Abstract
Pesticides based on the s-triazine ring structure are widely used in cultivation of food crops. Cleavage of the s-triazine ring is an important step in the mineralization of s-triazine compounds and hence in their complete removal from the environment. Cyanuric acid amidohydrolase cleaves cyanuric acid (2,4,6-trihydroxy-s-triazine), which yields carbon dioxide and biuret; the biuret is subject to further metabolism, which yields CO2 and ammonia. The trzD gene encoding cyanuric acid amidohydrolase was cloned into pMMB277 from Pseudomonas sp. strain NRRLB-12227, a strain that is capable of utilizing s-triazines as nitrogen sources. Hydrolysis of cyanuric acid was detected in crude extracts of Escherichia coli containing the cloned gene by monitoring the disappearance of cyanuric acid and the appearance of biuret by high-performance liquid chromatography (HPLC). DEAE and hydrophobic interaction HPLC were used to purify cyanuric acid amidohydrolase to homogeneity, and a spectrophotometric assay for the purified enzyme was developed. The purified enzyme had an apparent Km of 0.05 mM for cyanuric acid at pH 8.0. The enzyme did not cleave any other s-triazine or hydroxypyrimidine compound, although barbituric acid (2,4,6-trihydroxypyrimidine) was found to be a strong competitive inhibitor. Neither the nucleotide sequence of trzD nor the amino acid sequence of the gene product exhibited a significant level of similarity to any known gene or protein.
The s-triazine ring is a component of several widely used agricultural chemicals. In 1991 more than 80 million pounds of the s-triazine herbicides atrazine, cyanazine, and simazine were applied to corn, sorghum, and cotton in the United States alone (22). Biodegradation is an important mechanism of dissipation of s-triazine compounds in agricultural soils and in industrial wastewater produced during the manufacture of s-triazines. Previous studies have demonstrated that some degradation of atrazine occurs (2, 14, 25), although the biological portions of degradation that have been described involve primarily dealkylation of atrazine, not degradation of the triazine ring. Recently, complete mineralization of atrazine by several bacterial strains has been described (19, 23, 27).
Little is known about the biochemical mechanisms of s-triazine ring degradation. Workers in several labs have characterized enzymes that dechlorinate (9, 21) or N-dealkylate atrazine or related compounds (4, 24), but there have been no reports of characterization of the enzymes responsible for mineralization of the s-triazine ring. On the basis of studies of four s-triazine-degrading bacterial strains, Cook has postulated that most s-triazine degradation pathways converge at cyanuric acid and that this compound is the substrate for ring cleavage (7, 8). Cyanuric acid has been shown to be a central intermediate in the pathway of atrazine degradation in atrazine-degrading Pseudomonas cultures (24) and consortia (10). In several studies workers have observed hydrolytic cleavage of the s-triazine ring in bacteria (7) and fungi (16) during growth of the organisms on cyanuric acid as a nitrogen source. Cook et al. (7) assayed enzymes that catalyzed the conversion of cyanuric acid to biuret in the soluble portion of crude extracts of three bacteria (Pseudomonas sp. strains A and D and Klebsiella pneumoniae 99) that were able to utilize cyanuric acid as a source of nitrogen. These studies showed that the reaction was hydrolytic; however, no kinetic or physical properties of the enzymes were reported. Eaton and Karns (13) cloned the gene (trzD) for cyanuric acid amidohydrolase from these three strains of bacteria. All three trzD genes had the same restriction patterns, suggesting that there is extensive sequence identity. In this report I describe purification of the cyanuric acid amidohydrolase of these triazine-degrading bacteria from a strain of Escherichia coli containing the trzD gene of Pseudomonas sp. strain A and also describe some kinetic and physical characteristics of the enzyme, as well as the nucleotide sequence of the trzD gene.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Plasmid pJK204 was constructed by cloning a 2.0-kb HindIII-PstI fragment that contains the trzD gene of plasmid pRE458 (12), which was originally shown to contain the trzC and trzD genes of Pseudomonas sp. strain NRRLB-12227 (strain A of Cook and Hütter [6]), into pMMB277 (20). The trzD fragment was isolated from agarose gels by electroelution onto a type NA45 membrane (Schleicher and Schuell, Keene, N.H.) as recommended by the manufacturer. The isolated fragment was ligated to HindIII-PstI-digested pMMB277 with T4 ligase (BRL, Gaithersburg, Md.) and was transformed into transformation-competent E. coli DH5-α (BRL) as recommended by the manufacturer. Cells were plated onto Lennox broth (Gibco) that was solidified with 1.5% agar and contained chloramphenicol (34 μg/ml), 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal; 50 μg/ml), and 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Several white colonies were removed, and the presence of the proper insert was confirmed by restriction digestion and agarose gel electrophoresis of plasmid DNA prepared by the rapid boiling method of Holmes and Quigley (15).
