Abstract
Between April and December 1996, a serious outbreak of poliomyelitis occurred in Albania; almost 140 subjects were involved, and the episode presented an unusually high mortality rate (12%). During the outbreak, water samples from the Lana River in Tirana, Albania, and stool samples from two cases of paralytic poliomyelitis were collected and analyzed for the presence of polioviruses. Six polioviruses were isolated from the environmental and human samples, according to standard methods. All the samples were characterized by partial genomic sequencing of 330 bases across the 5′ untranslated region (5′-UTR) (nucleotide positions 200 to 530) and of 300 bases across the VP1 region (nucleotide positions 2474 to 2774). Comparison of these sequences with those present in data banks permitted the identification of environmental isolates Lana A and Lana B as, respectively, a Sabin-like type 2 poliovirus and an intertypic recombinant poliovirus (Sabin-like type 2/wild type 1), both bearing a G instead of an A at nucleotide position 481. The two other environmental polioviruses were similar to the isolates from the paralytic cases. They were characterized by a peculiar 5′-UTR and by a VP1 region showing 98% homology with the Albanian epidemic type 1 isolates reported by other authors. This study confirms the environmental circulation in Albania of recombinant poliovirus strains, likely sustained by a massive vaccination effort and by the presence in the environment of a type 1 poliovirus, as isolated from the Lana River in Tirana about 2 months before the first case of symptomatic acute flaccid paralysis was reported in this town.
The strategy for the global eradication of poliovirus requires an effective immunization program, an accurate environmental surveillance and assiduous epidemiological investigation program to monitor the circulation of wild and vaccine strains, and an increasing attention to aspects of policy, such as the recommendations of the World Health Organization’s International Office, in order to reduce the risk of adverse events following immunization (27). In 1988, the World Health Assembly projected the global eradication of poliomyelitis by the year 2000. Meanwhile, national immunization days were conducted in 65 countries in 1995, reaching 300 million children. Reported cases of poliomyelitis declined from 32,351 cases in 1988 to 7,024 in 1995 (6), and the complete eradication of the disease is now expected by 2005.
Despite improvements globally, the control of poliomyelitis in the Balkan area has been interrupted by bounded outbreaks, the most recent of which occurred in 1990 and 1991 in the Central Asian Republics and Central and Eastern Europe (the former Soviet Union) (22, 25). In 1996, a large outbreak occurred in Albania, resulting in 138 paralytic cases scattered throughout 27 districts of the country (10). This outbreak appears to be tied to a low level of immunity in the general population, poor personal hygiene in crowded families, and incorrect sewage disposal. More than 140,000 diarrheal diseases occur each year in Albania, and 10% of all the deaths of children under 5 years old are caused by diarrhea; at the moment, only 70% of the urban and 36% of the rural population of Albania has access to purified drinking water (26).
It has been demonstrated that inter- and intratypic recombination among different strains of poliovirus is frequent in vivo (5). The genetic variability affecting the RNA genome of the Sabin strains induces reversion towards those neurovirulent phenotypes mainly responsible for vaccine-associated paralytic poliomyelitis (VAPP), which is frequently associated with type 2 and type 3 strains of poliovirus and is seldom associated with type 1 viruses. Sabin poliovirus replication in the human gut could lead to reversions of the vaccine strains to pathogenic phenotypes by recombination during mixed in vivo infections (16). Events of recombination are relatively frequent for type 3 polioviruses, whereas types 2 and 1 are more stable (5, 12). Recently, Guillot et al. (14) have shown that several VAPP-causing strains classified as possible Sabin revertants were recombinations between wild-type and Sabin strains.
During the 1996 outbreak, personnel of the Hygiene Institute in Tirana had the opportunity to collect superficial water from the Lana River where it flows through Tirana. Four polioviruses were isolated from the environment, and two others were obtained from stool samples collected from people suffering from acute flaccid paralysis (AFP). Sequence analysis of the 330-bp amplicons originating from the 5′ untranslated region (5′-UTR) and from the 300-bp VP1 portion of the genomes classified the environmental isolate Lana B as an intertypic recombinant between the Sabin-like type 2 sequence tract furthest upstream and the wild-type type 1 sequence tract furthest downstream. The same method of sequence analysis for Lana A showed a 96% genetic similarity to the poliovirus type 2 Sabin strain (P712,ch,2ab), including the A→G mutation at nucleotide (nt) 481. The two other polioviruses (Lana C and D) and the two viruses isolated from human cases were similar, showing a VP1 region genetically linked to that of other type 1 Albanian isolates (10).
MATERIALS AND METHODS
Water sample.
