Abstract
The polyhydroxyalkanoic acid synthase gene from Chromobacterium violaceum (phaCCv) was cloned and characterized. A 6.3-kb BamHI fragment was found to contain both phaCCv and the polyhydroxyalkanoic acid (PHA)-specific 3-ketothiolase (phaACv). Escherichia coli strains harboring this fragment produced significant levels of PHA synthase and 3-ketothiolase, as judged by their activities. While C. violaceum accumulated poly(3-hydroxybutyrate) or poly(3-hydroxybutyrate-co-3-hydroxyvalerate) when grown on a fatty acid carbon source, Klebsiella aerogenes and Ralstonia eutropha (formerly Alcaligenes eutrophus), harboring phaCCv, accumulated the above-mentioned polymers and, additionally, poly(3-hydroxybutyrate-co-3-hydroxyhexanoate) when even-chain-length fatty acids were utilized as the carbon source. This finding suggests that the metabolic environments of these organisms are sufficiently different to alter the product range of the C. violaceum PHA synthase. Neither recombinant E. coli nor recombinant Pseudomonas putida harboring phaCCv accumulated significant levels of PHA. Sequence analysis of the phaCCv product shows homology with several PHA synthases, most notably a 48% identity with that of Alcaligenes latus (GenBank accession no. AAD10274).
Polyhydroxyalkanoic acids (PHAs) are carbon and energy reserve polymers produced in some bacteria when carbon sources are plentiful and another nutrient, such as nitrogen, phosphate, oxygen, or sulfur, becomes limiting. PHAs composed of monomeric units ranging from 3 to 14 carbons exist in nature. When the carbon source is exhausted, PHA is utilized by the bacterium (1, 29, 32). Some PHA polyesters have physical properties similar to those of polypropylene, making them a source of biodegradable plastic from renewable resources. Polymers of various compositions are produced, depending on the substrate specificity of the PHA synthase and the carbon source on which the bacterium is grown, as well as the metabolic pathways involved in the utilization of the carbon source. While homopolymers composed of 3-hydroxybutyric acid (3HB) are very brittle, mixtures possessing longer carbon backbones result in a more flexible polymer and, hence, a more marketable plastic. PHAs have potential applications in medicine and dentistry (1, 29), and a polymer composed of 3HB and 3-hydroxyvaleric acid (3HV) has been marketed under the trademark name BIO-POL (26). Another PHA with attractive physical properties is poly(3-hydroxybutyrate-co-3-hydroxyhexanoate), but at the present time there are only a few reports of bacteria that accumulate a copolymer composed entirely of 3HB and 3-hydroxyhexanoic acid (3HC) (4, 7, 14).
PHA production is best understood in Ralstonia eutropha (formerly Alcaligenes eutrophus [16, 17, 35]; for reviews see references 1 and 29). Poly(3-hydroxybutyrate) is synthesized from acetyl-coenzyme A (CoA) in a three-step pathway. The first reaction involves a PHA-specific 3-ketothiolase, encoded by phaARe, that condenses two acetyl-CoA molecules into acetoacetyl-CoA. The second reaction, which is the reduction of acetoacetyl-CoA to d-(−)-3-hydroxybutyryl-CoA, is catalyzed by an NADPH-dependent acetoacetyl-CoA reductase, encoded by phaBRe. The last reaction is catalyzed by PHA synthase, which is the product of the phaCRe gene. In this reaction, d-(−)-3-hydroxybutyrl-CoA is linked to an existing PHA molecule by the formation of an ester bond. In addition to the three-step pathway just described, different (d)-3-hydroxyacyl-CoA substrates may be used by the PHA synthase to construct PHAs of different monomeric compositions. These alternative substrates for PHA synthase could be provided by intermediates of other metabolic pathways, such as the fatty acid oxidation pathway, the fatty acid synthesis pathway, the methylmalonyl-CoA pathway, and the isoleucine-valine degradation pathway (20, 21, 33).
The focus of this research is the soil bacterium Chromobacterium violaceum, which has been known for quite some time to accumulate PHA (6, 30). C. violaceum is known to accumulate polymer composed primarily of 3HB and 3HV and can produce a homopolymer of 3HV when grown on valerate (30). Because of this ability to accumulate high levels of 3HV monomer, it is possible that this PHA synthase would also have enhanced ability to incorporate 3HC monomers. This paper is the first detailed genetic study of the C. violaceum PHA synthase and describes the cloning and molecular analysis of phaCCv, as well as attempts at expression of the cloned gene in Escherichia coli, Klebsiella aerogenes, Pseudomonas putida (phaC mutant) and R. eutropha (phaC mutant).
