Abstract
Allergic inflammation refers to a hyperimmune reaction that causes hypersensitivity responses such as hives, itchiness, runny nose, and cough due to specific allergens. Allergic diseases are known to be influenced by the diversity and distribution of intestinal microbiota, and Lactobacill is known to relieve allergic symptoms by modulating cytokines secreted by T helper type 1 (Th1)/Th2 cells. This study was designed to investigate the effects of Lactobacillus gasseri MG4247 and Lacticaseibacillus paracasei MG4272, MG4577, and MG4657 on levels of pro-inflammatory cytokines and proteins associated with allergic symptoms in RAW 264.7 macrophages, and RBL-2H3 mast cells, as well as their probiotic properties. MG4247, MG4272, and MG4577 significantly reduced tumor necrosis factor-α and interleukin (IL)-6 levels in LPS-induced RAW 264.7 macrophages, and markedly decreased IL-4, IL-5, and IL-13 levels and STAT6 phosphorylation in DNP-IgE/HSA sensitized RBL-2H3 mast cells. Furthermore, MG4247, MG4272, and MG4577 tolerated the acidic condition with pepsin and basic condition with bile salt, and showed a high adhesion rate (≥ 73.9%). In safety evaluation, MG4247, MG4272, and MG4577 showed no hemolytic or bile salt hydrolase activity and no cytotoxicity to HT-29 cells (≥ 96.7%). Hence, MG4272, MG4272, and MG4577 can be used as candidate probiotic strains to relieve cytokines associated with allergic inflammation.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12602-022-09950-4.
Keywords: Allergic symptom, Inflammation, Probiotics, Immune response
Introduction
Allergies are inflammatory diseases caused by the disorder of the immune system and hypersensitivity [1]. Allergies are caused by various allergens including food, insects, drugs, and the environment, and manifest as this allergic diseases such as rhinitis, anaphylaxis, asthma, and atopic dermatitis [2]. The prevalence of allergies is over 4 million worldwide, and it is increasing annually, making it a major global health concern [3]. Disruption of the normal immune response results in an imbalance between T helper type 1 (Th1) and T helper type 2 (Th2) cells [4]. In addition, macrophages participate in the immune response during allergic inflammation [5]. Macrophages secrete Th1 cytokines such as tumor necrosis factor α (TNFα), and interleukin (IL)-6, which are pro-inflammatory mediators of inflammation by allergen [5, 6]. Th2 cytokines, including IL-4, IL-5, and IL-13, are produced by the phosphorylation of signal transducer and activator of transcription 6 (STAT6) in mast cells, and sensitize allergens [4, 7]. Antihistamines have been used to treat of allergic symptoms; however, this class of drug is not ideal because of its adverse effects, such as dizziness, fatigue, and drowsiness [8]. Therefore, complementary and alternative treatments with minimal side effects are needed to improve allergic inflammation.
Probiotics, including lactic acid bacteria (LAB), are defined as living microorganisms that provide health benefits to the host [9]. Recently, probiotics have been reported to have various physiological roles and beneficial effects on various diseases such as gastrointestinal disorders, metabolic disorders, and allergic inflammation [10, 11]. Song et al. demonstrated that Lactiplantibacillus plantarum L67 suppressed the expression of pro-inflammatory cytokines in bisphenol A-exposed RBL-2H3 mast cells, and inflammatory factors in cadmium-treated RAW 264.7 macrophages [12]. Bifidobacterium longum IM55 and L. plantarum IM76 are reliving Th2 cytokine levels in house dust mite allergen and ovalbumin-induced allergic mouse models [13, 14]. Probiotics, including Lacticaseibacillus paracasei, Lactobacillus gasseri Lactobacillus acidophilus, and Lacticaseibacillus rhamnosus GG improve the condition of patients with allergic rhinitis by decreasing Th2 and pro-inflammatory cytokine levels [15, 16]. The anti-allergy response of probiotics must be investigated for further experiments based on animal models or clinical studies; additionally, more rapid screening by in vitro tests can save time and economically, and scientifically establish the beneficial effects of probiotics [17].
In this study, L. gasseri MG4247 and L. paracasei MG4272, MG4577, and MG4567, a species known to relieve cytokines in rhinitis patients, were investigated in RAW 264.7 macrophages and RBL-2H3 mast cells to evaluate the expression of pro-inflammatory and Th2 cytokines. In addition, LAB, which are thought to suppress allergic symptoms, were assessed for the probiotic properties including stability and safety in HT-29 intestinal epithelial cells.
Materials and Methods
Reagents and Apparatus
All the reagents were purchased from Sigma-Aldrich (St. Louis, MO, USA). De Man, Rogosa, and Sharpe (MRS) broth was purchased from BD Biosciences (Franklin Lakes, NJ, USA). RAW 264.7, RBL-2H3, and HT-29 cell lines were obtained from the Korean Cell Line Bank (Seoul, Korea). Dulbecco’s modified Eagle’s medium (DMEM), fetal bovine serum (FBS), penicillin–streptomycin (P/S), and phosphate-buffered saline (PBS) were purchased from Gibco (Gaithersburg, MD, USA). HiGene™ Total RNA Prep Kit (BIOFACT, Daejeon, Korea) was reverse-transcribed to cDNA using the SuperScript™ IV First-Strand Synthesis System (Invitrogen, Waltham, MA USA), and FastStart Essential DNA Green Master (Roche, Basel, Switzerland) was used for investigating mRNA expression. RIPA buffer was obtained from iNtRON (Seongnam-si, Gyeonggi-do, Korea) and contained phosphatase and protease inhibitor cocktails (GenDEPOT, Katy, TX, USA). The Pierce BCA Protein Assay Kit was purchased from Thermo Fisher Scientific (Waltham, MA, USA). Antibodies against STAT6, and phosphorylated STAT6 (p-STAT6) were purchased from Cell Signaling Technology (Danvers, MA, USA). Amersham™ Imager 600 (GE Healthcare, Chicago, IL, USA) and WesternBright ECL kit (Advansta, San Jose, CA USA) were used for protein image analysis.