Ten liters of superficial water from the Lana River in Tirana was collected and processed in two steps. The first step was performed in Albania. The sample was prefiltered through several layers of sterile gauze and ultraconcentrated by a Prep-scale ultrafiltration apparatus (Millipore) using a PTHL, 6-ft-2-in., 100,000-Da cutoff cartridge in polyethersulfone. The cartridge was washed with 5 liters of distilled water and preconditioned by filtering 500 ml of 3% beef extract, pH 7.0. The sample was concentrated at 1 lb/in2 income pressure, and the ultrafiltration stopped when the final volume of the sample reached 1.0 to 1.5 liters. The cartridge was completely emptied and washed with 100 to 120 ml of beef extract, pH 9.5. The ultraconcentrated sample and the washing buffer were collected together, the pH was adjusted to 7.0 to 7.2, and the mixture was frozen to be sent to Italy for the second step of processing. In Italy, the sample was reconcentrated by a similar ultrafiltration apparatus but using a smaller, PTHK, 1-ft-2-in., 100,000-Da cutoff cartridge in polyethersulfone. The sample was processed as before, and the final volume was less than 200 ml, including the 40 to 50 ml of washing buffer. The ultraconcentrated sample was reconcentrated by the polyethylene glycol 6000 precipitation, according to the method of Lewis and Metcalf (18), and the pellet was resuspended in 5 to 10 ml of sterile phosphate-buffered saline.
Poliovirus isolation by plaque assay.
Each sample was extracted twice with 30% chloroform, and the interface was extracted with 200 to 400 μl of cell culture media without fetal calf serum. The aqueous phases were collected together and added to a 20× solution of antibiotics (penicillin G, 100,000 U/ml; streptomycin, 120 mg/ml; kanamycin, 10 mg/ml; nystatin, 3.2 mg/ml) which was diluted at the time of the experiment. After 2 h at 37°C in a water bath, the sample was neutralized by using a mixture of polyclonal antibodies against all the known enteroviruses except for poliovirus (National Institute of Public Health and Environmental Hygiene, Bilthoven, The Netherlands) and kept for 2 h at 37°C in a water bath. An aliquot of the treated sample was used to infect a 2-day-old culture of Buffalo green monkey (BGM) cells, grown in a 90-mm-diameter petri dish. After 2 h at 37°C in a 5% CO2 atmosphere, the inoculum was discharged, and approximately 25 to 30 ml of minimum essential medium with Earle salts containing 2% fetal calf serum and 50% agar was added. The petri dishes were observed each day, and the single plaque was picked out and used to infect a 2-day-old monolayer growth in a 25-cm2 Falcon flask. When a complete cytopathic effect was observed, the flasks were frozen at −80°C. The presence of the virus was confirmed by a second passage on a 2-day-old monolayer of BGM cells, and individual viruses were classified as Lana A through D.
Virus isolation from patients.
Two polioviruses isolated from cases of AFP, collected in two distinct districts of Albania (Durres and Berat), were obtained directly from the Hygiene Institute in Tirana. The stool samples were collected in the first week after the patients were hospitalized, and the samples were passaged once on 2-day-old BGM cells. The cells were frozen at −80°C after a complete cytopathic effect was observed.
Genomic RNA extraction and RT-PCR test.
The supernatants of infected and mock-infected cells were clarified by low-speed centrifugation (1,400 × g for 15 min), and 100 μl of clarified supernatant was extracted by using a commercial kit based on the guanidinium thiocyanate method (Ultraspec; Bioteck). The RNA pellet was resuspended in 18 μl of nuclease-free water. Six microliters of RNA was combined with 4 μl of a universal buffer for reverse transcription (RT) (Promega), 1 μl of 50 mM random primer (Perkin-Elmer), 5 μl of water, and 0.8 μl of deoxynucleoside triphosphate (dNTP) at 100 mM (Promega), and the mixture was boiled at 95°C for 5 min and immediately placed in ice for at least 5 min. The RT step was performed by adding 7.4 μl of water, 20 U of RNasin (Promega), 4 U of avian leukosis virus reverse transcriptase (Promega), and water to a final volume of 20 μl. After 10 min at room temperature, the mixture was placed in a water bath at 37°C for 1 h. The first round of PCR was performed by using 5 μl of cDNA and 95 μl of a mixture containing 10 μl of 10× PCR buffer (Promega), 10 mM each dNTP (Promega), 4 U of Taq polymerase (Promega), 50 pmol each of sense and antisense primer, and water to a final volume of 100 μl per reaction. A nested PCR was performed for only the VP1 region in a 100-μl reaction volume by using 5 μl of the first PCR product in the same buffer as above but using internal sense and antisense primers. Negative and positive controls were included in each assay, and PCR products were electrophoresed through a 2% agarose gel in Tris-acetate-EDTA (TAE) buffer containing ethidium bromide. In the above PCRs, the cycles used were as follows: 2 min at 95°C; 30 cycles at 94°C for 1 min, 45°C for 1 min, and 72°C for 2 min; and a final elongation step at 72°C for 7 min. The primers used for the VP1 region were as follows (according to the nucleotide numbers of the wild-type poliovirus P1/Mahoney; accession no. V01149): external primers, nt 2402 to 2422 and nt 2881 to 2861 (3); internal primers, nt 2426 to 2446 and nt 2812 to 2792; and for 5′ noncoding region, nt 160 to 180 and nt 599 to 580.