MATERIALS AND METHODS
Bacterial culture conditions.
For routine maintenance, E. coli and K. aerogenes were grown at 37°C in Luria-Bertani medium (BBL, Cockeysville, Md.) while C. violaceum, R. eutropha, and P. putida were grown at 30°C in nutrient broth (Difco, Detroit, Mich.); both were supplemented with the appropriate antibiotic(s) as needed. The final concentrations of the antibiotics (Sigma, St. Louis, Mo.) were as follows: kanamycin, 50 μg/ml; chloramphenicol, 25 μg/ml; and tetracycline, 10 μg/ml. The minimal media used were M9 medium (2) for E. coli and K. aerogenes and a modified mineral salts medium (24) for C. violaceum, R. eutropha, and P. putida, containing 13.2 mM Na2HPO4 · 7H2O, 11 mM KH2PO4, 0.81 mM MgSO4 · 7H2O, 0.136 mM CaCl2, 0.047 mM NH4Cl, 1 ml of Ramsay’s trace element solution (19)/liter, and 5 mg of ferric ammonium citrate/liter. In all cases, the cultures were incubated in baffled flasks in an orbital shaker set at 180 rpm. The bacterial strains and plasmids used in this study are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Important features (reference or source) |
|---|---|
| C. violaceum | Source of phaC (DSM 30191) |
| E. coli XL1-Blue | F′ Tn10(Tetr); lac for blue/white screening (Stratagene) |
| E. coli S17-1 | Used for mating experiments (ATCC 47055) |
| E. coli DH5α | General strain for plasmid maintenance (Gibco-BRL) |
| K. aerogenes KC2671 | General Klebsiella strain (gift from R. Bender) |
| R. eutropha H16 PHB-4 | phaC negative mutant (DSM 541) |
| P. putida GpP104 | phaC negative mutant (gift from T. Mitsky, Ceregen) |
| pBBR1MCS-1 | Broad-host-range cloning vector; Cmr (11) |
| pAM | pBBR1MCS-1 + 6.3-kb C. violaceum insert |
| pJM9501 | pBBR1MCS-1 + kanamycin cassette |
| pCV7 | pJM9501 + 6.3-kb insert; opposite orientation to pAM |
| pCV8 | pJM9501 + 6.3-kb insert; same orientation as pAM |
| pJM9131 | Contains the pha pathway from R. eutropha; Kmr (34) |
| pUMS | Source of R. eutropha phaA and phaB genes; Tcr (25) |
| pBluescript II SK(+) | Sequencing vector; Apr (Stratagene) |
DNA manipulations.
All plasmid and mapping experiments were performed with standard techniques (2, 9) and commercially available enzymes (New England Biolabs, Beverly, Mass.; Boehringer Mannheim, Indianapolis, Ind.; Promega, Madison, Wis.; Gibco-BRL, Gaithersburg, Md.; and Stratagene, La Jolla, Calif.) and kits for gel purification and template purification (Qiagen, Chatsworth, Calif.).
Cloning of C. violaceum phaC gene.
A C. violaceum genomic library was constructed by ligation of 5- to 7-kb BamHI fragments into the BamHI site of pBBR1MCS-1 (12). The resulting plasmid library was used to transform XL1-Blue to chloramphenicol resistance (2). White colonies that resulted after growth on X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) (final concentration, 20 μg/ml; United States Biochemicals, Cleveland, Ohio) plates were further screened for phaCCv with a digoxigenin (DIG)-labeled PCR product corresponding to bases 1865 to 2430 (5′-CGCCGTGCATCAACAAGTA-3′ and 5′-TTGGTGGCGTCGCCGTTCCA-3′) of the R. eutropha phaC gene (11) and the Genius detection system (Boehringer Mannheim). By this procedure, plasmid pAM, containing a 6.3-kb insert, was identified and used for further studies.
Southern blot analyses.
In addition to probing with a phaCRe subfragment, the 6.3-kb insert DNA and subclones were similarly probed with DIG-labeled subfragments of phaARe and phaBRe, corresponding to bases 3323 to 3901 (5′-GCCGGCGGCCAGGAAAACAT-3′ and 5′-GGTCTTGCGGGGTCCACTCG-3′) and 4527 to 4932 (5′-CGTGGTGTTCCGCAAGATGA-3′ and 5′-GGACTCCTCCGACGACAACC-3′), respectively, of the published DNA sequence (11).
Nucleotide sequence analyses of phaCCv.
