Abstract
Background
Platinum based agents—cisplatin and carboplatin in combination with taxanes are used for the treatment of ovarian cancer (OC) patients. However, the majority of OC patients develop recurrent, platinum resistant disease that is uniformly fatal. Platinum treatment enriches for chemoresistant aldehyde dehydrogenase (ALDH) + ovarian cancer stem cells (OCSCs), which contribute to tumor recurrence and disease relapse. Acquired platinum resistance also includes metabolic reprograming and switching to oxidative phosphorylation (OXPHOS). Chemosensitive cells rely on glycolysis while chemoresistant cells have the ability to switch between glycolysis and OXPHOS, depending on which pathway drives a selective advantage for growth and chemoresistance. High expression of genes involved in OXPHOS and high production of mitochondrial ROS are characteristics of OCSCs, suggesting that OCSCs favor OXPHOS over glycolysis. Based on connections between OCSCs, chemoresistance and OXPHOS, we hypothesize that platinum treatment induces changes in metabolism that contribute to platinum-induced enrichment of OCSCs.
Methods
The effect of cisplatin on mitochondrial activity was assessed by JC1 staining and expression of OXPHOS genes by RT-qPCR. Cisplatin-induced changes in Sirtuin 1 (SIRT1) levels and activity were assessed by western blot. Small molecule inhibitors of mitochondrial complex I and SIRT1 were used to determine if their enzymatic activity contributes to the platinum-induced enrichment of OCSCs. The percentage of ALDH + OCSCs in OC cells and tumor tissue from xenograft models across different treatment conditions was analyzed using ALDEFLUOR assay and flow cytometry.
Results
We demonstrate that platinum treatment increases mitochondrial activity. Combined treatment of platinum agents and OXPHOS inhibitors blocks the platinum-induced enrichment of ALDH + OCSCs in vitro and in vivo. Furthermore, platinum treatment increases SIRT1 levels and subsequent deacetylase activity, which likely contributes to the increase in platinum-induced mitochondrial activity.
Conclusions
These findings on metabolic pathways altered by platinum-based chemotherapy have uncovered key targets that can be exploited therapeutically to block the platinum-induced enrichment of OCSCs, ultimately improving the survival of OC patients.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12967-022-03447-y.
Keywords: Ovarian cancer, Platinum, Chemoresistance, ALDH + cells, Cancer stem cells, OXPHOS, SIRT1
Background
Ovarian cancer (OC) is the 5th leading cause of cancer related deaths among women. The 5-year survival rate of OC patients has remained low for decades at 46% [1]. Surgical debulking followed by platinum and taxane-based chemotherapy are standard treatment modalities for OC patients [2]. The most common histologic sub-type of serous epithelial OC—high grade serous OC (HGSOC) initially responds well to platinum-based chemotherapy [3]. However, eventually patients with HGSOC relapse and develop a chemoresistant disease. Development of platinum resistance is the major obstacle in improving the survival of OC patients.
In the majority of solid tumors including OC, it has been firmly established that cancer stem cells (CSCs) are responsible for the development of chemoresistance and disease recurrence [4–6]. Aldehyde dehydrogenase (ALDH) is the most commonly used robust marker of CSCs [6], and ALDH + OCSCs have high tumor initiation capacity, enhanced ability to grow as spheroids and express high levels of stemness genes like BMI1, OCT4 and NOTCH3 [7, 8]. Furthermore, high expression and activity of the ALDH1A isoform strongly correlates with platinum resistant OC cells [8–10] and negatively correlates with survival of OC patients [11, 12]. Our group as well as others have demonstrated that treatment of OC cells with platinum-based agents results in enrichment of ALDH + OCSCs [7, 13, 14]. Collectively, the evidence suggests that platinum-induced enrichment of ALDH + OCSCs contributes to OC recurrence and relapse.
OCSCs are resistant to glucose deprivation and primarily rely on oxidative phosphorylation (OXPHOS) for energy requirements, which may also contribute to chemotherapy resistance [15–17]. Chemosensitive OC cells favor glycolysis while chemoresistant OC cells are able to switch between glycolysis and OXPHOS depending on which pathway provides selective advantage for growth and platinum resistance [16]. OCSCs exhibited high mitochondrial reactive oxygen species (ROS) production and high expression of enzymes involved in OXPHOS, implying that OCSCs preferentially utilize mitochondrial OXPHOS [15]. Lung and pancreatic CSCs also displayed higher membrane potential and lower glucose consumption rates compared to non-CSCs [18, 19]. Thus, there is strong evidence supporting altered metabolism as an important factor driving platinum resistance.
ALDH is a nicotinamide adenine dinucleotide (NAD +) dependent enzyme [20, 21]. We and others have demonstrated that platinum treatment results in an increase in expression of nicotinamide phosphoribosyltransferase (NAMPT)—a rate limiting enzyme in NAD + biosynthesis salvage pathway. The subsequent increase in cellular NAD + levels promotes ALDH + OCSC enrichment and contributes to platinum resistance [13, 14]. However, it is well-established that platinum resistance is a multifactorial phenomenon [22] and it is likely that additional metabolic pathways contribute to the enrichment of ALDH + OCSCs and chemoresistance.
In the present study, we examined the effect of platinum treatment of HGSOC cells on mitochondrial activity. We observed that mitochondrial membrane potential (∆ΨM) and expression of genes involved in mitochondrial OXPHOS increased 16 h after platinum treatment. Concomitantly, expression of genes in the glycolysis pathway was decreased by platinum treatment. Treatment of OC cells with OXPHOS inhibitors blocked the platinum-induced enrichment of ALDH + OCSCs, and increased deacetylase activity of sirtuin 1 (SIRT1) was required for platinum-induced enrichment of ALDH + OCSCs. In addition, we observed that SIRT1-regulated expression of mitochondrial transcription factor A (TFAM) likely contributed to the platinum mediated increase in mitochondrial activity. This first report demonstrating that OXPHOS inhibitors block platinum-induced enrichment of OCSCs supports further investigation into the role of mitochondrial OXPHOS in ovarian tumor recurrence.
Methods
Cell culture
HGSOC cell lines used in the study were maintained at 37 °C and 5% CO2 as described previously [23]. OVCAR5, OVCAR3 and OVSAHO cell lines were authenticated by ATCC in 2018. OVCAR5, OVCAR3 cells were cultured in DMEM 1X (Corning, #MT10013CV) and OVSAHO were cultured RPMI 1640 (Corning, #MT10040CV), containing 10% FBS (Gibco, #16000044) without antibiotics. All the cell lines used in the study were passaged less than 15 times. A 1.67 mM stock solution of cisplatin (Millipore Sigma, #232120) was made using 154 mM NaCl (Macron Fine chemicals, #7581-12) in water. All the HGSOC cell lines used in the study were treated with respective IC50 doses of cisplatin determined previously by Haley et al. using a 24 h treatment followed by a 72 h recovery period (OVCAR5 12 µM; OVSAHO 4 µM; OVCAR3 15 µM) [24]. Stock solutions of Rotenone (Sigma, #R8875-1G; 100 mM), IACS—010759—OXPHOS Inhibitor (MedChemExpress, #HY-112037; 10 mM), oligomycin (SelleckChem, #S1478; 10 mM), and SIRT1 inhibitor Ex-527 (Sigma, E7034; 10 mM) were made in DMSO. For all the experiments using these inhibitors, an equivalent amount of DMSO or inhibitors were added along with cisplatin and cells were incubated for 16 h at 37 °C and 5% CO2. All the treatment doses are specified in the figure legends.
JC-1 staining
OVCAR5 (1 × 106) and OVSAHO cells (1.5 × 106) were cultured in 100 mM plates for 24 h and treated with respective IC50 doses of cisplatin (OVCAR5 12 µM; OVSAHO 4 µM [24]) for 16 h. JC-1 staining was performed as per the manufacturer’s protocol (Thermo Fisher, #T3168). Briefly, cells were collected and washed with PBS and then resuspended in 1 ml PBS. Stock solutions of JC-1 were made in DMSO at a concentration of 5 mg/ml. Then JC-1 stain was added to cells at a final concentration of 2 µg/ml and cells were incubated at 37 °C, 5% CO2 for 30 min. Following incubation, the cells were filtered through 30 µm filters and analyzed by flow cytometry. For all the JC-staining experiments a no stain sample was used for compensation control. Analysis of JC-1 staining was performed using Flow Jo software (Becton, Dickinson & Company, RRID: SCR_008520). The mean red and green intensity of all cells in each biological replicate was determined using Flow Jo software. Then the ratio of red to green intensity was calculated for each biological replicate and the mean of ratio of red to green intensities of all 3 biological replicates was plotted in the graphs.
RNA isolation and reverse transcriptase—quantitative PCR (RT-qPCR)
Isolation of total RNA from cell pellets was performed using RNAeasy mini kit (Qiagen, #74104) as per the manufacturer’s protocol. Maxima first strand cDNA synthesis kit (Thermo Fisher, #K1642) for quantitative reverse transcription PCR was used to synthesize cDNA. FastStart Essential DNA green master (Roche, #06402712001) was used to perform RT-qPCR. Primers sequences of all candidate genes and their respective efficiencies are in Additional file 2: Table S1. Relative gene expression was calculated using the Pfaffl method to account for differences in primer efficiencies between the primers for the genes of interest and the housekeeping gene Actin B.