Preparation of cell extracts.
E. coli containing plasmid pJK204 was grown overnight in two 1-liter Lennox broth cultures containing chloramphenicol (34 μg/ml) and IPTG (0.1 mM) at 37°C with shaking. The cells were combined, harvested by centrifugation (6,000 × g, 4°C, 10 min), and suspended in 30 ml of ice-cold 25 mM potassium phosphate buffer (pH 7.0). The cells were lysed by passing suspended cells twice through a cold French pressure cell (15,000 lb/in2). The extract was centrifuged at 10,000 × g for 10 min at 4°C to remove the large debris, and the resulting supernatant was centrifuged at 105,000 × g for 2 h at 4°C to remove the membranes and other particulate material. The resulting supernatant was designated the crude soluble enzyme fraction and was used for further purification.
Enzyme purification.
The crude soluble enzyme fraction (33 ml) was applied at a rate of 2 ml/min to a DEAE 5PW high-performance liquid chromatography (HPLC) column (21.5 by 150 mm; Waters Associates, Milford, Mass.) that had been equilibrated with 25 mM potassium phosphate buffer (pH 7.0). The column was rinsed at a rate of 5 ml/min with the equilibration buffer until a stable baseline (optical density at 254 nm [OD254]) was obtained; then a linear 0 to 1 M NaCl gradient was run for 1 h. Fractions (5 ml) were collected, active fractions were identified as described below and pooled (10 ml recovered), and the ammonium sulfate concentration was adjusted to 1 M by adding 1.32 g of solid (NH4)2SO4. The enzyme preparation was centrifuged to remove precipitates that formed after (NH4)2SO4 was added, and the cleared supernatant, which contained all of the enzyme activity, was applied at a rate of 2 ml/min to a TSK-Phenyl 5PW (Tosoh Corp.) HPLC column (21.5 by 150 mm; HP-Genenchem, San Francisco, Calif.) that had been equilibrated with 25 mM potassium phosphate buffer (pH 7.0) containing 1 M (NH4)2SO4. The column was eluted with the starting buffer at a rate of 5 ml/min until a stable baseline (OD254) was obtained; then a decreasing linear 1 to 0 M (NH4)2SO4 gradient was run for 1 h. Fractions (5 ml) were collected, and the active fractions were identified and assayed as described below and then pooled.
Enzyme assays.
Active fractions were identified by mixing 5 μl of a fraction with 200 μl of 25 mM Tris-HCl (pH 8.0) containing 3 mM cyanuric acid in a 250-μl microcentrifuge tube that can be used as an insert in Waters autosampler vials. After overnight incubation the samples were examined to determine the loss of cyanuric acid by HPLC as described below. The reaction rates in crude extracts and column fractions were determined by mixing a sample of enzyme with 4 ml of 25 mM Tris-HCl (pH 8.0) containing 3 mM cyanuric acid in a Waters autosampler vial and immediately beginning a series of injections that were repeated every 5.5 min. The cyanuric acid concentration present at the time of each injection was calculated and plotted against the time of injection to determine the rate of cyanuric acid hydrolysis.
The rate of hydrolysis of cyanuric acid catalyzed by purified enzyme was measured spectrophotometrically by monitoring the decrease in absorbance at 220 nm due to cyanuric acid. The enzyme was mixed in a cuvette containing 1 ml of buffer (25 mM Tris-HCl pH 8.0 unless otherwise noted) supplemented with 0.15 mM (or less) cyanuric acid. The absorbance at 220 nm of cyanuric acid was linear in this range. An extinction coefficient of 6.283 OD220 units · ml · μmol−1 for cyanuric acid was determined empirically and was used to calculate the rate of cyanuric acid hydrolysis.
HPLC.
Cyanuric acid and biuret were separated on a C18 Resolve (Waters Associates) Radial-Pak HPLC column (8 by 100 mm; particle size, 5 μm; Waters Associates) by using an isocratic solvent system consisting of 5 mM octyltriethylammonium phosphate (Q-8 Ion-Pair Cocktail; Regis Chemical Co., Morton Grove, Ill.) in 5 mM potassium phosphate (final pH 6.8) at a flow rate of 2 ml/min. Biuret was detected at 200 nm and cyanuric acid was detected at 225 nm with a Waters model 490 multichannel spectrophotometric HPLC detector. Under these conditions biuret eluted at approximately 1.8 min, while cyanuric acid eluted at 3.5 min.
