Abstract
Domain-, class-, and subclass-specific rRNA-targeted probes were applied to investigate the microbial communities of three industrial and three laboratory-scale biofilters. The set of probes also included a new probe (named XAN818) specific for the Xanthomonas branch of the class Proteobacteria; this probe is described in this study. The members of the Xanthomonas branch do not hybridize with previously developed rRNA-targeted oligonucleotide probes for the α-, β-, and γ-Proteobacteria. Bacteria of the Xanthomonas branch accounted for up to 4.5% of total direct counts obtained with 4′,6-diamidino-2-phenylindole. In biofilter samples, the relative abundance of these bacteria was similar to that of the γ-Proteobacteria. Actinobacteria (gram-positive bacteria with a high G+C DNA content) and α-Proteobacteria were the most dominant groups. Detection rates obtained with probe EUB338 varied between about 40 and 70%. For samples with high contents of gram-positive bacteria, these percentages were substantially improved when the calculations were corrected for the reduced permeability of gram-positive bacteria when formaldehyde was used as a fixative. The set of applied bacterial class- and subclass-specific probes yielded, on average, 58.5% (± a standard deviation of 23.0%) of the corrected eubacterial detection rates, thus indicating the necessity of additional probes for studies of biofilter communities. The Xanthomonas-specific probe presented here may serve as an efficient tool for identifying potential phytopathogens. In situ hybridization proved to be a practical tool for microbiological studies of biofiltration systems.
Many industrial processes produce waste gases containing odorous organic or inorganic compounds, some of which may be toxic to human health (26). While the principle of filtering air with the help of microorganisms has been known for some time, Pomeroy (27) seems to be the first who applied biofiltration on a technical scale in the early 1950s. Biofiltration usually exhibits high removal efficiencies and lower investment and maintenance costs than physical and chemical treatment of waste gases. Biofilters are thus applied increasingly in a variety of industrial settings.
Extensive efforts have been made to optimize the technical aspects of biofilters. Comparably little is known about the biology of biofilters, although detailed biological information is necessary for long-term stability and further optimization of cleaning efficiencies. Most microbial studies of biofilters have used isolation techniques, which have led to the characterization of new species showing interesting physiological capabilities (16, 29). Molecular biological methods, in particular, are promising tools for studying the community level in biofilters. A PCR-based approach (32) and rRNA-targeted oligonucleotide probes (21) have been used to study waste gas cleaning systems containing artificial carrier materials. To our knowledge, the present study is the first to apply fluorescence in situ hybridization (FISH) to full-scale biofilters. In this study, the bacterial communities of three industrial and three laboratory-scale biofilters were characterized with oligonucleotide probes targeting domain-, class-, and subclass-specific regions of the 16S and 23S rRNAs.
A significant portion of the bacteria previously isolated from biofilters in our laboratory are members of the Xanthomonas branch of the class Proteobacteria, which forms a monophyletic group close to the root of the γ-Proteobacteria (22). Recently, the biofilter strains isolated in our laboratory were identified as two new genera and three new species of the Xanthomonas branch (9). Buchholz-Cleven et al. (7) isolated from a freshwater sediment a strain belonging to the Xanthomonas branch. They demonstrated that neither probe GAM42a, which is specific for the γ-Proteobacteria (19), nor probe BET42a hybridized with this strain. The present study confirms this finding on a larger scale, as none of the 18 tested representatives of the Xanthomonas branch hybridized with these probes. To allow in situ identification and enumeration of these bacteria, we thus designed, evaluated, and applied a new, 16S rRNA-targeted oligonucleotide probe (XAN818) for the detection of all sequenced members of the Xanthomonas branch.
MATERIALS AND METHODS
Cultivation and preparation of reference strains.
The reference strains used are listed in Table 1. The strains were obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH, Braunschweig, Germany (DSM); Laboratorium voor Microbiologie, Universiteit Gent, Ghent, Belgium (LMG); and our own collection (strains with prefixes B, L, and R). Except for strains DSM 20300T, DSM 20140T, and LMG11114, all DSM and LMG strains were cultivated according to the descriptions given by the collections. Those three strains and our strains were cultivated in nutrient broth (3 g of meat extract liter−1, 5 g of peptone liter−1, and 15 g of agar liter−1 were added for solid media) and harvested in the exponential phase (optical density at 578 nm, between 0.5 and 0.8). Pure cultures of members of the phylum Firmicutes (gram-positive bacteria) (5 ml) were fixed with 5 ml of ethanol (100%) and stored at −18°C. All other pure cultures (5 ml) of the reference strains (Table 1) were fixed with 15 ml of 4% paraformaldehyde (PFA) solution (3) for 24 h and centrifuged at 11,200 × g for 20 min. Pellets were washed twice in 20 ml of phosphate-buffered saline (10 mM Na2HPO4 · 2H2O adjusted to pH 7.4 with 10 mM Na2HPO4 · H2O; final concentration of NaCl, 130 mM) and centrifuged. Cells were then resuspended in 1:1 phosphate-buffered saline–ethanol and stored at −18°C.
TABLE 1.