ALDEFLUOR assay
OVCAR5 (1.0 × 105) and OVSAHO (1.5 × 106) were cultured in 100 mm dishes. Approximately 24 h after plating, cells were treated with cisplatin alone or in combination with DMSO or appropriate inhibitors and incubated at 37 °C, 5% CO2 for 16 h. After the incubation time ALDEFLUOR assay (Stem Cell Technologies, #01700) was performed as described previously [14].
Flow cytometry
LSR II Flow cytometer (BD Biosciences) was used for the analysis of both the JC-1 and ALDEFLUOR assay. JC1 J-monomers and J-aggregates was measured using 488 nm excitation and signal was detected using AF488 (green) and PE-A (red), respectively. Samples with no stain were analyzed for every experiment for compensation control. For all the ALDEFLUOR assays, 488 nm excitation was used, and the signal was detected using 530/30 filter to measure ALDH activity. To determine the percentage of ALDH positive cells in different conditions, the sample specific DEAB negative control was used. All the data analysis was done using FlowJo software (Becton, Dickinson & Company).
Generation of stable knockdown lines
For SIRT1 knockdown (Sigma, NM_012238, TRCN0000018981, TRCN0000018983) and empty vector (EV) TRC1, the lentiviral shRNA knockdown protocol from the RNAi consortium of the Broad Institute was used as described previously [23]. Briefly, OVCAR5 cells were transduced with appropriate amount of concentrated virus after titration along with 8 μg/ml polybrene. The following day media was changed and media containing 2.5 μg/ml puromycin was added for 3 days to select for infected cells. Approximately 3 days later cells were split and used for experiments.
Mitochondrial DNA (mtDNA) content analysis
1 × 106 OVCAR5 cells were plated and incubated at 37 °C and 5% CO2 for approximately 24 h. Then cells were treated with the IC50 dose of cisplatin (12 μM for OVCAR5). After 16 h treatment, cells were collected, DNA was isolated using DNAeasy blood and tissue kit (Qiagen, 69504) and quantified using a nanodrop. Mitochondrial DNA content relative to a nuclear DNA was quantified by qPCR and the Delta Delta Ct using primers that amplify mitochondrial DNA regions (NADH dehydrogenase sub-unit 1 and NADH dehydrogenase sub-unit 5) and a nuclear DNA (nDNA) region (β2—macroglobulin) as described previously [25–27]. See Additional file 2: Table S2 for primers.
Xenograft studies
All the animal studies were performed in accordance with the Association for Assessment and Accreditation of Laboratory Animal Care International and approved by Indiana University Bloomington Institutional Animal Care and Use Committee as described previously [14]. A power analysis performed using data from a pilot experiment indicated that 4 mice per group would provide 80% power to detect a difference in %ALDH + cells at p < 0.05. 2 × 106 OVCAR3 cells were injected subcutaneously in the flanks of NSG mice (IUSCCC In Vivo Therapeutics Core). Once the established tumors reached > 100 mm3, they were randomized into 3 different treatment groups and treated with vehicle alone or combination of carboplatin and vehicle or IACS-010759 (OXPHOS inhibitor). Carboplatin was administered once a week intraperitoneally at 50 mg/kg for 3 weeks. IACS-010759 was prepared in 10% DMSO and 90% corn oil. IACS-010759 was administered orally at 7.5 mg/kg 5 days a week for 3 weeks. At the end of the treatment schedule, mice were sacrificed and tumors were collected, disassociated and used for ALDEFLUOR assay as we have done previously [7, 14]. Tumor dissociation kit and gentle MACS dissociator (Miltenyi Biotech) was used to dissociate the tumors into single cells as per manufacturer’s protocol.
Correlation analysis using cBioportal
Correlation analysis of expression of SIRT1 and TFAM was performed using the cBioportal [28, 29] and RNA-seq data of ovarian cancer samples from The Cancer Genome Atlas [30].
Spheroid formation assay
1.5 × 103 cells pre-treated with DMSO or IACS alone or in combination with cisplatin (cisplatin, 6 μM; IACS—010,759, 1 μM) for 3 h were plated in 24-well low attachment plates (Corning, 3473) in stem cell media [DMEM-F12 (Corning, #10-017-CV) with 100 U Penicillin–Streptomycin, 0.4% BSA, 10 ng/ml bFGF (Invitrogen, #13256-029), 20 ng/ml EGF (Biolegend, 585,506), 5 μg Insulin (Sigma, 19278)]. The plates were incubated for 14 days at 37 °C, 5% CO2. Fresh stem cell media was supplemented every 3 days and the spheroids were imaged at 14 days. The percent area of spheroids in each well was calculated using ImageJ for each biological replicate and the graph was plotted using all the three replicates.
Antibodies
For western blot, the following antibodies were used: anti-pATMS1981 (Cell Signaling Technology (CST), #13050, 1:1000), anti-AcH4K16 (CST, #13534, 1:1000), anti-total H2A (CST, #12349, 1:1000), anti-SIRT1 (Santa Cruz, sc-74465, 1:1000), anti-HIF1a (CST, #36169,1:1000), anti-Myc (Abcam, ab32072,1:1000), anti-actin B (CST, #4970) and anti-vimentin (CST, #5741).
Statistics
The percentage of ALDH + cells across different conditions after ALDEFLUOR assay and percent area of spheroid assay was evaluated by one-way ANOVA with multiple comparisons using Graphpad prism and data is presented as ± SEM. Data of JC-1 staining, RT-qPCR and densitometry were evaluated by Student t test in Graphpad prism and excel.
Results
Platinum treatment increases mitochondrial OXPHOS activity
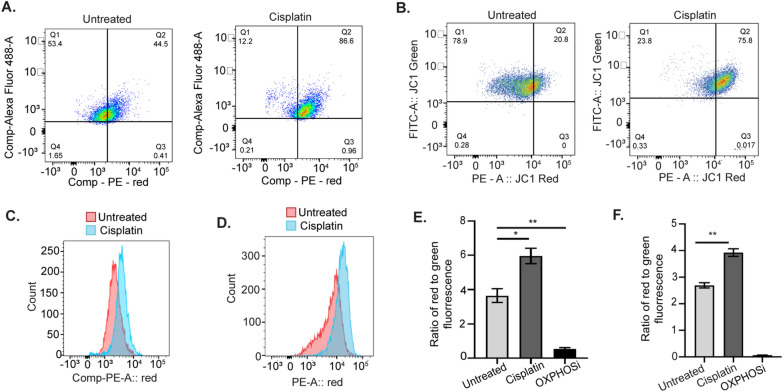
Development of platinum resistance in OC has been linked to increased reliance of cancer cells on OXPHOS [31]. Therefore, first we determined if mitochondrial membrane potential (∆ΨM) [32] increased after treatment of OC cells with cisplatin. To measure ∆ΨM, a lipophilic, cationic dye called JC-1 was used. JC-1 enters the mitochondria of healthy cells with normal ∆ΨM and spontaneously aggregates as red fluorescent J-aggregates; in contrast, in cells with disrupted ∆ΨM, JC-1 will not form aggregates and retains its green fluorescence [32]. Treatment of HGSOC cells—OVCAR5, OVSAHO with their respective IC50 doses of cisplatin for 16 h followed by JC-1 staining resulted in a shift in the population of cells from low red fluorescence to high red fluorescence (Fig. 1A, B, Additional file 1: Figure S1A), indicating an increase in mitochondrial J-aggregates after cisplatin treatment. A histogram of the red fluorescence for JC-1 staining in untreated and cisplatin treated OVCAR5 and OVSAHO cells confirmed that cisplatin treated cells shifted towards higher red fluorescence (Fig. 1C, D). Furthermore, the ratio of red to green fluorescence significantly increased after cisplatin treatment of both OVCAR5 and OVSAHO cells (Fig. 1E, F). Carbonyl cyanide m-chlorophenyl hydrazone (CCCP—OXPHOSi), which is known to reduce ∆ΨM [33], was used as an assay control and reduced the ratio of red to green fluorescence as expected (Fig. 1E, F). Treatment of OVCAR5 cells with cisplatin for 16 h had no effect on the mitochondrial DNA (mtDNA) content relative to nuclear DNA content (Additional file 1: Figure S1B), further supporting that the cisplatin-induced increase in OXPHOS was due to an increase in mitochondrial activity, not an increase in mitochondrial number.
Fig. 1.
