Abstract
The recent resurgence of deep learning (DL) has dramatically influenced the medical imaging field. Medical image analysis applications have been at the forefront of DL research efforts applied to multiple diseases and organs, including those of the lungs. The aims of this review are twofold: (i) to briefly overview DL theory as it relates to lung image analysis; (ii) to systematically review the DL research literature relating to the lung image analysis applications of segmentation, reconstruction, registration and synthesis. The review was conducted following the Preferred Reporting Items for Systematic Reviews and Meta-Analyses guidelines. 479 studies were initially identified from the literature search with 82 studies meeting the eligibility criteria. Segmentation was the most common lung image analysis DL application (65.9% of papers reviewed). DL has shown impressive results when applied to segmentation of the whole lung and other pulmonary structures. DL has also shown great potential for applications in image registration, reconstruction and synthesis. However, the majority of published studies have been limited to structural lung imaging with only 12.9% of reviewed studies employing functional lung imaging modalities, thus highlighting significant opportunities for further research in this field. Although the field of DL in lung image analysis is rapidly expanding, concerns over inconsistent validation and evaluation strategies, intersite generalisability, transparency of methodological detail and interpretability need to be addressed before widespread adoption in clinical lung imaging workflow.
Introduction
Respiratory diseases constitute significant global health challenges; five respiratory diseases are among the most common causes of death. 65 million people suffer from chronic obstructive pulmonary disease (COPD) and 339 million from asthma. 1,2 There are 1.8 million new lung cancer cases diagnosed annually and 1.6 million deaths worldwide, making it the most common and deadliest cancer on the planet. 3 Lung imaging is a critical component of respiratory disease diagnosis, treatment planning, monitoring and treatment assessment. Acquiring lung images, processing them and interpreting them clinically are crucial to achieving global reductions in lung-related deaths. Traditionally, the techniques employed to quantitatively analyse these images evolved from the disciplines of computational modelling and image processing; however, in recent years, deep learning (DL) has received significant attention from the lung imaging community.
DL is a subfield of machine learning that employs artificial neural networks with multiple deep or hidden layers. Whilst the fundamental theory was posited several decades ago, 4 DL gained international interest in 2012 when AlexNet, a type of neural network referred to as a convolutional neural network (CNN), won the ImageNet Large Scale Visual Recognition Challenge. That paper has been cited over 47,000 times and triggered a renaissance in DL research. 5 Subsequently, CNNs, and DL more generally, began to impact the medical imaging field profoundly. Development of fully convolutional networks such as V-Net and ConvNet demonstrated how deep-layered architectures could provide valuable functions in solving some of the field’s most critical applications, including common image analysis tasks. 6,7 Increased computational power due to the reduced cost of graphical processing units (GPUs) and publicly available annotated imaging data sets have since led to rapid developments and applications. 8
This review assesses the current literature on DL’s role in lung image analysis applications, discusses critical limitations for clinical adoption, and sets out a roadmap for future research.
Theory
Artificial neural networks
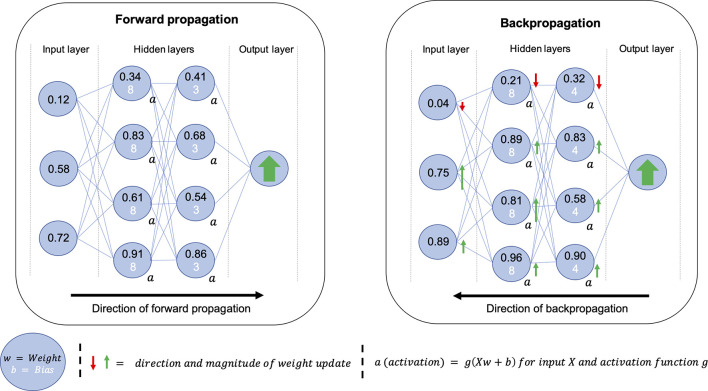
An artificial neural network (ANN), inspired by biological neurons, can be thought of as a series of connected nodes containing weights and biases which are combined using an activation function to produce an activation; the activation determines the strength of connections within the network. At the heart of DL is optimisation; an ANN learns by optimising weights and biases for a generalisable solution. This optimisation occurs in a two-step process of forward propagation and backpropagation. A basic diagram of an ANN with two hidden layers and generalised examples of forward propagation and backpropagation are shown in Figure 1. The use of hidden layers in the network allows more freedom for the weights and biases to be optimised. Forward propagation refers to the process of feeding an example to the network during training where the output of the neural network is compared to a desired output and a loss is calculated using a loss function. Backpropagation uses this loss to propagate changes in weights and biases throughout the network; thus by continually providing new examples, known as iterations, the model is optimised to approximate the function between the input and output domains. Figure 2 provides a glossary of the key technical terms used in this review.
Figure 1.
Simplified diagrams of the processes of forward propagation (left) and backpropagation (right) for a neural network with two hidden layers. The neural network is represented as a series of nodes, each of which contains a weight and bias. The weight and bias are combined using the activation function to produce an activation that impacts the strength of connections within the network. Once an input has been passed through the network, it is compared to a desired output, such as an expert segmentation of an anatomical region of interest, to produce a loss. This loss is used to propagate changes to weights and biases, hence, changing the strength of connections for the subsequent example. The continued repetition of this two-step process is known as network training.
Figure 2.
Glossary of key technical terms related to deep learning and image analysis. ANN, artificial neural network.
