Abstract
Elongation factor 4 (EF4/LepA) is a highly conserved guanosine triphosphatase translation factor. It was shown to promote back-translocation of tRNAs on post-translocational ribosome complexes and to compete with elongation factor G for interaction with pre-translocational ribosomes, thereby inhibiting the elongation phase of protein synthesis. Here, we report a crystal structure of EF4-GDP bound to the Thermus thermophilus ribosome with a P-site tRNA at 2.9 angstroms resolution. The C-terminal domain of EF4 reaches into the peptidyl transferase center and interacts with the acceptor stem of the peptidyl-tRNA in the P site. The ribosome is in an unusual state of ratcheting with the 30S subunit rotated clockwise relative to the 50S subunit, resulting in a remodeled decoding center. The structure is consistent with EF4 functioning either as a back-translocase or a ribosome sequester.
Summary:
In an EF4-ribosome complex the ribosome is in a clockwise ratcheted state which remodels the decoding center.
Protein biosynthesis by the ribosome is facilitated at multiple stages by translational guanosine triphosphatases (trGTPases) that catalyze the hydrolysis of guanosine 5’-triphosphate (GTP) to promote conformational changes leading to transitions between ribosome functional states (1). One of the critical and complex steps in protein synthesis is elongation, a unidirectional sequence of events catalyzed by elongation factor Tu (EF-Tu), which delivers an aminoacyl-transfer RNA (tRNA) to the ribosomal A site in response to the codon displayed on the messenger RNA (mRNA), and elongation factor G (EF-G) that promotes the movement of tRNAs in the A and P sites to the P and E sites, respectively, in a process called translocation. During these highly dynamic events, tRNAs transit through the ribosome with a concomitant counter-clockwise ratchet-like movement of the 30S subunit (2–4). Through multiple rounds of amino acid incorporation to the nascent peptide chain and tRNA translocation, the ribosome translates the genetic information in the mRNAs into proteins.
Under certain conditions, factors that are not required for normal elongation intervene in the elongation cycle to perform special functions (5–7). Elongation factor 4 (EF4), encoded by the lepA gene in Escherichia coli (8), is one such factor that is a highly conserved GTPase found in bacteria as well as in the mitochondria and chloroplasts of eukaryotes (9, 10). EF4 is known to have a ribosome-dependent GTPase activity (9, 11, 12) and to increase the rate of protein synthesis at high intracellular ionic strength (13), but has no effect on translational accuracy (13, 14). It also promotes tRNA back-translocation on post-translocation (POST) ribosomal complexes with tRNAs in the E and P sites (9, 11, 12, 15), thus reverting the reaction mediated by EF-G. An alternate model, based on single molecule Förster resonance energy transfer (smFRET) experiments, suggests that EF4 effectively competes with EF-G for binding to the pre-translocational (PRE) state of the ribosome with tRNAs in the A and P sites, thereby sequestering ribosomes in an intermediate state leading to a transient inhibition of the elongation cycle (16).
EF4 has six domains, of which four (I, II, III and V) are homologous to corresponding domains in EF-G (Fig. 1) (17). It differs from EF-G by having a short domain IV and lacking a G′ sub-domain, but possessing a conserved C-terminal domain (CTD) (17). The last 44 residues of the CTD are flexible and therefore not visible in the crystal structure of ribosome-free EF4 (17). A prior cryo-electron microscopy (cryo-EM) reconstruction showed that EF4 has the same binding site on the ribosome as other trGTPases and affects the conformation of the A-site tRNA (12). However, the low resolution of this cryo-EM map limits the conclusions that can be drawn from it about the structure and function of EF4 on the ribosome.
Fig. 1. The structure of EF4 bound to the ribosome.