Mitochondrial OXPHOS activity increases in response to cisplatin treatment in OC cells. Scatter plots of OVCAR5 (A) and OVSAHO (B) cells untreated or treated with 12 µM and 4 µM cisplatin, respectively, for 16 h followed by staining with JC-1 for 30 min and analysis by flow cytometry. Histogram of untreated and cisplatin treated OVCAR5 (C) and OVSAHO (D) cells after JC-1 staining as in A and B. Ratio of red to green fluorescence intensity in untreated and cisplatin treated OVCAR5 (E) and OVSAHO (F) cells after JC-1 staining as in A and B. The OXPHOS inhibitor carbonyl cyanide 3-chlorophenylhydrazone (OXPHOSi) was used as an assay control. Graphs display mean ratio of red to green fluorescence ± SEM (N = 3). Student t-test was used to calculate statistical significance. For all untreated versus cisplatin treated, P values *< 0.05, **< 0.005 and ***< 0.0005
Cisplatin treatment induces OXPHOS gene expression
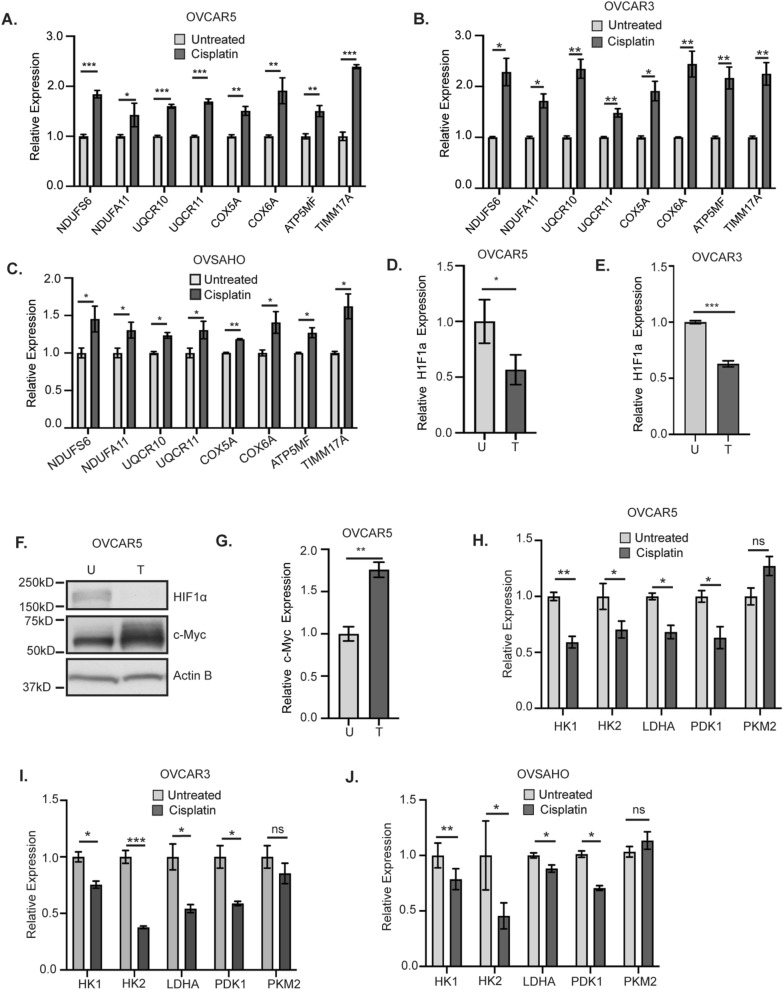
To determine if cisplatin treatment increased expression of genes of mitochondrial OXPHOS complexes, we performed RT-qPCR of untreated and 16 h cisplatin-treated cells. Increased expression of Complex I subunits—NADH ubiquinone oxidoreductase subunits S6, A11 (NDUFS6, NDUFA11); Complex III subunits—ubiquinol-cytochrome C reductase subunit X, XI (UQCR10, UQCR11); Cytochrome C oxidase complex subunits 5A, 6A (COX5A, COX6A); ATP synthase membrane subunit F (ATP5MF) and translocase of the inner mitochondrial membrane (TIMM17A) was observed in OVCAR5, OVCAR3, and OVSAHO cells after cisplatin treatment (Fig. 2A–C). Hypoxia-inducible factor 1 (HIF-1) is known to inhibit mitochondrial biogenesis and respiration by negatively regulating the activity of transcription factor c-Myc [34, 35]. Treatment of OVCAR5 and OVCAR3 cells with cisplatin for 16 h reduced the expression and protein levels of HIF1α (Fig. 2D–F, Additional file 1: Figure S2A). Additionally, cisplatin treatment increased c-Myc expression and protein levels in OVCAR5 cells (Fig. 2F, G) but not in OVCAR3 cells (Additional file 1: Figure S2A, B). HIF-1 regulates the expression of glycolytic genes like lactate dehydrogenase A (LDHA), Pyruvate dehydrogenase kinase (PDK1) and Pyruvate kinase isozyme M2 (PKM2) [36, 37]. We also examined the expression of hexokinase genes hexokinase 1 (HK1) and hexokinase 2 (HK2), which are involved in the first step in glycolysis [38]. Treatment of OVCAR5, OVCAR3, and OVSAHO cells with cisplatin for 16 h decreased expression of HK1, HK2, LDHA, and PDK1 but not PKM2 (Fig. 2H–J). Altogether, our findings suggest that platinum treatment increased ∆ΨM, increased expression of genes related to OXPHOS and decreased expression of genes involved in glycolysis.
Fig. 2.
Expression of genes involved in mitochondrial OXPHOS increases after cisplatin treatment. Gene expression of the indicated genes by RT-qPCR in OVCAR5 (A) OVCAR3 (B) and OVSAHO (C) cells untreated or treated with 12 µM, 15 µM or 4 µM cisplatin, respectively, for 16 h. Expression of HIF-1α in OVCAR5 (D) and OVCAR3 (E) cells treated as in A and C. F OVCAR5 cells were untreated (U) or treated with 12 µM cisplatin for 16 h (T). Cell lysates were collected and analyzed for indicated proteins by western blot. G c-Myc expression in untreated (U) or 12 µM cisplatin treated (T) OVCAR5 cells. Expression of the indicated glycolysis genes in OVCAR5 (H), OVCAR3 (I) and OVSAHO cells (J) treated as in A, B and C, respectively. Graphs display mean fold change ± SEM relative to untreated. Expression of all the genes was normalized to the house keeping gene Actin B. For all untreated versus cisplatin treated, P values *< 0.05, **< 0.005 and ***< 0.0005
OXPHOS is essential for the platinum-induced enrichment of ALDH + cells
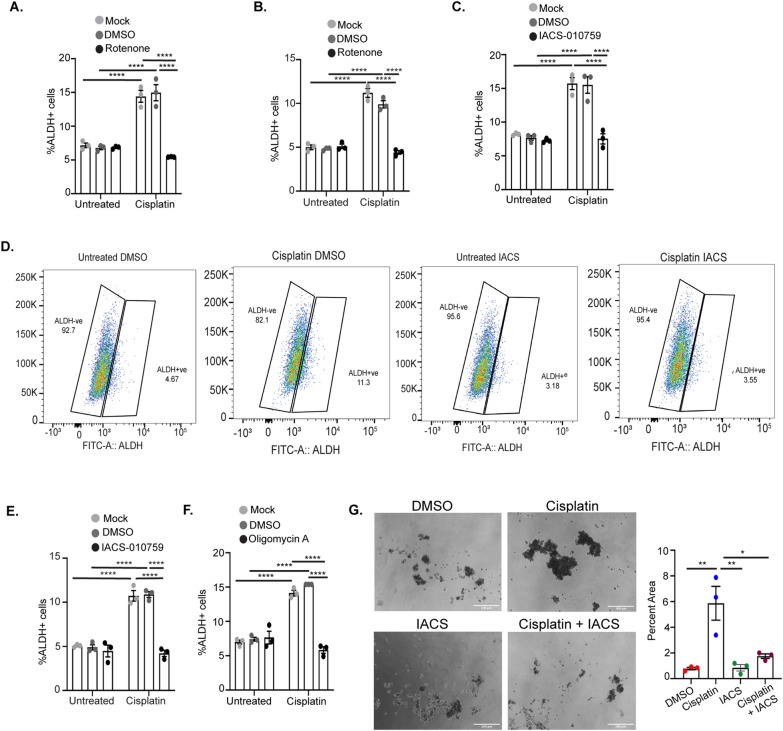
Platinum treatment has been previously demonstrated to enrich the ALDH + OCSC population [7, 13, 14]. To determine if the increase in OXPHOS in response to cisplatin treatment is essential for the platinum-induced enrichment of ALDH + cells, we treated OVCAR5 and OVSAHO cells with cisplatin alone or in combination with 5 µM Rotenone, a mitochondrial complex I inhibitor, or DMSO and performed the ALDEFLUOR assay. As expected, treatment of OVCAR5 and OVSAHO cells with cisplatin alone or in combination with DMSO increased the percentage of ALDH + OCSCs (Fig. 3A, B). Rotenone treatment blocked the platinum-induced increase in %ALDH + OCSCs without changing the basal percentage of ALDH + cells. IACS—010759 is a novel small molecule OXPHOS inhibitor, which like rotenone also inhibits complex I of the mitochondrial respiratory electron transport chain [39]. Treatment of OVCAR5 and OVSAHO cells with 1 µM IACS-010759 in combination with cisplatin abrogated the platinum-induced enrichment of ALDH + cells (Fig. 3C–E; Additional file 1: Figure S3A–C). IACS-010759 treatment did not affect the platinum-induced change in the expression of OXPHOS genes, c-Myc or HIF1α in OVCAR5 cells (Additional file 1: Figure S4A, B). Oligomycin inhibits the ATP synthase complex in the mitochondrial respiratory electron transport chain [40], and combination treatment of 1 µM oligomycin plus cisplatin blocked the platinum-induced increase in ALDH + cells (Fig. 3F). Several studies have established that cancer stem cells have the ability to grow in anchorage independent conditions forming spheroids [41, 42]. As previously demonstrated by our group, cisplatin treatment of OVCAR5 cells increased their ability to form spheroids under anchorage independent conditions compared to DMSO treated controls (Fig. 3G) [7]. Furthermore, treatment of OVCAR5 cells with IACS-010759 in combination with cisplatin blocked the cisplatin-induced increase in spheroid formation (Fig. 3G). Altogether, these data suggest that the increase in mitochondrial activity observed after cisplatin treatment is required for the platinum-induced enrichment of ALDH + OCSCs.