Preparation of Cell-Free Supernatants from LAB (SL)
All the LAB used in this study were obtained from MEDIOGEN (Jecheon, Korea). L. gasseri MG4247, and L. paracasei MG4272, MG4577, and MG4657 were isolated from Korean individuals. The selected strains were identified by the 16S rRNA gene sequencing (SolGent Co., Ltd. Korea). The DNA sequence was registered in the National Center for Biotechnology Information (NCBI) database using the Basic Local Alignment Search Tool (BLAST; NCBI Accession number-MG4247: MN069036.1, MG4272: MW947164.1, MG4577: MN833017.1, MG4657: ON025789). Based on BLAST, similarity of strains was confirmed through phylogenetic tree (Fig. S1). A phylogenetic tree was constructed with the neighbor-joining method using MEGA software ver. 10.0 (https://www.megasoftware.net/).
To obtain SL, each LAB cultured in MRS broth at 37 °C for 18 h in an anaerobic chamber were centrifuged (4000 × g) for 15 min at 4 °C. The obtained SLs were filtered using a 0.2-μm polytetrafluoroethylene (PTFE) membrane (ADVANTEC, Tokyo, Japan), and stored at −70 °C for the next experiment.
Cell Culture
Raw 264.7 macrophages and HT-29 cells were grown at 37 °C, 5% CO2 in RPMI medium containing 10% FBS and 1% P/S. RBL-2H3 basophilic leukemia mast cells were grown at 37 °C, 5% CO2 in Minimum Essential Medium (MEM) containing 15% FBS and 1% P/S. Cells were sub-cultured at 70–80% confluency.
Cell Viability
Cell viability was measured using the 3-(4,5-Dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assay [18]. Cells (3 × 104 cells/well) were seeded in 96 well plates and treated with SL for 24 h. MTT solution (0.1 mg/mL) was added to each well, and it was cultured for 1 h. After incubation, the formazan crystals in each well were dissolved in 150 µL DMSO. Absorbance was measured at 550 nm using a microplate reader.
Preparation of Total RNA
RAW 264.7 macrophages were seeded at 3 × 105 cells/well in 6-well plates and incubated for 24 h. The cells were treated with 100 ng/mL of lipopolysaccharide (LPS; Escherichia coli O111:B4) with or without samples diluted to 5% (v/v) in serum-free media (SFM) for 24 h. RBL-2H3 mast cells were seeded at 3 × 105 cells/well in 6-well plates and treated with 110 ng/mL monoclonal anti-dinitrophenyl antibody (DNP-IgE) for 1 h. The cells were incubated with SL, diluted to 5% (v/v) in SFM, for 1 h, and then with 25 ng/mL albumin, dinitrophenyl (DNP-HSA) for 1 h. Total RNA of RAW 264.7 and RBL-2H3 cells was isolated using HiGene™ Total RNA Prep Kit following the manufacturer’s protocols. Total RNA from RAW 264.7 macrophages and RBL-2H3 mast cells was isolated using the HiGene™ Total RNA Prep Kit.
Quantitative Real-Time Polymerase Chain Reaction
Total RNA from RAW 264.7 macrophages and RBL-2H3 mast cells was reverse-transcribed to cDNA using the SuperScript™ IV First-Strand Synthesis System. qPCR was performed using a LightCycler®96 Instrument (Roche) and the FastStart Essential DNA Green Master. The primer sequences are listed in Table 1. Acquisition on the SYBR green was recorded at the end of the extension step. The melting peaks of target genes were obtained with temperatures ranging from 81 to 90 °C (Fig. S2). All data were acquired using the LightCycler®96 software, and relative mRNA was normalized using the house-keeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Fig. S3). The expression levels were analyzed using the 2−ΔΔCT method [19].
Table 1.
Primer sequences of genes used in this study
| Gene | Sequence (from 5′ to 3′) | ||
|---|---|---|---|
| Sense | Antisense | ||
| RAW 264.7 macrophages (mouse) | IL-6 | TACCACTTCACAAGTCGGAGGC | CTGCAAGTGCATCATCGTTGTTC |
| TNF-α | GGTGCCTATGTCTCAGCCTCTT | GCCATAGAACTGATGAGAGGGAG | |
| GAPDH | TCTCCCTCACAATTTCCATCC | GGGTGCAGCGAACTTTATTG | |
| IL-4 | TCCTTACGGCAACAAGGAAC | GTGAGTTCAGACCGCTGACA | |
| RBL-2H3 mast cells (rat) | IL-5 | GATGAGGCTTCCTGTTCCTACT | TGACAAGTTTTGGAATAGTATTTCC |
| IL-13 | AACAGCAGCATGGTATGGAGCG | TGGGTCCTGTGGATGGCATTGC | |
| GAPDH | ATGGGAAGCTGGTCATCAAC | GTGGTTCACACCCATCACAA | |
IL-1β interleukin 1 beta, IL-6 interleukin 6, TNF-α tumor necrosis factor alpha, IL-4 interleukin 4, IL-5 interleukin 5, IL-13 interleukin 13, GAPDH glyceraldehyde-3-phosphate dehydrogenase
Protein Extraction and Western Blotting
The total protein in RBL-2H3 mast cells was extracted using RIPA lysis buffer containing phosphatase and protease inhibitors [20]. The extracted proteins were quantified using the Pierce BCA Protein Assay Kit. Western blotting was performed as previously described [21]. Briefly, 10 μg of protein was loaded onto 8% Tris–HCl gels for sodium dodecyl sulfate-poly acrylamide gel electrophoresis. The proteins were transferred to nitrocellulose membrane (Millipore, Middlesex County, MA, USA), washed, and blocked used by 5% skim milk in PBS-Tween buffer (PBST, pH 7.4) for 1 h. After blocking, the membranes were incubated with primary antibodies (STAT6, and pSTAT6, 1:1000) overnight. The membranes were then incubated with horseradish peroxidase-conjugated secondary antibody (1:10,000 in PBST) for 1 h. Protein expression was analyzed using Amersham™ Imager 600 with the WesternBright ECL kit.