(A) Overview of EF4, P-site tRNA (magenta), E-site tRNA (orange) and mRNA (cyan) bound to the 70S ribosome. The 50S and 30S subunits are shown in blue and yellow, respectively. (B) Cartoon representation of EF4 shown in its ribosome-bound conformation. Each domain of EF4 is colored as in panel A. Insets: Two views of the unbiased Fobs – Fcalc difference Fourier map of the helix-turn-helix of the CTD and the CCA-end of the P-site tRNA with the attached phenylalanine residue contoured at 3σ obtained after initial refinement with an empty ribosome as a starting model.
Structure determination of GTPase translation factors bound to the ribosome has remained a challenge (18). Using a new approach to crystallize trGTPases on the ribosome that covalently links the N-terminal domain of ribosomal protein L9 to the trGTPase (fig. S1) (19), we report a 2.9 Å resolution crystal structure of a complex between the T. thermophilus 70S ribosome and EF4 with GDP bound. The structure shows that the CTD of EF4 reaches into the peptidyl transferase center (PTC) of the 50S subunit and reveals details of the interactions of the CTD with the acceptor stem of the peptidyl-tRNA in the P site and other elements of the PTC. In contrast to the counter-clockwise ratcheting motion of the 30S subunit previously observed during EF-G-mediated tRNA translocation (4, 20–22), the 30S subunit in our EF4-ribosome complex is rotated clockwise which results in a remodeled decoding center.
Initial phase of the diffraction data obtained by using a model of an empty ribosome showed clear unbiased electron density for the mRNA, Phe-tRNAPhe in the P site, EF4 with its CTD near the acceptor stem of the Phe-tRNAPhe in the P site and for the phenylalanine residue attached to the P-site tRNA in the difference Fourier map (Fig. 1, A and B) (19). Residual electron density was observed in the E site, where a tRNAPhe was modeled (Fig. 1A). The overall conformation of EF4-GDP bound to the ribosome seen in our structure is in agreement with the cryo-EM reconstruction reported for the complex of EF4-GDPNP with the ribosome (Fig. 1, fig. S2 and movie S1) (12) and is similar to that seen in the crystal structure of ribosome-free EF4 (17). Binding of EF4 to the ribosome affects the position of its domain V which is displaced by more than 13 Å and rotated by ~30° to avoid a steric collision with the sarcin-ricin loop (SRL) of the 50S subunit (fig. S3). EF4 contacts both ribosomal subunits (Fig. 1A) and is held in position by interactions that are analogous to the ones previously reported for EF-G in complex with the ribosome (figs. S4 and S5) (23).
The terminal residues of the CTD form two α-helices (α1 and α2) connected by a short turn, resulting in a helix-turn-helix (HTH) module that occupies the A site of the 50S subunit and reaches into the PTC where it interacts with the P-site tRNA acceptor stem and 23S rRNA (Fig. 1, A and B and movie S1). Part of the CTD domain was built de novo into our electron density map (19) because of the absence of residues 556-599 in the crystal structure of the E. coli EF4 (17). The universally conserved nucleotide A2602 of 23S rRNA in the PTC, which is important for the peptide bond formation and release reactions (24), is sandwiched by the A76 ribose of the P-site tRNA and residue Arg569 of EF4 on one side, and by residues Lys561 and Cys562 on the other side (Fig. 2A). This is in agreement with data showing that EF4 bound to the ribosome can be cross-linked to a linezolid derivative antibiotic that binds near A2602 in the PTC (25). Helix α2 of the CTD is rich in basic residues (17), consistent with its location surrounded by RNA. Several of its positively charged residues interact with the phosphate backbone of the acceptor stem of the P-site tRNA (Fig. 2, A and B). The base of nucleotide A73 of the P-site tRNA stacks with the aliphatic moiety of the Lys572 side chain, while its sugar and Watson-Crick edges interact with residues Thr568 and Glu575, respectively, thereby gripping tightly nucleotide A73 (Fig. 2B). Finally, additional interactions form between the HTH module of the CTD and ribosomal protein L16 (Fig. 2C). These interactions collectively position the conserved Tyr563 residue in the PTC such that it can form hydrogen bonds with A2602 and the carbonyl oxygen of the phenylalanine residue attached to the P-site tRNA (Fig. 2A and movie S1), likely protecting the peptidyl-tRNA from premature hydrolysis. This observation is consistent with data showing that EF4 decreases the reactivity of the P-site tRNA with puromycin in both the pre- and post-translocation ribosomal complexes (9, 11, 12, 16).