Fig. 3.
Mitochondrial OXPHOS inhibitors in combination with cisplatin block the platinum-induced increase in percent ALDH + cells. Percent ALDH + OVCAR5 (A) and OVSAHO cells (B) untreated or treated with 12 µM or 4 µM cisplatin, respectively, alone or in combination with DMSO or 5 µM Rotenone for 16 h followed by ALDEFLUOR assay. Percentage ALDH + OVCAR5 (C) and OVSAHO (D, E) cells treated with cisplatin alone or in combination with DMSO or 1 µM IACS-010759 for 16 h followed by ALDEFLUOR assay. D Shows gates used to determine ALDH + cells for one biological replicate of OVSAHO cells treated with DMSO or IACS in combination with cisplatin. F Percent ALDH + OVCAR5 cells treated with cisplatin alone or in combination with DMSO or 1 µM oligomycin for 16 h followed by ALDEFLUOR assay. Graphs display mean ± SEM percent ALDH + cells in N = 3 biological replicates. G Spheroid formation assay in OVCAR5 cells pre-treated for 3 h with DMSO or 1 μM IACS-010759 alone or in combination with 6 μM cisplatin and cultured for 14 days. For all untreated versus cisplatin treated, P values *< 0.05, **< 0.005, ***< 0.0005, ****< 0.0001
SIRT1 contributes to the platinum-induced enrichment of ALDH + cells
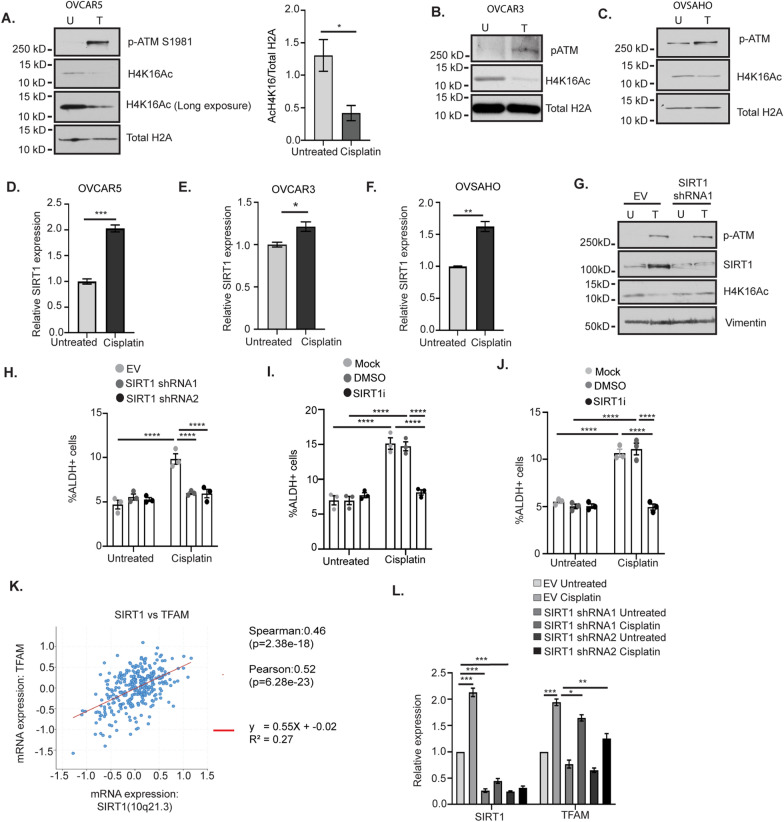
SIRT1, a NAD + -dependent deacetylase of acetylated histone 4 lysine 16 (H4K16Ac) and non-histone proteins, has been implicated in the regulation of metabolic processes [43, 44]. Cellular NAD + levels increase and regulate the deacetylase activity of SIRT1 after platinum treatment [13, 14, 45]. Therefore, we hypothesized that SIRT1 activity contributes to the observed cisplatin-induced metabolic changes that result in ALDH + OCSC enrichment. Cisplatin treatment for 16 h reduced the levels of H4K16Ac relative to untreated in OVCAR5, OVCAR3 and OVSAHO cells (Fig. 4A–C). Phosphorylation of ATM (p-ATM) at serine 1981 was used as a control for platinum-induced DNA damage response and we observed an expected increase in pATM after platinum treatment in all cell lines (Fig. 4A–C). Treatment of OVCAR5, OVCAR3, and OVSAHO cells with the respective IC50 doses of cisplatin for 16 h also resulted in increased SIRT1 mRNA levels (Fig. 4D–F) and increased SIRT1 protein levels in OVCAR5 cells (Fig. 4G). Furthermore, SIRT1 knockdown (KD) using shRNA rescued the platinum-induced decrease in H4K16Ac (Fig. 4G), indicating that SIRT1 likely deacetylates H4K16 in response to cisplatin treatment.
Fig. 4.
Platinum-induced increase in SIRT1 activity contributes to the enrichment of ALDH + cells. OVCAR5 (A), OVCAR3 (B) and OVSAHO (C) were untreated (U) or treated with respective IC50 dose of cisplatin (T) for 16 h (OVCAR5—12 μM, OVCAR3—15 μM and OVSAHO—4 μM). Cell lysates were collected and analyzed by western blot. A Graph displays mean ± SEM densitometric analysis of N = 3 biological replicates of H4K16ac relative to total H2A. Relative expression of SIRT1 in OVCAR5 (D), OVCAR3 (E) and OVSAHO (F) cells treated as in A, B and C, respectively. G OVCAR5 cells infected with empty vector (EV) or SIRT1 viral shRNA followed by treatment as in A. Lysates were collected and analyzed by western blot for the indicated proteins. H OVCAR5 cells infected EV or SIRT1 viral shRNA, untreated or treated as in A followed by ALDEFLUOR assay. Graph shows mean ± SEM percent ALDH + cells in N = 3 biological replicates. Percent ALDH + OVCAR5 (I) and OVSAHO (J) cells treated with cisplatin alone or in combination with DMSO or 3 µM Ex-527 for 16 h followed by ALDEFLUOR assay. Graph depicts mean ± SEM percent ALDH + cells in N = 3 biological replicates. (K) Correlation analysis of SIRT1 and TFAM expression using RNA-seq data from TCGA OC patient dataset. L Relative expression of SIRT1 and TFAM in OVCAR5 cells infected with EV or SIRT1 viral shRNA and treated as in A. Graph displays mean relative expression ± SEM to the untreated. Gene expression was normalized to the house keeping gene Actin B. For all comparisons, P—*< 0.05, **< 0.005, ***< 0.0005, ****< 0.0001
Next, to determine if SIRT1 plays a role in the platinum-induced enrichment of OCSCs, we combined SIRT1 KD or inhibition with platinum treatment. SIRT1 KD using two different shRNAs in OVCAR5 cells followed by cisplatin treatment blocked the platinum-induced increase in ALDH + cells without altering the baseline percentage of ALDH + cells (Fig. 4H). Additionally, inhibiting SIRT1 by treating cells with the SIRT1 inhibitor Ex-527 (3 µM) abrogated the platinum-induced increase in ALDH + cells in both OVCAR5 and OVSAHO cells (Fig. 4I, J).
SIRT1 has been shown to promote OXPHOS and mitochondrial biogenesis in adipocytes and hepatocellular carcinoma cells by regulating the expression of mitochondrial transcription factor A (TFAM) [46–48]. TFAM is important for proper transcription of mitochondrial and OXPHOS genes [49, 50]. To test the hypothesis that SIRT1 promoted the platinum-induced increase in ALDH + cells by promoting the increase in mitochondrial OXPHOS, we first analyzed the correlation between SIRT1 and TFAM expression in publicly available RNA-sequencing datasets from OC patients [30]. SIRT1 expression was positively correlated with TFAM expression (Fig. 4K). Additionally, treatment of OVCAR5 and OVCAR3 cells with cisplatin increased expression of TFAM (Fig. 4L, Additional file 1: Figure S4C). Next, we determined if the expression of TFAM is dependent on SIRT1 in response to cisplatin treatment in OC cells. SIRT1 KD using two different shRNAs resulted in a modest but significant reduction in the platinum-induced increase in TFAM expression (Fig. 4L). Altogether, these data suggest that SIRT1 activity plays a key role in platinum-induced enrichment of ALDH + cells in OC.