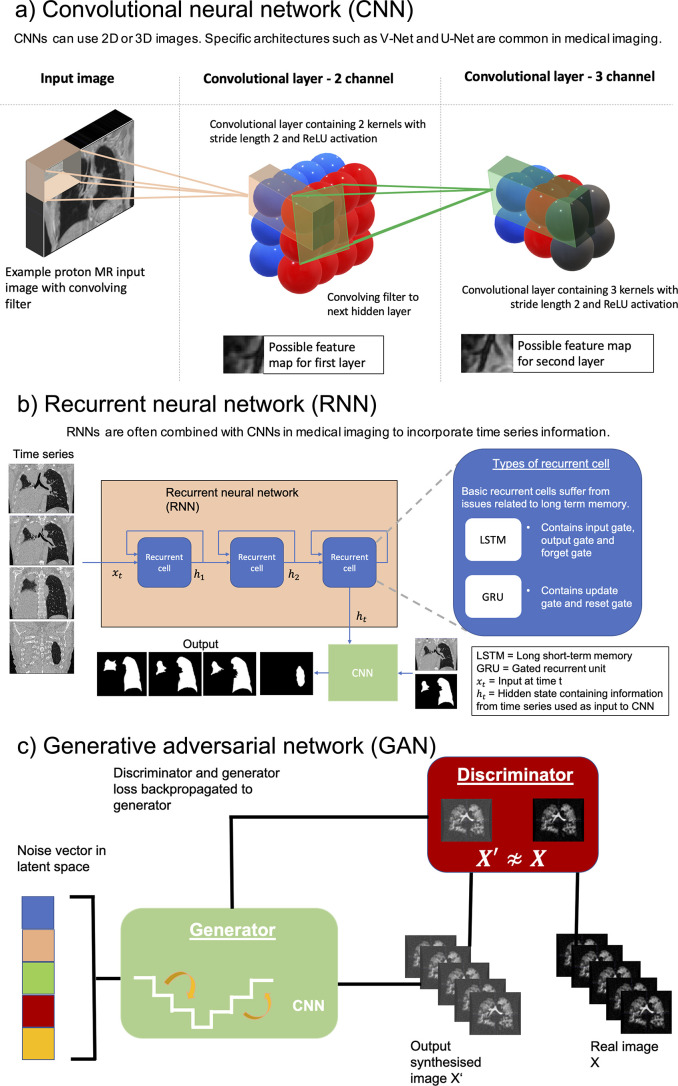
The structure of a DL network is known as an architecture. In the medical imaging field, three key architectures, namely, CNNs, recurrent neural networks (RNNs) and generative adversarial networks (GANs) are particularly prevalent. These structures are outlined in Figure 3. Understanding specific architectures such as V-Nets and GANs requires an in-depth understanding of complex linear algebra and matrix manipulation and is beyond this review’s scope; the interested reader is directed to several excellent papers on the subject. 6,9,10
Figure 3.
Illustration of three common types of deep learning architectures used in medical imaging: (a) CNN), (b) RNN and (c) GAN. In the lung image analysis examples given, the CNN and RNN are used for image segmentation while the GAN is used for image synthesis. CNN, convolutional neural network; GAN, generative adversarial network; RNN, recurrent neural network.
Preprocessing
Before images are fed into a neural network, they are frequently processed, often by accentuating differences between foreground and background voxels, to enhance performance and/or reduce training time. DL theory suggests that in high-dimensional matrices, local minima are very unlikely; instead, saddle points are more common due to the improbable likelihood that every dimension produces a minimum at the same location. These techniques can decrease the likelihood that the algorithm reaches a shallow saddle point, thereby causing slower optimisation. This is achieved through regularisation techniques and limiting outlier intensities. Cropping is regularly used to restrict the processing to voxels within the patient, 11 or coarse, manually drawn bounding boxes. 12 Table 1 summarises commonly used preprocessing techniques in the DL lung image analysis literature. In CNNs, other techniques such as batch normalisation, have been shown to reduce training time, acting as secondary regularisation techniques to minimise outliers and improve performance. 62,63
Table 1.