Fig. 2. Interactions of the EF4-CTD in the PTC.
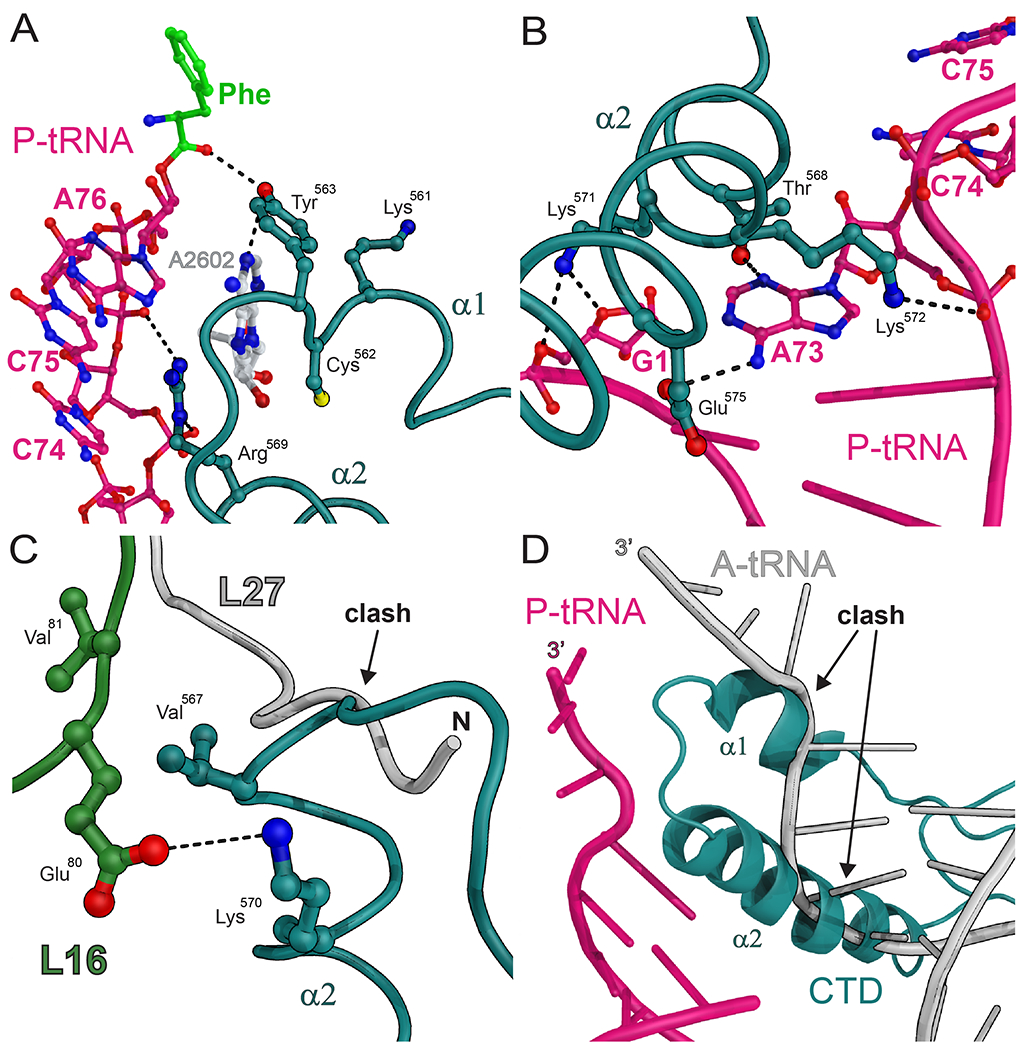
(A) Interactions between the EF4-CTD (deep teal), the CCA-end of the P-site tRNA (magenta) with its attached phenylalanine residue (green) and A2602 of 23S rRNA (white). (B) Interactions between helix α2 of the EF4-CTD (deep teal) and the acceptor stem of the P-site tRNA (magenta). Nucleotide A73 of the P-site tRNA is entrapped by residues Lys572, Thr568 and Glu575. (C) The helix-turn-helix (HTH) of the EF4-CTD (deep teal) sterically interferes with the usual location of the N-terminal region of ribosomal protein L27 (gray) (27). The stacking interactions between residues Val567 of EF4 and Val81 of ribosomal protein L16, and the salt bridge that forms between Lys570 of EF4 and Glu80 of protein L16 provide additional stabilization of HTH in the PTC. (D) The location of the EF4-CTD (deep teal) is not compatible with the acceptor stem of a tRNA bound in the classical A/A conformation (gray) (27).
