Abstract
The potential of terminal-restriction fragment length polymorphism (T-RFLP) and the detection of operational taxonomic units (OTUs) by capillary electrophoresis (CE) to characterize marine bacterioplankton communities was compared with that of denaturing gradient gel electrophoresis (DGGE). A protocol has been developed to optimize the separation and detection of OTUs between 20 and 1,632 bp by using CE and laser-induced fluorescence detection. Additionally, we compared T-RFLP fingerprinting to DGGE optimized for detection of less abundant OTUs. Similar results were obtained with both fingerprinting techniques, although the T-RFLP approach and CE detection of OTUs was more sensitive, as indicated by the higher number of OTUs detected. We tested the T-RFLP fingerprinting technique on complex marine bacterial communities by using the 16S rRNA gene and 16S rRNA as templates for PCR. Samples from the Northern and Middle Adriatic Sea and from the South and North Aegean Sea were compared. Distinct clusters were identifiable for different sampling sites. Thus, this technique is useful for rapid evaluation of the biogeographical distribution and relationships of bacterioplankton communities.
Fingerprinting techniques are valuable tools for the characterization of complex bacterial communities in the environment (1, 15). Different strategies have been developed over the last decade to characterize marine bacterial communities that escape, to a large extent, traditional cultivation techniques. These approaches include restriction fragment length polymorphism (RFLP) characterization of bacterial isolates (12, 24, 25) and complex bacterial communities (1) on horizontal or vertical gel electrophoresis. Visualization of RFLP bands is usually obtained by fluorescent labeling of all fragments with dyes such as ethidium bromide. High-resolution polyacrylamide gel electrophoresis is required to resolve the high number of bands resulting from RFLP (1).
Denaturing gradient gel electrophoresis (DGGE) is an alternative fingerprinting technique by which DNA fragments obtained after enzymatic amplification from complex microbial communities are separated according to their sequence information (15, 16). The separation of the different DGGE bands depends on the melting behavior of the PCR product and not on the size of the fragment.
Recently, terminal-restriction fragment length polymorphism (T-RFLP) has been shown to be an automated and sensitive approach to the characterization of bacterial assemblages of known composition by using capillary electrophoresis (CE) with laser-induced fluorescence (LIF) detection (2). This technique uses a PCR in which one of the primers is fluorescently labeled. After amplification, the PCR product is digested with one or more restriction enzymes, generating fragments with different lengths, depending on the DNA sequence of the bacteria analyzed and on the specificity of the enzyme. The T-RFLP pattern can be used to define the number of operational taxonomic units (OTUs) present in a complex bacterial community. It has been shown that for a bacterial strain only the end-labeled fragment (OTU) is detected (2). For mixed microbial communities, this has the advantage that each end-labeled fragment is specific for OTUs which have a restriction site at the same location, making the interpretation of complex communities easier. This approach was used for fingerprinting of complex bacterial communities (9) on polyacrylamide sequencing gels and recently for the characterization of archaea in the guts of marine fish with a capillary electrophoresis sequencer (23).
In this study, we compare T-RFLP and DGGE analyses of complex marine bacterial communities. Both methods were optimized for sensitive and accurate determination of OTUs from marine samples. It has been shown that the 16S rRNA can be used to characterize metabolically active bacteria, since active cells contain more rRNA than do nonactive cells (7, 19). Therefore, we used both the 16S rRNA gene (rDNA) and 16S rRNA in order to obtain insights into the structure of the total and the active bacterial communities, respectively. Finally, we applied this optimized T-RFLP fingerprinting technique and compared complex marine bacterial community samples from the upper mixed water column collected at different sites in the Mediterranean Sea.
MATERIALS AND METHODS
Collection of samples and isolation of bacterial strains.
Samples of natural bacterial communities were taken in the Middle Adriatic Sea (4 km off the coast of Ancona, Italy), in the Northern Adriatic Sea (5 km off the coast of Rovinj, Croatia), and at three stations in the North and two stations in the South Aegean Sea (Greece) (Table 1). Water temperatures ranged from 13°C (samples from the Aegean Sea) to 22°C (samples collected in the Middle Adriatic Sea). Three to 5 liters of seawater was filtered through GF/C filters (Whatman, Maidstone, England) to remove larger particles and phytoplankton. Subsequently, bacteria were concentrated onto Sterivex GV filtration units (Millipore, Bedford, Mass.) at a pressure of <105 Pa. Sterivex filters were immediately frozen and stored at −80°C until further processing in the laboratory.
TABLE 1.