The 6.3-kb BamHI insert was cloned in both orientations into pBluescript II SK(+), and subclones were generated from these plasmids by ligating fragments from a partial HincII digest into pBluescript II SK(+) cut with HincII. One subclone contained an approximately 3.0-kb insert that exhibited homology with the phaCRe probe. A series of overlapping subclones was generated for this plasmid (10), and the sequence was determined by a modified Sanger reaction (23) with the ThermoSequenase primer cycle sequencing kit (Amersham Corp., Arlington Heights, Ill.), prelabeled primers (LiCor, Lincoln, Nebr.), and an automated LiCor sequencer linked to an IBM OS/2 Warp computer. Sequence comparisons and alignments were performed with the Basic Local Alignment Search Tool (BLAST; National Center for Biotechnology Information) and the ClustalW Multiple Sequence Alignment Program (Baylor College of Medicine).
Enzyme assays.
Cells were grown in Luria-Bertani medium containing the appropriate antibiotics at 30°C with aeration. The cells were harvested by centrifugation (3,000 rpm in a Varafuge F centrifuge; Fisher Scientific), and the resulting cell pellets were stored frozen at −70°C at least overnight. The frozen pellets were thawed on ice prior to resuspension in 1/10 the original volume in lysis buffer (20 mM potassium phosphate [pH 7.2], 5 mM MgCl2, 1 mM EDTA [pH 8.0], 1 mM dithiothreitol, 9.2% glycerol). Upon resuspension, the cells were subjected to sonication on ice (four 10-s bursts with a microtip; Artek Sonicator). The resulting crude cell extract was centrifuged for 3 min in a microcentrifuge (16,000 × g), and the supernatant was used for enzyme and total-protein assays. PHA synthase, 3-ketothiolase, and NADPH-dependent acetyl-CoA reductase activities were measured by previously described methods (22, 25, 34). The total-protein concentration was determined with commercially available kits (Bio-Rad, Richmond, Calif.). For these experiments E. coli DH5α(pJM9131), which contains the R. eutropha PHA operon, was used as a positive control. One unit is defined as 1 μmol of substrate utilized per min.
Construction of pJM9501, pCV7, and pCV8.
Plasmid pJM9501 was constructed by inserting the kanamycin cassette into the SalI site of pBBR1MCS-1. For expression studies, the 6.3-kb C. violaceum insert was cloned into the BamHI site of pJM9501 in both orientations, resulting in pCV7 and pCV8.
PHA accumulation in heterologous hosts with the cloned phaCCv gene.
Plasmid pCV7 or pCV8 was introduced into E. coli DH5α(pUMS) and K. aerogenes KC2671(pUMS) by electroporation and into P. putida GpP104 and R. eutropha PHB-4 by the S17.1 mating technique (9). For E. coli and K. aerogenes, it was necessary to provide phaARe and phaBRe carried on pUMS in order for PHA accumulation to occur (27). For the expression studies, all bacterial strains were grown for 2 days at 30°C with aeration in the nitrogen-free minimal medium described above supplemented with 0.1% nutrient broth, the appropriate antibiotics, and a carbon source listed in Table 2. The final concentrations of the carbon sources were as follows: four-carbon to six-carbon fatty acids, 0.2%; higher-molecular-weight fatty acids (myristic acid, palmitic acid, and stearic acid), 5 mM (0.1 to 0.2%); and sugars and gluconate, 0.5%. Stock solutions of myristic acid, palmitic acid, and stearic acid were prepared by dissolving them in 10% Brij 58 and neutralizing the solution to pH 7.0 as described previously (28). The cells were harvested by centrifugation and washed in sterile saline, and the resulting pellet was lyophilized. PHA accumulation and composition were measured by methanolysis and gas chromatography of lyophilized samples as described previously (27).
TABLE 2.