The position occupied by the CTD of EF4 is not compatible with the simultaneous binding of the acceptor stem of a tRNA in the classical A/A conformation (Fig. 2D) or with the usual location of the N-terminal region of the ribosomal protein L27 (Fig. 2C), a protein that has been shown to increase the efficiency of peptide bond formation (26). The absence of electron density for the first eight residues of protein L27 in the electron density map suggests that they are disordered.
In this complex, the 30S subunit is rotated clockwise by ~5° (viewed from the solvent side) with respect to the 50S subunit (which we refer to as a clockwise ratchet-like movement) relative to the canonical state of the ribosome without any bound factors (Fig. 3A and movie S2) (27). This is in contrast with the previously reported counter-clockwise ratcheting motion of the 30S subunit observed in the structures of the ribosome in complex with EF-G, release factor 3 (RF3) and the ribosome recycling factor (RRF) (Fig. 3B) (4, 20–22, 28–31). During the clockwise ratchet-like movement of the ribosome, the platform and h44 of the small subunit remain essentially static, while the head, the shoulder and the spur undergo the largest movements. Inter-subunit bridges formed between protein S13 in the 30S head with H38 (B1a) and protein L5 (B1b) are both shifted by more than 4 Å, while bridge B8 formed between h14 in the 30S body and protein L14 is shifted by ~7 Å (fig. S6). Although the 30S subunit undergoes a clockwise ratcheting motion, the Phe-tRNAPhe remains in the classical P/P state as observed in the cryo-EM reconstruction of EF4-GDPNP bound to a non-rotated ribosome (12).
Fig. 3. Clockwise ratchet-like movement of the 30S subunit in the 70S ribosome.

(A) Alignment of the EF4-ribosome complex (yellow) on that of a ribosome in the classical state (green) (27) by superimposing their 23S rRNA reveals a clockwise ratcheting motion of the 30S subunit. (B) In contrast to the clockwise rotation observed here upon EF4 binding (yellow), the binding of EF-G-GTP to the ribosome stabilizes a counter-clockwise ratcheted state of the 30S subunit (orange) (21). In A and B, the 30S subunit in the classical state is green and the direction of the 30S subunit rotation relative to the 50S subunit (gray) is indicated by the curved arrows. The rotation angle between the classical state 30S (black dashed line) and the rotated state (black full line) is indicated.
In standard decoding, three universally conserved nucleotides in the 30S subunit, A1492 and A1493 in h44, and G530 in h18, change conformation and closely interact with the minor groove of the anticodon stem-loop (ASL) in the A site, thereby stabilizing the A-site tRNA in the decoding center (Fig. 4B) (27, 32). The clockwise ratchet-like movement of the ribosome causes an opening of the space between the subunits on the side where EF4 binds (Fig. 4A). A superimposition of our structure on that of a ribosome in the classical state (27) shows that the G530-loop (h18) and ribosomal protein S12 in the shoulder of the 30S shift away from the inter-subunit space by ~4 Å (Fig. 4, A and C, and movie S2), effectively perturbing the conformation of the decoding center. The decoding center in the clockwise ratcheted structure was superimposed on that of an “open” 30S subunit (33) and revealed that the G530-loop in our complex is displaced by ~2 Å further away from the A site (fig. S7), thereby showing that EF4 induces a greater degree of opening of the decoding center than any factor observed thus far.