Descriptions of samples collecteda
| Sample | Sampling date (day/mo/yr) | Latitude/longitude | Depth (m) | Sample collection depth (m) | chl a (μg liter−1) | No. of OTUs
|
|||
|---|---|---|---|---|---|---|---|---|---|
| CfoI | MvnI | RsaI | DGGE | ||||||
| an1 | 12/8/95 | 43°39.00/13°35.00 | 20 | 0.5 | 0.75 | 39 | 35 | 32 | NDb |
| an2 | 13/08/95 | 43°39.00/13°35.00 | 20 | 0.5 | 1.02 | 42 | 39 | 42 | ND |
| an3 | 14/08/95 | 43°39.00/13°35.00 | 20 | 0.5 | 0.85 | 38 | 51 | 45 | ND |
| an4 | 15/08/95 | 43°39.00/13°35.00 | 20 | 0.5 | 1 | 41 | 42 | 36 | ND |
| an50 | 6/9/95 | 43°39.00/13°35.00 | 20 | 0.5 | 0.79 | 32 | 31 | 28 | ND |
| an60 | 11/9/95 | 43°39.00/13°35.00 | 20 | 0.5 | 1.55 | 35 | 49 | 39 | ND |
| an816 | 24/09/95 | 43°39.00/13°35.00 | 20 | 0.5 | 1.35 | 26 | 44 | 43 | ND |
| msb1 | 8/3/98 | 36°04.50/25°17.00 | 1,740 | 10 | 0.17 | 44 | 42 | 39 | 35 |
| msb1rt | 8/3/98 | 36°04.50/25°17.00 | 1,740 | 10 | 0.17 | 42 | 44 | 38 | ND |
| msb3 | 7/3/98 | 36°00.00/23°53.60 | 1,190 | 10 | 0.18 | 44 | 42 | 39 | 35 |
| mnb1 | 14/03/98 | 40°14.90/25°12.00 | 1,300 | 10 | 0.64 | 43 | 38 | 36 | 33 |
| mnb1rt | 14/03/98 | 40°14.90/25°12.00 | 1,300 | 10 | 0.64 | 51 | 46 | 42 | ND |
| mnb2 | 15/03/98 | 40°04.65/24°36.20 | 960 | 10 | 0.63 | 44 | 39 | 38 | 33 |
| mnb2rt | 15/03/98 | 40°04.65/24°36.20 | 960 | 10 | 0.63 | 58 | 47 | 46 | ND |
| mnb3 | 13/03/98 | 39°13.45/25°00.00 | 820 | 10 | 0.17 | 40 | 42 | 40 | 36 |
| mnb4 | 19/03/98 | 39°47.10/25°31.50 | 95 | 10 | 0.37 | 41 | 39 | 33 | 28 |
| ro1 | 2/5/95 | 45°05.00/13°33.00 | 25 | 0.5 | 0.75 | 46 | 35 | 39 | ND |
| ro2 | 5/5/95 | 45°05.00/13°33.00 | 25 | 0.5 | 0.8 | 46 | 30 | 45 | ND |
| ro3 | 6/5/95 | 45°05.00/13°33.00 | 25 | 0.5 | 1 | 36 | 34 | 36 | ND |
| ro11 | 9/5/95 | 45°05.00/13°33.00 | 25 | 0.5 | 1.2 | 47 | 30 | 44 | ND |
Samples from the upper mixed water column were collected in the Middle Adriatic Sea off Ancona (an; Italy), in the South (msb) and North (mnb) Aegean Sea (Greece), and in the Northern Adriatic Sea off Rovinj (ro; Croatia). The number of OTUs found when the sample was digested with the respective tetrameric restriction enzyme (CfoI, MvnI, or RsaI) and in the DGGE gel is shown. For each sample, the patterns from the three tetrameric restriction digests were combined and used for UPGMA analysis (see Fig. 5).
ND, not determined.
Marine bacterial strains (MS-01, MS-02, MS-20, MS-21, MS-22, and MS-23) were isolated from marine snow aggregates collected in the Northern Adriatic Sea (3 km off the coast of Rovinj, Croatia). Marine snow was collected as described by Müller-Niklas et al. (14), and bacterial strains were isolated (11). Two milliliters of a liquid culture grown overnight at 20°C was collected by centrifugation at 3,200 × g for 20 min and washed twice with STE buffer (100 mM NaCl, 50 mM Tris-HCl [pH 7.4], 1 mM EDTA). Cell pellets were stored at −80°C until nucleic acid extraction.
Extraction of nucleic acids.
Isolated strains and natural bacterial communities were resuspended in 2 ml of lysis buffer (400 mM NaCl, 750 mM sucrose, 20 mM EDTA, 50 mM Tris-HCl, pH 9.0) and incubated with lysozyme (final concentration, 1 mg ml−1; Merck, Darmstadt, Germany) at 37°C for 30 min. Sodium dodecyl sulfate (final concentration, 1% [wt/vol]; Sigma, St. Louis, Mo.) and proteinase K (final concentration, 100 μg ml−1; Boehringer Mannheim Biochemicals B.V., Almere, The Netherlands) were added, and the samples were incubated at 55°C for 120 min. The lysate was checked under a microscope for complete lysis of the bacterial cells. Subsequently, the lysate was extracted with an equal volume of phenol-chloroform-isoamyl alcohol (25:24:1) and an equal volume of chloroform-isoamyl alcohol (24:1). The nucleic acids were precipitated (5), resuspended in 200 μl of diethylpyrocarbonate-treated water, and stored at −80°C.
DNA purification.
RNA was removed from the nucleic acid extracts by incubation with DNase-free RNase (final concentration, 100 μg ml−1; Pharmacia Biotech, Uppsala, Sweden) at 55°C for 30 min. DNA was further purified by using the Qiaex II Extraction Kit (Qiagen, Hilden, Germany) and following the manufacturer’s recommendations. Integrity of DNA was checked on 1% (wt/vol) agarose gels. DNA concentrations were determined fluorometrically in a DynaQuant fluorometer as recommended by the manufacturer (Pharmacia Biotech). DNA concentrations ranged between 35 and 60 ng μl−1.
RNA purification.
DNA was removed from 20 μl of nucleic acid extracts by using 4 U of RNase-free DNase (Pharmacia Biotech) as recommended by the manufacturer and extracted and precipitated as described above for the nucleic acids. The pellets were rinsed with 70% (vol/vol) ethanol and subsequently resuspended in 30 μl of diethylpyrocarbonate-treated water. Each RNA preparation was checked for remaining traces of undigested DNA in 50-μl PCR mixtures.
cDNA synthesis for reverse transcription-PCR.