Summary of common pre-processing techniques used for lung image analysis tasks, including values prevalent in the literature
| Preprocessing technique | Description | Modality | Literature values | References |
|---|---|---|---|---|
| Thresholding | The process of constraining the pixel values of an image to be between predefined values. | CT, MRI | CT intensity: [-1000, 700 HU] MRI intensity: [0,667] |
Wang et al. (2018), 13 Sousa et al. (2019), 14 Javaid et al. (2018), 15 Hofmanninger et al. (2020), 16 Jiang et al. (2019), 17 Tahmasebi et al. (2018), 18 Z. Zhong et al. (2019), 19 Zhou et al. (2019), 20 Park et al. (2019), 21 Gerard et al. (2019), 22 Yun et al. (2019), 23 Eppenhof & Pluim (2019), 24 Fu et al. (2020), 25 Jiang et al. (2020), 26 De Vos et al.(2019), 27 Stergios et al. (2018), 28 Ren et al. (2019) 29 |
| Normalisation and whitening | The process of transforming the distribution of image pixels to some distribution which is standardised across images. | CT, MRI, X-ray | Normalisation: [0,1] Mean/variance ≈ 0 |
Wang et al. (2018), 13 Liu et al. (2019), 30 Javaid et al. (2018), 15 Hofmanninger et al. (2020), 16 Akila Agnes et al. (2018), 31 Novikov et al. (2018), 32 Gaal et al. (2020), 33 Jiang et al. (2019), 17 Tahmasebi et al. (2018), 18 Zhou et al. (2019), 20 Hatamizadeh et al. (2019), 34 Sandkühler et al. (2019), 35 Rajchl et al. (2017), 36 Sentker et al. (2018), 37 Fletcher and Baltas (2020), 38 Jiang et al. (2020), 26 De Vos et al.(2019), 27 Galib et al. (2019), 39 Ferrante et al. (2018), 40 Stergios et al. (2018), 28 Beaudry et al. (2019), 41 Duan et al. (2019), 42 Liu et al. (2020), 43 Ren et al. (2019), 29 Olberg et al. (2018) 44 |
| Denoising | The process of removing noise from images in order to improve their quality. | CT, MRI | Gaussian, adaptive patch-based | J.Xu & Liu (2017), 45 Zha et al. (2019), 46 Tustison et al. (2019) 47 |
| Bias correction | A technique to correct for the low-frequency bias field that corrupts MR images. | HP gas MRI, MRI | N3/N4 bias correction | Tustison et al. (2019), 47 Zha et al. (2019), 46 Rajchl et al. (2017) 36 |
| Cropping | Cropping refers to the process of removing unwanted outer pixels or voxels of an image prior to being inputted to the network. This includes cropping by manually-defined regions of interest or external body masks. Cropping is commonly used to reduce computational cost and/or eliminate the influence of background voxels. | CT, MRI, X-ray, PET | Cropping to body mask, specific organ or manually-defined region. | Negahdar et al. (2018), 12 Soans & Shackleford (2018), 48 Zhu et al. (2019), 49 Hofmanninger et al. (2020), 16 Zha et al. (2019), 46 Hooda et al. (2018), 50 Mittal et al. (2018), 51 Jiang et al. (2018), 11 Zhao et al. (2019), 52 Zhou et al. (2019), 20 Moriya et al. (2018), 53 Kalinovsky et al. (2017), 54 Sandkühler et al. (2019), 35 Anthimopoulos et al. (2019), 55 Gao et al. (2016), 56 Rajchl et al. (2017), 36 C. Wang et al. (2019), 57 Juarez et al. (2019), 58 Juarez et al. (2018), 59 Eppenhof & Pluim (2019), 24 Sentker et al. (2018), 37 Fletcher and Baltas (2020), 38 Blendowski & Heinrich (2019), 60 Zhong et al. (2019), 61 Liu et al. (2020), 43 Olberg et al. (2018) 44 |
HU, Hounsfield unit; PET, Positron emission tomography.
Modalities included are those for which the pre-processing techniques have been used in the reviewed studies. This is not an exhaustive list of pre-processing techniques used.
Validation
Validation is used to evaluate the performance of trained DL networks and assess their generalisability to non-experimental settings. The goal is to develop a validation strategy that best represents the situation in which the algorithm is to be deployed.
Evaluation metrics
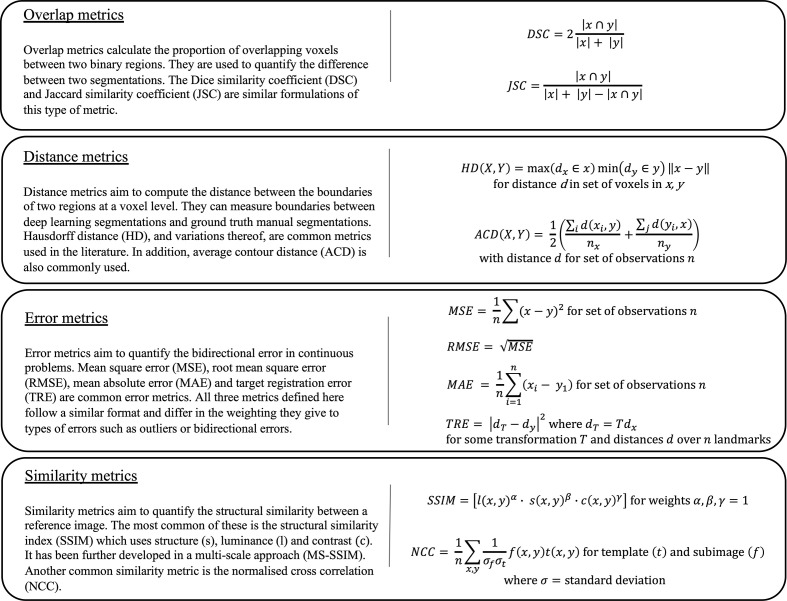
It is imperative to evaluate the performance of DL algorithms accurately. Evaluation metrics can be categorised into overlap, distance, error and similarity metrics and are summarised in Figure 4.
Figure 4.
Overview of four key categories of evaluation metrics (overlap, distance, error and similarity) used to evaluate the performance of deep learning methods in medical image analysis. Each category contains brief descriptions and mathematical formulations for some common metrics. In these equations, ‘x’ and ‘y’ denote the prediction and target of any deep learning task, respectively.
Validation techniques
Aside from the training set, an internal validation set is commonly used for tuning DL parameters to improve performance. A testing set is then used to provide an unbiased evaluation of performance on unseen data. In this review, validation sets used throughout the training phase are counted as training sets as the network has previously seen these images before testing. Therefore, the data split is the percentage of the total data used for training and internal validation vs that used for testing. Maintaining completely separate testing sets is somewhat uncommon in the literature and represents the ideal form of validation. 22,23,64 Validating on external multicentre data sets that have not been used for training should be the gold-standard in ensuring comparison between methods and generalisability. 65 However, this is uncommon as single-centre data sets, split into training and testing sets, are frequently used. To make the validation process more robust and generalisable, specific techniques are applied, such as k-fold cross-validation. In fourfold cross-validation, the datas et is randomly partitioned into a 75/25% training/testing split; this process is repeated with four different 25% blocks. Another approach is leave-one-out cross-validation which uses all of the data for training except one case for testing and repeats until all cases have been evaluated.