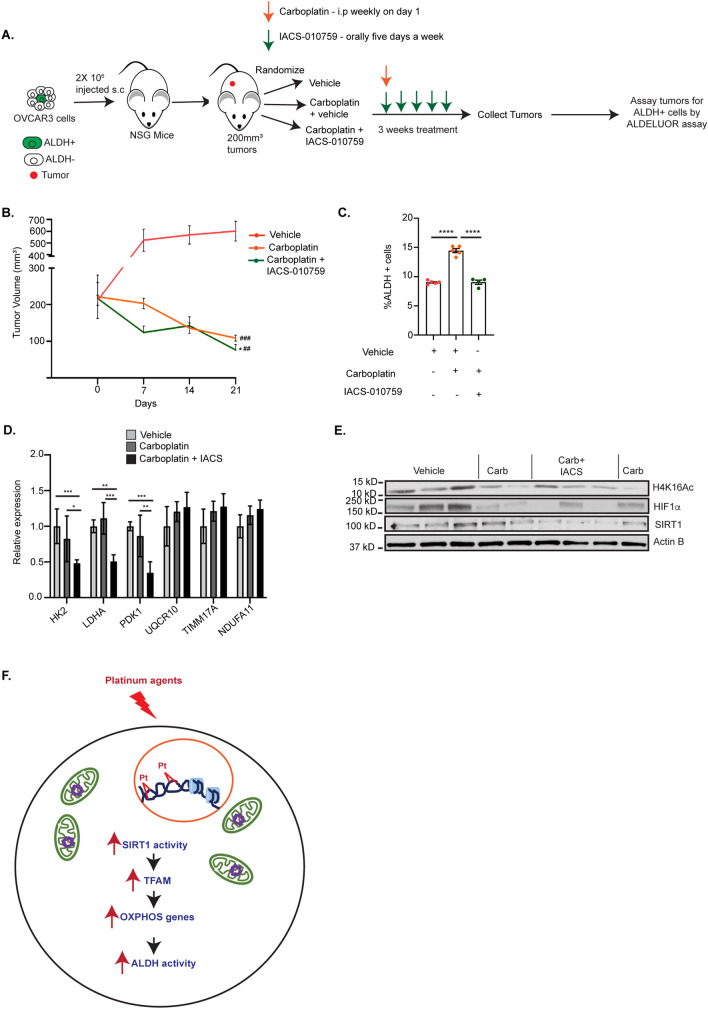
OXPHOS inhibition blocks the platinum-induced enrichment of ALDH + cells in vivo
We and others have demonstrated that platinum treatment results in enrichment of ALDH + OCSCs in vitro and in vivo [7, 13, 51, 52]. Furthermore, we demonstrated that treatment of OC cells with OXPHOS inhibitors in combination with cisplatin blocked the platinum-induced enrichment of ALDH + cells in vitro without affecting the basal ALDH percentage (Fig. 3A–F). Therefore, it was of interest to determine if OXPHOS inhibition blocks the platinum mediated increase in ALDH + OCSCs in vivo in mouse xenografts. As shown in Fig. 5A, 2 million OVCAR3 cells were injected subcutaneously in the flanks of NSG mice and after establishment of tumors, randomized mice received vehicle alone or a combination of carboplatin + vehicle or OXPHOS inhibitor IACS-010759. As expected, platinum treatment significantly reduced the tumor volume compared to the vehicle only group (Fig. 5B). In the group treated with carboplatin + IACS-010759, tumor volume was less than the group treated with carboplatin alone at the end of the study (Fig. 5B). We observed the expected increase in the percentage of ALDH + cells in the carboplatin + vehicle compared to the vehicle only group (Fig. 5C), consistent with previous studies by us and others [7, 13, 51, 52]. The combination treatment of IACS-010759 + carboplatin abrogated the platinum-induced enrichment of ALDH + cells in vivo (Fig. 5C). Somewhat unexpectedly, the expression of glycolysis genes HK2, LDHA and PDK1 decreased in tumors from mice treated with carboplatin in combination with IACS whereas the expression of OXPHOS genes did not change in any treatment group (Fig. 5D). SIRT1 protein levels in whole cell lysates from tumor xenografts were variable across treatment groups (Fig. 5E). However, consistent with our in vitro data, H4K16Ac levels were decreased in xenografts from mice treated with carboplatin alone or in combination with IACS compared to vehicle alone suggesting increased SIRT1 activity following carboplatin treatment (Fig. 5E). Tumors from carboplatin and carboplatin plus IACS treated mice also had reduced levels of HIF1α compared to vehicle alone (Fig. 5E). However, the reduction in tumor size in carboplatin treated groups make this result difficult to interpret. Overall, our data suggests that inhibiting OXPHOS using the complex I mitochondrial inhibitor IACS during carboplatin treatment blocks platinum mediated enrichment of ALDH + OCSCs in vivo.
Fig. 5.
OXPHOS inhibition blocks the platinum-induced enrichment of ALDH + cells in vivo. A 2 × 106 OVCAR3 cells were injected s.c in 6–7 weeks old NSG mice. Once the tumors were > 100 mm3, mice were randomized into three groups and treated with vehicle alone or combination of vehicle + carboplatin or IACS-010759 for 3 weeks as indicated. At the end of the study, tumors were collected, dissociated into single cells and the ALDEFLUOR assay was performed. B Tumor volumes were measured using a digital caliper through 3 weeks of treatment by the same investigator throughout the study. N = 4–5 mice per group. *P relative to carboplatin + vehicle. #P relative to vehicle. C Percentage of ALDH + cells in dissociated xenograft tumor samples using ALDEFLUOR assay. D Expression of glycolysis and OXPHOS genes in tumor xenografts. Graph indicates mean ± SEM in the different treatment groups. N = 4–5 mice per group. For all treated with vehicle alone versus carboplatin + vehicle or IACS-010759, P values *< 0.05, **< 0.005, ***< 0.0005, ****< 0.0001. E Western blot of the indicated proteins in whole cell lysates from tumor xenografts. F Model for how platinum treatment of HGSOC cells results in increased mitochondrial activity that contributes to the platinum-induced enrichment of ALDH + OCSCs. Platinum induces an increase in SIRT1 activity that increases TFAM levels, which in turn likely increases expression of OXPHOS genes that leads to an increase in mitochondrial activity
Discussion
Metabolic reprogramming is a distinctive characteristic of chemoresistant OC cells and likely occurs as a consequence of metabolic adaptation to the tumor microenvironment [16]. Metabolic reprogramming provides adenosine triphosphate [ATP] and precursors for macromolecular biosynthesis to meet the energy requirements for rapid proliferation and survival of tumor cells [53]. In addition to promoting tumorigenesis, alterations in cellular metabolism foster resistance to chemotherapy in OC and other cancer types [54–56]. The predominant mechanism of cytotoxicity of chemotherapeutic agents is inducing DNA damage and oxidative stress. Metabolic reprogramming enables tumor cells to adapt and manage pharmacological insults and oxidative stress [53]. The precise mechanism of how metabolic reprogramming promotes chemoresistance in OC is not understood. Several studies have established that ALDH + CSCs contribute to the development of chemoresistance in OC [7, 13, 14]. Our group and others have demonstrated that platinum treatment results in enrichment of ALDH + OCSCs [7, 13, 14, 52]. Persistence of these ALDH + OCSCs promotes tumor relapse and recurrence of chemoresistant disease contributing to high fatality rates among OC patients. Here, we demonstrate for the first time a role for OXPHOS and SIRT1 in the platinum-induced enrichment of ALDH + OCSCs.
There has been a long-standing debate about whether CSCs use glycolysis or OXPHOS for survival [57]. The metabolic pathway utilized by CSCs depends on which pathway provides a selective advantage in the tumor microenvironment. Here, we demonstrate that cisplatin treatment elevates expression of mitochondrial OXPHOS genes concurrently with an increase in percentage of ALDH + cells. Furthermore, OXPHOS inhibitors in combination with cisplatin or carboplatin block the enrichment of ALDH + cells in vitro and in vivo, suggesting that the platinum mediated increase in mitochondrial activity is required for the enrichment of ALDH + cells (Fig. 5F). The effect of platinum or mitochondrial complex I inhibitor IACS—010759 on other stem cell markers in OC like CD133, CD44 and others requires further investigation. However, our finding that IACS blocks cisplatin-induced spheroid formation suggests that IACS-010759 blocks platinum mediated enrichment of OCSCs. These findings agree with Viale et al. that pancreatic CSCs depend on OXPHOS for survival [19, 58]. It is likely that OXPHOS generates ROS to regulate metabolic plasticity of OCSCs, as recently suggested by Luo and Wicha [59].