Transcription of RNA into cDNA was performed with first-strand reaction mix beads (Pharmacia Biotech) in accordance with the manufacturer’s protocol by adding 1 μl of pd(N)6-primer (0.2 μg μl−1; Pharmacia Biotech). The cDNA was digested with DNase-free RNase in accordance with the DNA purification protocol to remove residual RNA and purified with the Qiaquick PCR Purification Kit (Qiagen). One microliter of cDNA was used per 50-μl PCR mixture.
PCR for T-RFLP.
The primers used for PCR were 27F-FAM (5′-AGA GTT TGA TCC TGG CTC AG-3′) and 1492R (5′-GGT TAC CTT GTT ACG ACT T-3′), which give a 1,503-bp product of the 16S rDNA (6). 27F-FAM was 5′ end labeled with phosphoramidite fluorochrome 5-carboxyfluorescein (5′ 6-FAM), which was synthesized by Eurogentec (Searing, Belgium). Each 50-μl PCR mixture contained both primers at 0.2 μM, 50 mM KCl, 10 mM Tris-HCl (pH 9.0), 1.5 mM MgCl2, 200 μM each deoxynucleoside triphosphate (Pharmacia Biotech), and 2.5 U of Taq polymerase (Pharmacia Biotech). Samples were amplified by using the following protocol: an initial denaturation step of 94°C for 3 min, followed by 30 cycles of denaturation at 94°C for 1 min, annealing at 55°C for 1 min, and extension at 72°C for 1 min. Cycling was completed by a final extension at 72°C for 7 min. The PCR products were purified with the Qiaquick PCR Purification Kit (Qiagen).
Restriction digests.
Each digest contained 8 μl of a cleaned PCR product, 10 U of a tetrameric restriction enzyme (i.e., the 4-bp cutter, CfoI, RsaI, or MvnI; Boehringer Mannheim Biochemicals B.V.), and the respective restriction buffer at 1× (ShuRE/CUT buffer; Boehringer Mannheim Biochemicals) and was filled up to a final volume of 50 μl with autoclaved Milli-Q water. Incubation was done at 37°C for 6 h. Restriction digests were desalted with the Qiaquick Nucleotide Removal Kit (Qiagen).
T-RFLP analysis.
Separation was performed at 3 kV for 75 min in a polyacryloylaminoethoxyethanol-coated 44-cm capillary (inside diameter, 75 μm; Bio-Rad, Hercules, Calif.). A replaceable 0.75% (wt/vol) hydroxypropyl methyl cellulose (H-4649; Sigma) running buffer in TBE (89 mM Tris-HCl, 89 mM boric acid, 1 mM EDTA, pH 8.3) was used. Samples were injected electrokinetically at 10 kV for 20 s. 5′ 6-FAM-labeled fragments were detected at a 520-nm wavelength by a Biofocus 3000 capillary electrophoresis apparatus (Bio-Rad) equipped with a Biofocus LIF2 detector (488-nm argon-ion laser and 594-nm helium-neon laser). A mixture of Low Range Standard and 100 bp Molecular Ruler (both Texas red labeled; Bio-Rad) was added to each sample at a final concentration of 5 fmol μl−1 and served as an internal size standard (Texas red-labeled fragments were detected at 630 nm).
T-RFLP analysis of patterns from complex marine bacterial communities.
We used three separate restriction digests for complex bacterial communities to obtain the fingerprinting information from different enzymes per sample. The sizes of 5′ 6-FAM-labeled fragments were determined by comparison with the internal Texas red size standards using a second-order polynomial regression. A binary matrix according to the presence or absence of aligned fragments (+ for the presence of an OTU and − for the absence of an OTU) was further analyzed by using the beta version of PAUP 4.0 (Sinauer Associates Inc. Publishers). The UPGMA (unweighted pair group with mathematical averages) method was applied with the site distance matrix method described by Nei and Li (18) to determine similarities between T-RFLP fingerprints. The same software and settings were used when the banding pattern of DGGE gels was UPGMA analyzed.
PCR for DGGE.
The primers used for PCR were DGGE341F (GM5F) (5′-CGC CCG CCG CGC CCC GCG CCC GGC CCG CCG CCC CCG CCC CCC TAC GGG AGG CAG CAG-3′) and 907R (5′-CCG TCA ATT CCT TTG AGT TT-3′) (17). Each 50-μl reaction mixture contained both primers at 20 pM, 50 mM KCl, 10 mM Tris-HCl (pH 9.0), 1.5 mM MgCl2, each deoxynucleoside triphosphate at 10 mM (Pharmacia Biotech), 1 U of Taq polymerase (Pharmacia Biotech), and 1 μl of template DNA. The optimal template concentration was determined by using serial dilutions (1/10, 1/20, 1/50, 1/100, and 1/200) and subsequent amplification of 1 μl as template DNA in a PCR. The first dilution lacking unspecific by-products as visualized after agarose gel electrophoresis was chosen as the optimal template concentration. The cycling program consisted of an initial denaturation step of 94°C for 5 min, followed by 10 cycles of denaturation at 95°C for 1 min, annealing at 66°C (decreasing in each cycle by 1°C) for 1 min, and an elongation step of 72°C for 3 min. We performed another 15 cycles of 95°C for 1 min, 56°C for 1 min, and elongation at 72°C for 3 min. Each run ended with a final elongation step of 72°C for 5 min.
DGGE analysis.