Nucleotide sequence determination.
Before sequencing, PCR products were purified by using a QIAquick column purification kit (Genenco) according to the manufacturer’s instructions. DNA sequencing was performed by the cycle sequencing method (Amplicycle kit; Perkin-Elmer) and by automatic DNA sequencing (Applied Biosystem model 37OA; Perkin-Elmer). In the latter procedure, about 500 ng of PCR product DNA and 0.8 pmol of one of the same primers used in the amplification reaction were employed in each sequencing reaction. Both strands of the amplified fragments were sequenced to confirm the nature of the product obtained.
Data analysis.
Sequence data were analyzed and compared to the accessible sequence data banks by the Genetics Computer Group (7) and BLAST software packages (1). Multiple alignments were performed by the CLUSTAL V program (15). Sequence relatedness between the reference strains and our isolates was analyzed by distance-determining methods provided by the PHYLIP software package (9). The Expect values are also reported (an Expect value is a BLAST indicator determining the statistical significance of the number of alignments found and indicates the number of times one might expect to see such a match merely by chance). The lower the Expect value, the better is the match. The reference strains introduced into the analysis were as follows: gi/3334777-8-9/emb/AJ00796-7-8/POV7966-7-8, gi/61252/emb/V01149/POLIO1B (P1/Mahoney), gb/L76402/POL5UTRF-15/Hong Kong/81, gi/61257/emb/V01150/POLIOS1 (P1/Sabin), gi/61127/emb/X00595/PIPOLS2 (P2/Sabin, P712,ch,2ab), gi/61114/emb/X01076/PIPO3119 (P3/119), gi/332895/emb/K01392/POL3L37 (P3L37), and gb/L76413/POL5-UTRN-50/URSS/87.
RESULTS AND DISCUSSION
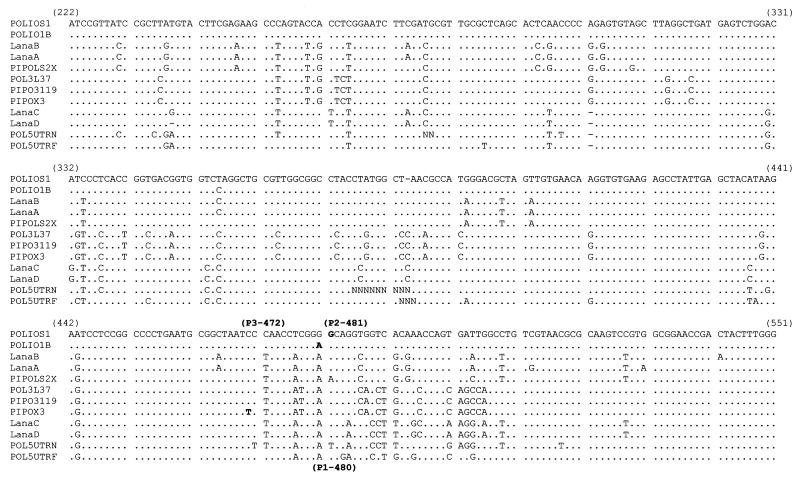
During the outbreak, in June 1996 and 2 months before the first case of AFP was detected in Tirana, water samples from the Lana River in Tirana were collected and partially concentrated at the Hygiene Institute of Tirana. The sampling point for all the withdrawals was the same. The preliminary assay for cultivable enteroviruses showed a viral concentration of 103 to 104 PFU/liter. It has to be emphasized that the Lana River collects not only superficial and rain waters but also untreated wastewater from Tirana and its suburbs. Furthermore, each year in Albania, more than 140,000 people are hospitalized with gastroenteritis. In the same period, the Hygiene Institute of Tirana sent us two unclassifiable polioviruses isolated from two cases of AFP. The first case involved a 22-year-old subject vaccinated with monovalent oral vaccine, and the second case involved a 17-year-old subject vaccinated with trivalent oral vaccine. Both subjects received several doses of vaccine. The extraction of isolated genomic RNAs was successfully achieved by using the Ultraspec kit (Bioteck), based on guanidium isothiocyanate. Intratypic differentiation of human and environmental polioviruses by restriction fragment length polymorphism assays was abandoned due to the difficulty of amplifying genome isolates in RT-PCRs driven by the primers previously described (11). Therefore, we reinforced the yield of amplicons by nested PCR by using another set of primers to amplify the same portion of the genome, extending from nt 2426 to 2812. Another amplicon of the genome region extending into the 5′-UTR from nt 160 to 599, produced in order to read nt 480, 481 and 472, indicated the presence of markers of attenuation for vaccine strains type 1, 2, and 3, respectively. Automated sequence analyses were performed according to standard methods. Figure 1 shows the sequence alignment of 5′-UTRs from wild-type, vaccine poliovirus reference strains and the corresponding regions of our isolates. Among the environmental isolates, Lana A showed a 96% genetic similarity