Organisms used and results of FISH specificity tests
| Species | Strain | FISH results obtained with the following probea:
|
|||
|---|---|---|---|---|---|
| EUB338 | BET42a | GAM42a | XAN818 | ||
| Acidiphilium acidophilum | DSM 700T | + | − | − | − |
| Agrobacterium tumefaciens | DSM 30147 | + | − | − | − |
| Gluconobacter oxydans subsp. oxydans | DSM 3503T | + | − | − | − |
| Paracoccus alkenifer | DSM 11593T | + | − | − | − |
| Paracoccus solventivorans | DSM 11592 | + | − | − | − |
| Alcaligenes faecalis | DSM 30033 | + | + | − | − |
| Beggiatoa alba | DSM 1416 | + | − | + | − |
| Pseudomonas pseudoalcaligenes | DSM 50188T | + | − | + | − |
| Pseudomonas putida | DSM 291T | + | − | + | − |
| Bacillus subtilis | DSM 10T | + | − | − | − |
| Corynebacterium glutamicum | DSM 20300T | + | − | − | − |
| Microbacterium laevaniformans | DSM 20140T | + | − | − | − |
| Pedobacter heparinus | DSM 2366T | + | − | − | − |
| Luteimonas mephitis | DSM 12574T | + | − | − | + |
| Luteimonas mephitis | B1959/26 | + | − | − | + |
| Luteimonas mephitis | B1962/27.1 | + | − | − | + |
| Luteimonas mephitis | R514 | + | − | − | + |
| Luteimonas mephitis | R515 | + | − | − | + |
| Pseudoxanthomonas broegbernensis | DSM 12573T | + | − | − | + |
| Stenotrophomonas maltophilia | LMG 11114 | + | − | − | + |
| Stenotrophomonas maltophilia | DSM 50170T | + | − | − | + |
| Stenotrophomonas nitritireducens | DSM 12575T | + | − | − | + |
| Stenotrophomonas nitritireducens | B1910/29.1 | + | − | − | + |
| Stenotrophomonas nitritireducens | L16 | + | − | − | + |
| Stenotrophomonas nitritireducens | L60 | + | − | − | + |
| Xanthomonas cucurbitae | LMG 690T | + | − | − | + |
| Xanthomonas hyacinthi | LMG 739T | + | − | − | + |
| Xanthomonas oryzae pv. oryzae | LMG 5047T | + | − | w | + |
| Xanthomonas pisi | LMG 847T | + | − | − | + |
| Xanthomonas sacchari | LMG 471T | + | − | − | + |
| Xylella fastidiosa | DSM 10026T | + | − | − | + |
Fluorescence signal was positive (+), negative (−), or weak (w).
Design of probes specific for the Xanthomonas branch.
Specific target sequences for the 16S rRNA of bacteria of the Xanthomonas branch were identified by use of the program package ARB (35). The most recent data set available for ARB (release 6pubmrz97) was supplemented with sequences obtained from the EMBL database. The accession numbers of these sequences are as follows: Xanthomonas, X95917, X95918, X95919, X95920, X95921, X95922, X99299, Y10754, Y10755, Y10756, Y10757, Y10758, Y10759, Y10760, Y10761, Y10762, Y10763, Y10764, Y10765, and Y10766; Nevskia, AJ001010 and AJ001011; Stenotrophomonas, AJ012229, AJ012230, X95923, X95925, and U62646; Luteimonas, AJ012228; Pseudoxanthomonas, AJ012231; denitrifying Fe(II)-oxidizing bacterium BrG3, U51103; and iron-oxidizing lithotroph, AF012541. The specificities of the target sequences obtained were also checked with the PROBE MATCH tool of the Ribosomal Database Project (17).
Optimization of probe specificity and FISH tests.
For FISH, 3 μl from each fixed sample was spotted onto precleaned (washed in 1% HCl and rinsed with 70% ethanol) and gelatin-coated [0.075% gelatin–0.01 CrK(SO4)2] slides. The slides were then dried at 42°C for 10 min. Following dehydration in 50, 80, and 100% ethanol for 3 min each, the samples were covered with 8 μl of hybridization buffer (19) and 1 μl of probe (50 ng/μl). Oligonucleotides were synthesized and fluorescently labeled with Cy3 (Amersham) at the 5′ end by Interactiva Biotechnologie GmbH (Ulm, Germany). The rRNA-targeted oligonucleotides used are summarized in Table 2.
TABLE 2.
Data on probes used in the present study
| Probe | Target (positions)a | Sequence (5′→3′) | Applied stringency (formamide concn [%]) | Source or reference |
|---|---|---|---|---|
| XAN818 | Xanthomonas branch 16S rRNA (818–835) | CAACATCCAGTTCGCATC | 10 | This study |
| EUB338 | Bacterial 16S rRNA (338–355) | GCTGCCTCCCGTAGGAGT | 10 | 4 |
| EUK516 | Eucaryal 18S rRNA (502–516) | ACCAGACTTGCCCTCC | 20 | 4 |
| ARCH915 | Archaeal 16S rRNA (915–934) | GTGCTCCCCCGCCAATTCCT | 20 | 33 |
| ALF968 | α-proteobacterial 16S rRNA (968–986) | GGTAAGGTTCTGCGCGTT | 35 | 23 |
| BET42a | β-proteobacterial 23S rRNA (1027–1043) | GCCTTCCCACTTCGTTT | 35 | 19 |
| CF319a | Cytophaga-Flavobacterium cluster of CFB phylum 16S rRNA (319–336) | TGGTCCGTGTCTCAGTAC | 35 | 18 |
| GAM42a | γ-proteobacterial 23S rRNA (1027–1043) | GCCTTCCCACATCGTTT | 35 | 19 |
| HGC69a | Actinobacterial (Firmicutes with high DNA G+C content) 23S rRNA (1901–1918) | TATAGTTACCACCGCCGT | 20 | 30 |
| LGC354a | Firmicutes with low DNA G+C content 16S rRNA (354–371) | TGGAAGATTCCCTACTGC | 35 | 20 |
| LGC354b | Firmicutes with low DNA G+C content 16S rRNA (354–371) | CGGAAGATTCCCTACTGC | 35 | 20 |
| LGC354c | Firmicutes with low DNA G+C content 16S rRNA (354–371) | CCGAAGATTCCCTACTGC | 35 | 20 |
| PLA46 | Planctomycetal 16S rRNA (46–63) | GACTTGCATGCCTAATCC | 35 | 24 |
| NON338 | Negative control | ACTCCTACGGGAGGCAGC | 10 | 38 |
E. coli numbering (6).
A formamide gradient of 0 to 70% in hybridization buffer was used to assess the optimal stringency for probe XAN818. The formamide concentrations used for the other hybridizations are given in Table 2. Samples were hybridized at 46°C for 90 min in isotonically equilibrated humid chambers. Samples were subsequently treated with a posthybridization wash as described by Manz et al. (19) at 48°C for 15 min. Sodium chloride concentrations in the washing buffer were adjusted by use of the formulae of Lathe (14). Slides were rinsed briefly with high-pressure liquid chromatography (HPLC) water (Riedel), air dried, and mounted in Vectashield (Vector Laboratories Inc., Burlingame, Calif.).
