (n = 23) and Kumasi (n = 10). Subsequent sampling in 2009 showed the seroprevalences to be more similar to each other (18–35%); again the difference was not significant between the locations (P = 0·07).
Overall, seroprevalence in mature bats was significantly greater than in either juvenile (OR 4·3, 95% CI 1·9–9·5) or SI bats (OR 6·1, 95% CI 3·5–10·7) (Table 1). Sexually mature females were more likely to be seropositive than sexually mature males (OR 1·6, 95% CI 1·1–2·3). Seroprevalence in sexually mature adults fluctuated between 23% (95% CI 15–33) and 49% (95% CI 39–52), with no significant difference between the proportion of seropositive sexually mature adults between sampling occasions over the 4-year period (P = 0·7) (Fig. 1).
Table 1.
Results of binomial logit link generalized linear models showing the explanatory variables for being LBV seropositive from 1169 bats. Data from neonates (n = 2) were excluded
| Coefficient estimate | s.e. | Z value | Pr(>|z|) | |
|---|---|---|---|---|
| Intercept | −2·014 | 0·328 | −6·146 | <0·001 |
| Age | ||||
| Sexually immature | −0·365 | 0·379 | −0·962 | 0·34 |
| Sexually mature | 1·45 | 0·327 | 4·433 | <0·001 |
| Sex: Female | 0·482 | 0·15 | 3·217 | 0·001 |
Fig. 1.
The trend in sexually mature adult E. helvum seroprevalence of anti-Lagos bat virus antibody in Accra, Ghana. Data from male and female animals are combined. Ninety-five percent confidence limits are shown.
Seroprevalence increased with age (Fig. 2). This relationship was maintained when age was categorized by 2- or 3-year periods (not shown). The force of infection (λ) was calculated to be lower for younger (4–6 years) (0·15, 95% CI 0·05–0·25) and older (⩾10 years) (0·16, 95% CI 0·13–0·22) adult bats than for bats aged 1–3 years (0·29, 95% CI 0·27–0·3) and 7–9 years (0·41, 95% CI 0·30–0·55).