PHA accumulation in C. violaceum and in heterologous hosts containing the cloned phaC gene
| Strain | Carbon source | % PHAa | Polymer composition (mol%)
|
||||
|---|---|---|---|---|---|---|---|
| 3HB | 3HV | 3HC | 3HH | 3HO | |||
| C. violaceum | Glucose | 38 | 98 | 2 | Trb | ||
| Fructose | 26 | 92 | 8 | Tr | |||
| Gluconate | 2 | 100 | |||||
| Butyric acid | Tr | 100 | |||||
| Valeric acid | 28 | 12 | 88 | ||||
| Hexanoic acid | 9 | 68 | 32 | Tr | |||
| Heptanoic acid | 15 | 5 | 95 | Tr | |||
| Octanoic acid | 4 | 78 | 22 | ||||
| Nonanoic acid | 22 | 10 | 90 | ||||
| Decanoic acid | 14 | 90 | 9 | Tr | |||
| Myristic acid | 41 | 97 | 3 | Tr | |||
| Palmitic acid | 41 | 99 | Tr | Tr | |||
| Stearic acid | 50 | 99 | 1 | Tr | |||
| K. aerogenes(pCV8) | Glucose | Tr | 100 | ||||
| K. aerogenes(pCV8, pUMS) | Glucose | 50 | 100 | Tr | |||
| Fructose | 40 | 100 | Tr | Tr | |||
| Butyric acid | 5 | 100 | |||||
| Valeric acid | 18 | 89 | 11 | ||||
| Hexanoic acid | 13 | 98 | Tr | 1 | |||
| Heptanoic acid | 28 | 48 | 52 | Tr | Tr | ||
| Octanoic acid | 12 | 93 | 1 | 6 | Tr | ||
| Nonanoic acid | 26 | 46 | 54 | Tr | Tr | ||
| Decanoic acid | 10 | 87 | 1 | 12 | Tr | ||
| Myristic acid | 8 | 95 | 5 | ||||
| Palmitic acid | 12 | 97 | 3 | ||||
| Stearic acid | 10 | 98 | 2 | ||||
| R. eutropha PHB-4(pCV8) | Gluconate | 81 | 100 | Tr | |||
| Fructose | 78 | 100 | Tr | ||||
| Butyric acid | 81 | 98 | Tr | 2 | |||
| Valeric acid | 31 | 19 | 81 | Tr | |||
| Hexanoic acid | 75 | 88 | Tr | 11 | Tr | ||
| Heptanoic acid | 55 | 68 | 27 | Tr | 5 | ||
| Octanoic acid | 68 | 92 | Tr | 8 | |||
| Nonanoic acid | 59 | 87 | 10 | Tr | 3 | ||
| Decanoic acid | 83 | 95 | Tr | 5 | |||
| Myristic acid | 31 | 86 | 2 | 10 | 2 | ||
| Palmitic acid | 41 | 89 | 4 | 6 | 1 | ||
| Stearic acid | 35 | 85 | 10 | 4 | 1 | ||
Yield = PHA weight/cell dry weight.
Tr, trace; less than 0.45 mol%.
Nucleotide sequence accession number. The complete nucleotide sequence of the 2,946-bp C. violaceum DNA fragment can be accessed under GenBank accession no. AF061446.
RESULTS
Cloning of the C. violaceum PHA synthase and activity in E. coli.
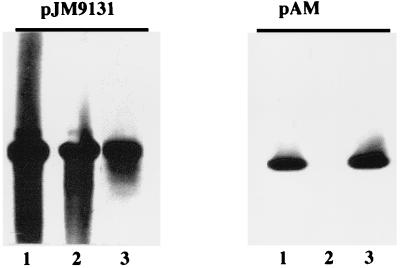
A plasmid library containing C. violaceum DNA was screened with a DIG-labeled PCR product corresponding to a conserved region of the R. eutropha phaC gene. Plasmid pAM, containing a 6.3-kb BamHI C. violaceum insert, was found to have sequences homologous to the phaCRe and the phaARe probes but did not react with the phaBRe probe (Fig. 1). To assay enzyme activity, E. coli DH5α harboring pAM or pJM9131 was cultured in rich medium in the absence of glucose. Under these conditions, E. coli DH5α containing pAM exhibited 3-ketothiolase (2,349 U/g of protein) and PHA synthase (35 U/g of protein) levels that were comparable to those of E. coli DH5α containing the R. eutropha PHA operon on pJM9131 (5,456 and 17 U/g of protein for 3-ketothiolase and PHA synthase, respectively). NADPH-dependent acetoacetyl-CoA reductase activity was detected in E. coli DH5α harboring pJM9131 (454 U/g of protein) but was not detected in E. coli DH5α(pAM).
FIG. 1.
Southern blot analysis. Plasmid DNA was digested with BamHI prior to electrophoresis and transfer to nylon filters. The filters were probed with the following DIG-labeled subfragments (see Materials and Methods): lanes 1, phaARe; lanes 2, phaBRe; and lanes 3, phaCRe. pJM9131 is a positive control carrying the R. eutropha PHA operon.
Construction of pCV7 and pCV8.
Because of the natural resistance of P. putida GpP104 to chloramphenicol (15) and in order to increase the versatility of the broad-host-range vector, it was necessary to construct pJM9501 by inserting a kanamycin cassette into pBBR1MCS-1. The 6.3-kb C. violaceum insert was cloned into the BamHI site of pJM9501 in both orientations. A SacII digest revealed that pCV8 has the same orientation as pAM while pCV7 has the opposite orientation. These plasmids were introduced into E. coli DH5α, K. aerogenes, and the phaC mutants of P. putida and R. eutropha for further expression studies.