Molecular weight determination.
The apparent molecular weight of the native enzyme was determined by HPLC by using a Waters Protein-Pak 300SW column (7.8 by 300 mm). A 200-μl sample of the pooled phenyl column fractions was injected, and the column was eluted with 25 mM potassium phosphate at a flow rate of 1 ml/min. The column was calibrated with horse spleen apoferritin (molecular mass, 443 kDa), sweet potato amylase (200 kDa), yeast alcohol dehydrogenase (150 kDa), and bovine erythrocyte carbonic anhydrase (29 kDa).
SDS-PAGE.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) of protein samples was performed by the method of Laemmli (18) by using a 10% acrylamide resolving gel and a 5% stacking gel in a mini-gel format. The gels were stained with Coomassie brilliant blue.
Protein assays.
The protein concentrations in crude extracts were determined by the method of Bradford (5) by using a Bio-Rad protein assay kit and immunoglobulin G as the standard. The concentrations in purified materials were determined by using the spectrophotometric assay of Kalb and Bernlohr (17).
DNA sequencing.
Plasmid pRE479 was prepared by ligating the 2.0-kb HindIII-PstI fragment containing the trzD gene into pUC19 that had been cut with the same enzymes. Colonies containing the proper insert were isolated and characterized as described above for pJK204. Plasmid pRE479 was isolated from E. coli by using mini plasmid purification kits (Qiagen, Inc., Valencia, Calif.). Sequencing was performed by workers at the University of Maryland DNA Sequencing Facility, Center for Agricultural Biotechnology, College Park, who used ABI automated sequencers and custom primers. DNA sequences were assembled by using the DNAStar software package.
BLASTN searches (1) of the GenBank nucleic acid database and BLASTP searches (1) of the SwissProt protein database and the GenBank database for sequences that are similar to sequence of the trzD gene and the sequence of the TrzD protein were conducted online at the NCBI web site (22a).
Chemicals.
Cyanuric acid was obtained from Eastman Chemicals, Rochester, N.Y. Biuret was obtained from Baker Chemical Co., Phillipsburg, N.J. Barbituric acid was obtained from Sigma Chemical Co., St. Louis, Mo. All other hydroxypyrimidines were obtained from Aldrich Chemical Co., Milwaukee, Wis. Protein standards were obtained from Sigma Chemical Co. or Boehringer Mannheim Co., Indianapolis, Ind.
Nucleotide sequence accession number.
The nucleotide sequence of the 2-kb PstI-HindIII fragment containing trzD and the amino acid sequence of TrzD have been submitted to the GenBank Nucleotide Database under accession no. AF086815.
RESULTS
Purification of cyanuric acid amidohydrolase.
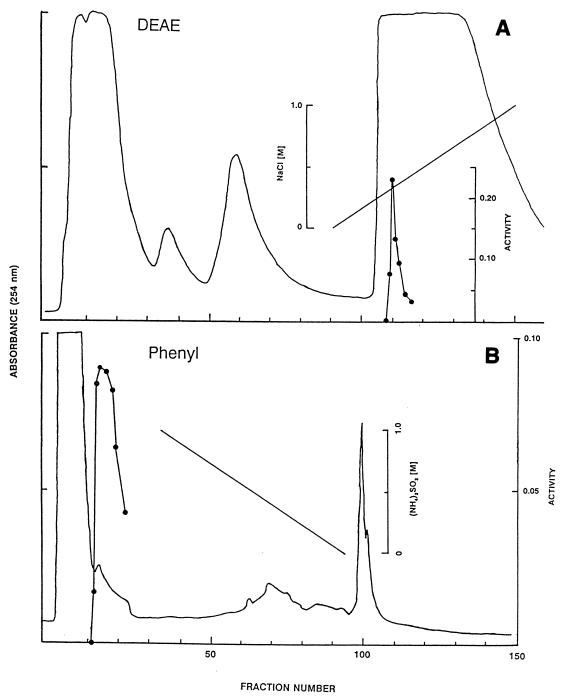
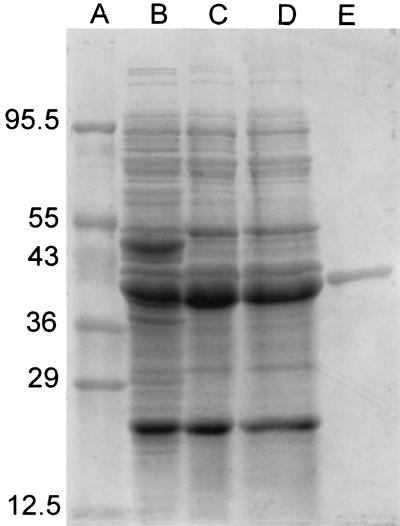
A summary of the results of purification of cyanuric acid amidohydrolase from E. coli DH5α containing the cloned trzD gene is shown in Table 1. The combination of DEAE chromatography and phenyl chromatography resulted in 30-fold purification of the enzyme. Figure 1 shows the elution profiles for cyanuric acid amidohydrolase on both of these columns. When the pooled phenyl fraction was examined by SDS-PAGE, a single band at a relative molecular mass of approximately 40,000 Da was observed, indicating that the enzyme had been purified to homogeneity (Fig. 2). The relative molecular mass of the active enzyme was determined to be approximately 185 kDa by SW300 gel filtration chromatography. This suggests that four of the 40-kDa subunits are combined to form the active enzyme.