Hemolytic and Bile Salt Hydrolase Activity
Hemolysis of each LAB strains was determined according to the method described in previous report [22]. LAB was cultured in 5% sheep blood on tryptic soy agar to assess the colonies; alpha (green colony), beta (clean zone), or gamma (no change). BSH activity of LAB was measured in MRS agar containing 0.5% (w/v) taurodeoxycholic acid (sodium salt) hydrate and 0.037% CaCl2 at 37 °C, for 48 h [23]. If LAB exhibits BSH activity, a precipitation zone surrounding the colony appears.
Simulated Gastrointestinal Tract
The survival rate of LAB in the simulated gastrointestinal tract was evaluated using a previously reported method [24]. After culturing the LAB for 18 h, they were washed twice with PBS (pH 7.4) and resuspended to 0.95–1.05 at 600 nm. Then, 1 mL of the solution was mixed with 9 mL of PBS (pH 2.5) containing 0.3% pepsin (simulated gastric fluid, SGF) and incubated at 37 °C with gentle shaking at 200 rpm. After 2 h, the pellet was obtained using centrifugation at 4000 rpm for 5 min. Subsequently, 10 mL (pH 7.4) of 1% pancreatin and 1% bile salt (stimulated intestinal fluid, SIF) solutions were added to the pellet and incubated at 37 °C for 2.5 h with gentle shaking at 200 rpm. The number of viable LAB was counted using MRS agar, and the survival rate was calculated as the percentage of viable cells compared to the initial number of cells.
Adhesion to HT-29 Intestinal Epithelial Cells
The adhesion of LAB to HT-29 cells was evaluated as previously described [22]. HT-29 cells (2 × 105 cells/mL) were seeded in 12-well plates and incubated at 37 °C and 5% CO2 for 24 h. LAB strains were cultured in MRS broth at 37 °C for 24 h. The LAB strains were resuspended at 1 × 108 CFU/mL in DMEM without FBS and P/S, and treated with HT-29 cells. After 2 h, the HT-29 cells were washed twice with PBS to detach non-adherent LAB. The number of viable LAB was measured by counting on MRS agar and was calculated as log CFU/mL.
Statistical Analysis
Results are expressed as the mean ± standard error of the mean (SEM). Statistical significance was analyzed using the Student’s t-test, and the Duncan’s multiple range tests with the IBM® Statistical Package for the Social Sciences Statistics (ver. 21.0 for Window, IBM, Armonk, NY, USA). Statistical significance was set at P < 0.05.
Results
Inhibition of Pro-inflammatory Cytokine Expression by L. gasseri and L. paracasei SL in LPS-Induced RAW 264.7 Macrophages
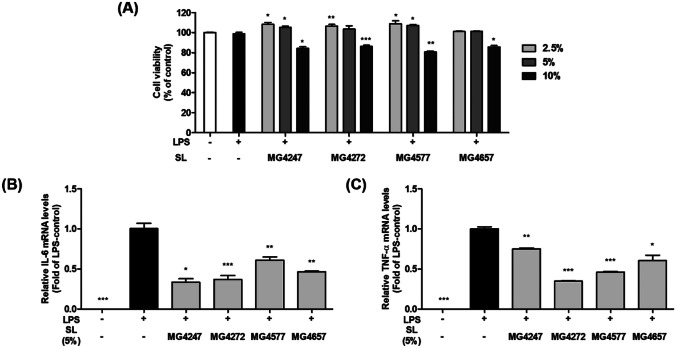
In order to establish a concentration that does not show cytotoxicity, MTT assay was performed with 2.5, 5, and 10% of SL treating in RAW 264.7 macrophages. The 2.5 and 5% SL in SFM exhibited no cytotoxicity (≥ 100%) on RAW 264.7 macrophages (Fig. 1a). With these results, SL 5% was chosen for subsequent experiments. To verify that SL affected pro-inflammation signaling, we examined mRNA expressions of IL-6 and TNFα, which lead to hyperimmune responses in LPS-induced RAW 264.7 macrophages (Fig. 1b and c). IL-6 and TNF-α mRNA levels in LPS-induced macrophages were significantly higher than those in untreated cells. IL-6 and TNF-α mRNA expressions were significantly downregulated by 0.33–0.61 fold and 0.35–0.75 fold in LPS-induced RAW 264.7 macrophages treated with SL, respectively.
Fig. 1.