For optimization of the stringency for probe XAN818, fluorescence intensities of reference strains hybridized with EUB338 and XAN818 were detected by use of a Zeiss Axiovert 10 microscope equipped with a 50-W super-pressure mercury lamp and an HQ-Cy3 filter set (AF Analysentechnik, Tübingen, Germany). Images were recorded with a charge-coupled device camera (SenSys; Photometrics Ltd., Tucson, Ariz.) and analyzed with the image analysis software package IPlab (Scanalytics, Fairfax, Va.). Camera parameters were kept constant for all measurements. Intensity threshold values were adjusted to mark the Cy3-labeled cells (37). Probe-conferred signal intensities of the cells were divided by the cell area to determine the intensities independently of cell size (25). Values of background fluorescence were subtracted from the fluorescence intensity values obtained. Mean values of signal intensities were normalized to the signal intensity obtained with probe EUB338 to correct for different contents of ribosomes (25). Between 200 and 400 cells were examined per replicate, except for Beggiatoa alba DSM 1416, for which 10 filaments were examined per replicate.
For FISH tests of reference strains (Table 1), hybridized and washed microorganisms were counterstained with 10 μl of 4′,6-diamidino-2-phenylindole (DAPI; 1 mg liter−1) at 4°C for 10 min in the dark and rinsed with HPLC water. DAPI- and probe-conferred fluorescence signals were examined by use of a Zeiss Axioskop epifluorescence microscope equipped with filter set no. 01 (Zeiss) and the HQ-Cy3 filter set.
Sample collection and physicochemical analyses.
Samples of biofilter material were collected from three industrial and three experimental biofilters. Samples Hamm A and B were collected from two sites of a biofilter used for hexane waste gas treatment in an oil mill (Hamm, Germany). Samples Sage A and B were collected from two sites of a biofilter used for waste gas treatment in an industrially operated chicken farm (Sage, Germany). Paderborn A and B were kindly provided by H.-J. Warnecke, Universität-Gesamthochschule Paderborn, Paderborn, Germany, from styrene-treated experimental biofilters. Sample Alberta was derived from a hexane-treated experimental biofilter and was kindly supplied by K. Budwill, Alberta Research Council, Vegreville, Alberta, Canada. Sample Eversburg was collected from a biofilter used for waste gas treatment in a municipal wastewater treatment plant (Osnabrück-Eversburg, Germany). The biofilter material consisted of crushed tree roots (Sage, Hamm, and Eversburg), a mixture of crushed wood and bark compost (Paderborn A), peat (Alberta), and porous clay (Paderborn B). Soil samples were taken just below the litter layer of a forest consisting mainly of Fagus sylvatica. The freshwater sample was collected from the upper 20 cm of a pond close to the University of Osnabrück. For pH measurements, 10 g of fresh filter material and soil were stirred in 25 ml of 1 M KCl for 30 min before being analyzed with a pH electrode. Between 40 and 50 g of fresh filter material and soil were dried at 80°C to assess the water content.
Direct enumeration and FISH of biofilter and environmental samples.
For direct counts, 10 g of fresh filter material and soil were stirred in 100 ml of Ringer solution (0.9% NaCl, 0.042% KCl, 0.024% NaHCO3) for 30 min. Subsequently, 5 ml of the solution was fixed with 15 ml of PFA solution (see above). Prior to direct enumeration, fixed samples were diluted in sterile 0.9% NaCl solution to obtain about 100 cells per microscopic field. After samples were stained with DAPI (final concentration, 2 mg liter−1) (28) for 5 min, 5-ml subsamples were filtered on black polycarbonate membrane filters (pore size, 0.2 μm; Nuclepore) by application of gentle vacuum pressure (≈2 kPa). Filters were mounted in melting-point bath oil (M-6884; Sigma). At least 400 DAPI-stained cells were counted by examining at least 20 randomly chosen microscopic fields.
Biofilter and soil samples were hybridized, washed, and counterstained with DAPI as described for the reference strains. Bacterial detection rates calculated from EUB338-positive counts relative to DAPI counts were corrected for different fixation procedures when high proportions (>10%) of members of the Firmicutes (gram-positive bacteria) were present. The differences between percentages of ethanol- and PFA-fixed samples for probes HGC69a, LGC354a, LGC354b, and LGC354c were thus added to the percentages of PFA-fixed samples for probe EUB338. When PFA-fixed samples yielded higher percentages of LGC354a-, LGC354b-, and LGC354c-labeled cells than ethanol-fixed samples, these (negative) differences were not taken into account for the correction of eubacterial counts. The freshwater sample was directly enumerated as described by Friedrich et al. (10) and hybridized as described by Glöckner et al. (12). Probe-positive counts were determined relative to DAPI counts. The number of DAPI-stained cells counted varied between 400 and 700 cells per replicate. At least 20 randomly chosen fields were examined per replicate. Between two and four replicates were counted per sample.
Statistical analysis.
For the analysis of relationships between variables, Pearson product-moment correlation coefficients were calculated. When the assumptions of normality and equal variance did not apply, the data were log10 transformed prior to calculating regressions. Percentages were transformed by arcsine √x/100 before statistical analysis.
RESULTS
Design and optimization of probe XAN818.
The rRNA data set of the software package ARB used for the probe design in the present study contained 5,138 small-subunit rRNA sequences of at least 1,400 residues (plus 2,778 sequences of shorter lengths). The alignment contained 34 nearly complete sequences of members of the Xanthomonas branch. Potential target regions for a probe specific for the Xanthomonas branch were positions 92 to 109, 818 to 835, 861 to 878, 1143 to 1160, and 1428 to 1445, based on Escherichia coli 16S rRNA numbering (6). Only positions 861 to 878 proved to be inaccessible, as found by whole-cell hybridization, among the targets that we tested (the latter four regions). Positions 818 to 835 were best suited for a probe specifically targeting bacteria of the Xanthomonas branch, since all sequenced members have retained this target sequence and nontarget organisms had a minimum of two mismatches (Table 3).