Expression of C. violaceum PHA synthase in heterologous hosts.
Expression of the C. violaceum phaC gene product was measured by the accumulation of PHA in the heterologous hosts grown in minimal medium containing one of several carbon sources (Table 2). No significant accumulation was seen in E. coli DH5α(pUMS) or P. putida harboring either of the plasmids (data not shown). However, R. eutropha(pCV7), R. eutropha(pCV8), K. aerogenes(pCV7, pUMS), and K. aerogenes(pCV8, pUMS) accumulated significant amounts of polymer. Because the yields and polymer compositions were similar for K. aerogenes and R. eutropha harboring either pCV7 or pCV8, only strains containing pCV8 are shown (Table 2). For K. aerogenes and R. eutropha, addition of IPTG (isopropyl-β-d-thiogalactopyranoside) to a final concentration of 1 mM had no effect on either the yield or the composition of the polymer (data not shown), suggesting that the lac promoter contained on the plasmid vector was not instrumental in the expression of the C. violaceum phaC gene. R. eutropha(pCV8) had, for the most part, higher PHA yields than those seen for K. aerogenes and for the wild-type C. violaceum. In R. eutropha, PHA was generally composed of larger molar percentages of 3HV, 3HC, 3-hydroxyheptanoic acid (3HH), and 3-hydroxyoctanoic acid (3HO) when grown on fatty acids. As the length of the even-chain-length fatty acid carbon source increased, so did the molar percentage of 3HC incorporated into the PHA polymer for K. aerogenes harboring pCV8. This trend continued until myristic acid was used as the carbon source (Table 2).
DNA sequence analyses.
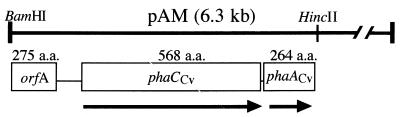
A mapping analysis of pAM revealed that phaCCv is located at the 5′ end of the 6.3-kb insert, closest to the lac promoter. To facilitate DNA sequencing of the PHA synthase, a 3.0-kb BamHI/HincII fragment resulting from a HincII partial digest at that end was constructed in pBluescript II SK(+) and unidirectional deletions were made from either end (10). Sequence analysis (with MacVector) of this 2,946-bp C. violaceum DNA fragment revealed that this subclone contains three putative open reading frames (ORFs) (Fig. 2). The translational product from the first ORF (designated OrfA) did not align significantly with any protein sequence in the GenBank database. A BLAST alignment of the translational product of the second ORF (nucleotides 741 to 2445) revealed that the amino acid sequence exhibited a very high homology with a PHA synthase from Alcaligenes latus (8) (BLAST score, 524; 48% identity) but was also quite similar to that from another Alcaligenes strain (13) and more than 20 other PHA synthases (with BLAST scores higher than 300). It was therefore designated phaCCv. The translational product of the third ORF exhibited homology with 10 PHA-specific thiolases (with BLAST scores above 200), most notably with the 3-ketothiolase from R. eutropha (17) (BLAST score, 315; 69% identity). It was therefore designated phaACv.
FIG. 2.
Molecular organization and orientation with respect to pAM of a 2,946-bp BamHI/HincII subfragment containing the phaCCv gene and sequences for orfA and phaACv genes. The arrows indicate the orientations of the genes. OrfA and phaCCv are partial ORFs. The number of amino acids (a.a.) encoded by each ORF is indicated.
DNA sequences upstream of the putative start of translation (base 741) were examined for homology to known prokaryotic control regions. A number of putative control sequences were detected, including possible ς70 promoters (TTGACA; −35) at bases 642 and 610 and a possible −24/12 promoter at base 582. Shine-Dalgarno consensus sequences were detected at bases 426, 727, and 2506 for orfA, phaCCv, and phaACv, respectively.