to P712,ch,2ab with a mutation at nt 481 (A→G). The two other viruses isolated from the environment (Lana C and D) and the two viruses isolated from cases of AFP showed almost the same nucleotide sequence (98% homology). At the same time, when submitted to the accessible data banks, these viruses revealed 5′-UTR sequences peculiarly homologous for type 3 and type 1 AFP isolates. The isolate sequences showed the highest level of statistical significance (i.e., had the lowest Expect value as calculated by the BLAST program [17]) for the type 3 isolate gb/L76413/POL-5UTRN-50/URSS/87 (Expect value: e−128) and showed a lower statistical significance for the isolate gb/L76402/POL5UTRF-15/Hong Kong/81 (Expect value: e−113) (Fig. 1). For the next lowest score, we have to move downward to an Expect value of 8e−94, identified by other poliovirus serotypes. At nt 480 and 472, our isolates carried bases A and C, respectively, characteristic of wild-type strains 1 and 3. According to the sequence comparison of 5′-UTRs shown in Fig. 1, it is not possible to definitely link isolates Lana C, Lana D, AFP6, and AFP8 to an independent poliovirus strain or to link them with certainty to country-circulating strains, because of the exiguity of 5′-UTR sequences present in the accessible data banks despite the large number of isolates reported in the literature from the many epidemics which have occurred in the Central Asian Republics and Central and Eastern Europe. For conciseness, the 5′-UTR sequences of human isolates AFP6 and AFP8 are not reported in the sequence alignment shown in Fig. 1 because they were identical to isolates Lana C and Lana D, except for a T→C transition at nt 375 and an A→C transversion at nt 435. This identity between environmental and human isolates, despite their isolation from geographically distinct areas, indicates a common viral source.
FIG. 1.
Nucleotide differences among reference strains of all three serotypes and the isolates from this study. The reverted bases characteristic of each serotype are shown (boldface) and numbered (within parentheses) according to the relative reference numeration.
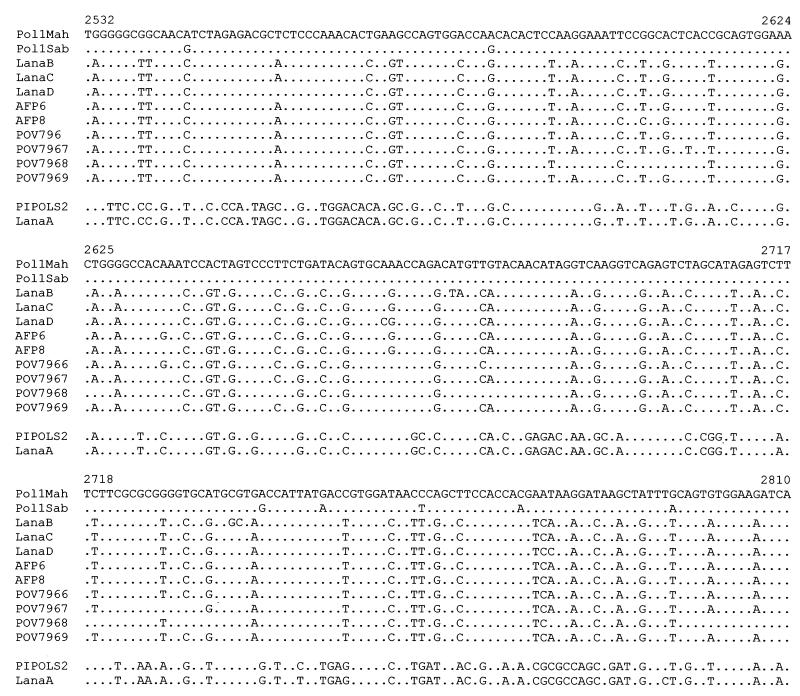
To further define our isolates at the molecular level, we compared the sequences of their 300-nt VP1 regions (nt 2474 to 2774) with those of other strains related to the Albanian outbreak (10) and with those of the wild and vaccine reference strains. In Fig. 2, the nucleotide alignment of the mentioned sequences shows 99% homology between Lana A and P712,ch,2ab with three nucleotide substitutions resulting in synonymous codons, thus yielding the total amino acid identity between the two sequences. The other environmental isolates, Lana B, Lana C, and Lana D, as well as the two human isolates, AFP6 and AFP8, showed 98% nucleotide homology (Expect value: e−154), with the type 1 POV796/7/8/9 (data bank entry). As shown in Fig. 2, the percentage of nucleotide homology between Lana B, C, and D, AFP6/8, and the wild, type 1 Mahoney reference strain consistently decreases to 80% (Expect value: 8e−23). Most of these nucleotide mutations resulted in synonymous codons. Therefore, seven amino acid changes (polyprotein amino acids: nt 599, A→S; nt 610, A→S; nt 614, T→A; nt 645, V→I; nt 673, P→S; nt 678, N→S; and nt 684, A→S) characterized 95% amino acid homology with the Mahoney reference strain. Lana B accumulated three further amino acid substitutions (nt 643, H→L; nt 664, C→W; and nt 665, V→L). In this regard, it has to be said that Lana B has been finally characterized as a recombinant Sabin-like type 2/wild-type 1 poliovirus (in 5′-UTR and VP1 regions, respectively) that presumably challenged the forces of selection in a such a way as to induce a genetic variation different than that of the original wild-type strain.