Fig. 4. Conformational changes in the decoding center of the 30S subunit.

(A) Side-view of the EF4-ribosome complex. The clockwise ratchet-like movement of the 30S subunit widens the space between the subunits on the side where EF4 binds (black arrow), effectively displacing the G530-loop (h18) and ribosomal protein S12 (dashed black box) away from the decoding center. (B and C) Close-up view of the dashed black box in panel A. (B) In the non-ratcheted ribosome (27), the close packing interactions between the G530-loop and the codon-anticodon mini-helix stabilize the tRNA in the A site (27, 32). (C) In the EF4-bound state, the clockwise ratcheting of the 30S subunit displaces the G530-loop and protein S12 away from h44, thereby widening the decoding center. For reference, the white mRNA, tRNA, h44, including nucleotides A1492 and A1493, are modeled from panel B. The latter nucleotides are disordered in the EF4 complex.
The structure illustrates how two closely related trGTPases, EF4 and EF-G, modulate the overall conformation of the functional centers of the ribosome in a completely different manner to perform their functions; both induce a ratchet-like movement of the 30S subunit albeit in opposite direction. The conformational malleability of the ribosome, exemplified by the ratcheting of the 30S subunit and remodeling of the decoding center, supports the growing consensus that the ribosome is a highly flexible particle that functions through multiple distinct conformations modulated by a multitude of protein factors at different stages of translation.
EF4 was first proposed to catalyze the reverse reaction mediated by EF-G that is, to promote back-translocation of tRNAs on POST ribosomal complexes from the E and P sites to the P and A sites, respectively (9, 11, 12, 15). One study suggests that EF4 binds with higher affinity to the POST state of the ribosome than the PRE state and the EF4-dependent GTP hydrolysis has a higher turnover rate with the POST than with the PRE ribosomes (12). The clockwise ratchet-like movement of the 30S subunit upon EF4 binding in our complex and the conformational changes observed in the decoding center could be arranged to facilitate the back-translocation function of EF4, since the ratcheting of the ribosome in the opposite direction upon EF-G-GTP binding leads to tRNA translocation (4, 20–22). In the EF4 complex, the widening of the decoding center may facilitate the re-accommodation of a back-translocated tRNA in the A site. Therefore, this structure could represent a state prior to the back-movement of tRNAs; however, it is unclear whether the anticodon or the acceptor stem of the P-site tRNA would move backward first since it remains in the classical P/P position in our complex.
Recently, the back-translocation function of EF4 has been challenged by a smFRET study which suggests that EF4 preferentially targets ribosomes in the PRE instead of the POST state and effectively competes with EF-G by sequestering ribosomes in an intermediate state between the PRE and POST states (16). The formation of this intermediate, which provides transient inhibition of the elongation cycle (16), can also occur upon EF4 interaction with the POST state, albeit at a much slower rate (11, 16). This argues against the back-translocation being the main function of EF4 (16). The interactions between the CTD of EF4 and the Phe-tRNAPhe in the P site of our complex, presumably protecting the phenylalanine residue attached the P-site tRNA from hydrolysis, seem to agree with the proposed sequestered state of the ribosome (16). However, in order for the CTD to reach the PTC, an A-site tRNA in the classical A/A conformation would have to be displaced due to steric overlaps (Fig. 2D). In this regard, the re-opening of the decoding center achieved through the clockwise ratchet-like movement of the 30S subunit could assist the displacement of the A-site tRNA by weakening the codon-anticodon interactions in the A site.
