Abstract
Among the relatively few established human prostate cancer cell lines, LNCaP cells are unique in their ability to model key stages of prostate cancer progression. Analyses of LNCaP cells and their derivatives have been invaluable for elucidating important translational aspects of prostate tumorigenesis, metastasis, and drug response, particularly in the context of androgen receptor signaling. Here we present major highlights from a wealth of literature that has exploited LNCaP cells and their derivatives to inform on prostate cancer progression and androgen response for improving the treatment of prostate cancer patients.
Prostate cancer is the most common cancer malignancy and a leading cause of cancer related deaths in men. Nowadays, most newly diagnosed cases are locally invasive and relatively indolent, and therefore do not require active intervention. However, for men with advanced prostate cancer the situation is more dire. The first line of defense is androgen-deprivation therapy (ADT), due to the essential role of the androgen receptor (AR) for all aspects of normal prostate cancer function, as well as all stages of prostate cancer progression (Fig. 1). Men who undergo ADT initially respond to treatment, however in most cases their tumors ultimately recur as castration-resistant prostate cancer (CRPC), so called because of its continued dependence on AR even in the absence of androgens. CRPC is often metastatic (mCRPC), with the primary site being the bone, and frequently lethal. Further treatment, including with second-generation anti-androgens, can give rise to even more aggressive disease variants, which tend to be AR-negative and can acquire features of neuroendocrine prostate cancer (NEPC). These aggressive variants are highly metastatic to bone as well as soft tissues, and have even worse disease outcomes than mCRPC.
Figure:
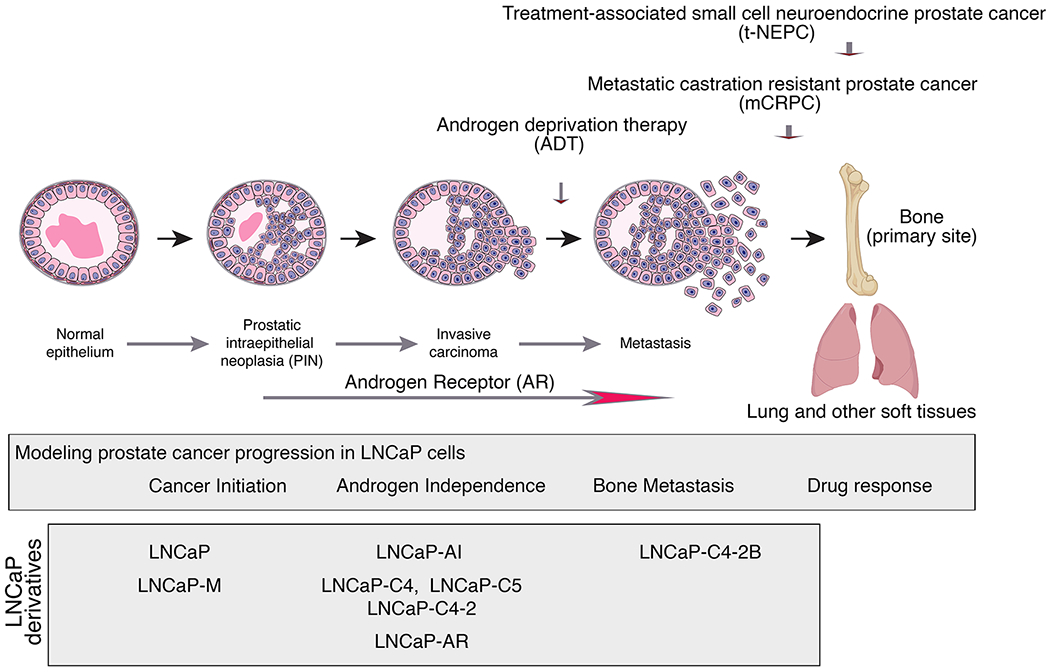
Schematic of prostate cancer progression showing the disease stages and the contribution of LNCaP cells and its derivatives to studying the disease stages.
Hence, to properly study prostate cancer, it is essential to model disease progression from the earliest to most advanced stages; however, this has proven extremely challenging to accomplish in vitro. The classic report by Horoszewicz and colleagues describes the characterization of the LNCaP prostate cancer cell line (1), which to this day remains one of the few and perhaps the only human cell line that can model a wide range of stages of prostate cancer progression, including the transition from androgen-dependence to castration-resistance, bone metastasis, drug sensitivity, and even the acquisition of NEPC features (Fig. 1).