Fig. 2.
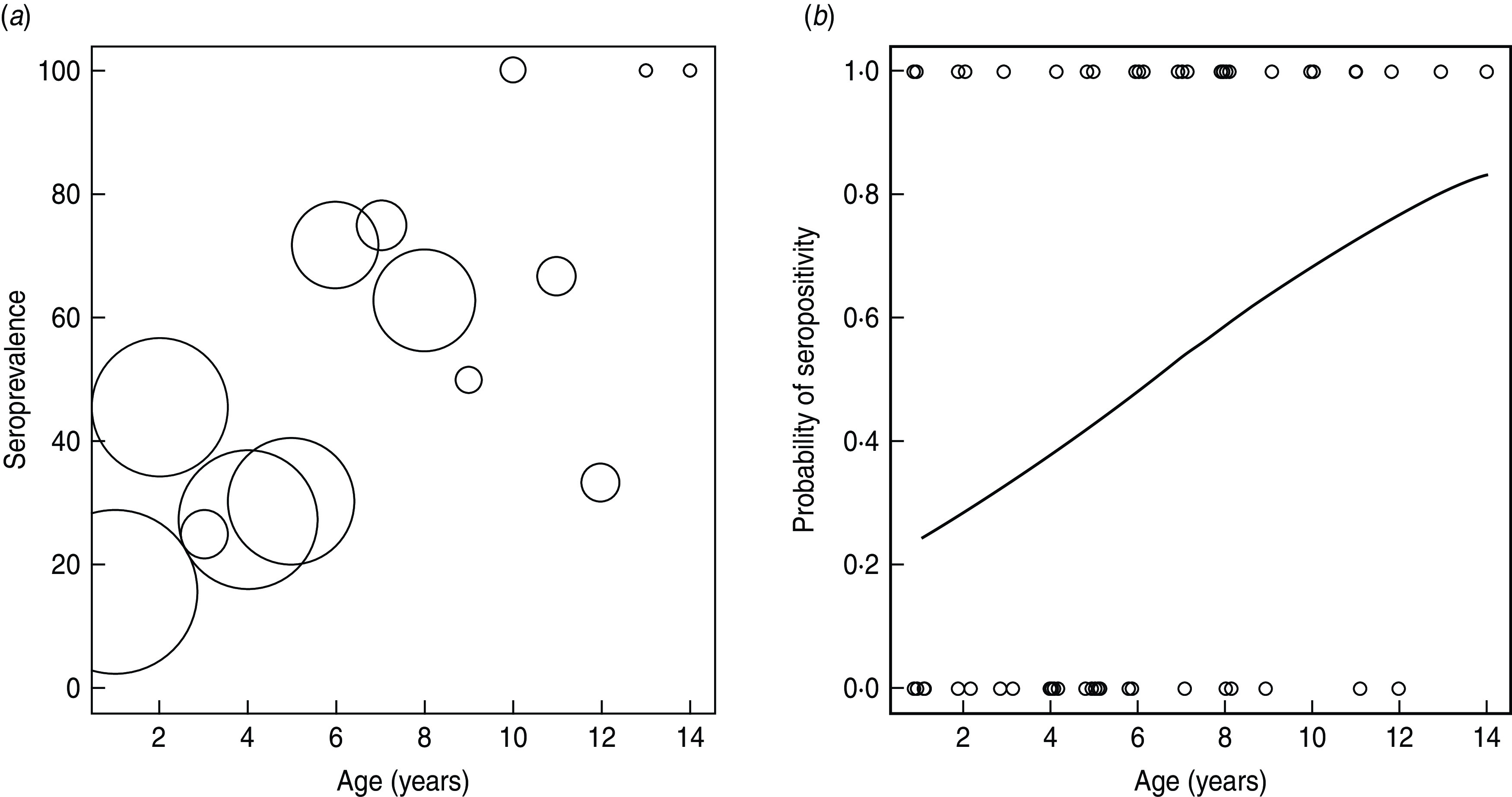
(a) The age-specific seroprevalence of anti-Lagos bat virus (LBV) antibody in 88 E. helvum from Ghana. Data from male and female animals are combined. Numbers of individuals are shown by bubble diameter, ranging in sample size from 11 (age 2 and 4 years) to 1 (age 13 and 14 years). (b) Predicted probability of being LBV seropositive in relation to age.
Contrary to the sample of categorically aged bats (Table 1), the best fitting GLM using fully age-specific data did not find sex to be an explanatory factor for being seropositive (Table 2), although the sample size was much smaller.
Table 2.
Results of binomial logit link generalized linear models showing the explanatory variables for being LBV seropositive in 88 bats of known age derived from tooth cementum annuli analysis [also see Fig. 2(a, b)]
| Coefficient estimate | s.e. | Z value | Pr(>|z|) | |
|---|---|---|---|---|
| Intercept | −4·251 | 0·449 | −2·934 | 0·003 |
| Age (yr) | 0·208 | 0·074 | 2·805 | 0·005 |
Virus prevalence
Total RNA was extracted successfully from all organ tissues tested from the 68 bats collected during field trips in late January and early February (from hereon January) and late March and early April (from hereon March) 2008, but Lyssavirus-specific RNA was not detected using either RT–qPCR assay or hnRT–PCR. The 54 brain samples collected in March 2009 and tested with RT–qPCR only were also negative for viral RNA. All samples were 18S host RNA positive, all negative controls were negative and all positive controls were positive, suggesting extraction and PCR reactions were successful. The seroprevalence in the sample of bats tested for cerebral lyssavirus infection using PCR was 42% (95% CI 33–51, Supplementary Table S2).