TABLE 3.
Difference alignment of the target sequence of probe XAN818 and sequences of exemplary matching target organisms and nontarget organisms
| Species | EMBL accession no. | No. of mismatches | Sequencea |
|---|---|---|---|
| Luteimonas mephitis | AJ012228 | 0 | GCCCUAAAC==================GGAGCAAUC |
| Pseudoxanthomonas broegbernensis | AJ012231 | 0 | GCCCUAAAC==================GGUUCAACU |
| Stenotrophomonas africae | U62646 | 0 | GCCCUAAAC==================GGUGCAAUU |
| Stenotrophomonas maltophilia | X95923 | 0 | GCCCUAAAC==================GGUGCAAUU |
| Stenotrophomonas nitritireducens | AJ012229 | 0 | GCCCUAAAC==================GGUGCAAUU |
| Xanthomonas campestris | X95917 | 0 | GCCCUAAAC==================GGUGCAAUU |
| Xylella fastidiosa | M26601 | 0 | GCCCUAAAC==================AGUGCAAAU |
| Beggiatoa alba | L40994 | 2 | GCCCUAAAC====a=====A=======GGAGAGACU |
| Acidiphilium acidophilum | M79399 | 3 | GCUGUAAAC====u=Ug==========GGN |
| Gluconobacter oxydans subsp. oxydans | X73820 | 3 | GCUGUAAAC====u=Ug==========GGAAACUUA |
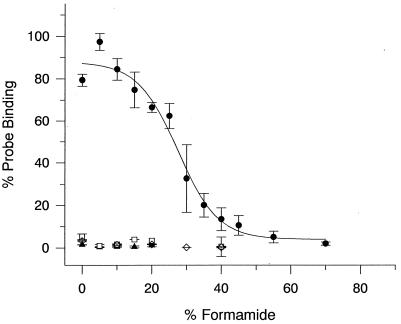
The optimum hybridization stringency in order to discriminate between target and nontarget organisms was evaluated by use of a formamide concentration gradient in the hybridization buffer (33). The effect of increased formamide concentration on the fluorescence intensity conferred by Cy3-labeled probe XAN818 hybridized with a target organism, Stenotrophomonas nitritireducens DSM 12575T, and three nontarget organisms is shown in Fig. 1. Probe binding to the target organism decreased at formamide concentrations above 10 to 15%, attaining background intensities at about 50% formamide. Probe-conferred fluorescence of the nontarget organisms was in the background range, even at 0% formamide. Nontarget organisms can thus be discriminated without the addition of formamide to the hybridization buffer. However, since there were no significant intensity losses, a concentration of 10% formamide was used for all subsequent hybridizations. FISH tests with reference strains confirmed a previous finding with an isolate of the Xanthomonas branch (7) that representatives of this lineage are detected by neither probe GAM42a nor probe BET42a (Table 1). Probe XAN818, presented here, hybridized with all members of the branch tested, confirming the results of the database retrieval.
FIG. 1.
Quantification of binding of probe XAN818 relative to that of probe EUB338 measured as fluorescence intensity. Different stringencies were obtained by varying the concentration of formamide in the hybridization buffer. The target sequence of probe XAN818 and base changes in nontarget organisms are shown in Table 3. Symbols: ●, S. nitritireducens DSM 12575T; ◊, B. alba DSM 1416; □, A. acidophilum DSM 700T; ▴, G. oxydans subsp. oxydans DSM 3503T. Error bars show standard deviations.
Physicochemical and community analyses.
H+ concentrations varied by more than 3 orders of magnitude in the different biofilter materials, with the pHs being 3.0 in sample Hamm A and 7.5 in sample Paderborn B (Table 4). The water content showed little variation among the organic materials, with an average of 72.6% and a coefficient of variation of 5.8%. Cell concentrations, as determined by direct counts with DAPI, varied between 5.5 × 109 and 7.8 × 1010 per g of dry weight among the organic biofilter materials, whereas counts of only 2.2 × 109 were determined for Paderborn B, which contained porous clay as the filter material (Table 4).
TABLE 4.
Physicochemical properties and cell concentrations of the samples
| Sample | pH | Water content (%) | Cell concna |
|---|---|---|---|
| Alberta | 7.0 | 76.5 | 7.8 × 1010 (1.1 × 1010) |
| Eversburg | 7.4 | 78.2 | 1.5 × 1010 (3.5 × 108) |
| Hamm A | 3.0 | 65.5 | 7.2 × 109 (1.9 × 109) |
| Hamm B | 4.2 | 71.0 | 5.5 × 109 (4.6 × 108) |
| Paderborn A | 4.1 | 74.0 | 1.4 × 1010 (1.6 × 109) |
| Paderborn B | 7.5 | 44.0 | 2.2 × 109 (3.4 × 107) |
| Sage A | 4.5 | 72.3 | 1.4 × 1010 (1.2 × 109) |
| Sage B | 6.7 | 70.4 | 2.2 × 1010 (6.4 × 108) |
| Soil A | 4.1 | 41.0 | 3.2 × 109 (2.9 × 108) |
| Soil B | 3.5 | 54.4 | 4.3 × 109 (3.2 × 108) |
| Freshwater pond | 7.5 | 6.1 × 106 (6.4 × 105) |
Reported as mean (standard deviation) for three replicates and given in counts per gram of dry weight for all but the freshwater pond sample, reported as counts per milliliter.
Treatment with Ringer solution removed considerable amounts of particles from the biofilter material in the size range of <10 μm to a few hundred micrometers. Most of the bacteria present in the preparations were attached to these particles. This result indicates that varying affinities of different taxa for particulate matter will not affect the results obtained after the treatment of biofilter material with Ringer solution.