TABLE 1.
Purification of cyanuric acid amidohydrolase
| Stage of purification | Sp act (μmol · min−1 · mg of protein−1)a | % Recoveryb | Fold puri-fication |
|---|---|---|---|
| Crude soluble fraction | 3.69 | ||
| DEAE | 19.43 | 46 | 5.3 |
| Phenyl | 109.47 | 28 | 29.7 |
Determined by the HPLC assay as described in the text.
Thirty-three milliliters of crude soluble fraction containing 4,700 IU of cyanuric acid amidohydrolase and 1.27 g of protein was applied to the DEAE column.
FIG. 1.
HPLC purification of cyanuric acid amidohydrolase on DEAE (A) and phenyl (B) columns. The continuous solid lines indicate the absorbance at 254 nm of the material eluting from the columns. The diagonal lines indicate the salt gradients in the eluting buffers. The solid circles show the rate of cyanuric acid hydrolysis catalyzed by 5 μl of each DEAE fraction or 10 μl of each phenyl fraction, as measured by the HPLC reaction. Rates are expressed as micromoles of cyanuric acid converted to biuret per minute.
FIG. 2.
SDS-PAGE of cyanuric acid amidohydrolase at various stages of purification. Lane A, protein standards (Diversified Biotech, Newton Centre, Mass.); lanes B through D, 10 μl of crude soluble fractions, 10 μl of pooled DEAE fractions, and 10 μl of pooled DEAE fractions after ammonium sulfate addition, respectively; lane E, 10 μl of pooled phenyl fractions. The numbers on the left indicate the sizes of the protein standards (in kilodaltons).
Development of spectrophotometric assay.
The HPLC assay for cyanuric acid amidohydrolase was found to be too insensitive for determining Km values and for other kinetic analyses, so attempts were made to develop a spectrophotometric assay for the enzyme. The UV spectrum of cyanuric acid in Tris-HCl buffer had a maximum absorbance peak at 216 nm. Addition of purified cyanuric acid amidohydrolase resulted in a steady decrease in this absorbance. A wavelength of 220 nm was chosen for the spectrophotometric assay since the background absorbance due to Tris was minimal at this wavelength. A plot of the absorbance at 220 nm of cyanuric acid in 25 mM Tris-HCl was linear for cyanuric acid concentrations of 0.015 to 0.15 mM (data not shown). When cyanuric acid amidohydrolase was added to solutions containing 0.15 mM cyanuric acid in Tris-HCl buffer, the absorbance decreased linearly, indicating that this method was a viable way to assay for cyanuric acid hydrolysis. No hydrolysis of cyanuric acid was observed in the absence of enzyme.
Effects of divalent cations on cyanuric acid hydrolysis.
Extensive dialysis of crude extracts of cyanuric acid amidohydrolase preparations against buffer containing 1 mM EDTA did not adversely affect the rate of cyanuric acid hydrolysis. Addition of either Mg2+ or Mn2+ ions at a concentration of 1 mM to reaction mixtures as either sulfate or chloride salts had no effect on the rate of cyanuric acid hydrolysis, while addition of Co2+, Cu2+, or Fe2+ at a concentration of 1 mM was slightly inhibitory. Addition of 1 mM Zn2+ reduced the reaction rate 100-fold. Thus, it appears that divalent cations are not required for cyanuric acid hydrolysis by cyanuric acid amidohydrolase, although the possibility that a tightly bound metal ion is present in the enzyme cannot be ruled out. No metal analysis of the enzyme was performed directly.