Platinum resistant OC cells display elevated expression of the deacetylase SIRT1 [60, 61]. We demonstrate that SIRT1 levels and activity increase after platinum treatment. NAD + functions as a co-factor for several enzymes like ALDH and SIRT1 [21, 62], and the increase in SIRT1 activity and expression in response to cisplatin treatment may be due to platinum-induced increase in cellular NAD + levels [13, 14]. The dependency of the platinum mediated increase in ALDH + cells on SIRT1 may be due to enhanced regulation of the platinum-induced metabolic switch to oxidative metabolism and improved antioxidant defenses by SIRT1, both of which could promote survival of CSCs [45, 63]. SIRT1 supports mitochondrial function in muscle cells after exercise or starvation by regulating TFAM [64, 65], and we show that SIRT1 KD reduces the platinum-induced increase in TFAM expression. We hypothesize that the SIRT1-mediated increase in TFAM leads to an increase in OXPHOS gene expression leading to an increase in OXPHOS and ALDH activity (Fig. 5F). However, the precise mechanism of how SIRT1 contributes to the increase in ALDH + cells and regulates TFAM in response to cisplatin treatment requires further investigation.
Persistence of ALDH + OCSCs is a major obstacle in improving the survival rate of platinum resistant, recurrent ovarian cancer patients. Our group as well as others have demonstrated various therapeutic strategies to target this OCSC population thereby providing preclinical evidence to improve prognosis of OC patients with chemoresistant disease [7, 13, 52, 66]. Even though the lack of verification of our findings in patient derived tumor samples is a limitation of our study, this work has furthered our understanding of how metabolic alterations caused by platinum based chemotherapeutic agents can potentially be exploited to target ALDH + OCSCs. Although the involvement of OXPHOS in platinum resistance has been previously demonstrated [31, 67], our study has elucidated how platinum treatment reprograms the metabolism of OC cells resulting in enrichment of ALDH + OCSCs. Furthermore, our data demonstrated how this alteration in metabolism can be targeted using the small molecule inhibitor IACS-010759 to block the persistence of the platinum resistant ALDH + OCSC population. Our study supports further investigation into exploiting OXPHOS inhibitors like IACS-010759 to target the ALDH + OCSC population.
Conclusions
The persistence of ALDH + cells after chemotherapy is a major cause of disease recurrence and relapse in OC [4]. We have demonstrated that cisplatin treatment of OC cells results in an increase in mitochondrial membrane potential. Our in vitro and in vivo data demonstrate that combined treatment with an OXPHOS inhibitor and platinum blocks the platinum-induced enrichment of ALDH + cells. Our findings reinforce the need for additional preclinical and clinical investigations aimed at exploiting OXPHOS inhibitors to delay tumor recurrence and improve survival in OC.
Supplementary Information
Additional file 1: Figure S1. Mitochondrial DNA (mtDNA) content does not increase in response to cisplatin treatment. A Plots showing gates used to determine single cells of OVCAR5 cells untreated, OXPHOSi or 16 h cisplatin after JC-1 staining. B OVCAR5 cells were treated with IC50 dose of cisplatin for 16 h. DNA was isolated and qPCR was performed using two primer sets for different regions in the mitochondrial DNA (mtDNA) NADH dehydrogenase sub-unit 1 (ND1) and NADH dehydrogenase sub-unit 5 (ND5) relative to beta 2 microglobulin in the genomic DNA. Graph depicts mean ± SEM of mtDNA relative to nuclear DNA of N = 3 biological replicates, P values *< 0.05, **< 0.005, ***< 0.0005. Figure S2. Platinum treatment reduces HIF1α protein levels. A Western blot of HIF1α, c-Myc and Actin B in OVCAR3 cells treated with IC50 dose of cisplatin (15 µM) for 16 hours. B c-Myc expression by RT-qPCR in OVCAR3 cells untreated (U) or treated with IC50 dose (15 µM) cisplatin for 16 h. Figure S3. Treatment of OC cells with mitochondrial complex I inhibitor in combination with cisplatin blocks platinum-induced enrichment of ALDH+ cells. A, B Plots showing gates used to determine single cells in OVSAHO cells treated with cisplatin alone or in combination with DMSO or 1 µM IACS-010759 for 16 h after ALDELUOR assay. C Plots showing gates used to determine ALDH+ cells using DEAB controls for one biological replicate of OVSAHO cells. Figure S4. IACS treatment does not alter platinum-induced gene expression changes. Expression of OXPHOS genes (A) and c-Myc, HIF1α (B) in OVCAR5 cells treated with DMSO or IACS-010759 (1 μM) in combination with cisplatin (12 μM) for 16 h. C TFAM expression in OVCAR3 cells untreated or treated with IC50 dose of cisplatin (15 μM) for 16 h.
Additional file 2: Table S1. qRT-primer sequences. Table S2. Primers for mitochondrial DNA (mtDNA) content.
Acknowledgements
We thank Christiane Hassel and the Indiana University Flow Cytometry facility for their assistance.
Abbreviations
- ∆ΨM
Mitochondrial membrane potential
- ALDH
Aldehyde dehydrogenase
- ATM
Ataxia telangiectasia mutated
- ATP5MF
ATP synthase membrane subunit F
- COX5A
Cytochrome C oxidase subunit 5A
- COX6A
Cytochrome C oxidase subunit 6A
- CSCs
Cancer stem cells
- DMSO
Dimethyl sulfoxide
- HGSOC
High grade serous ovarian cancer
- HIF1α
Hypoxia-inducible factor 1
- HK1
Hexokinase 1
- HK2
Hexokinase 2
- KD
Knockdown
- LDHA
Lactate dehydrogenase A
- mtDNA
Mitochondrial DNA
- NAD +
Nicotinamide adenine dinucleotide
- NAMPT
Nicotinamide phosphoribosyltransferase
- NDUFA11
NADH ubiquinone oxidoreductase subunit 11
- NDUFS6
NADH ubiquinone oxidoreductase subunit 6
- OC
Ovarian cancer
- OCSCs
Ovarian cancer stem cells
- OXPHOS
Oxidative phosphorylation
- PDK1
Pyruvate dehydrogenase kinase
- PKM2
Pyruvate kinase isozyme M2
- RT-qPCR
Reverse transcriptase-quantitative polymerase chain reaction
- ROS
Reactive oxygen species
- shRNA
Short hairpin RNA
- SIRT1
Sirtuin 1
- TFAM
Mitochondrial transcription factor A
- TIMM17A
Translocase of the inner mitochondrial membrane
- UQCR10
Ubiquinol- cytochrome C reductase subunit X
- UQCR11
Ubiquinol-cytochrome C reductase subunit XI
Author contributions
SS conceived, designed, analyzed all the experiments, wrote the original draft of the manuscript, edited and proofread the manuscript. TTV, TXM, RS, TDH and WW assisted in data acquisition. AHG performed some validation experiments and proofread the manuscript. KPN provided resources, funding acquisition, edited and proofread the manuscript. HMOH conceived, assisted with experimental design, supervised the study, and provided resources, funding acquisition, edited and proofread the manuscript. All authors read and approved the final manuscript.
Funding
This research was funded in part by the Ovarian Cancer Research Alliance (Grant Number 458788 to HMOH and KPN), the Ovarian Cancer Alliance of Greater Cincinnati and through the IU Simon Comprehensive Cancer Center P30 Support Grant (P30CA082709). SS was supported by the Doane and Eunice Dahl Wright Fellowship generously provided by Ms. Imogen Dahl.
Availability of data and materials
All the data generated in the manuscript and materials used in the study are available upon reasonable request.
Declarations
Ethics approval and consent to participate
The animal study was approved by Indiana University Bloomington Institutional Animal Care and Use Committee and was performed in adherence with the Association for Assessment and Accreditation of Laboratory Animal Care International.