LNCaP cells were established from a lymph node metastasis of a Caucasian patient with metastatic prostate cancer (1). Further characterization has shown that LNCaP cells are highly aneuploid but are wild-type for TP53 (2). Most notably, LNCaP cells express AR, albeit a mutated version which has broader steroid-binding specificity than wild-type AR. LNCaP cells also express downstream targets of AR activity, including prostate specific antigen (PSA), a well-known marker of prostate cancer, and NKX3.1, a homeoprotein that has essential transcriptional regulatory functions in prostatic epithelial cells and is expressed in benign prostate and well-differentiated adenocarcinoma.
For reasons that remain unclear, it has been exceedingly difficult to establish cell lines from human prostate cancer, which is also true of 3-D organoid and to a lesser extent patient-derived xenograft models. Consequently, relatively few human prostate cancer cell lines have been established, and even fewer are commonly used. The classic and most widely utilized human prostate cancer cell lines are DU145, PC3, and LNCaP cells (2); among these, only LNCaP cells express AR and are androgen-responsive. This feature is undoubtedly related to why LNCaP cells have a unique capacity to model prostate cancer progression.
For the most part, this has been accomplished by the generation of a series of derivatives from the original LNCaP cell line, which model various aspects of prostate cancer progression (Fig. 1). Seminal work from Leland Chung and colleagues led to the generation of a series of LNCaP cell lines that model hormone-dependent (the LNCaP-M line) and castration-resistant (the LNCaP-C4, LNCaP-C4-2 and LNCaP-C5 lines) prostate cancer. These derivatives were established from tumors that arose following implantation of the original LNCaP cell line, together with a bone stromal cell line, into intact or castrated host mice (3). Analyses of the resulting hormone-dependent and castration-resistant LNCaP derivatives provided some of the first evidence of the importance of the tumor microenvironment for prostate cancer progression, particularly following androgen deprivation. Further studies from Chung and colleagues led to the generation of additional LNCaP derivatives that develop osteoblastic bone metastases, which reinforced the notion that the microenvironment plays an integral role in bone metastasis (4). Notably, these LNCaP derivatives are still widely used for studying castration-resistance and bone metastasis, and for modeling the role of the tumor stroma in vivo.
As noted above, LNCaP cells are among the few prostate cancer cell lines that express AR, albeit a mutated version. This important feature spurned a plethora of studies that cumulatively have elucidated the essential role of AR for castration-resistant prostate cancer and led to the development of novel therapies for treatment of patients with advanced disease. In groundbreaking work, Charles Sawyers and colleagues developed a derivative of the LNCaP cell line that expresses exogenous wild-type AR in addition to the endogenous mutated version (LNCaP-AR cells), and showed that gain of AR expression is sufficient for the transition from an hormone-responsive to an castration-resistant state (5).
This transformative observation led to a paradigm shift in the treatment of CRPC based on targeting of AR. In particular, Sawyers and colleagues used the LNCaP-AR cells to evaluate the concept that antagonizing AR could inhibit castration-resistant tumor growth, and by evaluating a series of novel anti-androgens in LNCaP-AR cells, identified enzalutamide as an optimal compound (6). Enzalutamide is now an FDA approved standard of care for treatment of men with advanced prostate cancer, highlighting how these observations revolutionized the treatment of men with prostate cancer. Sawyers and colleagues subsequently showed that enzalutamide resistance in LNCaP-AR-derived tumors can sometimes occur through induction of glucocorticoid receptor, thereby bypassing the AR blockade (7), thus providing further translational insights into mechanisms of tumor growth in the absence of androgens.
As noted above, men who undergo treatment with second-generation anti-androgens can develop more aggressive variants of mCRPC, including features of NEPC. These treatment resistant variants are thought to arise via lineage plasticity, in which a cell can change its identity from one committed phenotype to another; indeed, lineage plasticity is now widely appreciated to play a critical role in prostate tumorigenesis and drug resistance. Notably, this important translational insight was initially demonstrated in LNCaP cells, which were shown to give rise to NEPC-like cells following androgen deprivation (8). Indeed, many subsequent papers have established the plasticity of LNCaP cells to convert to NEPC-like cells as an adaptive mechanism for resistance to second-generation anti-androgens.