Kinetic constants and substrate specificity.
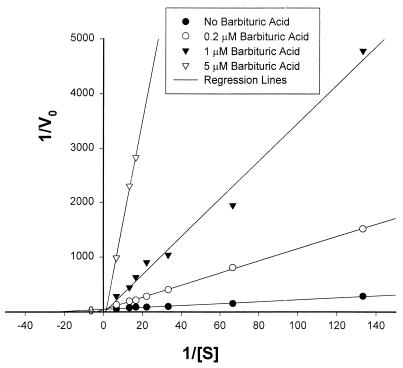
A Km of 50 μM for cyanuric acid and a turnover rate of 15,000 μmol of cyanuric acid min−1 (μmol of enzyme)−1 were obtained for purified cyanuric acid amidohydrolase at pH 8.0 in 25 mM Tris-HCl at 30°C from a linear regression analysis of Woolf plots (11). Several compounds that are structurally related to cyanuric acid were tested as possible substrates for cyanuric acid amidohydrolase. Ammeline (2,4-diamino-6-hydroxy-s-triazine) and ammelide (2-amino-4,6-dihydroxy-s-triazine) were not cleaved by the enzyme when they were added to a reaction mixture at a concentration of 2 mM. A number of pyrimidine compounds were tested as substrates at a concentration of 2 mM; these compounds included uracil (2,4-dihydroxypyrimidine), 5,6-dihydrouracil, cytosine (4-amino-2-hydroxypyrimidine), 2,4,5-trihydroxypyrimidine, and barbituric acid (2,4,6-trihydroxypyrimidine), and none of them was transformed by the enzyme as determined by either HPLC or spectrophotometric assays. When the compounds listed above were tested at a concentration of 0.05 mM to determine whether they were able to inhibit hydrolysis of cyanuric acid by cyanuric acid amidohydrolase, only barbituric acid was found to have any inhibitory effect. As shown in Fig. 3, barbituric acid is a potent competitive inhibitor of cyanuric acid hydrolysis. A Ki of less than 0.1 μM for barbituric acid was calculated from the intercepts of the Lineweaver-Burk plots shown in Fig. 3.
FIG. 3.
Lineweaver-Burke plots showing competitive inhibition of cyanuric acid hydrolysis by barbituric acid.
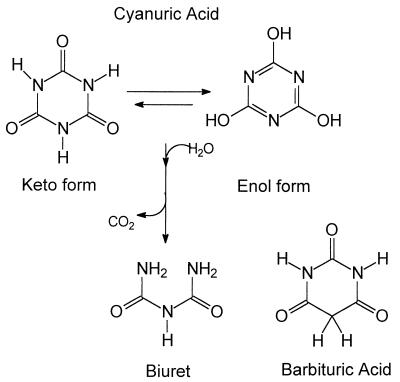
The reaction catalyzed by cyanuric acid amidohydrolase is shown in Fig. 4, as is the structure of barbituric acid. The optimum pH for cyanuric acid hydrolysis by cyanuric acid amidohydrolase was between 8.0 and 8.5, and the optimum temperature for the reaction was between 45 and 50°C (data not shown).
FIG. 4.
Chemical structures of barbituric acid (triketo tautomer) and cyanuric acid (triketo and enol forms) and the reaction catalyzed by cyanuric acid amidohydrolase which results in the formation of biuret.
Identification of reaction product.
The product resulting from hydrolysis of cyanuric acid by the purified enzyme was identified as biuret by HPLC and by examining the comparative UV spectra at pH 8 and 13. Biuret has very little UV absorbance at pH 8 but a large absorbance peak at pH 13 (λmax, 216 nm).