Cell viability and pro-inflammatory cytokines expression of LPS-induced RAW 264.7 macrophages treated with SL. A Cell viability was assessed using the MTT assay in RAW 264.7 macrophages pretreated with SL (2.5, 5, and 10%) for 1 h and then LPS (0.1 μg/mL) with SL treatment for 24 h. Data has no statistical significance. The mRNA expression of pro-inflammatory cytokines, B IL-6, and C TNF-α, was evaluated using qPCR. Gene expression was normalized with GAPDH. RAW 264.7 macrophages were pretreated with LPS (0.1 μg/mL) and treated with SL for 3 h. Data are presented as the means ± SEM (n = 3). Statistical Significance was determined by one-way analysis of variance according to Student’s t-test; **p < 0.01, ***p < 0.001 vs. non-treated control
Reduction of IL-4, IL-5, and IL-13 Cytokines by SL from L. gasseri and L. paracasei in DNP-IgE Sensitized RBL-2H3 Mast Cells
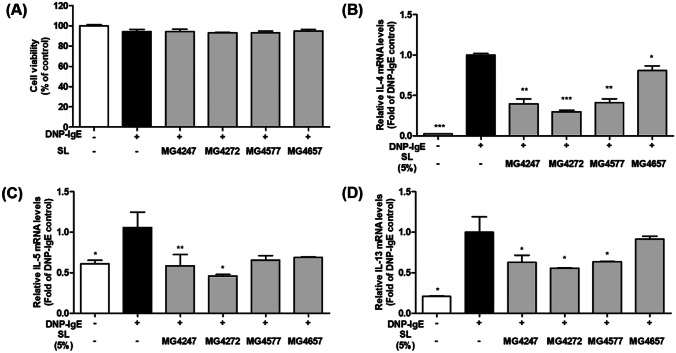
The 5% SL in SFM showed no cytotoxicity (≥ 93%) in RBL-2H3 cells stimulated with DNP-IgE/HSA (Fig. 2a). As with Raw 264.7 macrophage, SL 5% was chosen for subsequent experiments. To verify whether SL might affect the cytokine expression in mast cells, IL-4, IL-5, and IL-13 mRNA expression was examined in DNP-IgE/HSA-sensitized RBL-2H3 mast cells (Fig. 2b–d). Exposure to DNP-IgE/HSA increased IL-4 (47.10-fold), IL-5 (1.66-fold), and IL-13 (4.84-fold) mRNA expression compared with that in non-sensitized RBL-2H3 mast cells; however, treatment with SL significantly decreased the mRNA expression of all cytokines (IL-4, 0.30–0.81 fold; IL-5, 0.46–0.69 fold; and IL-13, 0.76–0.91 fold), except for IL-13, in MG4657 cells. Therefore, MG4247, MG4272, and MG4577 were selected for the subsequent studies.
Fig. 2.
Effect of SL on viability and cytokines levels in DNP-IgE/HSA-sensitized RBL-2H3 mast cells. A Cell viability was assessed using the MTT assay in RBL-2H3 cells treated with anti-DNP-IgE (110 ng/ml) for 1 h, followed by stimulation with SL (5%) for 24 h. The mRNA expression of cytokines, B IL-4, C IL-5, and D IL-13, was evaluated using qPCR in DNP-IgE/HSA induced RBL-2H3 mast cell. Each gene expression was measured in RBL-2H3 cells treated with anti-DNP-IgE (110 ng/ml) for 1 h, followed by stimulation with SL (5%) for IL-4 and IL-13 for 1 h, and IL-5 (3 h), and then with 25 ng/mL DNP-HSA antigen (25 ng/ml). Gene expression was normalized with GAPDH. Data are presented as the mean ± SEM (n = 3). Statistical significance was determined by one-way analysis of variance according to Student’s t-test; **p < 0.01, ***p < 0.001 vs. non-treated control
Effect on STAT6 Phosphorylation by SL from L. gasseri and L. paracasei in DNP-IgE Sensitized RBL-2H3 Cells
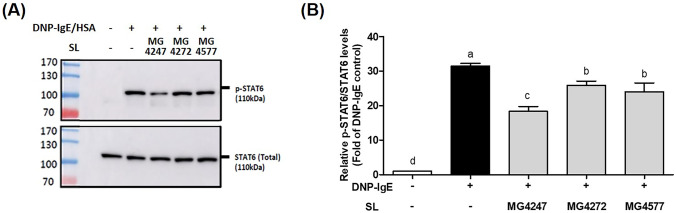
Western blotting was performed to determine whether the LAB candidates affected STAT6 phosphorylation, which leads to the secretion of IL-4, IL-5, and IL-13 in DNP-IgE/HSA-sensitized RBL-2H3 mast cells (Fig. 3a). Exposure of RBL-2H3 mast cells to DNP-IgE/HSA markedly upregulated the expression of p-STAT6/STAT6 (31.49-folds) compared to that in untreated mast cells. SL treatment of RBL-2H3 mast cells induced with DNP-IgE/HSA reduced the expression of p-STAT6/STAT6 (18.36 to 25.91-fold) (Fig. 3b).
Fig. 3.
Effects of SL on STAT6 phosphorylation in DNP-IgE/HSA-sensitized RBL-2H3 mast cell. RBL-2H3 cells were treated with anti-DNP-IgE (110 ng/ml) for 1 h, followed by stimulation with SL (5%) for 1 h, and then with DNP-HSA antigen (25 ng/ml). A STAT6 and p-STAT6 levels were detected by western blotting. B The graphs show STAT6 protein expression relative to no phosphorylation. Data are presented as the mean ± SEM (n = 3). Different letters on each column indicate statistical significance
Safety of L. gasseri MG4247, L. paracasei MG4272, and MG4577 as probiotics
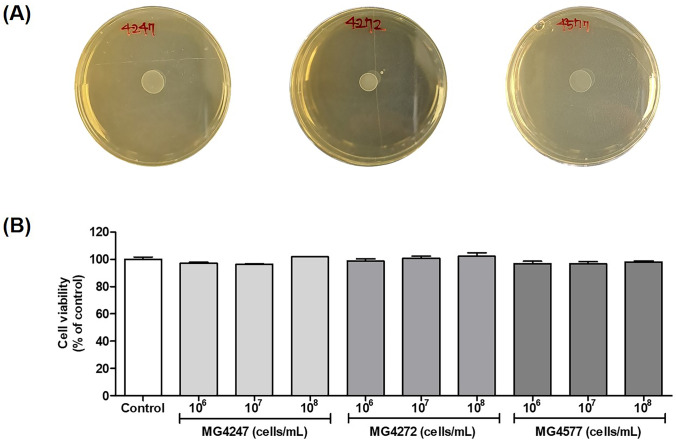
To investigate the safety of MG4247, MG4272, and MG4577, which alleviated the effects of cytokines associated with allergic symptoms, hemolysis, BSH, and cytotoxicity on intestinal epithelial cells were measured. MG4247, MG4272, and MG4577 did not show β-hemolysis (data not shown), indicating no hemolytic activity on the host. In terms of BSH activity, MG4247, MG4272, and MG4577 showed no precipitation (Fig. 4a). Moreover, MG4247, MG4272, and MG4577 (1 × 106–108 cells/mL) were not significantly toxic, indicating that none of the LAB damaged the intestinal epithelium after adherence (Fig. 4b).