Methods
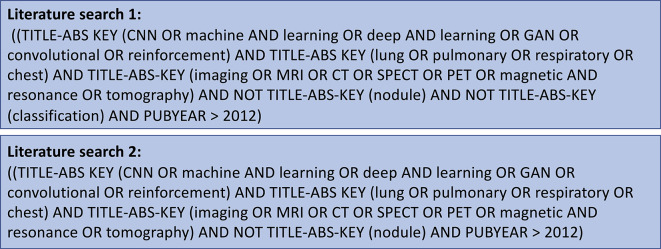
The protocol for this literature review was performed using the preferred reporting items for systematic reviews and meta-analyses (PRISMA)-statement. 66 The literature search was conducted on 1 April 2020 using multiple databases (Web of Science, Scopus, PubMed) and aimed to identify studies written in English published between 1 January 2012, the same year that the seminal AlexNet paper was published, 5 and the date of the search. The search strategy is defined in Figure 5. Further studies that met the selection criteria were identified by handsearching references and through the authors’ input.
Figure 5.
The search strategy used on Scopus, Web of Science and PubMed to identify relevant studies for inclusion in the review. Further studies that met the selection criteria were identified by handsearching references and through the authors’ input.
Several recent reviews have focussed primarily on DL-based lung classification and detection 67–69 ; accordingly, this review was limited in scope to the lung image analysis applications of segmentation, registration, reconstruction and synthesis. Both published peer-reviewed scientific papers and conference proceedings were included due to recent developments in the field.
Results and discussion
Study selection
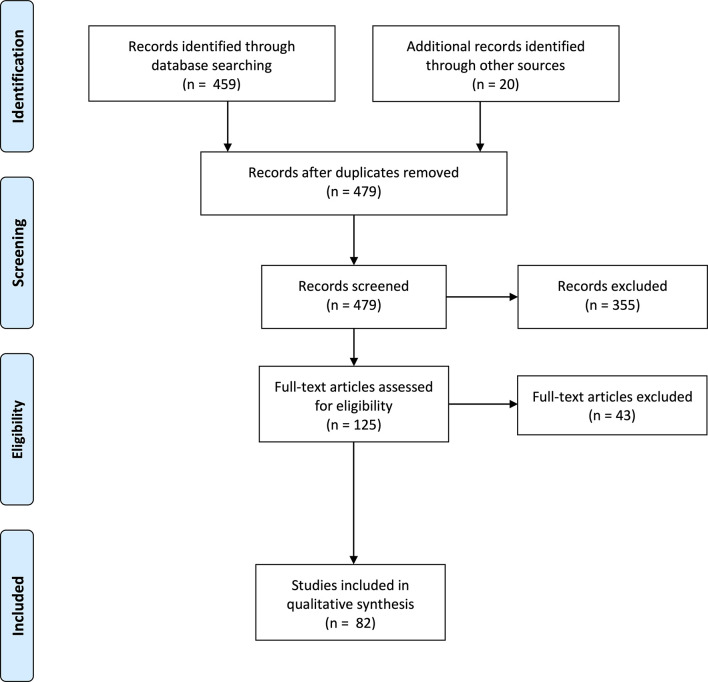
479 non-overlapping papers were retrieved. 355 papers were excluded due to not meeting the eligibility criteria. In particular, many papers focused on classification or used traditional machine learning techniques beyond this review’s scope. Upon reviewing the remaining papers, 82 studies were included for analysis. The PRISMA flowchart is shown in Figure 6.
Figure 6.
PRISMA flowchart of studies identified, screened, assessed for eligibility and included in the literature review analysis. PRISMA, preferred reporting items for systematic reviews and meta-analyses.
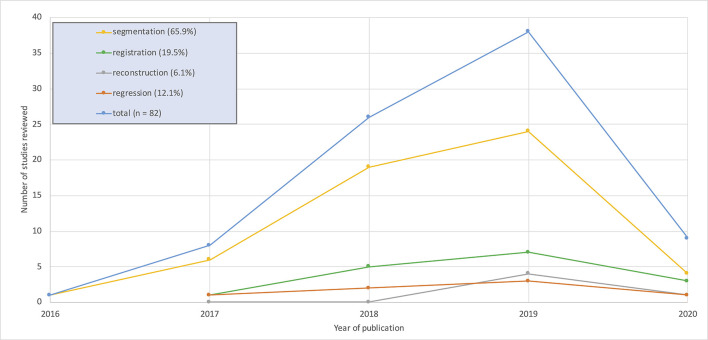
No studies that met the inclusion criteria were published before 2016 with the majority appearing since 2018. Image segmentation applications accounted for 65.9% of the studies reviewed. The remaining 34% are divided between synthesis, reconstruction and registration applications. Full details are shown in Figure 7.
Figure 7.
Graphical overview of the number of studies per year for the four image analysis applications considered in this review. 2020 values calculated up to 1 April 2020.