Consent for publication
Not applicable.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Torre LA, Trabert B, DeSantis CE, Miller KD, Samimi G, Runowicz CD, et al. Ovarian cancer statistics, 2018. CA Cancer J Clin. 2018;68(4):284–296. doi: 10.3322/caac.21456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ozols RF. Treatment goals in ovarian cancer. Int J Gynecol Cancer. 2005;15(Suppl 1):3–11. doi: 10.1111/j.1525-1438.2005.15351.x. [DOI] [PubMed] [Google Scholar]
- 3.Vaughan S, Coward JI, Bast RC, Jr, Berchuck A, Berek JS, Brenton JD, et al. Rethinking ovarian cancer: recommendations for improving outcomes. Nat Rev Cancer. 2011;11(10):719–725. doi: 10.1038/nrc3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lupia M, Cavallaro U. Ovarian cancer stem cells: still an elusive entity? Mol Cancer. 2017;16(1):64. doi: 10.1186/s12943-017-0638-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gupta PB, Chaffer CL, Weinberg RA. Cancer stem cells: mirage or reality? Nat Med. 2009;15(9):1010–1012. doi: 10.1038/nm0909-1010. [DOI] [PubMed] [Google Scholar]
- 6.Muralikrishnan V, Hurley TD, Nephew KP. Targeting aldehyde dehydrogenases to eliminate cancer stem cells in gynecologic malignancies. Cancers. 2020 doi: 10.3390/cancers12040961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y, Cardenas H, Fang F, Condello S, Taverna P, Segar M, et al. Epigenetic targeting of ovarian cancer stem cells. Cancer Res. 2014;74(17):4922–4936. doi: 10.1158/0008-5472.CAN-14-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Landen CN, Jr, Goodman B, Katre AA, Steg AD, Nick AM, Stone RL, et al. Targeting aldehyde dehydrogenase cancer stem cells in ovarian cancer. Mol Cancer Ther. 2010;9(12):3186–3199. doi: 10.1158/1535-7163.MCT-10-0563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Condello S, Morgan CA, Nagdas S, Cao L, Turek J, Hurley TD, et al. beta-Catenin-regulated ALDH1A1 is a target in ovarian cancer spheroids. Oncogene. 2015;34(18):2297–2308. doi: 10.1038/onc.2014.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Januchowski R, Wojtowicz K, Sterzyska K, Sosiska P, Andrzejewska M, Zawierucha P, et al. Inhibition of ALDH1A1 activity decreases expression of drug transporters and reduces chemotherapy resistance in ovarian cancer cell lines. Int J Biochem Cell Biol. 2016;78:248–259. doi: 10.1016/j.biocel.2016.07.017. [DOI] [PubMed] [Google Scholar]
- 11.Silva IA, Bai S, McLean K, Yang K, Griffith K, Thomas D, et al. Aldehyde dehydrogenase in combination with CD133 defines angiogenic ovarian cancer stem cells that portend poor patient survival. Cancer Res. 2011;71(11):3991–4001. doi: 10.1158/0008-5472.CAN-10-3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuroda T, Hirohashi Y, Torigoe T, Yasuda K, Takahashi A, Asanuma H, et al. ALDH1-high ovarian cancer stem-like cells can be isolated from serous and clear cell adenocarcinoma cells, and ALDH1 high expression is associated with poor prognosis. PLoS ONE. 2013;8(6):e65158. doi: 10.1371/journal.pone.0065158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nacarelli T, Fukumoto T, Zundell JA, Fatkhutdinov N, Jean S, Cadungog MG, et al. NAMPT inhibition suppresses cancer stem-like cells associated with therapy-induced senescence in ovarian cancer. Cancer Res. 2020;80(4):890–900. doi: 10.1158/0008-5472.CAN-19-2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sood R, Xiao S, Sriramkumar S, Hassel C, Nephew KP, O’Hagan HM. Novel epigenetic-metabolic inhibitor combination treatment blocks platinum-induced ovarian cancer stem cell enrichment. Biorxiv. 2021 doi: 10.1101/2021.03.18.435878. [DOI] [Google Scholar]
- 15.Pasto A, Bellio C, Pilotto G, Ciminale V, Silic-Benussi M, Guzzo G, et al. Cancer stem cells from epithelial ovarian cancer patients privilege oxidative phosphorylation, and resist glucose deprivation. Oncotarget. 2014;5(12):4305–4319. doi: 10.18632/oncotarget.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dar S, Chhina J, Mert I, Chitale D, Buekers T, Kaur H, et al. Bioenergetic adaptations in chemoresistant ovarian cancer cells. Sci Rep. 2017;7(1):8760. doi: 10.1038/s41598-017-09206-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghoneum A, Gonzalez D, Abdulfattah AY, Said N. Metabolic plasticity in ovarian cancer stem cells. Cancers. 2020 doi: 10.3390/cancers12051267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ye XQ, Li Q, Wang GH, Sun FF, Huang GJ, Bian XW, et al. Mitochondrial and energy metabolism-related properties as novel indicators of lung cancer stem cells. Int J Cancer. 2011;129(4):820–831. doi: 10.1002/ijc.25944. [DOI] [PubMed] [Google Scholar]
- 19.Viale A, Pettazzoni P, Lyssiotis CA, Ying H, Sanchez N, Marchesini M, et al. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature. 2014;514(7524):628–632. doi: 10.1038/nature13611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kiefer FW, Orasanu G, Nallamshetty S, Brown JD, Wang H, Luger P, et al. Retinaldehyde dehydrogenase 1 coordinates hepatic gluconeogenesis and lipid metabolism. Endocrinology. 2012;153(7):3089–3099. doi: 10.1210/en.2011-2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Y, Thompson DC, Koppaka V, Jester JV, Vasiliou V. Ocular aldehyde dehydrogenases: protection against ultraviolet damage and maintenance of transparency for vision. Prog Retin Eye Res. 2013;33:28–39. doi: 10.1016/j.preteyeres.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stewart DJ. Mechanisms of resistance to cisplatin and carboplatin. Crit Rev Oncol Hematol. 2007;63(1):12–31. doi: 10.1016/j.critrevonc.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 23.Sriramkumar S, Matthews TD, Ghobashi AH, Miller SA, VanderVere-Carozza PS, Pawelczak KS, et al. Platinum-induced ubiquitination of phosphorylated H2AX by RING1A is mediated by replication protein A in ovarian cancer. Mol Cancer Res. 2020;18(11):1699–1710. doi: 10.1158/1541-7786.MCR-20-0396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haley J, Tomar S, Pulliam N, Xiong S, Perkins SM, Karpf AR, et al. Functional characterization of a panel of high-grade serous ovarian cancer cell lines as representative experimental models of the disease. Oncotarget. 2016;7(22):32810–32820. doi: 10.18632/oncotarget.9053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maiuri AR, Li H, Stein BD, Tennessen JM, O'Hagan HM. Inflammation-induced DNA methylation of DNA polymerase gamma alters the metabolic profile of colon tumors. Cancer Metab. 2018;6:9. doi: 10.1186/s40170-018-0182-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rooney JP, Ryde IT, Sanders LH, Howlett EH, Colton MD, Germ KE, et al. PCR based determination of mitochondrial DNA copy number in multiple species. Methods Mol Biol. 2015;1241:23–38. doi: 10.1007/978-1-4939-1875-1_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Machado TS, Macabelli CH, Sangalli JR, Rodrigues TB, Smith LC, Meirelles FV, et al. Real-time PCR quantification of heteroplasmy in a mouse model with mitochondrial DNA of C57BL/6 and NZB/BINJ strains. PLoS ONE. 2015;10(8):e0133650. doi: 10.1371/journal.pone.0133650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cancer Genome Atlas Research N Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474(7353):609–15. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matassa DS, Amoroso MR, Lu H, Avolio R, Arzeni D, Procaccini C, et al. Oxidative metabolism drives inflammation-induced platinum resistance in human ovarian cancer. Cell Death Differ. 2016;23(9):1542–1554. doi: 10.1038/cdd.2016.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sivandzade F, Bhalerao A, Cucullo L. Analysis of the mitochondrial membrane potential using the cationic JC-1 dye as a sensitive fluorescent probe. Bio Protoc. 2019 doi: 10.21769/BioProtoc.3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Linsinger G, Wilhelm S, Wagner H, Hacker G. Uncouplers of oxidative phosphorylation can enhance a Fas death signal. Mol Cell Biol. 1999;19(5):3299–3311. doi: 10.1128/mcb.19.5.3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang H, Gao P, Fukuda R, Kumar G, Krishnamachary B, Zeller KI, et al. HIF-1 inhibits mitochondrial biogenesis and cellular respiration in VHL-deficient renal cell carcinoma by repression of C-MYC activity. Cancer Cell. 2007;11(5):407–420. doi: 10.1016/j.ccr.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 35.Sutphin PD, Giaccia AJ, Chan DA. Energy regulation: HIF MXIes it up with the C-MYC powerhouse. Dev Cell. 2007;12(6):845–846. doi: 10.1016/j.devcel.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 36.Luo W, Hu H, Chang R, Zhong J, Knabel M, O'Meally R, et al. Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell. 2011;145(5):732–744. doi: 10.1016/j.cell.2011.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wheaton WW, Chandel NS. Hypoxia. 2. Hypoxia regulates cellular metabolism. Am J Physiol Cell Physiol. 2011;300(3):C385–93. doi: 10.1152/ajpcell.00485.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.DeWaal D, Nogueira V, Terry AR, Patra KC, Jeon SM, Guzman G, et al. Hexokinase-2 depletion inhibits glycolysis and induces oxidative phosphorylation in hepatocellular carcinoma and sensitizes to metformin. Nat Commun. 2018;9(1):446. doi: 10.1038/s41467-017-02733-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Molina JR, Sun Y, Protopopova M, Gera S, Bandi M, Bristow C, et al. An inhibitor of oxidative phosphorylation exploits cancer vulnerability. Nat Med. 2018;24(7):1036–1046. doi: 10.1038/s41591-018-0052-4. [DOI] [PubMed] [Google Scholar]