In addition to the wealth of information that has been gleaned by studying advanced prostate cancer using LNCaP cells, these cells have also been invaluable for modeling early-stage disease, particularly for investigating external influences that promote cancer initiation, such as oxidative stress and inflammation. As an example, in our recent publication we modeled cancer initiation in LNCaP cells and showed that, following oxidative stress, NKX3.1 becomes localized to mitochondria where it protects against oxidative stress and thereby disease progression (9). Notably, oxidative stress has the opposite effect on AR in these cells, suggesting that analyses of LNCaP cells can provide insights into the coordinate actions of regulators that protect cells during early stages of cancer progression.
In related work, Gelmann and colleagues have studied the relationship of DNA damage and cancer initiation in LNCaP cells. They have shown that NKX3.1 protects against DNA damage (10), which is due to the ability of NKX3.1 to protect against common genomic rearrangements and to interactions with key DNA repair genes. Considering that genes that protect against DNA damage are now known to be dysregulated in advanced prostate cancer, we envision that the potential for LNCaP cells as a model for analyzing DNA repair in prostate cancer will open exciting possibilities for future studies.
As should be evident from this brief overview, LNCaP cells have proven to be a powerful model to study many aspects of prostate tumorigenesis, as well as a vital and unique resource to the field of prostate cancer. On the other hand, it is equally important to recognize that a large body of work and many important conclusions have been based on analysis of a single cell model and its derivatives. Notably, LNCaP cells were derived from a Caucasian man, whereas we know that African American men develop more aggressive prostate cancer that may not be reflected by studying LNCaP cells. So, while LNCaP cells have been an excellent resource, we continue to need addition cell line models to ensure that we provide the most insightful and translationally accurate information that is reflective of the population that is most susceptible to the disease.
Acknowledgements:
We are grateful to Phil Watson and Michael Shen for their comments. Work in the CAS laboratory is supported by funding from the NIH (R01 CA173481, R01 CA183929, and P01 CA265768).
Footnotes
Competing interests: The authors have no competing interests to declare.
References
- 1.Horoszewicz JS, Leong SS, Kawinski E, Karr JP, Rosenthal H, Chu TM , et al. LNCaP model of human prostatic carcinoma. Cancer Res 1983;43:1809–18 [PubMed] [Google Scholar]
- 2.van Bokhoven A, Varella-Garcia M, Korch C, Johannes WU, Smith EE, Miller HL , et al. Molecular characterization of human prostate carcinoma cell lines. Prostate 2003;57:205–25 [DOI] [PubMed] [Google Scholar]
- 3.Wu HC, Hsieh JT, Gleave ME, Brown NM, Pathak S, Chung LW. Derivation of androgen-independent human LNCaP prostatic cancer cell sublines: role of bone stromal cells. Int J Cancer 1994;57:406–12 [DOI] [PubMed] [Google Scholar]
- 4.Wu TT, Sikes RA, Cui Q, Thalmann GN, Kao C, Murphy CF , et al. Establishing human prostate cancer cell xenografts in bone: induction of osteoblastic reaction by prostate-specific antigen-producing tumors in athymic and SCID/bg mice using LNCaP and lineage-derived metastatic sublines. Int J Cancer 1998;77:887–94 [DOI] [PubMed] [Google Scholar]
- 5.Chen CD, Welsbie DS, Tran C, Baek SH, Chen R, Vessella R , et al. Molecular determinants of resistance to antiandrogen therapy. Nat Med 2004;10:33–9 [DOI] [PubMed] [Google Scholar]
- 6.Tran C, Ouk S, Clegg NJ, Chen Y, Watson PA, Arora V , et al. Development of a second-generation antiandrogen for treatment of advanced prostate cancer. Science 2009;324:787–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arora VK, Schenkein E, Murali R, Subudhi SK, Wongvipat J, Balbas MD , et al. Glucocorticoid receptor confers resistance to antiandrogens by bypassing androgen receptor blockade. Cell 2013;155:1309–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shen R, Dorai T, Szaboles M, Katz AE, Olsson CA, Buttyan R. Transdifferentiation of cultured human prostate cancer cells to a neuroendocrine cell phenotype in a hormone-depleted medium. Urol Oncol 1997;3:67–75 [DOI] [PubMed] [Google Scholar]
- 9.Papachristodoulou A, Rodriguez-Calero A, Panja S, Margolskee E, Virk RK, Milner TA , et al. NKX3.1 Localization to Mitochondria Suppresses Prostate Cancer Initiation. Cancer Discov 2021;11:2316–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bowen C, Gelmann EP. NKX3.1 activates cellular response to DNA damage. Cancer Res 2010;70:3089–97 [DOI] [PubMed] [Google Scholar]