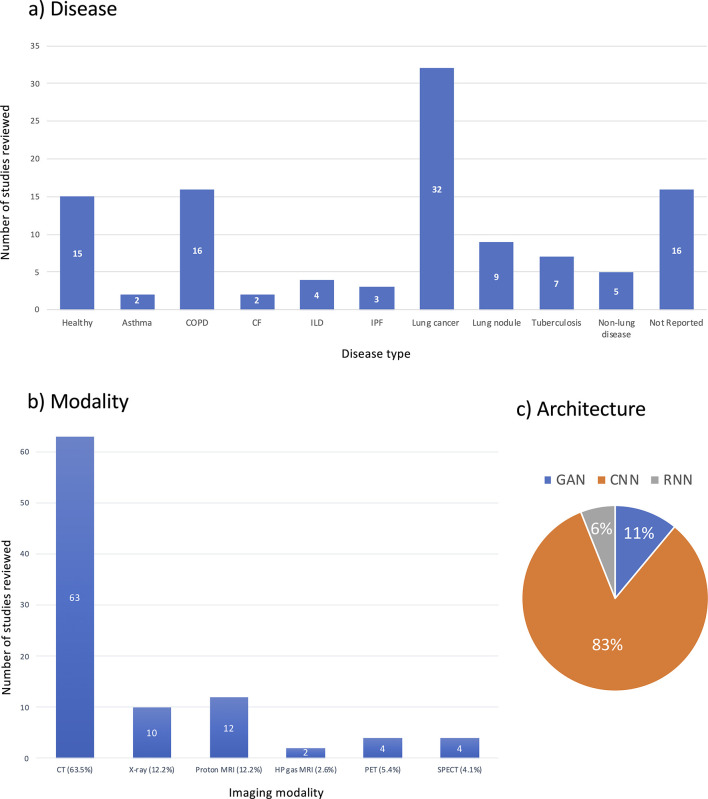
The majority of studies reviewed used structural imaging modalities (87.8%), with most using CT (63.5%). Functional lung imaging studies only constitute 12.1% of the reviewed studies and are spread across PET, SPECT and hyperpolarised gas MRI. Graphical summaries of the studies reviewed with respect to disease present in patient cohorts, imaging modality and architecture are shown in Figure 8.
Figure 8.
Graphical overview of breakdown of deep learning lung image analysis studies reviewed by (a) disease present in patient cohorts, (b) imaging modality and (c) architecture. Absolute numbers of papers are provided in (a, b).
Segmentation
Image segmentation is the process of partitioning an image into one or more segments that encompass anatomical or pathological specific regions of interest (ROIs), such as the lungs, lobes, or a tumour. Studies describing DL-based segmentation applications of pulmonary ROIs are summarised in Table 2.
Table 2.
Summary of reviewed studies on deep learning for lung image segmentation. The entries are arranged alphabetically by pulmonary region of interest (ROI), followed by modality
| Study | Modality | ROI | Disease | Number of subjects | Dimentionality | Architecture | Pre-processing |
Percentage data split
(training*/testing) |
Performance |
|---|---|---|---|---|---|---|---|---|---|
| Wang et al. (2018) 13 | CT | Whole lung | COPD, IPF | 575 | 2D | ResNet-101 | Clipped −1000 to +1000 HU, Normalisation [0,1] | 5-fold CV | DSC = 0.988 ± 0.012 ASD = 0.562±0.52 mm |
| Dong et al. (2019) 70 | CT | Whole lung | Lung cancer | 35 | 3D | U-Net-GAN | LOOCV | DSC = 0.97±0.01 HD95 = 2.29±2.64 mm MSD = 0.63±0.63 mm |
|
| Liu et al. (2019) 30 | CT | Whole lung | NR | 100 | 2D | SegNet | Class grouping, Normalisation [−1000,800] | 40/60 | DSC = 0.98 |
| Lustberg et al. (2018) 71 | CT | Whole lung | Lung cancer | 470 | NR | CNN | 95/5 | DSC = 0.99±0.01 Median HD = 0.4±0.2 cm |
|
| Negahdar et al. (2018) 12 | CT | Whole lung | Multiple | 83 | 3D | V-Net | Bounding box for lung, cropped to bounding box | 58/42 | DSC(n = 12)=0.983±0.002 DSC(n = 23)=0.990±0.002 |
| Soans & Shackleford (2018) 48 | CT | Whole lung | Lung cancer | 422 | 3D | CNN with spatial constraints | ROI extraction for organ localisation | 71/29 | ROC(Left)=0.954 ROC(right)=0.949 |
| Soliman et al. (2018) 72 | CT | Whole lung | NR | 95 | 3D | Deep-CNN | Post-processed hole filling | LOOCV | DSC = 0.984±0.068 HD95 = 2.79±1.32 mm PVD = 3.94±2.11% |
| Sousa et al. (2019) 14 | CT | Whole lung | Lung lesion | 908 | 3D | Modified V-Net | Clipped [−1000, 400 HU] | 98/2 | ASD = 0.576 mm DSC = 0.987 |
| X. Zhou et al. (2017) 73 | CT | Whole lung | NR | 106 | 2D/3D | FCN VGG16 | Transfer learning from ImageNet ILSVRC‐2014 | 95/5 | JSC = 0.903±0.037 |
| Zhu et al. (2019) 49 | CT | Whole lung | Lung Cancer | 66 | 3D | U-Net | Cropping to ROI | 55/45 | DSC = 0.95±0.01 MSD = 1.93±0.51 mm HD95 = 7.96±2.57 mm |
| Gerard et al. (2018) 74 | CT | Whole lung | COPD, IPF | 1749 | 3D | Course-Fine ConvNet | Transfer learning from COPDGene and SPIROMICS, fine-tuned on animal model | 92/8 | JSC = 0.99 ASD = 0.29 mm |
| Javaid et al. (2018) 15 | CT | Whole lung | Lung cancer | 13 | 2D | Dilated U-Net | Only axial slices selected, clipped −1000 to 3000 HU, Normalisation [0,1] | 94/6 | DSC = 0.99 ± 0.01 HD ≈ 4.5 mm |
| J. Xu & Liu (2017) 45 | CT | Whole lung | NR | 20 | 2D | MFCNN | gaussian denoising | 50/50 | DSC = 0.754 |
| Hu et al. (2020) 75 | CT | Whole lung | NR | 75 | 2D | Mask R-CNN +k-means | NR | DSC = 0.973 ±0.032 | |
| Hofmanninger et al. (2020) 16 | CT | Whole lung | Multiple | 266 | 2D | U-Net | Body mask, Clipped [−1024, 600 HU], Normalisation [0,1] | 87/13 | DSC = 0.98 ±0.03 HD95 = 3.14 ±7.4 mm MSD = 0.62 ±0.93 |
| Xu et al. (2019) 76 | CT | Whole lung | Lung cancer, COPD | 224 | 2D | one layer CNN | Post-processed hole filling | 8-fold CV | DSC = 0.967 ±0.001 HD = 1.44±0.04 mm |
| Tustison et al. (2019) 47 | HP gas MRI Proton MRI |
Functional lung Whole lung |
NR NR |
113 268 |
2D 3D |
U-Net U-Net |
Template-based data augmentation, N4 bias correction, denoising | 65/35 77/23 |