Assuming 100% PCR sensitivity, random sampling, and given a population size of over 10 000 in the 2008 dry season, the upper 95% confidence level of cerebral infection was 4·2%. For March 2009, the upper 95% confidence level was 7·1%. Similarly, combining the 122 brains over the 15-month study period, the upper 95% confidence limit was 3·1%.
Overall, 796 oropharyngeal swabs were tested for Lyssavirus RNA using RT–qPCR (Supplementary Table S3). None was virus RNA positive. The proportion of individual oropharyngeal swabs in which host 18S RNA was detected was 80%, and upper prevalence estimates from the population samples were adjusted to reflect this. Adjusted upper 95% confidence limits of virus prevalence in oral swabs was 1–9% in each year. The overall seroprevalence in the sample of bats tested using PCR for salivary excretion of Lyssavirus was 32·3% (95% CI 29–36, n = 710), with serological results not available for 86/796 samples (Supplementary Table S3).
Capture–mark–recapture analysis
Radiotelemetry allowed detection of bats from Accra in both of the other colonies visited: one in Kumasi in 2008 and two in Tanoboase in 2009, demonstrating a degree of colony connectivity. The overall seroprevalence for LBV antibodies in this sample was 25% (23/91 tested). Seropositivity did not significantly affect survival over the year (likelihood ratio test statistic 0·28, P = 0·58), thus providing no evidence for differential survival rates between seropositive and seronegative bats.
DISCUSSION
This longitudinal study of wild E. helvum bats demonstrated endemic LBV infection with horizontal transmission, as did the analysis of age-specific seroprevalence. Significantly greater seroprevalence was detected in mature bats than in juvenile or sexually immature bats. Interestingly, the λ estimations suggest λ is greater for young (1–3 years) and middle-aged (7–9 years) bats than for young adult (4–6 years) or older (⩾10 years) bats; however, data for older bats were sparse. Whether this is real, or due to increasing disease-induced mortality in the younger or older adults (and thus removing seropositive animals from the population) requires further study. Seroprevalence was significantly different between the sexes, with females being more likely to be seropositive, but the difference was small. Sampling of this population, as for many wildlife studies, may not have been truly random and age may have confounded the effect of sex if the sexually mature females sampled from the colony were older than the sexually mature males. Future studies should determine if there is significant population structure within the colony and if capture bias caused this apparent effect.
Similarly, while not significant at the P = 0·05 level, differences among colonies in Ghana were detected and may reflect differing infection dynamics and/or differing age structures within the different colonies. Further studies are required to clarify this. More generally, the high seroprevalence of anti-LBV antibodies in the Ghanaian E. helvum colonies, which is consistent with those reported for Kenyan and Nigerian colonies [11, 20], provides clear evidence that the bats are exposed to the infection. Possible mechanisms to explain the presence of anti-LBV VNAs in E. helvum include infection, with seroconversion and recovery, or that animals seroconvert and remain latently infected (although latent infection has yet to be detected). Neither of these hypotheses excludes the possibility that a proportion of animals might die from central nervous infection, as supported by current knowledge from virus isolations [11]. The sample sizes are small for the capture–recapture analyses, and our conclusions must therefore be cautious; however, our initial telemetry data suggest that seropositive bats survive, on average, at the same rate as seronegative bats, suggesting that infection-associated mortality in adults is low. It also possibly suggests that latent infection followed by death from rabies does not occur in LBV-seropositive bats. Certainly, no LBV was detected from healthy bat brains (n = 122) during the study.