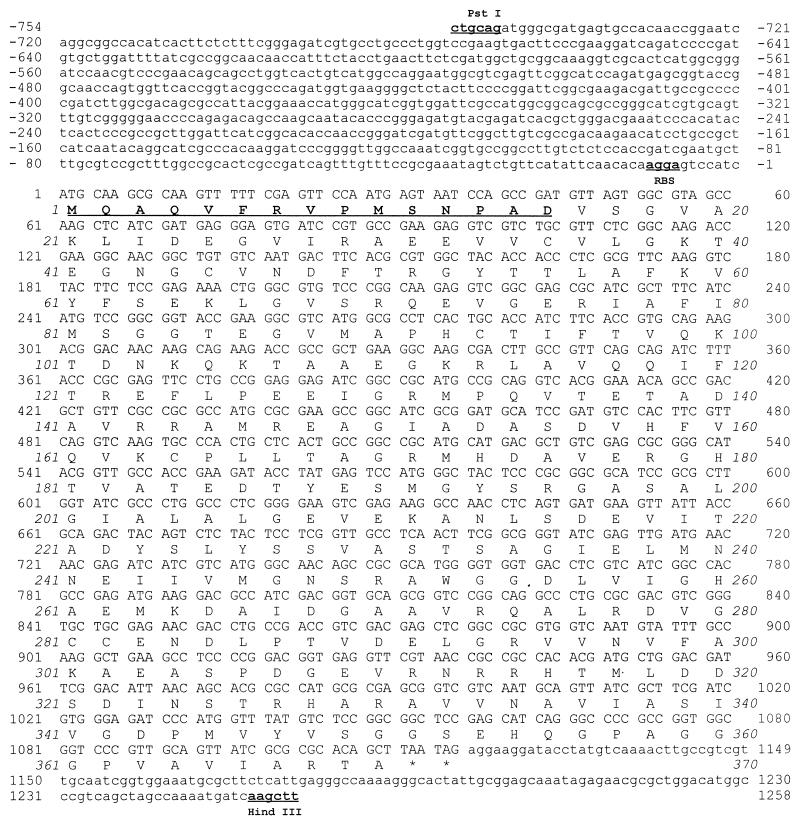
DNA sequence.
The DNA sequence of the 2.0-kb PstI-HindIII fragment containing the trzD gene is shown in Fig. 5. The total size of the fragment is 2,012 bp. The coding region for cyanuric acid amidohydrolase begins 754 bp from the PstI site at the 5′ end of the fragment and is preceded by a potential ribosome binding site (AGGA) 11 bp upstream. The first 15 residues at the amino terminus of the purified protein were determined to be Met-Gln-Ala-Gln-Val-Phe-Arg-Val-Pro-Met-Ser-Asn-Pro-Ala-Asp, which matches the DNA sequence; this indicates that the mRNA is translated from its own initiation sites so that the enzyme is not produced in E. coli as a fusion protein with the β-galactosidase gene present on the cloning vector. The molecular mass predicted from the DNA sequence (39.4 kDa) also agrees well with the 40-kDa subunit size observed in polyacrylamide gels. A BLASTN search of the trzD base sequence against the GenBank nucleic acid database revealed no significant similarity with any other DNA sequence. Likewise, a BLASTP search of the hypothetical TrzD protein’s amino acid sequence against the SwissProt database and GenBank data revealed no significant similarity with any known protein.
FIG. 5.
Sequence of the 2-kb PstI-HindIII fragment from pRE479 containing the trzD gene. The amino acids whose identities were confirmed by N-terminal amino acid sequencing of purified cyanuric acid amidohydrolase are underlined and in boldface type. A putative ribosome binding site (RBS) upstream of the coding region is underlined and in boldface type. Stop codons are indicated by asterisks.
The 2.0-kb PstI-HindIII fragment contains an open reading frame from nucleotide positions −754 to 35 that codes for a hypothetical protein with homology to YlbE and YahG of E. coli, hypothetical proteins whose functions are not known. From nucleotide position 1130 to the end of the sequence there is an open reading frame that codes for a hypothetical 43-amino-acid peptide that exhibits some similarity to the amino terminus of ArcC, the product of the carbamate kinase gene of E. coli. Since the structure of carbamoyl phosphate, the product of the carbamate kinase reaction, is somewhat similar to the structure of biuret, it is possible that this open reading frame is the start of a gene involved in biuret metabolism.
DISCUSSION
In this report I describe purification and properties of an enzyme that is responsible for hydrolytic cleavage of the s-triazine ring, which makes mineralization of the carbon and nitrogen contained in s-triazines possible. The first report of this enzyme was that of Cook and coworkers (7), who assayed and partially purified cyanuric acid amidohydrolase (EC 3.5.2) from Pseudomonas strain D (= NRRLB-12228). In that study the enzyme was not completely purified, nor were kinetic constants, pH optima, or temperature optima reported.