DGGE was performed as described by Muyzer et al. (16) using a DCode Universal Mutation Detection System (Bio-Rad). The gels were cast discontinuously; i.e., the gradient gel was cast until it was 1 cm below the end of the comb teeth and overlaid with water-saturated isobutanol (Fluka, Buchs, Switzerland) until polymerization was finished (after 3 h). A polyacrylamide gel without urea or formamide was cast on top of the gradient gel in which the comb was inserted. DGGE gels were run at a constant voltage of 75 V at 60°C for 18 h. The gels were poststained with GelStar (FMC Bioproducts, Rockland, Maine) in accordance with the manufacturer’s instructions and acquired on a Fluor-S MultiImager (Bio-Rad) with an excitation wavelength of 302 nm and an emission filter wavelength of 520 nm. Generally, three images with integration times of 1, 2, and 3 min were taken to obtain one optimally illuminated image, one oversaturated image, and one undersaturated image of the same gel. Schematic drawings of the band patterns were made by combining the information from all of the images acquired from one gel and aligning them with the originals, thereby increasing the information retrievable from one gel.
RESULTS AND DISCUSSION
Optimization of CE running conditions.
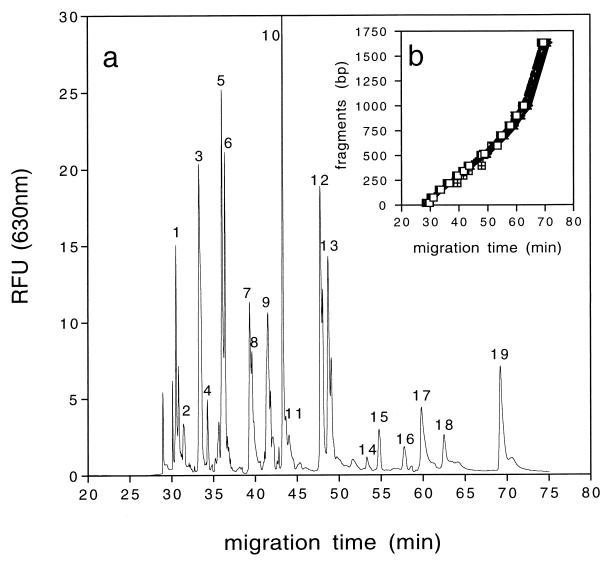
CE running conditions were optimized for the separation of double-stranded DNA fragments with sizes between 20 and 1,632 bp. The highest resolution was found with 0.75% hydroxypropyl methyl cellulose–TBE buffer and a voltage of 3 kV run for 75 min. We used a Low Range Standard (Bio-Rad) for sizing of the PCR products and a mixture of two commercially available size markers (Texas red-labeled Low Range Standard and 100 bp Molecular Ruler from Bio-Rad) for sizing of T-RFLP fragments. In our protocol, the 220- and 221-bp fragments showed a difference in migration time of 23 ± 8 s (n = 30) while the difference between the 504- and 517-bp fragments was 52 ± 6 s (n = 30, Fig. 1a). The applied running conditions resulted in a constant migration of double-stranded DNA, as indicated by almost identical slopes (Fig. 1b; second-order polynomial; n = 20), and in correct sizing of the PCR product (Fig. 2a; 1,509 ± 8 bp; n = 32). The resolution decreased with increasing fragment size (3), but still, less than 5 bp of separation was obtained for fragments of up to 1,000 bp.
FIG. 1.
(a) Migration behavior of the Texas red standard mixture when run at 3 kV for 75 min: 1, 75 bp; 2, 100 bp; 3, 154 bp; 4, 200 bp; 5, 220 bp; 6, 221 bp; 7, 296 bp; 8, 300 bp; 9, 344 bp; 10, 396 bp; 11, 400 bp; 12, 500 and 504 bp; 13, 517 bp; 14, 600 bp; 15, 700 bp; 16, 800 bp; 17, 900 bp; 18, 1,000 bp; 19, 1,632 bp. (b) A second-order polynomial regression was applied to determine the unknown fragments with these standards (n = 20). Unlabeled small peaks are unspecific peaks due to slight impurities or partial degradation.
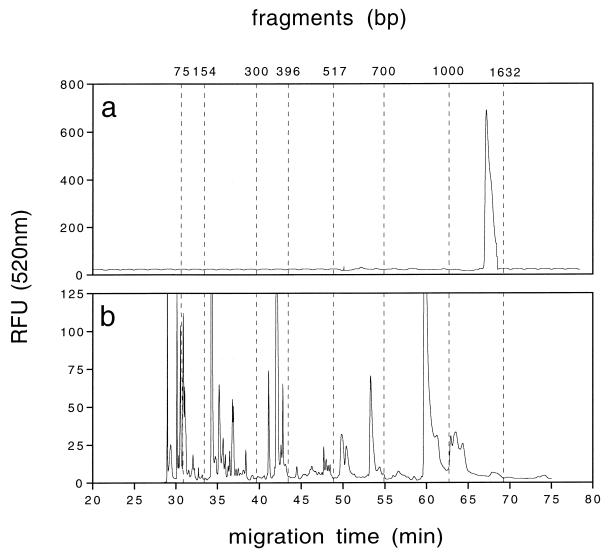
FIG. 2.
(a) Screening for 16S rDNA PCR products from the South Aegean Sea (msb1). The calculated length of the 16S rDNA product is 1,503 ± 8 bp (n = 32). (b) T-RFLP fingerprinting of a complex marine bacterial community from the South Aegean Sea (msb1) cut with CfoI for 6 h. Thirty-two of 44 T-RFLP OTUs were found between 20 and 517 bp. The largest fragments were ≈1,000 bp.