FIG. 2.
Alignment of 278-nt region (nt 2532 to 2810) coding for VP1. The comparison is with gi/61252/emb/V01149/POLIO1B, gi/61257/emb/V01150/POLIOS1, gi/3334777-8-9/emb/AJ00796-7-8/POV7966-7-8, and gi/61127/emb/X00595/PIPOLS2.
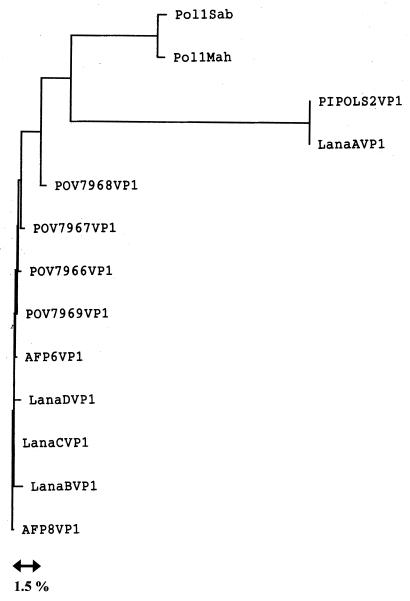
We also generated a dendrogram of the nucleotide sequence relationship between the isolates examined in this study (Fig. 3). The three branches represent essentially different genotypes that diverge by more than 15% and form groups composed of the type 2 Sabin strain, the environmental and human Albanian isolates, and poliovirus type 1 reference strains, respectively. Conversely, the high percentage of genetic identity within each cluster could account for a direct transmission linkage between different isolates.
FIG. 3.
Dendrogram showing sequence relationship among Albanian isolates and data bank reference strains. The tree is based on 278 nt of the VP1-coding region. Pol1Sab, poliovirus type 1 strain Sabin; Pol1Mah, poliovirus type 1 strain Mahoney; PIPOLS2VP1, poliovirus type 2 strain Sabin; POV7966, -7, -8, -9, human poliovirus cases from Albania (10); AFP6 and -8, human poliovirus cases (this study); LanaA, -B, -C, and -D, environmental poliovirus isolated from the Lana River (this study).
To summarize, for the environmental isolate Lana A, the sequence analysis of both the 5′-UTR and VP1 regions showed 96% genetic similarity to the poliovirus type 2 Sabin strain, including an A→G mutation at nt 481. Furthermore, the mutations observed in the VP1 region of Lana A were silent, producing a protein identical to that produced by reference strain P712,ch,2ab, thus undoubtedly characterizing it as a Sabin-like poliovirus. Interestingly, the characterization of the environmental isolate Lana B defined a recombinant genome showing a 5′-UTR region identical to that of P712,ch,2ab except for nt 481 (which was G, as in the wild type) and a portion of the VP1 sequence which showed 98% homology with the type 1 strains isolated from humans in Albania reported by Fiore et al. (10) and 80% homology with the Mahoney type 1 poliovirus. All the other isolates were strongly linked by their VP1 regions to the type 1 strains isolated in the Albanian epidemics by Fiore et al. (10). The 5′-UTRs of those isolates were characterized by a pattern of similarity with the poliovirus sequences gb/L76413/POL5-UTRN-50/URSS/87 and gb/L76402/POL5UTRF-15/Hong Kong/81 peculiar among the sequences contained in the accessible data banks.