Fig. 4.
Safety tests of LAB as probiotics. A BSH activity of L. gasseri MG4247 and L. paracasei MG4272 and MG4577. B Cell viability of HT-29 intestinal epithelial cells treated with L. gasseri MG4247 and L. paracasei MG4272 and MG4577 (1 × 106, 107, and 108 cells/mL). Data are presented as the mean ± SEM (n = 3). Data has no statistical significance
Stability of L. gasseri MG4247, L. paracasei MG4272, and MG4577 in Stimulated GIT
The viability of L. gasseri MG4247 L. paracasei MG4272, and MG4577, which exhibited an anti-allergy effect in this study, was determined using artificial GIT (Table 2). Initial L. gasseri MG4247, L. paracasei MG4272, and MG4577 numbers at OD600 were determined as 8.03, 8.92, and 9.66 Log CFU/mL. In stimulated SGF (at pH 2.5 with 0.3% pepsin), all strains survived ≥ 75%. In stimulated SIF with surviving strains in SGF, L. gasseri MG4247, L. paracasei MG4272, and MG4577 showed 96.18, 87.18, and 73.90% viability.
Table 2.
Tolerance of stimulated GIT to L. gasseri MG4247, L. paracasei MG4272, and MG4577
| Strains | Initial counts (Log CFU/mL) | SGF (Log CFU/mL) | SIF (Log CFU/mL) | Survival rate (%) |
|---|---|---|---|---|
| L. gasseri MG4247 | 8.03 ± 0.01 | 7.85 ± 0.02 | 7.73 ± 0.04 | 96.18 ± 0.51 |
| L. paracasei MG4272 | 8.92 ± 0.01 | 7.96 ± 0.01 | 7.77 ± 0.01 | 87.18 ± 0.11 |
| L. paracasei MG4577 | 9.66 ± 0.03 | 7.25 ± 0.03 | 7.14 ± 0.01 | 73.90 ± 0.50 |
Simulated gastric fluid (SGF) contained 0.3% pepsin at pH 2.5, stimulated intestinal fluid (SIF) contained 1% pancreatin and 1% bile salt at pH 7.4
Data are presented as the mean ± standard deviation (duplicate)
Adhesion of L. gasseri MG4247, L. paracasei MG4272, and MG4577 to HT-29 Intestinal Epithelial Cells
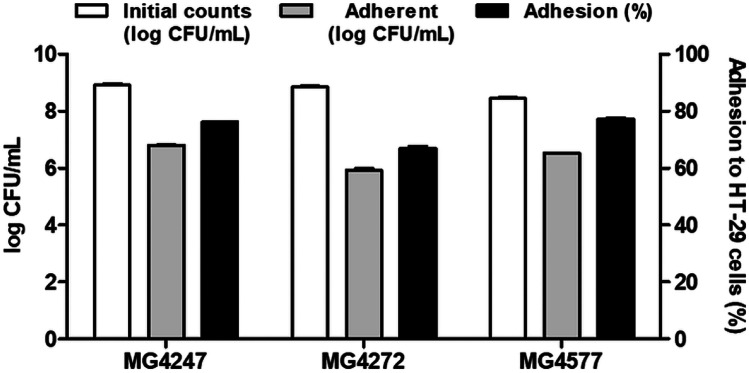
To determine the ability to adhere to intestinal epithelial cells, LAB attachment to HT-29 cells was determined using plate counting (Fig. 5). As a result of expressing the attached L. gasseri MG4247, L. paracasei MG4272, and MG4577 as a percentage in proportion to the initial counts, all strain were confirmed to be 66% or more adherent to HT-29 intestinal epithelial cells.
Fig. 5.
Adhesion of L. gasseri MG4247, L. paracasei MG4272, and MG4577 to HT-29 intestinal epithelial cells. Data are presented as the mean ± SEM (n = 3)
Discussion
Allergic inflammatory responses are stimulated by various environmental factors, including pollen, food, and climatic conditions If allergic responses are untreated, they can lead to bronchitis, pneumonia, asthma, or chronic obstructive pulmonary disease (COPD) [25]. Hence, allergic inflammation should be treated before it progresses to COPD or other diseases. Probiotics not only strengthen the host’s gut immunity but also attenuate inflammatory and allergic diseases [11]. In patients with allergic diseases, the gut microbiota is altered; in comparison with healthy individuals, in patients with allergic diseases, the relative abundance of Lactobacillus and Bifidobacterium is reduced, whereas the abundance of pathogens such as Escherichia coli, Clostridium difficile, and Staphylococcus aureus is increased [26]. In particular, metabolites such as short-chain fatty acids produced by probiotics regulate allergic inflammation through the G protein-coupled receptors in intestinal epithelial cells [27]. Therefore, consuming probiotics with potential as therapeutic agents for allergy symptoms can be a more effective alternative to using the native gut microbiota. When LAB is cultured in MRS, which is the most used to confirm the metabolite difference of LAB, metabolite giving beneficial effect to human is present in the supernatant of MRS medium [28]. It has been reported that various metabolites such as SCFA are produced when probiotics are taken and stabilized in the intestine in human [29]. In addition, different metabolites, even belonging to the same species, are produced in each strain [28]. This study assessed the anti-allergy effects of SL from LAB on pro-inflammatory cytokine levels in macrophages and mast cells. Moreover, we selected LAB as functional probiotic candidates and evaluated their stability and safety as probiotics.