Most experimental studies of lyssaviruses report that not all cerebrally infected individuals secrete lyssaviruses in their saliva [37–40]. Further knowledge of virus excretion in infected bats is required because, while saliva-positive animals typically have virus-positive brains, there are now two studies reporting salivary viral secretion in healthy bats [37, 41]. Both cerebral and oral swab prevalences were estimated to be less than 9% during the current study. Assuming there were no false negatives, there are several possible explanations for failure to detect viral nucleic acid. First, the actual prevalence might have been lower than the detectable prevalence, a function of sample size. Sample sizes of 149 and 299 would be required to have a 95% probability of detecting at least one positive animal based on a true prevalence of 2% and 1%, respectively, at each sampling event. However, overall we tested 796 oropharyngeal swabs, which, in theory, could detect a prevalence of ∼0·5% with 95% confidence given certain assumptions. Therefore we suspect viral prevalence is low in the colony. Second, sampling may have been undertaken in a location and/or time when virus prevalence, which may vary seasonally [19], was low. Near-term pregnant females migrate in March to unknown destinations [26, 27]. The influx of new susceptible individuals into these locations may cause an increase in virus prevalence in these colonies and therefore future studies should sample from these currently unidentified locations. Third, infection may be present in another, as-yet untested, tissue (possibly in an immunoprivileged site). However, in vitro and in vivo studies have shown LBV to be neurotropic (or at least isolated from animal brains). Although initial viral replication may occur at the primary inoculation site [42], studies, including a hamster model using LBV and MOKV, suggest that axons alone support the generation of progeny virus [43, 44]. An E. helvum specimen examined from Kenya by Kuzmin and others had widespread dissemination of LBV virus in nerves and nerve bundles, submandibular salivary gland ganglia and acini, with the majority of virus detected in the brain [11]. Moreover, LBV has been isolated from the brains of a range of animals including bats [14–16] and has been experimentally shown to have similar mortality rates to RABV [45].
In summary, this study answers several key questions regarding LBV–E. helvum interactions. Although the results here clearly demonstrate that LBV infection is endemic in the population of E. helvum found in Ghana, further work, including experimental infection studies, is required to capture the dynamics of LBV transmission and pathogenesis in a reservoir host. Our current understanding of bat ecology and Lyssavirus epidemiology is not sufficient to explain maintenance of lyssaviruses in any tropical bat species. Future studies should therefore aim to understand the drivers of infection dynamics within this species, including seasonality and migration.
Supplementary Material
Supplementary information supplied by authors.
ACKNOWLEDGEMENTS
We thank the three anonymous reviewers for their constructive comments. We thank the Executive Director, Wildlife Division, and the Director of Veterinary Services, Ghana for their commitment to this project. We acknowledge T. J. McKinley, University of Cambridge, and the Ecology and Evolution of Infectious Diseases ecology workshop (2011) for help with statistical analyses, and Dave Selden (AHVLA) for technical support. D.T.S.H. and K.S.B are funded by The Wellcome Trust, J.L.N.W. is supported by the Alborada Trust, A.A.C. is supported by a Royal Society Wolfson Research Merit Award and A.R.F. is funded by the UK Department for Environment, Food and Rural Affairs (Defra) by grant SEV3500. The Cambridge Infectious Diseases Consortium, the Research and Policy for Infectious Disease Dynamics programme of the Science and Technology Directorate, Department of Homeland Security, Fogarty International Center, National Institutes of Health, and The Lubee Foundation Luis F. Bacardi Bat Conservation and Research Fund provided further support for the study.
Supplementary material
For supplementary material accompanying this paper visit https://doi.org/10.1017/S0950268812000167.
click here to view supplementary material
DECLARATION OF INTEREST
None.