DSC (HP gas)=0.92 DSC (Proton) = 0.94 |
| Akila Agnes et al. (2018) 31 | LDCT | Whole lung | NR | 220 | 2D | CDWN | Normalised [mean = 0] | 91/9 | DSC = 0.95 ± 0.03 JSC = 0.91 ± 0.04 |
| Zha et al. (2019) 46 | UTE proton MRI | Whole lung | Healthy, CF, asthma | 45 | 2D | CED (U-Net and autoencoder) | Denoising, bias field correction, body mask | 5-fold CV | DSC (right) = 0.97±0.015 DSC (left) = 0.96±0.012 |
| Hwang & Park (2017) 77 | X-ray | Whole lung | Healthy, lung nodules | 247 | 2D | U-Net | 2-fold CV | DSC = 0.980±0.008 JSC = 0.961±0.015 ASD (mm) = 0.675±0.122 ACD (mm) = 1.237±0.702 |
|
| Souza et al. (2019) 78 | X-ray | Whole lung | Healthy, Tuberculosis | 138 | 2D | ResNet-18 with FC layer | Scaled to same input size, post processing erosion, dilation, filtering | 73/27 | DSC = 0.936 JSC = 0.881 |
| Dai et al. (2018) 64 | X-ray | Whole lung | Healthy, Tuberculosis, lung nodules | 385 | 2D | SCAN (structure correcting adversieral network) | Scaled to same input size | 85/15 | IoU = 94.7±0.4% DSC = 0. 973 ± 0.02 |
| C. Wang (2017) 79 | X-ray | Whole lung | Healthy, lung nodules | 247 | 2D | Multi task U-Net | Scaled to same input size, post processing hole filling | NR | JSC = 0.959 ± 0.017 AD = 1.29 ± 0.80 mm |
| Novikov et al. (2018) 32 | X-ray | Whole lung | Healthy, lung nodules | 247 | 2D | InvertedNet + All-dropout | Normalised [mean = 0, SD = 0] | 3-fold CV | DSC = 0.974 JSC = 0.949 |
| Hooda et al. (2018) 50 | X-ray | Whole lung | Healthy, Tuberculosis, lung nodules | 385 | 2D | FCN-8+dropout | Scaled to same input size, random cropping | 75/25 | DSC = 0.959 |
| Mittal et al. (2018) 51 | X-ray | Whole lung | Healthy, Tuberculosis, lung nodules | 385 | 2D | LF-SegNet | Scaled to same input size, random cropping | 48/52 | DSC = 0.951 |
| Gaal et al. (2020) 33 | X-ray | Whole lung | Healthy, Tuberculosis, lung nodules | 1047 | 2D | Adversarial attention U-Net | Scaled to same input size, CLAHE, Normalisation [−1,1] | 24/76 | DSC = 0.962±0.04 |
| Chen et al. (2019) 80 | CT | Lung tumour | Lung cancer | 134 | 3D | HSN (2D + 3D CNN) | 78/22 | DSC = 0.888±0.033 | |
| Jiang et al. (2018) 11 | CT, MRI | Lung tumour | Lung cancer | 400 CT (377) MRI (23) |
2D | Tumour aware semi-supervised Cycle-GAN | Scaled to same input size, Image synthesis from CT to MRI, body mask | 98/2 | DSC = 0.63 ± 0.24 HD95 = 11.65±6.53 |
| Jiang et al. (2019) 17 | CT, MRI | Lung tumour | Lung cancer | 405 CT (377) MRI (28) |
2D | Tumour aware pseudo MR and T2w MR U-Net | Scaled to same input size, Image synthesis from CT to MR, Clipped [−1000,500 HU] and [0,667], Normalised [−1, 1] | 95/5 | DSC = 0.75±0.12 HD95 = 9.36±6.00 mm VR = 0.19±0.15 |
| Tahmasebi et al. (2018) 18 | MRI | Lung tumour | Lung cancer | 6 | 2D | Adapted FCN | Rescaled 10–95% of intensities, Normalisation [0,1] | 5-fold CV | DSC = 0.91 ± 0.03 HD = 2.88 ± 0.86 mm RMSE = 1.20 ± 0.34 |
| Z. Zhong et al. (2019) 19 | FDG PET, CT | Lung tumour | Lung cancer | 60 PET (60) CT (60) |
3D | DFCN Co-Seg U-Net | Scaled to same input size, Clipped [−500,200 HU] and [0.01,20] | 80/20 | DSC (CT) = 0.861±0.037 DSC (PET) = 0.828±0.087 |
| Zhao et al. (2019) 52 | PET, CT | Lung tumour | Lung cancer | 84 PET (84) CT (84) |
3D | V-Net +feature fusion | Cropped to ROI | 57/43 | DSC = 0.85±0.08 VE = 0.15±0.14 |
| Zhou et al. (2019) 20 | CT | Lung tumour | NR | 1350 | 3D | P-SiBA | Transfer learning from ImageNet ILSVRC‐2014, Cropped to ROI, Rescaled by +1000 HU and dividing by 3000 and Normalisation [0,1] | NR | DSC = 0.809 ± 0.12 HD = 7.612 ± 5.03 mm vs = 0.883 ± 0.13 |
| Moriya et al. (2018) 53 | Micro CT | Lung tumour | Lung cancer | 3 | 3D | JULE CNN + k-means | Body mask, patch extraction | NMI = 0.390 | |
| Imran et al. (2019) 81 | CT | Lobes | COPD, ILD | 563 | 3D | Progressive dense V-Net | 48/52 | DSC (n = 84)=0.939±0.02 DSC (n = 154)=0.950±0.007 DSC (n = 55)=0.934 |
|
| Park et al. (2019) 21 | CT | Lobes | COPD | 196 | 3D | U-Net | Clipped [-1024,–400 HU] | 80/20 | DSC = 0.956 ± 0.022 JSC = 0.917 ± 0.031 MSD = 1.315 ± 0.563 HSD = 27.89±7.50 |