That it is possible to separate T-RFLP fragments under denaturing conditions which differ by only 2 bp has been previously shown by using sequencing gels (9) or a capillary electrophoresis sequencer (23) for fragments of up to 600 bp. Although our samples were not run under denaturing conditions, comparable resolution was still obtained. With our CE separation buffer, a nearly linear regression was found for fragments of up to 1,000 bp (Fig. 1b). The largest fragments obtained with the three enzymes used in this study were about 1,000 bp and were therefore included in the analysis. However, more than 70% of the T-RFLP fragments (for all of the T-RFLP samples analyzed) were within the 20- to 517-bp range. The high accuracy of separation of the T-RFLP fragments is obtained by the addition of the size standard to each sample. Although the size markers showed unspecific smaller peaks (Fig. 1a) due to slight impurities and/or partial degradation, correct sizing of the fragments was achieved. These impurities had no effect on correct sizing and determination of OTUs because no bleeding through to the 520-nm detection channel was observed. Another crucial problem for accurate separation is efficient desalting of the sample. For example, we were not able to resolve the 220- and 221-bp fragments of the size standard when the salt concentration in the sample was higher than 50 mM.
T-RFLP analysis.
Optimized T-RFLP fingerprinting under nondenaturing conditions was first tested with bacteria isolated from marine snow (Table 2) and digested with CfoI and RsaI. Each of the randomly chosen marine snow isolates produced a specific OTU when the template DNA (50 ng/50-μl PCR mixture) from the respective strain was subjected to a PCR (Table 2). Additionally, Escherichia coli was used as a positive control in the PCR, giving a specific T-RFLP fragment of 359 bp. To test for potential bias between strains in a complex mixture of DNA templates, all six DNA templates from the marine snow isolates (10 ng of each template) were combined in a single 50-μl PCR mixture (Table 2). All of the OTUs of the six strains were identified again in a single PCR mixture and characterized with the information obtained from the runs when template DNA from each isolate was added separately (Table 2). Figure 2b shows a T-RFLP chromatogram obtained when the 16S rDNA PCR product from a complex bacterial community (msb1) was digested with CfoI. The pattern in Fig. 2b shows large peaks and a large number of smaller peaks, which were also included in the analysis (Table 1). Of the 44 OTUs detected in this sample, 32 were smaller than 517 bp, 10 were in the size range of 517 to 1,000 bp, and 2 were larger than 1,000 bp.
TABLE 2.
T-RFLP OTU fragment sizesa
| Isolate | Mean OTU fragment size (bp) ± SD
|
|
|---|---|---|
| Single template | Mixture of all 6 templates | |
| MS-01 | 352 ± 3 | 352 ± 3 |
| MS-02 | 213 ± 2 | 208 ± 2 |
| MS-20 | 223 ± 2 | 222 ± 2 |
| MS-21 | 115 ± 4 | 118 ± 4 |
| MS-22 | 326 ± 1 | 326 ± 1 |
| MS-23 | 582 ± 5 | 583 ± 5 |
| E. coli | 359 ± 1 | NIb |
Shown are sizes of T-RFLP OTU fragments from six marine strains digested with two restriction enzymes (CfoI and RsaI) and sizes of T-RFLP OTU fragments of each marine snow isolate (single template; n = 8) and T-RFLP OTUs when all six templates were present in one PCR mixture (combination of all six templates; n = 4).
NI, not included in reaction mixture.
The migration behavior did not change with the amount of DNA applied to the capillary, for either the Texas red size marker or the 5′ 6-FAM-labeled fragments, making correct sizing of the different OTUs independent of the relative intensity. This guarantees correct size determination of highly abundant, as well as less abundant, OTUs. The high sensitivity of the CE-LIF system makes it crucial to check PCR products for artifacts (primer dimers or nonspecific PCR products). This potential contamination cannot be detected by conventional gel electrophoresis (data not shown). Only PCR products showing a single peak were used for subsequent reactions. We successfully determined PCR products after five cycles of amplification with the CE-LIF system. Lower cycle numbers, however, prohibit the detection of less abundant templates after the PCR. To avoid exclusion of less abundant members of the community by low cycle numbers, we used 30 cycles, which could cause bias in the PCR (8, 21). Because we interpreted our results not quantitatively but rather qualitatively (presence or absence of OTUs), a bias in the PCR cannot be excluded but should not influence the interpretation of our data. In addition, bacteria can express more than one rRNA operon (20), which has to be considered for quantitative studies. Therefore, making assumptions about the quantitative abundance of a specific OTU seems to be possible only if PCR bias and the number of operons are considered.
Despite the high resolution of T-RFLP, there is the possibility that two different species could have the same restriction site in the 16S rDNA, resulting in an identical peak. Therefore, we performed three independent restriction digests using different enzymes to increase the information for a single sample. With T-RFLP fingerprinting, only the end-labeled fragments are detected; therefore, we cannot make any assumptions about how often a restriction enzyme actually cuts the 16S rDNA. This somehow decreases the information obtainable from the whole RFLP pattern produced but makes interpretation of complex communities easier when it is assumed that every fragment is representative of a single OTU. The restriction enzymes used here were selected according to Moyer et al. (13) and gave the best resolution for 106 bacterial clones from 10 tetrameric restriction enzymes tested in a computer simulation.
DGGE analysis.
Staining with a highly sensitive nucleic acid stain (GelStar; FMC Bioproducts) and use of the improved acquisition method for DGGE gels presented here are advantageous for the characterization of less intense OTUs. Three separate integration times were chosen to detect all of the bands of various intensities. Our lower threshold was the band still detectable with the longest integration time. DGGE has the advantage that separation is dependent not on the size but rather on the melting behavior of the PCR product, making DGGE more discriminating than when a restriction fragment is used. Additionally, bands of interest can be excised and used for subsequent sequencing reactions. We are currently developing a protocol in which isolated clones are redetected in the T-RFLP fingerprint of the complex bacterial community to allow subsequent screening and sequencing also for T-RFLP. However, DGGE depends on the primers used because one of the primers has a poly(GC) clamp (40 bp long) which makes PCRs more accessible to primer-dimer formation. Other than with T-RFLP, in which every sample has its own inline size standard, the lack of appropriate size standards for DGGE makes gel-to-gel comparisons difficult.