REFERENCES
- 1.Badrane H, Tordo N. Host switching in Lyssavirus history from the Chiroptera to the Carnivora orders. Journal of Virology 2001; 75: 8096–8104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kuzmin IV, et al. Phylogenetic relationships of Irkut and West Caucasian bat viruses within the Lyssavirus genus and suggested quantitative criteria based on the N gene sequence for lyssavirus genotype definition. Virus Research 2005; 111: 28–43. [DOI] [PubMed] [Google Scholar]
- 3.Kuzmin IV, et al. Bat lyssaviruses (Aravan and Khujand) from Central Asia: phylogenetic relationships according to N, P and G gene sequences. Virus Research 2003; 97: 65–79. [DOI] [PubMed] [Google Scholar]
- 4.Kuzmin IV, et al. Shimoni bat virus, a new representative of the Lyssavirus genus. Virus Research 2010; 149: 197–210. [DOI] [PubMed] [Google Scholar]
- 5.Badrane H, et al. Evidence of two Lyssavirus phylogroups with distinct pathogenicity and immunogenicity. Journal of Virology 2001; 75: 3268–3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hanlon CA, et al. Efficacy of rabies biologics against new lyssaviruses from Eurasia. Virus Research 2005; 111: 44–54. [DOI] [PubMed] [Google Scholar]
- 7.Weyer J, et al. Cross-protective and cross-reactive immune responses to recombinant vaccinia viruses expressing full-length lyssavirus glycoprotein genes. Epidemiology and Infection 2008; 136: 670–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daoust PY, Wandeler AI, Casey GA. Cluster of rabies cases of probable bat origin among red foxes in Prince Edward Island, Canada. Journal of Wildlife Diseases 1996; 32: 403–406. [DOI] [PubMed] [Google Scholar]
- 9.Leslie MJ, et al. Bat-associated rabies virus in skunks. Emerging Infectious Diseases 2006; 12: 1274–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayman DTS, et al. Antibodies against Lagos Bat Virus in Megachiroptera from West Africa. Emerging Infectious Diseases 2008; 14: 926–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuzmin IV, et al. Lagos bat virus in Kenya. Journal of Clinical Microbiology 2008; 46: 1451–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wright E, et al. Virus neutralising activity of African fruit bat (Eidolon helvum) sera against emerging lyssaviruses. Virology 2010; 408: 183–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuzmin IV, et al. Possible emergence of West Caucasian bat virus in Africa. Emerging Infectious Diseases 2008; 14: 1887–1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Markotter W, et al. Isolation of Lagos bat virus from water mongoose. Emerging Infectious Diseases 2006; 12: 1913–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Markotter W, et al. Lagos bat virus, South Africa. Emerging Infectious Diseases 2006; 12: 504–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mebatsion T, Cox JH, Frost JW. Isolation and characterization of 115 street rabies virus isolates from Ethiopia by using monoclonal antibodies: identification of 2 isolates as Mokola and Lagos bat viruses. Journal of Infectious Diseases 1992; 166: 972–977. [DOI] [PubMed] [Google Scholar]
- 17.Mallewa M, et al. Rabies encephalitis in malaria-endemic area, Malawi, Africa. Emerging Infectious Diseases 2007; 13: 136–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knobel DL, et al. Re-evaluating the burden of rabies in Africa and Asia. Bulletin of the World Health Organization 2005; 83: 360–368. [PMC free article] [PubMed] [Google Scholar]
- 19.George DB, et al. Host and viral ecology determine bat rabies seasonality and maintenance. Proceedings of the National Academy of Sciences USA 2011; 108: 10208–10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dzikwi AA, et al. Evidence of Lagos bat virus circulation among Nigerian fruit bats. Journal of Wildlife Diseases 2010; 46: 267–271. [DOI] [PubMed] [Google Scholar]
- 21.Kamins AO, et al. Uncovering the fruit bat bushmeat commodity chain and the true extent of bat hunting in Ghana, West Africa. Biological Conservation (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boulger LR, Porterfield JS. Isolation of a virus from Nigerian fruit bats. Transactions of the Royal Society of Tropical Medicine and Hygiene 1958; 52: 421–424. [DOI] [PubMed] [Google Scholar]