The gene encoding the enzyme studied in this report originated in Pseudomonas strain A (= NRRLB-12227) of Cook et al. (7); this gene was shown by Eaton and Karns (13) to have a restriction pattern identical to that of the gene from Pseudomonas strain D for 12 enzymes that cut within the cloned region. Thus, the enzyme described here is the same enzyme that was described by Cook et al. The gene was cloned into E. coli and overproduced by expression from the tac promoter of vector pMMB277. This provided a way to rapidly grow large amounts of cells with high initial specific activity. The combination of HPLC on DEAE columns and HPLC on phenyl columns resulted in a pure enzyme preparation that was suitable for amino-terminal sequence analysis. This analysis confirmed that the enzyme was produced from its own translation start site and was not the result of a fusion with the β-galactosidase protein encoded by the vector. Thus, this protein is probably authentic cyanuric acid amidohydrolase as it is produced in the native host, although the possibility that some processing of the protein might occur in the native host that does not occur in E. coli cannot be ruled out.
Cyanuric acid amidohydrolase has a fairly low Km for the substrate cyanuric acid (50 μM) and a high turnover rate (15,000 mol of substrate · mol of enzyme−1 · min−1). The enzyme appears to have a much higher affinity for the competitive inhibitor barbituric acid, which is a structural analog of cyanuric acid (Fig. 4). Cyanuric acid exists in various tautomeric forms in aqueous solution, and the diketo tautomer predominates at physiological pH values (26). Binding of cyanuric acid at the active site of the enzyme may stabilize cyanuric acid in the transition state, which encourages hydrolysis. Barbituric acid also exists in a partially tautomerized form in aqueous solution; however, substitution of a pyrimidine ring for the s-triazine ring changes the pKa of transitions such that the monoketo form of barbituric acid predominates at physiological pH values (3). A comparison of the chemistries of barbituric acid and cyanuric acid and the interactions of these acids with the active site of the enzyme may provide some clue as to the mechanism of reaction of cyanuric acid amidohydrolase.
The extensive use of triazine herbicides in agriculture worldwide lends importance to the study of the means by which these compounds are removed from the environment and their carbon and nitrogen are recycled. The cleavage of the s-triazine ring is an important step in this process. The observation that barbituric acid is a potent inhibitor of this cleavage may provide a useful tool to scientists studying the dissipation of s-triazines in the environment and to scientists studying the metabolism of s-triazines in plants, animals, and microbes. Because of the lack of similarity between the trzD sequence and other sequences in various databases, the evolutionary origin of this enzyme cannot be predicted. Sadowsky et al. (24) noted that the triazine-degrading proteins AtzA, AtzB, and AtzC exhibit amino acid similarities that indicate that they are members of an amidohydrolase superfamily that includes AdeC of E. coli and PyrC of Bacillus subtilis. These enzymes are all metal-binding hydrolases that act on nitrogenous heterocyclic ring substrates. Although cyanuric acid amidohydrolase cleaves an amide bond like AtzB and AtzC, the fact that it does not seem to require any metal ions for activity and the fact that it exhibits no significant amino acid sequence similarity with AtzC, AtzA, and AdeC indicate that it is a member of a different amidohydrolase family than these proteins. It would be interesting to use the cloned trzD gene and barbituric acid to study hydrolysis of cyanuric acid at the gene and enzyme levels in members of recently described atrazine-degrading bacterial cultures.
ACKNOWLEDGMENTS
I thank Richard Eaton of the U.S. Environmental Protection Agency, Gulf Breeze, Fla., for providing pRE479 and for helpful discussions. Excellent technical assistance with protein purification was provided by Donald Wiggins.
REFERENCES
- 1.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Behki R M, Kahn S U. Degradation of atrazine by Pseudomonas. J Agric Food Chem. 1986;34:746–749. [Google Scholar]
- 3.Bojarski J T, Mokrosz J L, Barton H J, Paluchowska M H. Recent progress in barbituric acid chemistry. Adv Heterocycl Chem. 1985;38:229–297. [Google Scholar]
- 4.Boundy-Mills K L, de Souza M L, Mandelbaum R T, Wackett L P, Sadowsky M J. The atzB gene of Pseudomonas sp. strain ADP encodes the second enzyme of a novel atrazine degradation pathway. Appl Environ Microbiol. 1997;63:916–923. doi: 10.1128/aem.63.3.916-923.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–252. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 6.Cook A M, Hütter R. s-Triazines as nitrogen sources for bacteria. J Agric Food Chem. 1981;29:1135–1143. [Google Scholar]