DISCUSSION
It is well known that certain experimental conditions may be manipulated in the biological synthesis of PHA to result in polymers of various compositions. One of these conditions is the choice of PHA synthase, the enzyme that incorporates (d)-3-hydroxyacyl-CoA substrates into the PHA polymer. We hypothesized that because the C. violaceum PHA synthase is much better at incorporating 3HV units into polymer than the R. eutropha PHA synthase (30), the broader substrate range might also allow it to be used to incorporate higher percentages of 3HC into the polymer in recombinant hosts. The findings show that this is the case for poly(3-hydroxybutyrate-co-3-hydroxyhexanoate) accumulation in K. aerogenes and R. eutropha, which incorporated 3HC to levels as high as 12 mol%. In particular, more 3HC was incorporated into the polymer as the size of the even-chain-length fatty acid carbon source increased for K. aerogenes(pUMS), until a backbone 10 carbons long was reached (Table 2). This finding may reflect the increased induction of fatty acid metabolism genes by the presence of longer-chain fatty acids (3, 5, 18). However, this effect was greatly decreased for myristic, palmitic, and stearic acids (14, 16, and 18 carbons long, respectively), since cultures containing these carbon sources accumulated less PHA and less 3HC than did the cultures grown on heptanoic through decanoic acids (Table 2). Perhaps this reflects a decreased ability of these organisms to take up the longer-chain fatty acids, a smaller flux of 3HB-3HC substrates within the cell, or the fact that the longer-chain fatty acids could not be solubilized into the medium at levels as high as those for heptanoic through decanoic acids (28).
Another condition that may be manipulated for PHA accumulation is the bacterial host used to synthesize PHA. In this study, K. aerogenes(pUMS) and R. eutropha used the same contingent of genes to synthesize PHA (phaA and phaB from R. eutropha and phaC from C. violaceum). Yet R. eutropha(pCV8) accumulated polymer that contained significant amounts of 3HH and 3HO when grown on odd-chain-length fatty acids and even-chain-length fatty acids, respectively, but K. aerogenes(pUMS, pCV8) was unable to do so. In addition, the composition of the PHA polymer in both K. aerogenes and R. eutropha differs from that accumulated by C. violaceum cultured in the same medium (Table 2). This suggests that R. eutropha and K. aerogenes have additional enzymes capable of synthesizing the substrates for the phaCCv gene product used in these experiments or that they harbor the same enzymes with different substrate specificities than those in C. violaceum. Because the phaB gene product is directly involved in synthesizing d-(−)-3-hydroxyacyl-CoA substrates, one possible candidate for an additional enzyme is an alternate ketoacyl-CoA reductase in R. eutropha. Alternatively, additional metabolic pathways could provide substrates for the PHA synthase. In other organisms, fatty acid metabolism, fatty acid oxidation, the methylmalonyl-CoA pathway, and the isoleucine-valine degradation pathway are involved in PHA accumulation (20, 21, 33).
It was also interesting to note that phaCCv was not expressed in either E. coli(pUMS) or P. putida. This is significant because most PHA synthases isolated previously that have been expressed in R. eutropha also show good expression in P. putida (which contains enzymes that provide substrates for the PHA synthase) (31, 33). Likewise, PHA synthase genes which have previously been shown to be expressed in K. aerogenes are usually also expressed in E. coli strains (reference 36 and unpublished data). The reason for this unusual difference in expression levels is not known.
DNA sequence analysis suggests that phaCCv and phaACv are arranged in an operon (Fig. 2). The 6.3-kb fragment does not appear to contain phaB, as indicated by Southern blot analysis (Fig. 1) and enzyme analysis (data not shown). Because it is not known whether the phaB gene is contained within the phaCCv-phaACv operon or is located elsewhere on the chromosome, it is unclear whether this gene arrangement resembles that seen in other bacteria harboring type I synthases (31) or represents a novel gene arrangement.
ACKNOWLEDGMENTS
This work was funded by a grant from the Monsanto Corporation located in St. Louis, Missouri.
We thank Anne Stangl and Ken Gonyer for technical assistance, Robert Bender and Timothy Mitsky for their generous donation of strains, and Henry Valentin, Ivor Knight, Jon Monroe, Brian Hall, and Ho-Gun Rhie for helpful discussions.