In allergic inflammation, the major cytokines that stimulate inflammation are Th1 and Th2 cytokines [4]. Th1 cytokines, especially TNF-α and IL-6, induce inflammation in macrophages [5]. LPS, which binds to the toll-like receptor 4 in the cell membrane, promotes the nuclear factor kappa-light-chain-enhancer of activated B cells signaling pathway. Because of this pathway, macrophages produce TNF-α- and IL-6 cytokines and Th1 cells undergo differentiated [30]. Our results showed that all LAB strains significantly decreased the mRNA expression of IL-6 and TNF-α. Th2 cytokines are known to cause allergic inflammation by increasing the production of immunoglobulin E (IgE) in B cells [31]. In this study, MG4247, MG4272, and MG4577 reduced the mRNA levels of Th2 cytokines, including IL-4, IL-5, and IL-13. Similarly, many Lactobacillus strains have shown anti-allergy effects by modulating Th2 cytokines in cells and animal model [32]. IL-4/13 cytokines, which are attached to the IL-4/13 receptor, induce STAT6 protein expression in mast cells [33]. JAKs, activated by IL-4/13 cytokines, phosphorylate STAT6, which abnormally secretes Th2 cytokines and initiates allergic inflammation [34]. The p-STAT6, which produces Th2 cytokines, was decreased by MG4247, MG4272, and MG4577 treatment. Comparing the difference between both LAB strains, it was confirmed that both strains significantly lowered Th1 cytokines; however, in Th2 cytokines, especially L. paracasei was significantly reduced. Considering our results, MG4247, MG4272, and MG4577 showed anti-allergy effects by regulating Th1 and Th2 cytokine levels in macrophages and mast cells.
The in vitro properties of certified probiotics include BSH activity, hemolysis, resistance to gastric acidity, and basic conditions with bile acid, and adherence to epithelial cells [35]. Therefore, stability and safety as probiotics of MG4247, MG4272, and MG4577 with anti-allergic effect were investigated. Probiotics have been reported to have various beneficial effects on the host when they reach the intestines after ingestion [36]. Therefore, it is necessary for the probiotics to have the ability to survive in the GIT. In this study, MG4247, MG4272, and MG4577 showed survival rates more than 73.90% in simulated GIT. Other commercial probiotics, including Lactobacillus and Bifidobacterium sp. exhibited 51.72–93.90% survival rate after passing the simulated GIT [36]. In particular, L. paracasei was reported to exhibit 37.60 to 79.88% viability in simulated GIT [37]. Our results demonstrate similar viability of LAB in artificial GIT compared to other probiotics. In addition, probiotics attached to the intestinal mucosa promote various beneficial effects by producing metabolites and stimulating the gut barrier in the host [38]. First, we established that MG4247, MG4272, and MG4577 were non-toxic and adherent to intestinal epithelial cells. The adhesion of MG4247, MG4272, and MG4577 to intestinal epithelial cells was more than 66%. Other probiotics, which were distributed recently, were reported to be 60.2% adherent to colon cells [39]. Compared to the results of cited literature, it is suggested that MG4247, MG4272, and MG4577 may have a higher adhesion rate in intestinal epithelial cells. Although it has been scientifically verified in vitro that L. gasseri MG4272, L. paracasei MG4247, and MG4577 down-expressed allergy-related cytokines, further studies in animal models sensitized by various allergens or/and clinical trials still need to be performed.
Conclusions
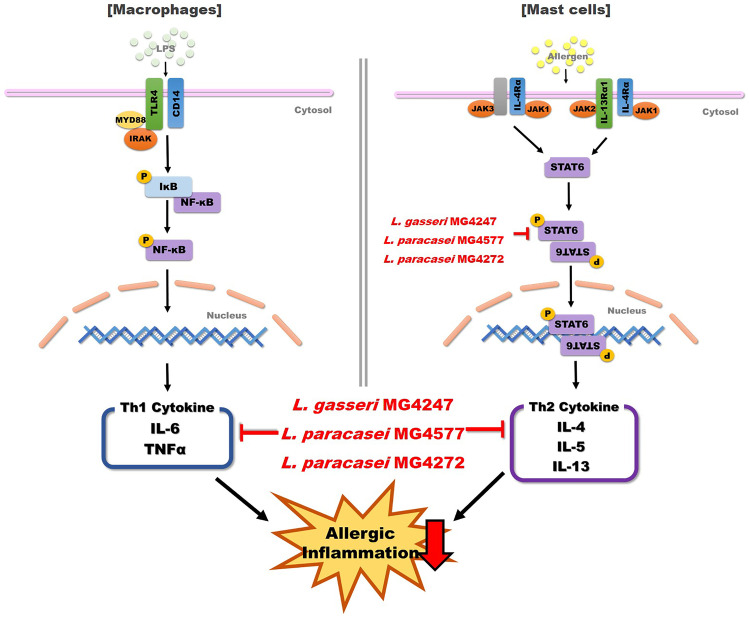
Our results suggest that LAB are effective in modulating allergic inflammation in macrophages and mast cells. In the present study, treatment with LAB reduced TNF-α and IL-6 expression levels in LPS-induced RAW 264.7 macrophages and the levels of Th2 cytokines, IL-4, IL-5, and IL-13, by reducing p-STAT6 protein expression in DNP-IgE/HSA-sensitized RBL-2H3 mast cells (Fig. 6). Among LAB, L. gasseri MG4272 and L. paracasei MG4247 and MG4577 can be qualified as probiotics because of their safety and stability in the gastric intestine of the host. Thus, L. gasseri MG4272, L. paracasei MG4247, and MG4577 secured have safety as a probiotic, and these strains decrease the Th1 cytokines production, and also suppress secretion of Th2 cytokines via reduction of pSTAT6 phosphorylation.
Fig. 6.
L. gasseri MG4247 and L. paracasei MG4577 reduced allergic symptoms via modulating phosphorylation of STAT6, Th1, and Th2 cytokines in macrophages, and mast cells
Supplementary Information
Below is the link to the electronic supplementary material.