- 7.Cook A M, Beilstein P, Grossenbacher H, Hütter R. Ring cleavage and degradative pathway of cyanuric acid in bacteria. Biochem J. 1985;231:25–30. doi: 10.1042/bj2310025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cook A M. Biodegradation of s-triazine xenobiotics. FEMS Microbiol Rev. 1987;46:93–116. [Google Scholar]
- 9.de Souza M L, Sadowsky M J, Wackett L P. Atrazine chlorohydrolase from Pseudomonas strain ADP: gene sequence, enzyme purification, and protein characterization. J Bacteriol. 1996;178:4894–4900. doi: 10.1128/jb.178.16.4894-4900.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Souza M L, Newcombe D, Alvey S, Crowley D, Hay A, Sadowsky M J, Wackett L P. Molecular basis of a bacterial consortium: interspecies catabolism of atrazine. Appl Environ Microbiol. 1998;64:178–184. doi: 10.1128/aem.64.1.178-184.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dowd J E, Riggs D S. A comparison of estimates of Michaelis-Menton kinetic constants from various linear transformations. J Biol Chem. 1965;240:860–869. [PubMed] [Google Scholar]
- 12.Eaton R W, Karns J S. Cloning and analysis of s-triazine catabolic genes from Pseudomonas sp. strain NRRLB-12227. J Bacteriol. 1991;173:1215–1222. doi: 10.1128/jb.173.3.1215-1222.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eaton R W, Karns J S. Cloning and comparison of the DNA encoding ammelide aminohydrolase and cyanuric acid amidohydrolase from three s-triazine-degrading bacterial strains. J Bacteriol. 1991;173:1363–1366. doi: 10.1128/jb.173.3.1363-1366.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giardina M C, Giardi M T, Filacchione G. Atrazine metabolism by Nocardia: elucidation of the initial pathway and synthesis of potential metabolites. Agric Biol Chem. 1982;46:1439–1445. [Google Scholar]
- 15.Holmes D S, Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981;114:193–199. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- 16.Jensen H L, Abdel-Ghaffer A S. Cyanuric acid as nitrogen source for microorganisms. Arch Mikrobiol. 1969;67:1–5. doi: 10.1007/BF00413674. [DOI] [PubMed] [Google Scholar]
- 17.Kalb V F, Bernlohr R W. A new spectrophotometric assay for protein in cell extracts. Anal Biochem. 1977;82:362–371. doi: 10.1016/0003-2697(77)90173-7. [DOI] [PubMed] [Google Scholar]
- 18.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 19.Mandelbaum R T, Allan D L, Wackett L P. Isolation and characterization of a Pseudomonas sp. that mineralizes the s-triazine herbicide atrazine. Appl Environ Microbiol. 1995;61:1451–1457. doi: 10.1128/aem.61.4.1451-1457.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morales V M, Backman A, Bagdasarian M. A series of wide host range low copy number vectors that allow direct screening for recombinants. Gene. 1991;97:39–47. doi: 10.1016/0378-1119(91)90007-x. [DOI] [PubMed] [Google Scholar]
- 21.Mulbry W W. Purification and characterization of an inducible s-triazine hydrolase from Rhodococcus coralinus. Appl Environ Microbiol. 1994;60:613–618. doi: 10.1128/aem.60.2.613-618.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.National Agricultural Statistics Service and Economic Research Service, U.S. Department of Agriculture. Agricultural chemical usage 1991: field crops summary. U.S. Washington, D.C: Department of Agriculture; 1992. [Google Scholar]
- 22a.National Center for Biotechnology Information. 24 February 1998, posting date. BLAST 2.0.4. [Online.] www.ncbi.nlm.nih.gov/BLAST/. [7 August 1998, last date accessed.]
- 23.Radosevich M, Traina S J, Hao Y-L, Tuovinen O H. Degradation and mineralization of atrazine by a soil bacterial isolate. Appl Environ Microbiol. 1995;61:297–302. doi: 10.1128/aem.61.1.297-302.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sadowsky M J, Tong Z, deSouza M, Wackett L P. AtzC is a new member of the amidohydrolase protein superfamily and is homologous to other atrazine-metabolizing enzymes. J Bacteriol. 1998;180:152–158. doi: 10.1128/jb.180.1.152-158.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shocken M J, Speedie M K. Physiological aspects of atrazine degradation by higher marine fungi. Arch Environ Contam Toxicol. 1984;13:707–714. [Google Scholar]
- 26.Smolin E M, Rapoport L. Cyanuric acid and derivatives. In: Weissberger A, editor. The chemistry of heterocyclic compounds, vol. 13. s-Triazines and derivatives. New York, N.Y: Interscience; 1959. pp. 17–48. [Google Scholar]
- 27.Yanze-Kontchou C, Gschwind N. Mineralization of the herbicide atrazine as a carbon source by a Pseudomonas strain. Appl Environ Microbiol. 1994;60:4297–4302. doi: 10.1128/aem.60.12.4297-4302.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]