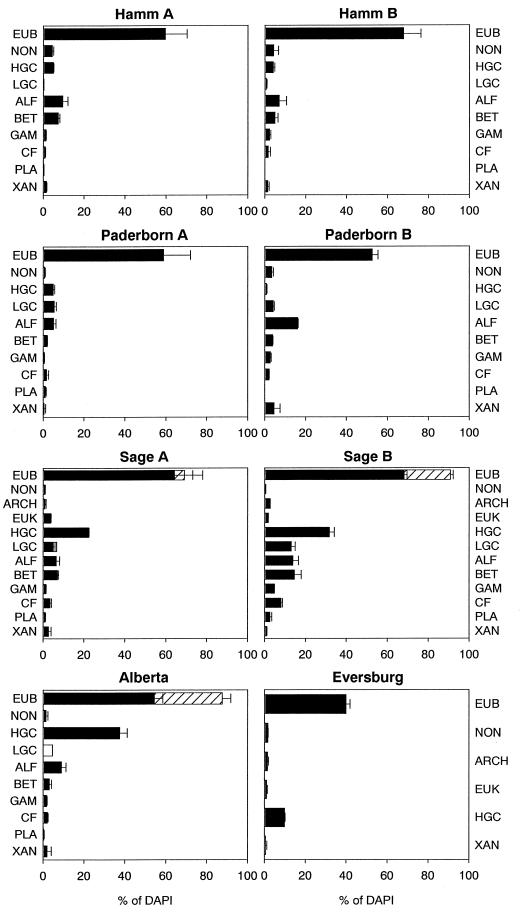
Bacterial detection rates obtained with probe EUB338 varied between 39.9% in sample Eversburg and 68.2% in sample Sage B (Fig. 2) and were similar to the rates reported for other systems (12, 13, 25, 34, 39). Probe EUB338 is usually applied in combination with PFA-fixed samples. However, ethanol is more suitable for the fixation of members of the Firmicutes (30). In samples with high proportions of members of the Firmicutes, bacterial detection rates may thus be underestimated when PFA is used as a fixative. In the present study, percentages of EUB338-positive bacteria were therefore corrected for samples with high proportions of bacteria belonging to the Firmicutes by hybridizing and enumerating ethanol- and PFA-fixed samples with probes LGC354a, LGC354b, LGC354c, and HGC69a (Table 5). Ethanol proved to be more effective than PFA for members of the Actinobacteria. The largest difference between ethanol- and PFA-fixed samples was found for the Alberta sample. Detection rates (relative to those obtained with DAPI) obtained with probe HGC69a were 37.5% for the ethanol-fixed sample and merely 4.3% for the PFA-fixed sample. In comparison, there were no consistent differences between the two fixatives for bacteria belonging to the Firmicutes with a low G+C content (Table 5). Whereas ethanol was more effective than PFA for sample Sage B, it was less effective for samples Alberta and Sage A. The positive differences between detection rates for ethanol- and PFA-fixed samples were then added to the proportions obtained with probe EUB338. This correction led to higher detection rates, particularly for samples Alberta and Sage B, for which 87.7 and 90.8% of all DAPI-stained cells, respectively, were thus identified as bacteria with probe EUB338 (Fig. 2).
FIG. 2.
Probe-specific counts, relative to direct enumeration with DAPI (percentages) (■). Percentages of EUB338 positive bacteria corrected for samples with high proportions of members of the Firmicutes (▨). Percentages of LGC obtained when PFA fixation was more efficient than ethanol fixation (□) (see Materials and Methods and Table 5). Percentages given are corrected for negative control NON338. Probe names are abbreviations of those shown in Table 2. Error bars indicate standard deviations for two to four replicates.
TABLE 5.
Percentages of cells labeled with probes HGC69a, LGC354a, LGC354b, and LGC354c after ethanol and PFA fixation, relative to DAPI counts
| Sample | %a of cells labeled with the following probe(s) after the indicated type of fixation:
|
|||||
|---|---|---|---|---|---|---|
| HGC69a
|
LGC354a, LGC354b, and LGC354c
|
|||||
| Ethanol | PFA | Difference | Ethanol | PFA | Difference | |
| Alberta | 37.5 (3.7) | 4.3 (1.0) | 33.2 | 0 (0) | 4.6 (0) | −4.6 |
| Sage A | 22.2 (0.3) | 17.3 (1.1) | 4.9 | 5.0 (0.9) | 6.6 (0.1) | −1.6 |
| Sage B | 31.7 (2.4) | 13.6 (1.2) | 18.1 | 13.1 (1.9) | 8.7 (0.8) | 4.4 |
Reported as mean (standard deviation) for two to four replicates.
The two sampling sites for the Hamm biofilter showed similar bacterial community patterns, with the α-Proteobacteria (ALF) being the most dominant group, followed by the β-Proteobacteria (BET) and the Actinobacteria (HGC) (Fig. 2). Members of the Cytophaga-Flavobacterium cluster (CF), γ-Proteobacteria (GAM), Xanthomonas branch (XAN), Firmicutes with low G+C DNA content (LGC), and Planctomycetes (PLA) varied between 0 and 2.3% (Fig. 2). Interestingly, proportions for members of the Xanthomonas branch were in the same range as those for members of the γ-Proteobacteria. The Paderborn biofilters used for styrene degradation differed with respect to the filter material used (see Materials and Methods). They also differed in terms of their bacterial community patterns. Whereas in Paderborn biofilter A ALF, HGC, and LGC were of similar importance, ALF dominated in biofilter B (Fig. 2). In this biofilter, members of the Xanthomonas branch were the second most dominant group, accounting for 4.5% of all DAPI-stained cells. The two sampling sites for the Sage biofilter used for waste gas treatment in an industrially operated chicken farm showed different detection rates but similar community patterns. The bacterial communities of both sites were dominated by HGC, BET, ALF, and LGC (Fig. 2). Members of the Xanthomonas group accounted for 2.6 and 0.9% at sites A and B, respectively, and were again in the range for members of the γ-Proteobacteria. Members of the Eucarya and Archaea were only of minor importance in terms of abundance, representing 0.9 to 3.6%, relative to DAPI counts. In the Alberta laboratory-scale biofilter used for hexane degradation, HGC was the most dominant group, followed by ALF, BET, and CF (Fig. 2). XAN and GAM accounted for 2.0 and 1.8%, respectively (Fig. 2).