Author Contribution
Chang-Ho Kang and Ji Yeon Lee contributed to the study conception and design. Data collection, investigation, and analysis were performed by Ji Yeon Lee, Ju-Hui Kang, and Ye-Rin Jung. The first draft of the manuscript was written by Ji Yeon Lee. Chang-Ho Kang contributed to supervision, and commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by Prof. Tae-Bong Kang, Konkuk University.
Data Availability
The authors declare that all data and materials support published claims and comply with field standards.
Declarations
Ethics Approval
This article does not contain any studies with human or animal subjects.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Conflict of Interest
J.Y. Lee and C.-H. Kang are currently employed by the MEDIOGEN Corporation, Republic of Korea. None of the other authors had any conflict of interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Hwang S-W, Sun X, Han J-H, Kim T-Y, Koppula S, Kang T-B, Hwang J-K, Lee K-H. Fermentation-mediated enhancement of ginseng’s anti-allergic activity against IgE-mediated passive cutaneous anaphylaxis in vivo and in vitro. J Microbiol Biotechnol. 2018;28:1626–1634. doi: 10.4014/jmb.1807.07057. [DOI] [PubMed] [Google Scholar]
- 2.Jenerowicz D, Silny W, Danczak-Pazdrowska A, Polanska A, Osmola-Mankowska A, Olek-Hrab K (2012) Environmental factors and allergic diseases. Ann Agric Environ Med 19 [PubMed]
- 3.Pawankar R, Canonica GW, Holgate ST, Lockey RF. Allergic diseases and asthma: a major global health concern. Curr Opin Allergy Clin Immunol. 2012;12:39–41. doi: 10.1097/ACI.0b013e32834ec13b. [DOI] [PubMed] [Google Scholar]
- 4.Yoshimoto T. The hunt for the source of primary interleukin-4: how we discovered that natural killer T cells and basophils determine T helper type 2 cell differentiation in vivo. Front Immunol. 2018;9:716. doi: 10.3389/fimmu.2018.00716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saradna A, Do DC, Kumar S, Fu Q-L, Gao P. Macrophage polarization and allergic asthma. Transl Res. 2018;191:1–14. doi: 10.1016/j.trsl.2017.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kany S, Vollrath JT, Relja B. Cytokines in inflammatory disease. Int J Mol Sci. 2019;20:6008. doi: 10.3390/ijms20236008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nam YK, Jin SC, Kim MH, Choi LY, Lee Y-B, Yang WM. Banhahubak-Tang tablet, a standardized medicine attenuates allergic asthma via inhibition of janus kinase 1 (JAK1)/signal transducer and activator of transcription 6 (STAT6) signal pathway. Molecules. 2020;25:2206. doi: 10.3390/molecules25092206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fein MN, Fischer DA, O’Keefe AW, Sussman GL. CSACI position statement: newer generation H 1-antihistamines are safer than first-generation H 1-antihistamines and should be the first-line antihistamines for the treatment of allergic rhinitis and urticaria. Allergy Asthma Clin Immunol. 2019;15:1–6. doi: 10.1186/s13223-019-0375-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim K-T, Kim J-W, Kim S-I, Kim S, Nguyen TH, Kang C-H. Antioxidant and anti-Inflammatory effect and probiotic properties of lactic acid bacteria Isolated from canine and feline feces. Microorganisms. 2021;9:1971. doi: 10.3390/microorganisms9091971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee E-S, Song E-J, Nam Y-D, Lee S-Y. Probiotics in human health and disease: from nutribiotics to pharmabiotics. J Microbiology. 2018;56:773–782. doi: 10.1007/s12275-018-8293-y. [DOI] [PubMed] [Google Scholar]
- 11.Lopez-Santamarina A, Gonzalez EG, Lamas A, Mondragon AdC, Regal P, Miranda JM (2021) Probiotics as a possible strategy for the prevention and treatment of allergies. a narrative review. Foods 10:701 [DOI] [PMC free article] [PubMed]
- 12.Song S, Lee S-J, Park D-J, Oh S, Lim K-T. The anti-allergic activity of Lactobacillus plantarum L67 and its application to yogurt. J Dairy Sci. 2016;99:9372–9382. doi: 10.3168/jds.2016-11809. [DOI] [PubMed] [Google Scholar]
- 13.Kim W-G, Kang G-D, Kim H, Han M, Kim D-H. Bifidobacterium longum IM55 and Lactobacillus plantarum IM76 alleviate allergic rhinitis in mice by restoring Th2/Treg imbalance and gut microbiota disturbance. Benef Microbes. 2019;10:55–67. doi: 10.3920/BM2017.0146. [DOI] [PubMed] [Google Scholar]
- 14.Kim J-K, Kim J-Y, Kim HI, Han MJ, Kim D-H. Bifidobacterium longum and Lactobacillus plantarum alleviate house dust mite allergen-induced allergic rhinitis by regulating IL-4, IL-5, and IL-10 expression. Food Agric Immunol. 2019;30:581–593. doi: 10.1080/09540105.2019.1608161. [DOI] [Google Scholar]
- 15.Steiner NC, Lorentz A (2021) Probiotic potential of Lactobacillus species in allergic rhinitis. Int Arch Allergy Immunol 1–12 [DOI] [PubMed]
- 16.Chen YS, Lin YL, Jan RL, Chen HH, Wang JY. Randomized placebo-controlled trial of lactobacillus on asthmatic children with allergic rhinitis. Pediatr Pulmonol. 2010;45:1111–1120. doi: 10.1002/ppul.21296. [DOI] [PubMed] [Google Scholar]
- 17.Lei W, Liu C, Pan L, Peng C, Wang J, Zhou H. Screening of probiotic Lactobacilli with potential anti-allergic activity based on hyaluronidase inhibition and degranulation of RBL-2H3 cells in vitro. LWT. 2021;140:110707. doi: 10.1016/j.lwt.2020.110707. [DOI] [Google Scholar]