With the acquisition protocol presented here, DGGE sensitivity can be increased considerably. With the nucleic acid stain used (GelStar; FMC Bioproducts), the DGGE detection threshold is restricted to 20 pg of double-stranded DNA, but when the LIF detector and 5′ 6-FAM-labeled fragments are used, 1 fg of double-stranded DNA can still be detected (Bio-Rad). The difference of 1 order of magnitude between the detection limits of double-stranded DNA could be responsible for the higher numbers of OTUs detected with the CE-LIF system.
Comparison of DGGE and T-RFLP.
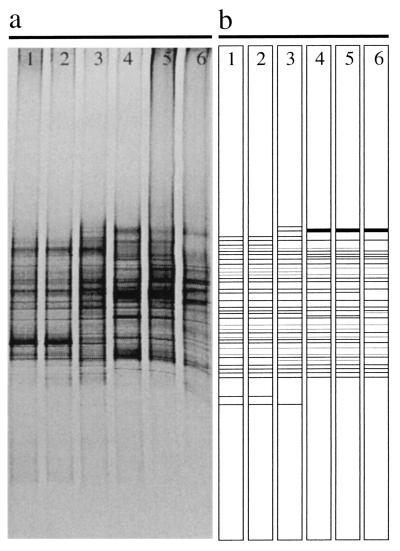
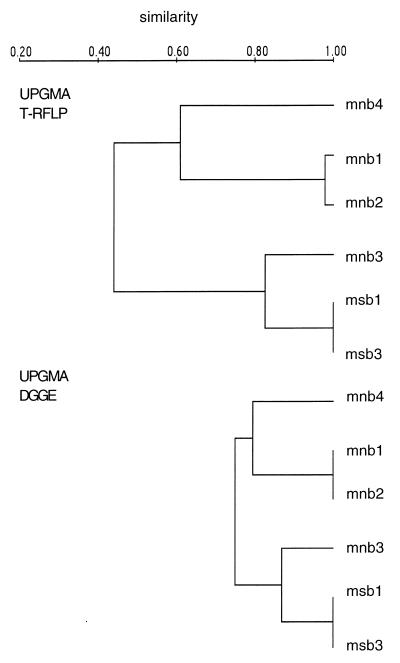
Figure 3a shows the banding pattern of the original DGGE gel integrated for 2 min, and Fig. 3b depicts the schematic banding pattern of this gel with less intense bands included. Based on the presence of individual DGGE bands, a matrix was established and compared to the matrix obtained by the T-RFLP approach. Both fingerprinting techniques showed the same clustering for the samples taken from the South and North Aegean Sea; only the branch lengths differed due to different numbers of OTUs obtained with the two fingerprinting techniques (Fig. 4). We found a larger number of OTUs with T-RFLP than with DGGE (Table 1). Similar results have been found for eukarya communities in activated sludge (10). Therefore, we only used T-RFLP for subsequent community analysis. The successful detection of less abundant OTUs is important for the determination of the actual bacterial diversity present in a certain habitat. The actual diversity of bacteria might be underestimated if the detection threshold were too high. Members of the bacterial community present in low abundance might still play an important role in the transformation of dissolved organic matter. Moreover, as soon as the environmental conditions (e.g., nutrient concentration or composition) change, these rare members of the bacterial community might outcompete others.
FIG. 3.
DGGE pattern from the samples taken in the Aegean Sea. (a) Banding pattern of the original gel stained with GelStar (FMC Bioproducts), acquired with a charge-coupled device camera over a 2-min exposure period. Longer integration times enhanced the signal from weak bands, thereby oversaturating stronger bands. (b) The DGGE gel was acquired with three different integration times, and the bands were drawn in a schematic diagram. Lanes: 1, msb3 (35 OTUs); 2, msb1 (35 OTUs); 3, mnb3 (36 OTUs); 4, mnb1 (33 OTUs); 5, mnb2 (33 OTUs); 6, mnb4 (28 OTUs).
FIG. 4.
UPGMA dendrograms from T-RFLP and DGGE analyses of PCR products of samples taken in the Aegean Sea. For the samples analyzed with T-RFLP, the numbers of OTUs from three separate restriction digests were pooled.
Comparison of PCR and reverse transcription-PCR products from microbial communities originating from different sites.