- 40.Ashton TM, McKenna WG, Kunz-Schughart LA, Higgins GS. Oxidative phosphorylation as an emerging target in cancer therapy. Clin Cancer Res. 2018;24(11):2482–2490. doi: 10.1158/1078-0432.CCR-17-3070. [DOI] [PubMed] [Google Scholar]
- 41.Bapat SA, Mali AM, Koppikar CB, Kurrey NK. Stem and progenitor-like cells contribute to the aggressive behavior of human epithelial ovarian cancer. Cancer Res. 2005;65(8):3025–3029. doi: 10.1158/0008-5472.CAN-04-3931. [DOI] [PubMed] [Google Scholar]
- 42.Zhang S, Balch C, Chan MW, Lai HC, Matei D, Schilder JM, et al. Identification and characterization of ovarian cancer-initiating cells from primary human tumors. Cancer Res. 2008;68(11):4311–4320. doi: 10.1158/0008-5472.CAN-08-0364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu TF, McCall CE. Deacetylation by SIRT1 reprograms inflammation and cancer. Genes Cancer. 2013;4(3–4):135–147. doi: 10.1177/1947601913476948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vaquero A, Scher M, Lee D, Erdjument-Bromage H, Tempst P, Reinberg D. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol Cell. 2004;16(1):93–105. doi: 10.1016/j.molcel.2004.08.031. [DOI] [PubMed] [Google Scholar]
- 45.Canto C, Auwerx J. Targeting sirtuin 1 to improve metabolism: all you need is NAD(+)? Pharmacol Rev. 2012;64(1):166–187. doi: 10.1124/pr.110.003905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Y, Xu S, Li J, Zheng L, Feng M, Wang X, et al. SIRT1 facilitates hepatocellular carcinoma metastasis by promoting PGC-1alpha-mediated mitochondrial biogenesis. Oncotarget. 2016;7(20):29255–29274. doi: 10.18632/oncotarget.8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Majeed Y, Halabi N, Madani AY, Engelke R, Bhagwat AM, Abdesselem H, et al. SIRT1 promotes lipid metabolism and mitochondrial biogenesis in adipocytes and coordinates adipogenesis by targeting key enzymatic pathways. Sci Rep. 2021;11(1):8177. doi: 10.1038/s41598-021-87759-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang J, Peng J, Kong D, Wang X, Wang Z, Liu J, et al. Silent information regulator 1 suppresses epithelial-to-mesenchymal transition in lung cancer cells via its regulation of mitochondria status. Life Sci. 2021;280:119716. doi: 10.1016/j.lfs.2021.119716. [DOI] [PubMed] [Google Scholar]
- 49.Kaufman BA, Durisic N, Mativetsky JM, Costantino S, Hancock MA, Grutter P, et al. The mitochondrial transcription factor TFAM coordinates the assembly of multiple DNA molecules into nucleoid-like structures. Mol Biol Cell. 2007;18(9):3225–3236. doi: 10.1091/mbc.E07-05-0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kanki T, Ohgaki K, Gaspari M, Gustafsson CM, Fukuoh A, Sasaki N, et al. Architectural role of mitochondrial transcription factor A in maintenance of human mitochondrial DNA. Mol Cell Biol. 2004;24(22):9823–9834. doi: 10.1128/MCB.24.22.9823-9834.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Y, Zong X, Mitra S, Mitra AK, Matei D, Nephew KP. IL-6 mediates platinum-induced enrichment of ovarian cancer stem cells. JCI Insight. 2018 doi: 10.1172/jci.insight.122360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang Y, Zhao G, Condello S, Huang H, Cardenas H, Tanner EJ, et al. Frizzled-7 identifies platinum-tolerant ovarian cancer cells susceptible to ferroptosis. Cancer Res. 2021;81(2):384–399. doi: 10.1158/0008-5472.CAN-20-1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen X, Chen S, Yu D. Metabolic reprogramming of chemoresistant cancer cells and the potential significance of metabolic regulation in the reversal of cancer chemoresistance. Metabolites. 2020 doi: 10.3390/metabo10070289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Han CY, Patten DA, Richardson RB, Harper ME, Tsang BK. Tumor metabolism regulating chemosensitivity in ovarian cancer. Genes Cancer. 2018;9(5–6):155–175. doi: 10.18632/genesandcancer.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Montopoli M, Bellanda M, Lonardoni F, Ragazzi E, Dorigo P, Froldi G, et al. “Metabolic reprogramming” in ovarian cancer cells resistant to cisplatin. Curr Cancer Drug Targets. 2011;11(2):226–235. doi: 10.2174/156800911794328501. [DOI] [PubMed] [Google Scholar]
- 56.Cardoso MR, Santos JC, Ribeiro ML, Talarico MCR, Viana LR, Derchain SFM. A metabolomic approach to predict breast cancer behavior and chemotherapy response. Int J Mol Sci. 2018 doi: 10.3390/ijms19020617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Peiris-Pages M, Martinez-Outschoorn UE, Pestell RG, Sotgia F, Lisanti MP. Cancer stem cell metabolism. Breast Cancer Res. 2016;18(1):55. doi: 10.1186/s13058-016-0712-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Viale A, Corti D, Draetta GF. Tumors and mitochondrial respiration: a neglected connection. Cancer Res. 2015;75(18):3685–3686. doi: 10.1158/0008-5472.CAN-15-0491. [DOI] [PubMed] [Google Scholar]
- 59.Luo M, Wicha MS. Targeting cancer stem cell redox metabolism to enhance therapy responses. Semin Radiat Oncol. 2019;29(1):42–54. doi: 10.1016/j.semradonc.2018.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403(6771):795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 61.Liang XJ, Finkel T, Shen DW, Yin JJ, Aszalos A, Gottesman MM. SIRT1 contributes in part to cisplatin resistance in cancer cells by altering mitochondrial metabolism. Mol Cancer Res. 2008;6(9):1499–1506. doi: 10.1158/1541-7786.MCR-07-2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guo X, Kesimer M, Tolun G, Zheng X, Xu Q, Lu J, et al. The NAD(+)-dependent protein deacetylase activity of SIRT1 is regulated by its oligomeric status. Sci Rep. 2012;2:640. doi: 10.1038/srep00640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Singh CK, Chhabra G, Ndiaye MA, Garcia-Peterson LM, Mack NJ, Ahmad N. The role of sirtuins in antioxidant and redox signaling. Antioxid Redox Signal. 2018;28(8):643–661. doi: 10.1089/ars.2017.7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chandrasekaran K, Anjaneyulu M, Choi J, Kumar P, Salimian M, Ho CY, et al. Role of mitochondria in diabetic peripheral neuropathy: influencing the NAD(+)-dependent SIRT1-PGC-1alpha-TFAM pathway. Int Rev Neurobiol. 2019;145:177–209. doi: 10.1016/bs.irn.2019.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ruderman NB, Xu XJ, Nelson L, Cacicedo JM, Saha AK, Lan F, et al. AMPK and SIRT1: a long-standing partnership? Am J Physiol Endocrinol Metab. 2010;298(4):E751–E760. doi: 10.1152/ajpendo.00745.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fang F, Munck J, Tang J, Taverna P, Wang Y, Miller DF, et al. The novel, small-molecule DNA methylation inhibitor SGI-110 as an ovarian cancer chemosensitizer. Clin Cancer Res. 2014;20(24):6504–6516. doi: 10.1158/1078-0432.CCR-14-1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nayak AP, Kapur A, Barroilhet L, Patankar MS. Oxidative phosphorylation: a target for novel therapeutic strategies against ovarian cancer. Cancers. 2018 doi: 10.3390/cancers10090337. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Mitochondrial DNA (mtDNA) content does not increase in response to cisplatin treatment. A Plots showing gates used to determine single cells of OVCAR5 cells untreated, OXPHOSi or 16 h cisplatin after JC-1 staining. B OVCAR5 cells were treated with IC50 dose of cisplatin for 16 h. DNA was isolated and qPCR was performed using two primer sets for different regions in the mitochondrial DNA (mtDNA) NADH dehydrogenase sub-unit 1 (ND1) and NADH dehydrogenase sub-unit 5 (ND5) relative to beta 2 microglobulin in the genomic DNA. Graph depicts mean ± SEM of mtDNA relative to nuclear DNA of N = 3 biological replicates, P values *< 0.05, **< 0.005, ***< 0.0005. Figure S2. Platinum treatment reduces HIF1α protein levels. A Western blot of HIF1α, c-Myc and Actin B in OVCAR3 cells treated with IC50 dose of cisplatin (15 µM) for 16 hours. B c-Myc expression by RT-qPCR in OVCAR3 cells untreated (U) or treated with IC50 dose (15 µM) cisplatin for 16 h. Figure S3. Treatment of OC cells with mitochondrial complex I inhibitor in combination with cisplatin blocks platinum-induced enrichment of ALDH+ cells. A, B Plots showing gates used to determine single cells in OVSAHO cells treated with cisplatin alone or in combination with DMSO or 1 µM IACS-010759 for 16 h after ALDELUOR assay. C Plots showing gates used to determine ALDH+ cells using DEAB controls for one biological replicate of OVSAHO cells. Figure S4. IACS treatment does not alter platinum-induced gene expression changes. Expression of OXPHOS genes (A) and c-Myc, HIF1α (B) in OVCAR5 cells treated with DMSO or IACS-010759 (1 μM) in combination with cisplatin (12 μM) for 16 h. C TFAM expression in OVCAR3 cells untreated or treated with IC50 dose of cisplatin (15 μM) for 16 h.
Additional file 2: Table S1. qRT-primer sequences. Table S2. Primers for mitochondrial DNA (mtDNA) content.
Data Availability Statement
All the data generated in the manuscript and materials used in the study are available upon reasonable request.