| Wang et al. (2018) 13 | CT | Lobes | COPD, IPF | 1280 | 3D | DenseNet | Clipped −1000 to +1000 HU, Normalisation [0,1] | 5-fold CV | DSC = 0.959±0.087 ASD = 0.873±0.61 mm |
| Hatamizadeh et al. (2019) 34 | CT | Lung lesion | NR | 87 | 3D | DALS CNN | Scaled to same input size, Normalisation [NR] | 90/10 | DSC = 0.869 ± 0.113 HD = 2.095 ± 0.623 mm |
| Kalinovsky et al. (2017) 54 | CT | Lung lesion | Tuberculosis | 338 | 2D | GoogLeNet CNN | Images cropped into four quadrants | 80/20 | IoU = 0.95 ROC = 0.775 |
| Gerard et al. (2019) 22 | CT | Lung fissure | COPD, Lung cancer | 5327 | 3D | Two Seg3DNets | Clipped [-1024,–200 HU], Linear rescaling | 30/70 | ASD = 1.25 SDSD = 2.87 |
| Sandkühler et al. (2019) 35 | MRI | Lung defect region | NR | 35 | 2D | GAE-LAE RNN with LCI Loss | Z-normalisation [−4,4], Lung mask, Normalisation [0,1], Histogram stretching | 80/20 | Qualitative evaluation - 42% images rated ‘very good’, 19% rated ‘perfect’ |
| Vakalopoulou et al. (2018) 82 | CT | ILD pattern | ILD | 46 | 2D | AtlasNet | 37/63 | DSC = 0.677 HD = 3.981 mm ASD = 1.274 mm |
|
| Anthimopoulos et al. (2019) 55 | CT | ILD pattern | ILD | 172 | 2D | FCN-CNN | Pre-computed lung mask | 5-fold CV | Accuracy = 81.8% |
| B. Park et al. (2019) 83 | CT | ILD pattern | COP, UIP, NSIP | 647 | 2D | U-Net | 88/12 | DSC = 0. 988 ± 0.006 JSC = 0.978 ± 0.011 MSD = 0.27 ± 0.18 mm HSD = 25.47 ± 13.63 mm |
|
| Gao et al. (2016) 56 | CT | ILD pattern | ILD | 17 | 2D | CNN based CRF unary classifier | Transfer learning from ImageNet, Pre-computed lung mask | Accuracy = 92.8% | |
| Suzuki et al. (2020) 84 | CT | Diffuse lung disease | NR | 372 | 3D | U-Net | 5-fold CV | DSC = 0.780±0.169 | |
| Wang et al. (2018) 85 | MRI | Foetal lung | NR | 18 | 2D | BIFSeg P-Net | Trained on different organs, Image specific fine-tuning | 66/33 | DSC = 0.854±0.059 |
| Rajchl et al. (2017) 36 | MRI | Foetal lung | Healthy, IUGR | 55 | 3D | DeepCut CNN + CRF | Bounding box for ROI, Bias correction, Normalisation [mean = 0], Transfer learning from LeNet | 5-fold CV | DSC = 0.749±0.067 |
| Edmunds et al. (2019) 86 | Cone-beam CT | Diaphragm | Lung cancer | 10 | 2D | Mask R-CNN | Scaled to same input size | 9-fold CV | Mean error = 4.4 mm |
| C. Wang et al. (2019) 57 | CT | Airways | NR | 38 | 3D | Spatial-CNN (U-Net) | Random cropping | 92/8 3-fold MCCV | DSC = 0. 887 ± 0.012 CO = 0.766 ± 0.06 |
| Juarez et al. (2019) 58 | CT | Airways | Lung cancer | 32 | 3D | U-Net GNN | Bounding box for ROI | 63/37 | DSC = 0.885 Airway completeness = 74% |
| Yun et al. (2019) 23 | CT | Airways | COPD | 89 | 2D | 2.5D CNN | Clipped [−700,700 HU] | 78/22 | Mean Branch detected = 65.7% |
| Juarez et al. (2018) 59 | CT | Airways | Healthy, CF, CVID | 24 | 3D | U-Net | Bounding box for ROI | 75/25 | DSC = 0.8 |
ACD, Average contour distance; AD, Average distance; ASD, Average surface distance; CDWN, Convolutional deep wide network; CE, Classification error; CF, Cystic fibrosis; CLAHE, Contrast limited adaptive histogram equalisation; CNN, Convolutional neural network; CO, Centreline overlap; COPD, Chronic obstructive pulmonary disorder; CV, Cross-validation; CVID, Common variable immunodeficiency disorders; DSC, Dice similarity coefficient; FDG, Fluorine-18‐fluorodeoxyglucose; GAN, Generative adversarial network; HD95, Hausdorff distance 95%; HD, Hausdorff distance; HSD, Hausdorff surface distance; HU, Hounsfield unit; ILD, Interstitial lung disease; IPF, Idiopathic pulmonary fibrosis; IUGR, Intrauterine growth restriction; IoU, Intersection over union; JSC, Jaccard similarity coefficient; LOOCV, Leave-one-out cross-validation; MAP, Mean average precision; MCCV, Monte carlo cross-validation; MSD, Mean surface distance; NMI, Normalised mutual information; NR, Not reported; NSIP, Nonspecific interstitial pneumonia; PVD, Percent ventilated defect; RMSE, Root mean square error; ROC, Receiver operating characteristic; ROI, Region of interest; SD, Standard deviation; SDSD, Standard deviation of surface distances; UIP, Usual interstitial pneumonia; VE, Volume error; VR, Relative volume ratio; VS, Volumetric similarity.
The entries are arranged alphabetically by pulmonary ROI, followed by modality.