REFERENCES
- 1.Anderson A J, Dawes E A. Occurrence, metabolism, metabolic role, and industrial uses of bacterial polyhydroxyalkanoates. Microbiol Rev. 1990;54:450–472. doi: 10.1128/mr.54.4.450-472.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. J. New York, N.Y: Wiley and Sons; 1987. [Google Scholar]
- 3.Black P N, DiRusso C C. Molecular and biochemical analyses of fatty acid transport, metabolism, and gene regulation in Escherichia coli. Biochim Biophys Acta. 1994;1210:123–145. doi: 10.1016/0005-2760(94)90113-9. [DOI] [PubMed] [Google Scholar]
- 4.Caballero K P, Karel S F, Register R A. Biosynthesis and characterization of hydroxybutyrate-hydroxycaproate copolymers. Int J Biol Macromol. 1995;17:86–92. doi: 10.1016/0141-8130(95)93522-y. [DOI] [PubMed] [Google Scholar]
- 5.Di Russo C C, Metzger A K, Heimert T L. Regulation of transcription of genes required for fatty acid transport and unsaturated fatty acid biosynthesis in Escherichia coli by FadR. Mol Microbiol. 1993;7:311–322. doi: 10.1111/j.1365-2958.1993.tb01122.x. [DOI] [PubMed] [Google Scholar]
- 6.Forsyth W G C, Hayward A C, Roberts J B. Occurrence of poly-β-hydroxybutyric acid in aerobic gram-negative bacteria. Nature. 1958;182:800–801. doi: 10.1038/182800a0. [DOI] [PubMed] [Google Scholar]
- 7.Fukui T, Doi Y. Cloning and analysis of the poly(3-hydroxybutyrate-co-3-hydroxyhexanoate) biosynthesis genes of Aeromonas caviae. J Bacteriol. 1997;179:4821–4830. doi: 10.1128/jb.179.15.4821-4830.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Genser K F, Renner G, Schwab H. Molecular cloning, sequencing and expression in Escherichia coli of the poly(3-hydroxyalkanoate) synthesis genes from Alcaligenes latus DSM1124. J Biotechnol. 1998;64:125–135. doi: 10.1016/s0168-1656(98)00093-5. [DOI] [PubMed] [Google Scholar]
- 9.Gerhardt P, editor. Methods for general and molecular bacteriology. Washington, D.C: American Society for Microbiology; 1994. [Google Scholar]
- 10.Henikoff S, Wallace J C, Brown J P. Finding protein similarities with nucleotide sequence databases. Methods Enzymol. 1990;183:111–132. doi: 10.1016/0076-6879(90)83009-x. [DOI] [PubMed] [Google Scholar]
- 11.Janes B, Hollar J, Dennis D. New biosynthetic biodegradable polymers of industrial interest from microorganisms. Amsterdam, The Netherlands: Kluwer Publishers; 1990. Molecular characterization of the poly-β-hydroxybutyrate biosynthetic pathway of Alcaligenes eutrophus H16; pp. 175–190. [Google Scholar]
- 12.Kovach M E, Phillips R W, Elzer P H, Roop II R M, Peterson K M. pBBR1MCS: a broad-host-range cloning vector. BioTechniques. 1994;16:800–802. [PubMed] [Google Scholar]
- 13.Lee I, Rhee Y H, Kim J-Y. Cloning and characterization of the genes involved in the biosynthesis of polyhydroxybutyrates in Alcaligenes sp. SH-69. GenBank accession no. U78047. 1996. [Google Scholar]
- 14.Liebergesell M, Mayer F, Steinbüchel A. Analysis of polyhydroxyalkanoate acid biosynthesis genes of anoxygenic phototrophic bacteria reveals synthesis of a polyester exhibiting an unusual composition. Appl Microbiol Biotechnol. 1993;40:292–300. [Google Scholar]
- 15.Mitsky, T. (Monsanto Corporation, St. Louis, Mo.). Personal communication.
- 16.Peoples O P, Sinskey A J. Poly-beta-hydroxybutyrate (PHB) biosynthesis in Alcaligenes eutrophus H16. Identification and characterization of the PHB polymerase gene (phbC) J Biol Chem. 1989;264:15298–15303. [PubMed] [Google Scholar]
- 17.Peoples O P, Sinskey A J. Poly-beta-hydroxybutyrate biosynthesis in Alcaligenes eutrophus H16. Characterization of the genes encoding beta-ketothiolase and acetoacetyl-CoA reductase. J Biol Chem. 1990;264:15293–15297. [PubMed] [Google Scholar]
- 18.Quail M A, Dempsey C E, Guest J R. Identification of a fatty acyl responsive regulator (FarR) in Escherichia coli. FEBS Lett. 1994;356:183–187. doi: 10.1016/0014-5793(94)01264-4. [DOI] [PubMed] [Google Scholar]