The proportions of bacteria that were detected with probe EUB338 and that could be assigned to taxa with the group-specific probes used varied greatly among biofilters and sites. Whereas in sample Sage B nearly all bacteria could be identified as members of the taxa examined, there were considerable proportions of unidentified bacteria among the other filter samples. These varied between 18% in Paderborn B and nearly 46% in Hamm B, relative to DAPI counts, indicating that significant percentages of other, nontargeted bacterial taxa were present in these samples. The correction of eubacterial detection rates by use of the results shown in Table 5 had a significant influence on the proportions of unidentified taxa. Before correction, the sum of the percentages derived from bacterial class- and subclass-specific probes was higher than that derived from probe EUB338 for sample Sage B and nearly equal to the latter for sample Alberta. This situation changed after the correction was taken into account: whereas in the Sage B sample nearly all bacteria could be identified as members of the taxa examined, 27.6% of bacteria belonged to unidentified taxa in the Alberta sample. Hence, the correction of eubacterial detection rates in samples with high concentrations of members of the Firmicutes, as used in the present study, may be crucial to derive meaningful data. Percentages of Planctomycetes in counts obtained with probe PLA46 were not subtracted from the counts obtained with probe EUB338, as these organisms generally do not hybridize with probe EUB338 (24).
Probes EUB338, NON338, and XAN818 were also applied in terrestrial and aquatic environments. Percentages of bacteria detected with probe EUB338 were 45.0% ± 0.2% and 50.4% ± 4.4% in soil samples A and B, respectively, and 55.9% ± 1.0% in the freshwater pond sample. Amounts of bacteria belonging to the Xanthomonas branch were fairly low in these samples, being 1.5 and 1.1% in soil samples A and B, respectively, and close to the detection limit in the freshwater pond sample.
There were no significant relationships between physicochemical (pH and water content) and bacterial parameters (total cell concentration, probe-specific percentages) of the biofilter data.
DISCUSSION
Design of probes specific for the Xanthomonas branch.
Our alignment demonstrated that positions 818 to 835 (E. coli numbering) are most appropriate for a probe targeting the 16S rRNA of members of the Xanthomonas branch. Among the four potential target regions that we examined, only positions 861 to 878 were inaccessible by whole-cell hybridization. Interestingly, Fuchs et al. (11) recently found good accessibility of positions 853 to 870 and 863 to 880 in cells of E. coli DSM 30083T. Since both probes, i.e., that used by Fuchs et al. (11) to target positions 863 to 880 and the one that we used to target positions 861 to 878, were self-complementary to the same extent, potential Watson-Crick pairings within the probes may not be the reason for the observed differences in accessibility. These results rather suggest that accessibility may vary among different taxa. The fluorescence intensity conferred by probe XAN818 was 97% that conferred by probe EUB338 at 5% formamide for Stenotrophomonas nitritireducens DSM 12575T (Fig. 1). This intensity was 5.7 times higher than that found for the same region in cells of E. coli DSM 30083T, which showed only 17% the fluorescence intensity obtained with probe EUB338 (11). The hybridization stringency applied by Fuchs et al. (11) was similar to that obtained at 0% formamide in the present study (for fluorescence values, see Fig. 1). Thus, the observed lower probe-conferred fluorescence in E. coli DSM 30083T may be due to probe-specific differences (e.g., probe quality, dissociation temperature, and so forth) and/or to different accessibilities of the same target site among the taxa examined. The latter could increase the taxonomic specificity of in situ hybridization beyond the level obtained by sequence differences. The results presented here stress the necessity to test every newly developed probe individually.
In situ identification of members of the Xanthomonas branch.
Methods for the in situ identification of bacteria of the Xanthomonas branch are interesting for several applications. Members of the genus Xanthomonas are sometimes difficult to identify, especially if the origin of the isolates is uncommon (5). In comparison to time-consuming methods based on isolation techniques, in situ hybridization facilitates identification procedures. Probe XAN818 may also be suited for the assessment of Xylella- and Xanthomonas-caused plant diseases. The identification of the causative microorganisms has been addressed by a number of studies using different techniques (1, 2, 15). However, the methods used do not allow in situ identification and enumeration, which are possible with FISH. Furthermore, probe XAN818 allows differentiation between bacteria of the genera Pseudomonas and Xanthomonas, which is sometimes a difficult task (5). Apart from its use for the phytopathogenic genera, probe XAN818 may be useful in clinical studies focusing on the identification of human pathogens such as Stenotrophomonas maltophilia (8, 31).
Tanner et al. (36) have recently shown that PCR may easily amplify contaminant DNA, resulting in biased results obtained by PCR-based assessments of bacterial diversity. These problems may arise because of the difficulty in preparing genomic DNA absolutely free from contaminating DNA and the exquisite sensitivity of PCR. Therefore, the occurrence of an organism indicated by a cloned rRNA sequence requires explicit detection of that organism in situ (36). Probe XAN818 may serve as a tool for such analyses if cloned rRNA sequences can be assigned to the Xanthomonas group, a contaminant of which was found by Tanner et al. (36) in their negative extraction controls.
Community analysis of biofilter samples.
No significant correlations were obtained from the statistical analysis of the parameters pH and water content versus bacterial parameters of the pooled data from the biofiltration systems. There were similar community patterns between sampling sites for the industrial biofilters Hamm and Sage. For samples from biofilter Hamm, detection rates also were similar. These differed for the two Sage biofilter sites (Fig. 2). The higher detection rates at Sage site B might be a result of the nearly neutral pH at this site, because higher acidity, as found at site A, might reduce overall bacterial activity, numbers of ribosomes and, thus, detection rates with rRNA-targeted probes. It can be expected that waste gas loading will be an important factor in determining the microbial communities of biofilters. The rather complex waste gas composition of a chicken farm (Sage) may thus result in a different community pattern than artificial waste gas containing the single compound hexane (Alberta) or styrene (Paderborn). Apart from waste gas composition, the filter material itself also may have an impact on the microbial community. The material of the Paderborn A biofilter consisted of crushed wood and bark compost, whereas the Paderborn B filter contained porous clay. Even though both biofilters were used for styrene degradation, the community patterns differed substantially (Fig. 2). Also, the pHs differed, being 4.1 and 7.5 at sites A and B, respectively. The different community patterns may be a consequence of the differences in the physical and/or chemical environment.