Our results show that during elongation EF4 can remodel the decoding center in an opposite way to that induced by the accommodation of an A-site tRNA. The re-opening of the A site on the 30S subunit may lead to back-translocation of the P-site tRNA on POST ribosome complexes or help displace the A-site tRNA on PRE complexes. Thus, EF4 appears to reverse the process performed by either EF-G or EF-Tu, thereby providing a structural basis for the previously proposed functions of EF4 as a back-translocase (9, 11, 12, 15) or a ribosome sequester (16). The high conservation of EF4 underlines the importance of the conformational reversibility of the ribosome during elongation which may provide an evolutionary advantage by ensuring that ribosomes do not stall during the elongation cycle. Structures of additional EF4-ribosome complexes, along with more directed experiments, will be essential to fully understand the role of EF4.
Supplementary Material
Acknowledgments
We thank P. Moore, S. Seetharaman, I. Lomakin and Y. Polikanov for valuable discussions and critical reading of the manuscript along with G. Blaha for help in designing and generating the mutant T. thermophilus strain. We thank A. Dahlberg and S. Gregory for providing the thermostable kanamycin gene and advice with genetic manipulations of T. thermophilus. We thank R. Grodzicki for preparing the Phe-tRNAPhe. We also thank the staffs of the Advanced Photon Source beamline 24-ID and the National Synchrotron Light Source beamlines X25 and X29 for help during data collection and the Center for Structural Biology facility at Yale University for computational support. This work was supported by NIH grant GM022778 (to T.A.S.). Coordinates and structure factors have been deposited in the Protein Data Bank with accession codes 4QJS and 4QJT.
Footnotes
References and Notes
- 1.Clementi N, Polacek N, Ribosome-associated GTPases: the role of RNA for GTPase activation. RNA Biol. 7, 521 (Sep-Oct, 2010). [DOI] [PubMed] [Google Scholar]
- 2.Julian P et al. , Structure of ratcheted ribosomes with tRNAs in hybrid states. Proc. Natl. Acad. Sci. U. S. A 105, 16924 (Nov 4, 2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Agirrezabala X et al. , Visualization of the hybrid state of tRNA binding promoted by spontaneous ratcheting of the ribosome. Mol. Cell 32, 190 (Oct 24, 2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frank J, Agrawal RK, A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature 406, 318 (Jul 20, 2000). [DOI] [PubMed] [Google Scholar]
- 5.Yoshizawa S, Bock A, The many levels of control on bacterial selenoprotein synthesis. Biochim. Biophys. Acta 1790, 1404 (Nov, 2009). [DOI] [PubMed] [Google Scholar]
- 6.Roberts MC, Update on acquired tetracycline resistance genes. FEMS Microbiol. Lett 245, 195 (Apr 15, 2005). [DOI] [PubMed] [Google Scholar]
- 7.Caldon CE, Yoong P, March PE, Evolution of a molecular switch: universal bacterial GTPases regulate ribosome function. Mol. Microbiol 41, 289 (Jul, 2001). [DOI] [PubMed] [Google Scholar]
- 8.March PE, Inouye M, Characterization of the lep operon of Escherichia coli. Identification of the promoter and the gene upstream of the signal peptidase I gene. J. Biol. Chem 260, 7206 (Jun 25, 1985). [PubMed] [Google Scholar]
- 9.Qin Y et al. , The highly conserved LepA is a ribosomal elongation factor that back-translocates the ribosome. Cell 127, 721 (Nov 17, 2006). [DOI] [PubMed] [Google Scholar]
- 10.Zhang D, Qin Y, The paradox of elongation factor 4: highly conserved, yet of no physiological significance? Biochem. J 452, 173 (Jun 1, 2013). [DOI] [PubMed] [Google Scholar]
- 11.Liu H, Pan D, Pech M, Cooperman BS, Interrupted catalysis: the EF4 (LepA) effect on back-translocation. J. Mol. Biol 396, 1043 (Mar 5, 2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Connell SR et al. , A new tRNA intermediate revealed on the ribosome during EF4-mediated back-translocation. Nat. Struct. Mol. Biol 15, 910 (Sep, 2008). [DOI] [PubMed] [Google Scholar]