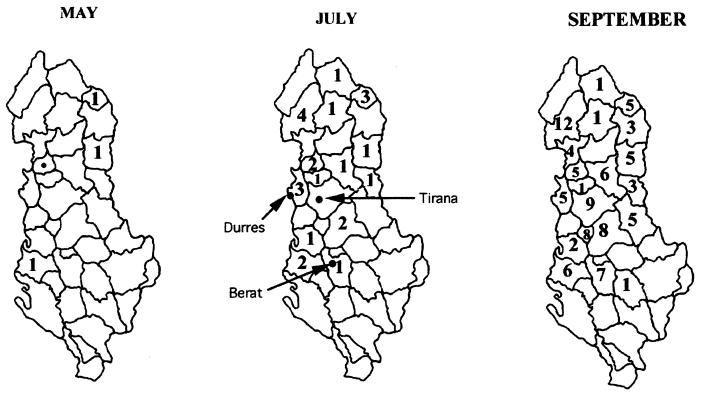
The outbreak of poliomyelitis started in May 1996 with the diagnosis of a 1-year-old child (VAPP; index case), 10 days after the first local, oral, vaccine dose. Almost 1 month later, three other human cases were identified in three different districts. A total of 138 cases were officially reported, and the mortality rate was unusually high: 12%. The paralytic poliomyelitis in Albania has been described in its epidemiological aspects by Prevots et al. (20). The index case was in the district of Lac, whereas the other three cases (in 2-, 29-, and 30-year-old subjects) were identified in three different districts located far away from the index case and each other. Albania is considered to be the poorest country in Europe, and, due to the present economic and health situations in that country, a constant massive movement of people from the rural and mountainous regions towards the coast is in evidence. In the rural areas, a high percentage (23.6%) of children are underweight in comparison with the children in urban areas (10.2%) (4). Consequently, the direction of migration is always from the country’s inner, mountainous regions towards the seaside of the country, which includes the country’s main cities. From an epidemiological point of view, the outbreak of poliomyelitis appears to have a multifocal origin, a view confirmed by the appearance in May of other cases of poliomyelitis in areas distant from the sites of the initial three cases. In July, the outbreak involved three large areas of the country, with a higher number of AFP diagnoses being made in the country’s northeast regions (mountain districts). During the following months, the three areas were confluent, and 27 of Albania’s 36 districts presented cases of AFP (Fig. 4). The cause of this outbreak was the low level of immunity of the people, as shown by Squarcione et al. (21), who found high levels of seronegativity towards one or more polioviruses in a large group of Albanian refugees. In particular, the seronegativity reached 40% for poliovirus type 3 and around 20% for poliovirus types 1 and 2. These data are in good agreement with the data reported from other developing countries. Several factors can determine this negative result: the low level of immunogenicity of the vaccine, the vaccination escape, the absence of a “cold chain” to avoid vaccine inactivation, the copresence of other enteric viruses in the gut, etc. Triki et al. (24) have documented that the only host-related factor of the antibody response toward the polio vaccine was the presence of other enteric viruses. In fact, infection with other enteric viruses can cause a state of diarrhea in which the mucosal structure of the infected person is altered, and, consequently, a more rapid clearance of the gastrointestinal tract and a reduced poliovirus immunization can occur (19). The mean age of the people suffering from AFP was 21, and this group included subjects vaccinated during the 1960s and 1970s with monovalent oral poliovirus vaccine by receiving just two doses of vaccine produced directly in Albania. Strebel et al. (22), analyzing cases of paralytic poliomyelitis in Romania, have shown that cases of VAPP were not influenced by a change in oral poliovirus vaccine manufacturers. Studies conducted in industrialized countries have shown 100% seroconversion in vaccinated infants, whereas the seroconversion rate is lower in developing countries, particularly for poliovirus types 3 and 1 (23). These data confirm those obtained by Green et al. (13), who found different levels of seroconversion in young adults involved in a 1988 outbreak in Israel. The appearance of different immunogenic strains of poliovirus in a country has been documented. Hovi et al. (16) described the appearance of a new variant of poliovirus type 3 which differed from the type 3 vaccine strains in both immunological and molecular characteristics; not much information about poliovirus strains in Albania is available, with the exception of strains isolated in the large outbreaks before the 1980s. From the early 1980s until 1995, only cases of VAPP have been reported, and no wild poliovirus has been isolated (8). Green et al. (13) have documented that the poliovirus involved in the 1988 outbreak in Israel was the descendant of a wild poliovirus present in Israel and Jordan since the early 1980s. In Albania, a complete surveillance system and maintenance of poliovaccine were organized by the World Health Organization only in the early 1990s.
FIG. 4.