The most commonly used biofiltration material, crushed tree roots, poses some difficulties for investigations of the microorganisms involved. The main reasons are the large number of different size classes, the morphological heterogeneity of the material, and the high organic content of the material. The results presented here demonstrate that FISH is a valuable tool for studying biofiltration systems, being both practical and reproducible. Our results also stress the necessity to account for the different efficiencies of formaldehyde and ethanol in permeabilizing the cell wall of bacteria of the phylum Firmicutes, particularly in systems with high concentrations of these bacteria. Detection rates with bacterium-specific probes can be significantly improved if these differences are corrected for. The results of the present study also show the presence of significant proportions of bacteria that can be identified with probe EUB338 but not with probes targeting major groups within the Bacteria. Therefore, identification and enumeration of the remaining taxa of the microbial community of biofilters should be an interesting avenue for future research.
REFERENCES
- 1.Albibi R, Chen J, Lamikanra O, Banks D, Jarret R L, Smith B J. RAPD fingerprinting Xylella fastidiosa Pierce’s disease strains isolated from a vineyard in North Florida. FEMS Microbiol Lett. 1998;165:347–352. [Google Scholar]
- 2.Alippi A M, Ronco L, Alippi H E. Bacterial leaf blight affecting Syngonium podophyllum in Argentina. Rev Argent Microbiol. 1994;26:133–138. [PubMed] [Google Scholar]
- 3.Amann R I, Krumholz L, Stahl D A. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990;172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amann R I, Ludwig W, Schleifer K-H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Auling G, Busse H-J, Pilz F, Webb L, Kneifel H, Claus D. Rapid differentiation, by polyamine analysis, of Xanthomonas strains from phytopathogenic pseudomonads and other members of the class Proteobacteria interacting with plants. Int J Syst Bacteriol. 1991;41:223–228. [Google Scholar]
- 6.Brosius J, Dull T J, Sleeter D, Noller H F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981;148:107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- 7.Buchholz-Cleven B E E, Rattunde B, Straub K L. Screening for genetic diversity of isolates of anaerobic Fe(II)-oxidizing bacteria using DGGE and whole-cell hybridization. Syst Appl Microbiol. 1997;20:301–309. [Google Scholar]
- 8.Denton M, Kerr K G. Microbiological and clinical aspects of infection associated with Stenotrophomonas maltophilia. Clin Microbiol Rev. 1998;11:57–80. doi: 10.1128/cmr.11.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finkmann, W., K. Altendorf, E. Stackebrandt, and A. Lipski. Characterization of N2O-producing Xanthomonas-like isolates from biofilters as Stenotrophomonas nitritireducens sp. nov., Luteimonas mephitis gen. nov., sp. nov., and Pseudoxanthomonas broegbernensis gen. nov., sp. nov. Int. J. Syst. Bacteriol., in press. [DOI] [PubMed]
- 10.Friedrich U, Schallenberg M, Holliger C. Pelagic bacteria-particle interactions and community-specific growth rates in four lakes along a trophic gradient. Microb Ecol. 1999;37:49–61. doi: 10.1007/s002489900129. [DOI] [PubMed] [Google Scholar]
- 11.Fuchs B M, Wallner G, Beisker W, Schwippl I, Ludwig W, Amann R. Flow cytometric analysis of the in situ accessibility of Escherichia coli 16S rRNA for fluorescently labeled oligonucleotide probes. Appl Environ Microbiol. 1998;64:4973–4982. doi: 10.1128/aem.64.12.4973-4982.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glöckner F O, Amann R, Alfreider A, Pernthaler J, Psenner R, Trebesius K, Schleifer K-H. An in situ hybridization protocol for detection and identification of planktonic bacteria. Syst Appl Microbiol. 1996;19:403–406. [Google Scholar]
- 13.Kalmbach S, Manz W, Szewzyk U. Dynamics of biofilm formation in drinking water: phylogenetic affiliation and metabolic potential of single cells assessed by formazan reduction and in situ hybridization. FEMS Microbiol Ecol. 1997;22:265–279. [Google Scholar]
- 14.Lathe R. Synthetic oligonucleotide probes deduced from amino acid sequence data. Theoretical and practical considerations. J Mol Biol. 1985;183:1–12. doi: 10.1016/0022-2836(85)90276-1. [DOI] [PubMed] [Google Scholar]
- 15.Leite R P, Minsavage G V, Bonas U, Stall R E. Detection and identification of phytopathogenic Xanthomonas strains by amplification of DNA sequences related to the hrp genes of Xanthomonas campestris pv. vesicatoria. Appl Environ Microbiol. 1994;60:1068–1077. doi: 10.1128/aem.60.4.1068-1077.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lipski A, Klatte S, Bendinger B, Altendorf K. Differentiation of gram-negative, nonfermentative bacteria isolated from biofilters on the basis of fatty acid composition, quinone system, and physiological reaction profiles. Appl Environ Microbiol. 1992;58:2053–2065. doi: 10.1128/aem.58.6.2053-2065.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maidak B L, Cole J R, Parker C T, Jr, Garrity G M, Larsen N, Li B, Lilburn T G, McCaughey M J, Olsen G J, Overbeek R, Pramanik S, Schmidt T M, Tiedje J M, Woese C R. A new version of the RDP (Ribosomal Database Project) Nucleic Acids Res. 1999;27:171–173. doi: 10.1093/nar/27.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manz W, Amann R, Ludwig W, Vancanneyt M, Schleifer K-H. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum cytophaga-flavobacter-bacteroides in the natural environment. Microbiology. 1996;142:1097–1106. doi: 10.1099/13500872-142-5-1097. [DOI] [PubMed] [Google Scholar]