- 13.Pech M et al. , Elongation factor 4 (EF4/LepA) accelerates protein synthesis at increased Mg2+ concentrations. Proc. Natl. Acad. Sci. U. S. A 108, 3199 (Feb 22, 2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shoji S, Janssen BD, Hayes CS, Fredrick K, Translation factor LepA contributes to tellurite resistance in Escherichia coli but plays no apparent role in the fidelity of protein synthesis. Biochimie 92, 157 (Feb, 2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamamoto H et al. , EF-G and EF4: translocation and back-translocation on the bacterial ribosome. Nat. Rev. Microbiol 12, 89 (Feb, 2014). [DOI] [PubMed] [Google Scholar]
- 16.Liu H et al. , The conserved protein EF4 (LepA) modulates the elongation cycle of protein synthesis. Proc. Natl. Acad. Sci. U. S. A 108, 16223 (Sep 27, 2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Evans RN, Blaha G, Bailey S, Steitz TA, The structure of LepA, the ribosomal back translocase. Proc. Natl. Acad. Sci. U. S. A 105, 4673 (Mar 25, 2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Selmer M, Gao YG, Weixlbaumer A, Ramakrishnan V, Ribosome engineering to promote new crystal forms. Acta Crystallogr. D Biol. Crystallogr 68, 578 (May, 2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Materials and methods are available as supporting material on Science Online.
- 20.Zhou J, Lancaster L, Donohue JP, Noller HF, Crystal structures of EF-G-ribosome complexes trapped in intermediate states of translocation. Science 340, 1236086 (Jun 28, 2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tourigny DS, Fernandez IS, Kelley AC, Ramakrishnan V, Elongation factor G bound to the ribosome in an intermediate state of translocation. Science 340, 1235490 (Jun 28, 2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pulk A, Cate JH, Control of ribosomal subunit rotation by elongation factor G. Science 340, 1235970 (Jun 28, 2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao YG et al. , The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science 326, 694 (Oct 30, 2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Youngman EM, Brunelle JL, Kochaniak AB, Green R, The active site of the ribosome is composed of two layers of conserved nucleotides with distinct roles in peptide bond formation and peptide release. Cell 117, 589 (May 28, 2004). [DOI] [PubMed] [Google Scholar]
- 25.Colca JR et al. , Cross-linking in the living cell locates the site of action of oxazolidinone antibiotics. J. Biol. Chem 278, 21972 (Jun 13, 2003). [DOI] [PubMed] [Google Scholar]
- 26.Maguire BA, Beniaminov AD, Ramu H, Mankin AS, Zimmermann RA, A protein component at the heart of an RNA machine: the importance of protein L27 for the function of the bacterial ribosome. Mol. Cell 20, 427 (Nov 11, 2005). [DOI] [PubMed] [Google Scholar]
- 27.Selmer M et al. , Structure of the 70S ribosome complexed with mRNA and tRNA. Science 313, 1935 (Sep 29, 2006). [DOI] [PubMed] [Google Scholar]
- 28.Zhou J, Lancaster L, Trakhanov S, Noller HF, Crystal structure of release factor RF3 trapped in the GTP state on a rotated conformation of the ribosome. RNA 18, 230 (Feb, 2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jin H, Kelley AC, Ramakrishnan V, Crystal structure of the hybrid state of ribosome in complex with the guanosine triphosphatase release factor 3. Proc. Natl. Acad. Sci. U. S. A 108, 15798 (Sep 20, 2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dunkle JA et al. , Structures of the bacterial ribosome in classical and hybrid states of tRNA binding. Science 332, 981 (May 20, 2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang W, Dunkle JA, Cate JH, Structures of the ribosome in intermediate states of ratcheting. Science 325, 1014 (Aug 21, 2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ogle JM et al. , Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 292, 897 (May 4, 2001). [DOI] [PubMed] [Google Scholar]
- 33.Wimberly BT et al. , Structure of the 30S ribosomal subunit. Nature 407, 327 (Sep 21, 2000). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