Poliomyelitis case distribution in Albania. ●, index case; numbers, number of poliomyelitis cases reported in the district.
Our data confirm the data from Green et al. (13) showing the presence of wild poliovirus types 1 and 3 (gb/L76413/POL5-UTRN-50/URSS/87) in a particular area of Albania.
Almost 700,000 doses of trivalent oral vaccine were distributed from May to June 1996, ensuring a large circulation of attenuated poliovirus in a country where the presence of poliovirus in previous years could be suspected. The possible rearrangement of poliovirus with still-present wild types has been well documented by Cammack et al. (5), and such rearrangement has been reported in an outbreak in Finland (16). This rearrangement event also appears to be a possible cause of the epidemic in Albania. It would be advisable to elaborate new immunization strategies for isolated and not regularly checked populations. In order to reduce cases of VAPP, preference should be given to the use of inactivated poliovirus vaccine, alone or followed by the oral vaccine, as recommended by the American Academy of Pediatrics (2).
ACKNOWLEDGMENTS
This investigation was developed under the “Control of Diarrhoea Diseases including Cholera” emergency program founded by UNICEF and the Italian Ministry of Foreign Affairs, and this work was supported by the Community of St. Egidio nongovernmental organization.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped blast and PSI-Blast: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.American Academy of Pediatrics Committee on Infectious Diseases. Poliomyelitis prevention: revised recommendations for use of inactivated and live oral poliovirus vaccines. Pediatrics. 1999;103:171–172. doi: 10.1542/peds.103.1.171. [DOI] [PubMed] [Google Scholar]
- 3.Balanant J, Guillot S, Candrea A, Delpeyroux F, Crainic R. The natural genomic variability of poliovirus analyzed by a restriction fragment length polymorphism assay. Virology. 1991;184:645–654. doi: 10.1016/0042-6822(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 4.Buonomo E, Marazzi M C, Mancinelli S, Hoxha D, Cenko F, Palombi L. Infant nutritional and health status, feeding practices in rural and urban Albania. Ann Ig. 1998;10:163–171. [PubMed] [Google Scholar]
- 5.Cammack N, Phillis A, Dunn G, Patel V, Minor P D. Intertypic genomic rearrangement of poliovirus strains in vaccinees. Virology. 1988;167:507–514. [PubMed] [Google Scholar]
- 6.Cochi S L, Hull H F, Sutter R W, Wilfert C M, Katz S L. Commentary: the unfolding story of global poliomyelitis eradication. J Infect Dis. 1997;175(Suppl. 1):S1–S3. doi: 10.1093/infdis/175.supplement_1.s1. [DOI] [PubMed] [Google Scholar]
- 7.Devereaux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the Vax. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diamanti E, Ibrahimi B, Tafaj F, Mezini E, Dodbiba A, Dobi V, Catone S, Genovese D, Simeoni P, Fiore L. Surveillance of suspected poliomyelitis in Albania, 1980–1995: suggestion of increased risk of vaccine associated poliomyelitis. Vaccine. 1998;16:940–948. doi: 10.1016/s0264-410x(98)80025-x. [DOI] [PubMed] [Google Scholar]
- 9.Felsenstein J. Evolution trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- 10.Fiore L, Genovese D, Diamanti E, Catone S, Ridolfi B, Ibrahimi B, Konomi R, Van der Avoort H G A M, Hovi T, Crainic R, Simeoni P, Amato C. Antigenic and molecular characterization of wild type 1 poliovirus causing outbreaks of poliomyelitis in Albania and neighboring countries in 1996. J Clin Microbiol. 1998;36:1912–1918. doi: 10.1128/jcm.36.7.1912-1918.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Furione M, Guillot S, Otelea D, Balanant J, Candrea A, Crainic R. Poliovirus with natural recombination genomes isolated from vaccine-associated paralytic poliomyelitis. Virology. 1993;196:113–120. doi: 10.1006/viro.1993.1468. [DOI] [PubMed] [Google Scholar]
- 12.Georgescu M M, Delpeyroux F, Crainic R. Tripartite genome organization of a natural type 2 vaccine/nonvaccine recombinant poliovirus. J Gen Virol. 1995;76:2343–2348. doi: 10.1099/0022-1317-76-9-2343. [DOI] [PubMed] [Google Scholar]