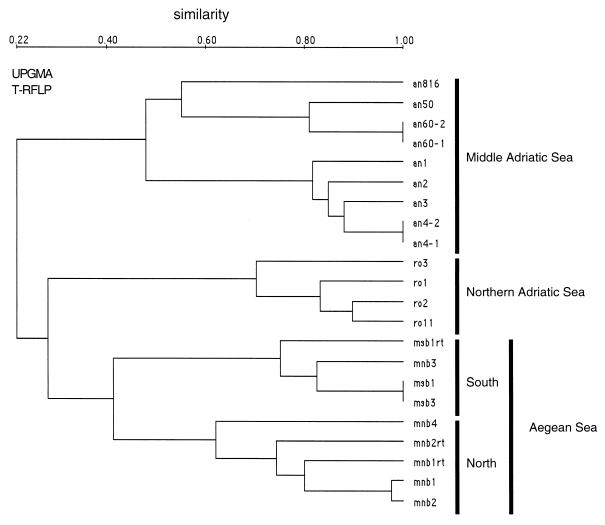
The T-RFLP patterns analyzed from the different sampling sites showed three distinct clusters, for the Middle Adriatic Sea, for the Aegean Sea, and for the Northern Adriatic Sea (Fig. 5). For the Middle Adriatic Sea, samples an1 to an4 were more related to each other than to an50, an60, and an816, which were taken 3 weeks later, indicating a change in the community structure. During sampling of an50 and an60, strong winds led to increased resuspension of the sediment, which might have caused this change in community structure. The second distinct cluster (from the Aegean Sea) is characterized by two distinct subclusters of samples from the North and the South Aegean Sea (Fig. 4 and 5). These two distinct subclusters reflect the hydrological and trophic differences between the two sampling sites (Table 1). Only one station in the North Aegean Sea (mnb3) showed chlorophyll a (chl a) concentrations (indicative of phytoplankton biomass) similar to those of the South Aegean Sea. These results indicate that under low-nutrient conditions (indicated here by low chl a concentrations), a different bacterial community exists than under the generally higher-nutrient conditions (indicated by higher chl a values) found in the North Aegean Sea. Another important factor which might be responsible for the separation between the sampling sites (North versus South Aegean Sea) is the influence of water from the Black Sea at the stations in the North Aegean Sea (26). The third distinct cluster identified with UPGMA, the samples collected in the Northern Adriatic Sea, showed a close relationship between the ro2 and ro11 samples, while the ro1 and ro3 samples were different, indicating fluctuations in community structure during the 1-week interval of sampling.
FIG. 5.
UPGMA dendrogram based on comparison of patterns from the upper mixed water column at different sampling sites. Designations: an, samples taken in the Middle Adriatic Sea off Ancona; msb, samples from the South Aegean Sea; mnb, samples from the North Aegean Sea; ro, samples taken in the Northern Adriatic Sea off Rovinj. Samples an4-1 and an4-2, as well as an60-1 and an60-2, are replicates from the same template. For samples msb1rt, mnb1rt, and mnb2rt, rRNA was reverse transcribed and used as the template in the PCR.
Metabolically active bacterial cells contain more rRNA than do dormant cells (7, 19). Therefore, rRNA has been suggested to be an indicator of the active bacterial community present, as indicated by the direct correlation found between rRNA content and the overall bacterial growth rate (4). We used the more sensitive T-RFLP technique to characterize complex marine bacterial communities on both the rDNA and the rRNA levels. The T-RFLP pattern of the community obtained when 16S rRNA (msb1rt) was used as a template was found to cluster within the samples taken from the South Aegean Sea but showed differences compared to the fingerprints obtained when 16S rDNA was used as a template (Fig. 5). The other 16S rRNA T-RFLP pattern (mnb1rt and mnb2rt) also differed from the corresponding rDNA T-RFLP pattern. Two rRNA samples expressed a higher number of OTUs (mnb1rt and mnb2rt), and one RNA sample (msb1rt) expressed an equal number of OTUs compared to the rDNA level (Table 1). A higher number of OTUs if rRNA is used as the template might be an indication of the presence of bacteria that are low in abundance but have a high rRNA content. The abundance of these bacteria seems to be too low to give an OTU signal on the rDNA level. Therefore, these active members of the bacterial community would escape characterization on the rDNA level. This also shows the high potential of the rRNA approach for detection of less abundant rRNA-containing bacteria in the environment. A similar conclusion was reached for sulfate-reducing bacterial populations by using DGGE (22).
Conclusions.
We have shown that T-RFLP fingerprinting has a slightly higher resolution than DGGE and is therefore useful for rapid and sensitive evaluation of the biogeographical distribution and relationships of bacterioplankton communities. Additionally, the rRNA approach as shown here in combination with T-RFLP fingerprinting is a first step toward a more sensitive characterization of bacteria on the rRNA level in the marine environment.
ACKNOWLEDGMENTS
We thank the captain and crew of the RV Aegaeo for their cooperation during sample collection and especially Tassos Tselepidis, Stella Psarra, Vivi Pitta, and Vassilis Zervakis for their help. Roberto Danovaro and Mauro Fabiano helped with the samples taken in the Middle Adriatic Sea, and for the samples from the Northern Adriatic Sea, the help of the staff from the Institute Ruder Boscovic at Rovinj, Croatia, was appreciated. We further thank Harry Witte for technical assistance and Hendrik Schaefer for critically reading a previous draft of the manuscript.
This research was supported by a grant from the European Union to G.J.H. (MAST-MTP II, MATER, no. MAS3-CT96-0051).