- 18.Bahuguna A, Khan I, Bajpai VK, Kang SC. MTT assay to evaluate the cytotoxic potential of a drug. Bangladesh J Pharmacol. 2017;12:115–118. doi: 10.3329/bjp.v12i2.30892. [DOI] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Kim T-Y, Kang J-H, Lee S-B, Kang T-B, Lee K-H. Down-regulation of pro-necroptotic molecules blunts necroptosis during myogenesis. Biochem Biophys Res Commun. 2021;557:33–39. doi: 10.1016/j.bbrc.2021.04.004. [DOI] [PubMed] [Google Scholar]
- 21.Mahmood T, Yang P-C. Western blot: technique, theory, and trouble shooting. N Am J Med. 2012;4:429. doi: 10.4103/1947-2714.100998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee JY, Kim H, Jeong Y, Kang C-H. Lactic acid bacteria exert a hepatoprotective effect against ethanol-induced liver injury in HepG2 cells. Microorganisms. 2021;9:1844. doi: 10.3390/microorganisms9091844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shehata M, El Sohaimy S, El-Sahn MA, Youssef M. Screening of isolated potential probiotic lactic acid bacteria for cholesterol lowering property and bile salt hydrolase activity. Ann Agric Sci. 2016;61:65–75. doi: 10.1016/j.aoas.2016.03.001. [DOI] [Google Scholar]
- 24.Qi X, Simsek S, Chen B, Rao J. Alginate-based double-network hydrogel improves the viability of encapsulated probiotics during simulated sequential gastrointestinal digestion: Effect of biopolymer type and concentrations. Int J Biol Macromol. 2020;165:1675–1685. doi: 10.1016/j.ijbiomac.2020.10.028. [DOI] [PubMed] [Google Scholar]
- 25.Allergy C (2021) Can allergies cause pneumonia? Columbia Allergy. If left untreated, these symptoms can lead to bronchitis or pneumonia, such as chronic obstructive pulmonary disease (COPD). Accessed Dec 26 2021
- 26.Fang Z, Li L, Zhang H, Zhao J, Lu W, Chen W (2021) Gut microbiota, probiotics, and their interactions in prevention and treatment of atopic dermatitis: a review. Front Immunol 2799 [DOI] [PMC free article] [PubMed]
- 27.McKenzie C, Tan J, Macia L, Mackay CR. The nutrition-gut microbiome-physiology axis and allergic diseases. Immunol Rev. 2017;278:277–295. doi: 10.1111/imr.12556. [DOI] [PubMed] [Google Scholar]
- 28.Moradi M, Molaei R, Guimarães JT. A review on preparation and chemical analysis of postbiotics from lactic acid bacteria. Enzyme Microb Technol. 2021;143:109722. doi: 10.1016/j.enzmictec.2020.109722. [DOI] [PubMed] [Google Scholar]
- 29.Markowiak-Kopeć P, Śliżewska K. The effect of probiotics on the production of short-chain fatty acids by human intestinal microbiome. Nutrients. 2020;12:1107. doi: 10.3390/nu12041107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu T, Zhang L, Joo D, Sun S-C. NF-κB signaling in inflammation. Signal Transduct Target Ther. 2017;2:1–9. doi: 10.1038/sigtrans.2017.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berker M, Frank LJ, Geßner AL, Grassl N, Holtermann AV, Höppner S, Kraef C, Leclaire MD, Maier P, Messerer DAC. Allergies–AT cells perspective in the era beyond the TH1/TH2 paradigm. Clin Immunol. 2017;174:73–83. doi: 10.1016/j.clim.2016.11.001. [DOI] [PubMed] [Google Scholar]
- 32.Hajavi J, Esmaeili SA, Varasteh AR, Vazini H, Atabati H, Mardani F, Momtazi-Borojeni AA, Hashemi M, Sankian M, Sahebkar A. The immunomodulatory role of probiotics in allergy therapy. J Cell Physiol. 2019;234:2386–2398. doi: 10.1002/jcp.27263. [DOI] [PubMed] [Google Scholar]
- 33.Sherman MA, Secor VH, Lee SK, Lopez RD, Brown MA. STAT6-independent production of IL-4 by mast cells. Eur J Immunol. 1999;29:1235–1242. doi: 10.1002/(SICI)1521-4141(199904)29:04<1235::AID-IMMU1235>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 34.Chen H, Jiang Z. The essential adaptors of innate immune signaling. Protein Cell. 2013;4:27–39. doi: 10.1007/s13238-012-2063-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Joint F (2002) Guidelines for the evaluation of probiotics in food, London, Ontario, Canad. April 30 and May 1, 2002 http://www.who.int/foodsafety/publications/fs_management/probiotics2/en/index.html
- 36.da Silva MN, Tagliapietra BL, do Amaral Flores V, dos Santos Richards NSP (2021) In vitro test to evaluate survival in the gastrointestinal tract of commercial probiotics. Curr Res Nutr Food Sci 4:320–325 [DOI] [PMC free article] [PubMed]
- 37.Bengoa AA, Zavala L, Carasi P, Trejo SA, Bronsoms S, de los Ángeles Serradell M, Garrote GL, Abraham AG (2018) Simulated gastrointestinal conditions increase adhesion ability of Lactobacillus paracasei strains isolated from kefir to Caco-2 cells and mucin. Food Res Int 103:462–467 [DOI] [PubMed]
- 38.Monteagudo-Mera A, Rastall RA, Gibson GR, Charalampopoulos D, Chatzifragkou A. Adhesion mechanisms mediated by probiotics and prebiotics and their potential impact on human health. Appl Microbiol Biotechnol. 2019;103:6463–6472. doi: 10.1007/s00253-019-09978-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Piatek J, Gibas-Dorna M, Olejnik A, Krauss H, Wierzbicki K, Zukiewicz-Sobczak W, Glowacki M. The viability and intestinal epithelial cell adhesion of probiotic strain combination-in vitro study. Ann Agric Environ Med. 2012;19:99–102. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all data and materials support published claims and comply with field standards.