- 13.Green M S, Handsher R, Cohen D, Melnik J L, Slepon R, Mendelsohn E, Danon Y L. Age differences in immunity against wild and vaccine strains of poliovirus prior to the 1988 outbreak in Israel and response to booster immunization. Vaccine. 1993;11:75–81. doi: 10.1016/0264-410x(93)90342-u. [DOI] [PubMed] [Google Scholar]
- 14.Guillot S, Caro V, Dahourou G, Cuervo N, Balanant J, Delpeyroux F, Crainic R. Looking for parents of the vaccine/wild (V/W) poliovirus recombinants. Presented at the 10th meeting of the European study group on the molecular biology of picornaviruses. Jena, Germany, 5 to 11 September 1998. 1998. [Google Scholar]
- 15.Higgins D G, Sharp P M. Clustal: a package for performing multiple sequence alignment on a microcomputer. Gene. 1988;73:237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- 16.Hovi T, Huovilainen A, Kuronen T, Poyry T, Salama N, Cantell K, Kinnunen E, Lapinleimu K, Roivainen M, Stenvik M, Silander A, Thoden C J, Salminen S, Weckstrom P. Outbreak of paralytic poliomyelitis in Finland: widespread circulation of antigenically alterated poliovirus type 3 in a vaccinated population. Lancet. 1986;21:1427–1432. doi: 10.1016/s0140-6736(86)91566-7. [DOI] [PubMed] [Google Scholar]
- 17.Karlin S, Altschul S F. Applications and statistics for multiple high-scoring segments in molecular sequences. Proc Natl Acad Sci USA. 1993;90:5873–5877. doi: 10.1073/pnas.90.12.5873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lewis G D, Metcalf T G. Polyethylene glycol precipitation for recovery of pathogenic viruses, including hepatitis A virus and human rotavirus from oyster, water, and sediment samples. Appl Environ Microbiol. 1988;54:1983–1988. doi: 10.1128/aem.54.8.1983-1988.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patriarca P A, Wright P F, Jacob J T. Factors affecting the immunogenicity of oral poliovirus vaccine in developing countries. Rev Infect Dis. 1991;13:926–939. doi: 10.1093/clinids/13.5.926. [DOI] [PubMed] [Google Scholar]
- 20.Prevots D R, Ciofi degli Atti M L, Sallabanda A, Diamanti E, Aylward R B, Kakariqqi E, Fiore L, Ylli A, Van der Avoort H, Sutter R W, Tozzi A E, Panei P, Schinaia N, Genovese D, Oblapenko G, Greco D, Wassilak S G F. Outbreak of paralytic poliomyelitis in Albania 1966: high attack rate among adults and apparent interruption of transmission following nationwide mass vaccination. Clin Infect Dis. 1998;26:419–425. doi: 10.1086/516312. [DOI] [PubMed] [Google Scholar]
- 21.Squarcione S, Germinario C, Iandolo E, Lo Caputo S, Bergamini F, Profeta M L, Greco D, Quarto M, Barbuti S. Seroimmunity to poliomyelitis in an Albanian immigrant population. Vaccine. 1992;10:853–856. doi: 10.1016/0264-410x(92)90049-p. [DOI] [PubMed] [Google Scholar]
- 22.Strebel P M, Aubert-Combiescu A, Ion-Nedelcu N, Biberi-Moroeanu S, Combiescu M, Sutter R W, Kew O M, Pallansch M A, Patriarca P A, Cochi S L. Paralytic poliomyelitis in Romania, 1984–1992. Evidence for a high risk of vaccine-associated disease and reintroduction of wild-virus infection. Am J Epidemiol. 1994;140:1111–1124. doi: 10.1093/oxfordjournals.aje.a117211. [DOI] [PubMed] [Google Scholar]
- 23.Sutter R W, Patriarca P A, Suleiman A J M, Pallansch M A, Zell E R, Malankar P G, Brogan S, Al-Ghassani A A K, El-Bualy M S. Paralytic poliomyelitis in Oman: association between regional differences in attack rate and variations in antibody responses to oral poliovirus vaccine. Int J Epidemiol. 1993;22:936–944. doi: 10.1093/ije/22.5.936. [DOI] [PubMed] [Google Scholar]
- 24.Triki H, Ould Mohamed M V, Ben Aissa R, Bouratbine A, Ben Ali Kamec M, Bouraoui S, Koubaa C, Zouari S, Mohsni E, Crainic R, Dellagi K. Influence of host related factors on the antibody response to trivalent oral poliovaccine in Tunisian infants. Vaccine. 1997;15:1123–1129. doi: 10.1016/s0264-410x(97)00001-7. [DOI] [PubMed] [Google Scholar]
- 25.World Health Organization. Expanded programme on immunization. Poliomyelitis outbreak, Bulgaria. Weekly Epidemiol Rec. 1992;67:336–337. [PubMed] [Google Scholar]
- 26.World Health Organization. European centre for environment and health: concern for Europe’s tomorrow. Stuttgart, Germany: World Health Organization; 1995. [Google Scholar]
- 27.World Health Organization. Surveillance of adverse events following immunisation. Geneva, Switzerland: World Health Organization; 1997. [Google Scholar]