- 23.Thomas DW. Annual migration of three species of West African fruit bats (Chiroptera: Pteropodidae). Canadian Journal of Zoology 1983; 61: 2266–2272. [Google Scholar]
- 24.Richter HV, Cumming GS. First application of satellite telemetry to track African straw-coloured fruit bat migration. Journal of Zoology 2008; 275: 172–176. [Google Scholar]
- 25.Richter HV, Cumming GS. Food availability and annual migration of the straw-colored fruit bat (Eidolon helvum). Journal of Zoology 2006; 268: 35–44. [Google Scholar]
- 26.Hayman DT, et al. Long-term survival of an urban fruit bat seropositive for ebola and lagos bat viruses. PLoS One 2010; 5(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hayman DTS, et al. Demography of straw-colored fruit bats in Ghana. Journal of Mammalogy (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Divljan A, Parry-Jones K, Wardle GM. Age determination in the grey-headed flying fox. Journal of Wildlife Management 2006; 70: 607–611. [Google Scholar]
- 29.Aubert M. Methods for the calculation of titres. In: Meslin FX, Kaplin MM, Koprowski H, eds. Laboratory Techniques in Rabies. Geneva: World Health Organisation, 1996, pp. 445–459. [Google Scholar]
- 30.Hayman DT, et al. A universal real-time assay for the detection of lyssaviruses. Journal of Virological Methods (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heaton PR, et al. Heminested PCR assay for detection of six genotypes of rabies and rabies-related viruses. Journal of Clinical Microbiology 1997; 35: 2762–2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DeFrees SL, Wilson DE. Eidolon helvum. Mammalian Species 1988; 312: 1–5. [Google Scholar]
- 33.Crawley MJ. Statistics: An Introduction Using R. Chichester: John Wiley and Sons, Ltd, 2005. [Google Scholar]
- 34.Anderson RM, May RM. Infectious Diseases of Humans. Oxford: Oxford University Press, 1991. [Google Scholar]
- 35.Arnason AN. The estimation of population size, migration rates and survival in a stratified population. Researches on Population Ecology 1973; 13: 97–113. [Google Scholar]
- 36.Schwarz CJ, Schweigert JF, Arnason AN. Estimating migration rates using tag-recovery data. Biometrics 1993; 49: 177–193. [Google Scholar]
- 37.Franka R, et al. Susceptibility of North American big brown bats (Eptesicus fuscus) to infection with European bat lyssavirus type 1. Journal of General Virology 2008; 89: 1998–2010. [DOI] [PubMed] [Google Scholar]
- 38.Freuling CM, et al. Experimental infection of Serotine bats (Eptesicus serotinus) with European bat lyssavirus type 1a (EBLV-1a). Journal of General Virology 2009; 90: 2493–2502. [DOI] [PubMed] [Google Scholar]
- 39.Johnson N, et al. Experimental study of European bat lyssavirus type-2 infection in Daubenton's bats (Myotis daubentonii). Journal of General Virology 2008; 89: 2662–2672. [DOI] [PubMed] [Google Scholar]
- 40.McColl KA, et al. Pathogenesis studies with Australian bat lyssavirus in grey-headed flying foxes (Pteropus poliocephalus). Australian Veterinary Journal 2002; 80: 636–641. [DOI] [PubMed] [Google Scholar]
- 41.Aguilar-Setien A, et al. Salivary excretion of rabies virus by healthy vampire bats. Epidemiology and Infection 2005; 133: 517–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schnell MJ, et al. The cell biology of rabies virus: using stealth to reach the brain. Nature Reviews Microbiology 2009; 8: 51–61. [DOI] [PubMed] [Google Scholar]
- 43.Murphy FA, et al. Comparative pathogenesis of rabies and rabies-like viruses: infection of the central nervous system and centrifugal spread of virus to peripheral tissues. Laboratory Investigation 1973; 29: 1–16. [PubMed] [Google Scholar]
- 44.Murphy FA, et al. Comparative pathogenesis of rabies and rabies-like viruses. Viral infection and transit from inoculation site to the central nervous system. Laboratory Investigation 1973; 28: 361–376. [PubMed] [Google Scholar]
- 45.Markotter W, et al. Lagos bat virus virulence in mice inoculated by the peripheral route. Epidemiology and Infection 2009; 137: 1155–11162. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information supplied by authors.
For supplementary material accompanying this paper visit https://doi.org/10.1017/S0950268812000167.
click here to view supplementary material