- 19.Manz W, Amann R, Ludwig W, Wagner M, Schleifer K-H. Phylogenetic oligodeoxynucleotide probes for the major subclasses of proteobacteria: problems and solutions. Syst Appl Microbiol. 1992;15:593–600. [Google Scholar]
- 20.Meier H, Amann R, Ludwig W, Schleifer K-H. Specific oligonucleotide probes for in situ detection of a major group of Gram-positive bacteria with low DNA G+C content. Syst Appl Microbiol. 1999;22:186–196. doi: 10.1016/S0723-2020(99)80065-4. [DOI] [PubMed] [Google Scholar]
- 21.Møller S, Pedersen A R, Poulsen L K, Arvin E, Molin S. Activity and three-dimensional distribution of toluene-degrading Pseudomonas putida in a multispecies biofilm assessed by quantitative in situ hybridization and scanning confocal laser microscopy. Appl Environ Microbiol. 1996;62:4632–4640. doi: 10.1128/aem.62.12.4632-4640.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moore E R B, Krüger A S, Hauben L, Seal S E, de Baere R, de Wachter R, Timmis K N, Swings J. 16S rRNA gene sequence analyses and inter- and intrageneric relationships of Xanthomonas species and Stenotrophomonas maltophilia. FEMS Microbiol Lett. 1997;151:145–153. doi: 10.1111/j.1574-6968.1997.tb12563.x. [DOI] [PubMed] [Google Scholar]
- 23.Neef A. Anwendung der in situ-Einzell-Identifizierung von Bakterien zur Populationsanalyse von komplexen mikrobiellen Biozönosen. Ph.D. thesis. Munich, Germany: Technische Universität München; 1997. [Google Scholar]
- 24.Neef A, Amann R, Schlesner H, Schleifer K-H. Monitoring a widespread bacterial group: in situ detection of planctomycetes with 16S rRNA-targeted probes. Microbiology. 1998;144:3257–3266. doi: 10.1099/00221287-144-12-3257. [DOI] [PubMed] [Google Scholar]
- 25.Neef A, Zaglauer A, Meier H, Amann R, Lemmer H, Schleifer K-H. Population analysis in a denitrifying sand filter—conventional and in situ identification of Paracoccus spp. in methanol-fed biofilms. Appl Environ Microbiol. 1996;62:4329–4339. doi: 10.1128/aem.62.12.4329-4339.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ottengraf S P P. Exhaust gas purification. In: Rehm H J, Reed G, editors. Biotechnology. Vol. 8. Weinheim, Germany: VCH-Verlagsgesellschaft; 1986. pp. 425–452. [Google Scholar]
- 27.Pomeroy, R. D. 1957. U.S. patent 2,793,096.
- 28.Porter K G, Feig Y S. The use of DAPI for identifying and counting aquatic microflora. Limnol Oceanogr. 1980;25:943–948. [Google Scholar]
- 29.Reichert K, Lipski A, Pradella S, Stackebrandt E, Altendorf K. Pseudonocardia asaccharolytica sp. nov. and Pseudonocardia sulfidoxydans sp. nov., two new dimethyl sulfide-degrading actinomycetes and emended description of the genus Pseudonocardia. Int J Syst Bacteriol. 1998;48:441–449. doi: 10.1099/00207713-48-2-441. [DOI] [PubMed] [Google Scholar]
- 30.Roller C, Wagner M, Amann R, Ludwig W, Schleifer K-H. In situ probing of Gram-positive bacteria with high DNA G+C content using 23S rRNA-targeted oligonucleotides. Microbiology. 1994;140:2849–2858. doi: 10.1099/00221287-140-10-2849. [DOI] [PubMed] [Google Scholar]
- 31.Rusin P A, Rose J B, Haas C N, Gerba C P. Risk assessment of opportunistic bacterial pathogens in drinking water. Rev Environ Contam Toxicol. 1997;152:57–83. doi: 10.1007/978-1-4612-1964-4_2. [DOI] [PubMed] [Google Scholar]
- 32.Sakano Y, Kerkhof L. Assessment of changes in microbial community structure during operation of an ammonia biofilter with molecular tools. Appl Environ Microbiol. 1998;64:4877–4882. doi: 10.1128/aem.64.12.4877-4882.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stahl D A, Amann R. Development and application of nucleic acid probes. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. Chichester, England: John Wiley & Sons Ltd.; 1991. pp. 205–248. [Google Scholar]
- 34.Stoffels M, Amann R, Ludwig W, Hekmat D, Schleifer K-H. Bacterial community dynamics during start-up of a trickle-bed bioreactor degrading aromatic compounds. Appl Environ Microbiol. 1998;64:930–939. doi: 10.1128/aem.64.3.930-939.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Strunk, O., O. Gross, B. Reichel, M. May, S. Hermann, N. Stuckman, B. Nonhoff, M. Lenke, A. Ginhart, A. Vilbig, T. Ludwig, A. Bode, K.-H. Schleifer, and W. Ludwig. March 1997, posting date. ARB: a software environment for sequence data. [Online.] Department of Microbiology, Technische Universität München, Munich, Germany. http://www.mikro.biologie.tu-muenchen.de. [June 1999, last date accessed.]
- 36.Tanner M A, Goebel B M, Dojka M A, Pace N R. Specific ribosomal DNA sequences from diverse environmental settings correlate with experimental contaminants. Appl Environ Microbiol. 1998;64:3110–3113. doi: 10.1128/aem.64.8.3110-3113.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trebesius K, Amann R, Ludwig W, Mühlegger K, Schleifer K-H. Identification of whole fixed bacterial cells with nonradioactive 23S rRNA-targeted polynucleotide probes. Appl Environ Microbiol. 1994;60:3228–3235. doi: 10.1128/aem.60.9.3228-3235.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wallner G, Amann R, Beisker W. Optimizing fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry. 1993;14:136–143. doi: 10.1002/cyto.990140205. [DOI] [PubMed] [Google Scholar]
- 39.Zarda B, Hahn D, Chatzinotas A, Schönhuber W, Neef A, Amann R I, Zeyer J. Analysis of bacterial community structure in bulk soil by in situ hybridization. Arch Microbiol. 1997;168:185–192. [Google Scholar]