- 19.Ramsay B A, Saracovan I, Ramsay J A, Marchessauld R H. Effect of nitrogen limitation on long-side-chain poly-β-hydroxyalkanoate synthesis by Pseudomonas resinovorans. Appl Environ Microbiol. 1992;58:744–746. doi: 10.1128/aem.58.2.744-746.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rhie H G, Dennis D R. Role of fadR and atoC(Con) mutations in poly(3-hydroxybutyrate-co-3-hydroxyvalerate) synthesis in recombinant pha+ Escherichia coli. Appl Environ Microbiol. 1995;61:2487–2492. doi: 10.1128/aem.61.7.2487-2492.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rhie H G, Dennis D. The function of ackA and pta genes is necessary for poly (3-hydroxybutyrate-co-3-hydroxyvalerate) synthesis in recombinant pha+ Escherichia coli. Can J Microbiol. 1995;41(Suppl. 1):200–206. doi: 10.1139/m95-188. [DOI] [PubMed] [Google Scholar]
- 22.Ritchie G A F, Senior P J, Dawes E A. The purification and characterization of acetoacetyl-coenzyme A reductase from Azotobacter beijerinckii. Biochem J. 1971;121:309–316. doi: 10.1042/bj1210309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schlegel H G, Kaltwasser H, Gottschalk G. Ein Submersverfahren zur Kultur wasserstoffoxydierender Bakterien: Wachstumsphysiologische Untersuchungen. Arch Mikrobiol. 1961;38:209–222. [PubMed] [Google Scholar]
- 25.Senior P J, Dawes E A. The regulation of poly-β-hydroxybutyrate metabolism in Azotobacter beijerinckii. Biochem J. 1973;134:225–238. doi: 10.1042/bj1340225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sherwood M. Bacterial plastic comes to market. Bio/Technology. 1983;1:388–389. [Google Scholar]
- 27.Slater S, Gallaher T, Dennis D. Production of poly-(3-hydroxybutyrate-co-3-hydroxyvalerate) in a recombinant Escherichia coli strain. Appl Environ Microbiol. 1992;58:1089–1094. doi: 10.1128/aem.58.4.1089-1094.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spratt S K, Ginsburgh C L, Nunn W D. Isolation and genetic characterization of mutants defective in propionate metabolism. J Bacteriol. 1981;146:1166–1169. doi: 10.1128/jb.146.3.1166-1169.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steinbüchel A. Polyhydroxyalkanoic acids. In: Byrom D, editor. Biomaterials: novel materials from biological sources. New York, N.Y: Stockton Press; 1991. pp. 123–213. [Google Scholar]
- 30.Steinbüchel A, Debzi E-M, Marchessault R H, Timm A. Synthesis and production of poly(3-hydroxyvaleric acid) homopolyester by Chromobacterium violaceum. Appl Microbiol Biotechnol. 1993;39:443–449. [Google Scholar]
- 31.Steinbüchel A, Hustede E, Liebergesell M, Pieper U, Timm A, Valentin H. Molecular basis for biosynthesis and accumulation of polyhydroxyalkanoate acids in bacteria. FEMS Microbiol Rev. 1992;103:217–230. doi: 10.1111/j.1574-6968.1992.tb05841.x. [DOI] [PubMed] [Google Scholar]
- 32.Steinbüchel A, Schlegel H G. Physiology and molecular genetics of poly(β-hydroxyalkanoic acid) synthesis in Alcaligenes eutrophus. Mol Microbiol. 1991;5:535–542. doi: 10.1111/j.1365-2958.1991.tb00725.x. [DOI] [PubMed] [Google Scholar]
- 33.Valentin H E, Dennis D. Metabolic pathway for poly(3-hydroxybutyrate-co-3-hydroxyvalerate) formation in Nocardia corallina: inactivation of mutB by chromosomal integration of a kanamycin resistance gene. Appl Environ Microbiol. 1996;62:372–379. doi: 10.1128/aem.62.2.372-379.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Valentin H E, Steinbüchel A. Application of enzymatically synthesized short-chain-length hydroxy fatty acid coenzyme A thioesters for assay of polyhydroxyalkanoic acid synthases. Appl Microbiol Biotechnol. 1994;40:699–709. [Google Scholar]
- 35.Yabuuchi E, Kosako Y, Yano I, Hotta H, Nishiuchi Y. Transfer of two Burkholderia and an Alcaligenes species to Ralstonia gen. nov.: proposal of Ralstonia pickettii (Ralston, Palleroni, and Doudoroff 1973) comb. nov., Ralstonia solanacearum (Smith 1896) comb. nov. and Ralstonia eutropha (Davis 1969) comb. nov. Microbiol Immunol. 1995;39:897–904. doi: 10.1111/j.1348-0421.1995.tb03275.x. [DOI] [PubMed] [Google Scholar]
- 36.Zhang H, Obias V, Gonyer K, Dennis D. Production of polyhydroxyalkanoates in sucrose-utilizing recombinant Escherichia coli and Klebsiella strains. Appl Environ Microbiol. 1994;60:1198–1205. doi: 10.1128/aem.60.4.1198-1205.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]