REFERENCES
- 1.Acinas S, Rodriguez-Valera F, Pedros-Alio C. Spatial and temporal variation in marine bacterioplankton diversity as shown by RFLP fingerprinting of PCR amplified 16S rDNA. FEMS Microbiol Ecol. 1997;24:27–40. [Google Scholar]
- 2.Avaniss-Aghajani E, Jones K, Chapman D, Brunk C. A molecular technique for identification of bacteria using small subunit ribosomal RNA sequences. BioTechniques. 1994;17:144–146. , 148–149. [PubMed] [Google Scholar]
- 3.Baba Y, Ishimaru N, Samata K, Tushako M. High-resolution separation of DNA restriction fragments by capillary electrophoresis in cellulose derivate solutions. J Chromatogr A. 1993;653:329–335. doi: 10.1016/0021-9673(93)83191-T. [DOI] [PubMed] [Google Scholar]
- 4.DeLong E F, Wickham G S, Pace N R. Phylogenetic stains: ribosomal RNA-based probes for the identification of single cells. Science. 1989;243:1360–1363. doi: 10.1126/science.2466341. [DOI] [PubMed] [Google Scholar]
- 5.Giovannoni S. The polymerase chain reaction. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. New York, N.Y: John Wiley & Sons, Inc.; 1991. pp. 177–204. [Google Scholar]
- 6.Lane D J. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. New York, N.Y: John Wiley & Sons, Inc.; 1991. pp. 115–176. [Google Scholar]
- 7.Lee S H, Kemp P F. Single-cell RNA content of natural marine planktonic bacteria measured by hybridization with multiple 16S rRNA-targeted fluorescent probes. Limnol Oceanogr. 1994;39:869–879. [Google Scholar]
- 8.Liesack W, Weyland H, Stackebrandt E. Potential risks of gene amplification by PCR as determined by 16S rDNA analysis of a mixed-culture of strict barophilic bacteria. Microb Ecol. 1991;21:191–198. doi: 10.1007/BF02539153. [DOI] [PubMed] [Google Scholar]
- 9.Liu W-T, Marsh T L, Cheng H, Forney L J. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl Environ Microbiol. 1997;63:4516–4522. doi: 10.1128/aem.63.11.4516-4522.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marsh T L, Liu W-T, Forney L J, Cheng H. Beginning a molecular analysis of eukaryal community in activated sludge. Water Sci Technol. 1998;37:455–460. [Google Scholar]
- 11.Martinez J, Smith D C, Steward G F, Azam F. Variability in ectohydrolytic enzyme activities of pelagic marine bacteria and its significance for substrate processing in the sea. Aquat Microb Ecol. 1996;10:223–230. [Google Scholar]
- 12.Moyer C L, Dobbs F C, Karl D M. Estimation of diversity and community structure through restriction fragment length polymorphism distribution analysis of bacterial 16S rRNA genes from a microbial mat at an active, hydrothermal vent system, Loihi Seamount, Hawaii. Appl Environ Microbiol. 1994;60:871–879. doi: 10.1128/aem.60.3.871-879.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moyer C L, Tiedge J M, Dobbs F C, Karl D M. A computer-simulated restriction fragment length polymorphism analysis of bacterial small-subunit rRNA genes: efficacy of selected tetrameric restriction enzymes for studies of microbial diversity in nature. Appl Environ Microbiol. 1996;62:2501–2507. doi: 10.1128/aem.62.7.2501-2507.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Müller-Niklas G, Schuster S, Kaltenböck E, Herndl G J. Organic content and bacterial metabolism in amorphous aggregations of the northern Adriatic Sea. Limnol Oceanogr. 1994;39:58–68. [Google Scholar]
- 15.Murray A E, Hollibaugh J T, Orrego C. Phylogenetic compositions of bacterioplankton from two California estuaries compared by denaturing gradient gel electrophoresis of 16S rDNA fragments. Appl Environ Microbiol. 1996;62:2676–2680. doi: 10.1128/aem.62.7.2676-2680.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muyzer G, DeWaal E C, Uitterlinden A G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muyzer G, Teske A, Wirsen C O, Jannasch H W. Phylogenetic relationships of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel electrophoresis of 16S rDNA fragments. Arch Microbiol. 1995;164:165–172. doi: 10.1007/BF02529967. [DOI] [PubMed] [Google Scholar]
- 18.Nei M, Li W-H. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA. 1979;76:5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poulsen L K, Ballard G, Stahl D A. Use of rRNA fluorescence in situ hybridization for measuring the activity of single cells in young and established biofilms. Appl Environ Microbiol. 1993;59:1354–1360. doi: 10.1128/aem.59.5.1354-1360.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmidt T M. Multiplicity of ribosomal RNA operons in prokaryotic genomes. In: De Bruijn F J, Lupski J R, Weinstock G M, editors. Bacterial genomes: physical structure and analysis. New York, N.Y: Chapman & Hall; 1997. pp. 221–229. [Google Scholar]
- 21.Suzuki M T, Giovannoni S J. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol. 1996;62:625–630. doi: 10.1128/aem.62.2.625-630.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Teske A, Wawer C, Muyzer G, Ramsing N B. Distribution of sulfate-reducing bacteria in a stratified fjord (Mariager Fjord, Denmark) as evaluated by most-probable-number counts and denaturing gradient gel electrophoresis of PCR-amplified ribosomal DNA fragments. Appl Environ Microbiol. 1996;62:1405–1415. doi: 10.1128/aem.62.4.1405-1415.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van der Maarel M J E C, Artz R R E, Haanstra R, Forney L J. Association of marine archaea with the digestive tracts of two marine fish species. Appl Environ Microbiol. 1998;64:2894–2898. doi: 10.1128/aem.64.8.2894-2898.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vaneechoutte M, Rossau R. Rapid identification of bacteria of the Comamonadaceae with amplified ribosomal DNA-restriction analysis (ARDRA) FEMS Microbiol Lett. 1992;93:227–234. doi: 10.1111/j.1574-6968.1992.tb05102.x. [DOI] [PubMed] [Google Scholar]
- 25.Weidner S, Arnold W, Puhler A. Diversity of uncultured microorganisms associated with the seagrass Halophila stipulacea estimated by restriction fragment length polymorphism analysis of PCR-amplified 16S rRNA genes. Appl Environ Microbiol. 1996;62:766–771. doi: 10.1128/aem.62.3.766-771.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zervakis, V., D. Georgopoulos, E. Papageorgiou, and V. Papadopoulos. Hydrological characteristics and circulation in the North Aegean throughout 1997–1998. Submitted for publication.