The training data set includes internal validation data.
CT segmentation
CT is the most common modality for clinical lung imaging due to superior spatial resolution, rapid scan times and widespread availability. This is reflected in the DL lung segmentation literature with the majority of studies to date focusing on CT. For whole-lung segmentation, 3D networks are often used, whereas in interstitial lung disease (ILD) pattern segmentation, only 2D networks have been applied to date. The application often dictates the use of 2D and 3D networks; segmentation of the whole lung leads to a volumetric 3D region in which features such as overall lung shape, or the position of the trachea can be encoded. In contrast, segmenting ILD patterns is often conducted on central 2D slices; hence, a 2D network may be more appropriate as, in this approach, no features are conserved between slices. 55,83
Across the CT papers reviewed, both the median and mode training/testing data splits were 80/20%, with many using k-fold cross-validation with less than 50 patients. Even as an independent testing set, using only 5–10 patients for testing limits generalisability. Moreover, some studies cite the number of images or 2D slices rather than the number of subjects. If data from the same subject are included in both the testing and training phases, it is likely that the algorithm has already seen a similar slice from the same patient as the individual data points are spatially correlated and do not strictly represent independent data points.
The Dice similarity coefficient (DSC) overlap metric is the most common evaluation metric used. Most studies tackling whole-lung segmentation report DSC values above 0.90, with some achieving values above 0.98. For other pulmonary ROIs, the highest DSC values reported are often lower (e.g. DSC (airways) ≈ 0.85). However, overlap metrics such as the DSC can be insensitive to errors in large volumes as the percent error is low compared to the overall pixel count. 87 Frequently, high DSC values are reported despite errors that require significant manual intervention before a segmentation is clinically useful. As the airways occupy smaller volumes, the DSC metric is more sensitive. In terms of Hausdorff-based distance metrics, whole-lung segmentation studies report HD95 values ≈10 mm; however, Dong et al 70 report a HD95 as low as 2.249 ± 1.082 mm averaged across both lungs. The lack of a standardised evaluation metric can make direct comparisons between different methods challenging.
Image segmentation is challenging to evaluate. Currently, manual segmentations by expert observers are used as the gold-standard; however, it is well-known that expert segmentations are susceptible to interobserver variability. 88 Often, only one observer segments all the images in a training data set; hence, if a different observer segments the testing images, the algorithm may not perform as expected. This poses problems for widespread generalisation if certain biases in segmentation are preserved as there is no clear ‘true’ expert segmentation; therefore, differences in DL segmentations and expert segmentations may not be solely the result of DL errors. Most expert segmentations are conducted using semi-automatic software and image editing tools; the tools given to the user can convey a propensity for features, such as smooth lung borders, which may, in fact, be inaccurate. In other anatomical sites such as the liver, a DSC of 0.95 was obtained by DL; the interobserver variability for the DL approach was 0.69% compared to 2.75% for manual expert observers. 89 The low degree of interobserver variability in DL segmentations may be a positive step towards consistent segmentations between institutions. Using multiple expert segmentations and averaging the error may reduce interobserver variability effects; however, this is unlikely to be widely adopted due to the time required. In addition, medical imaging grand challenges can provide diverse data from multiple institutions with corresponding expert segmentations, limiting the extent of individual researcher bias.
MRI segmentation
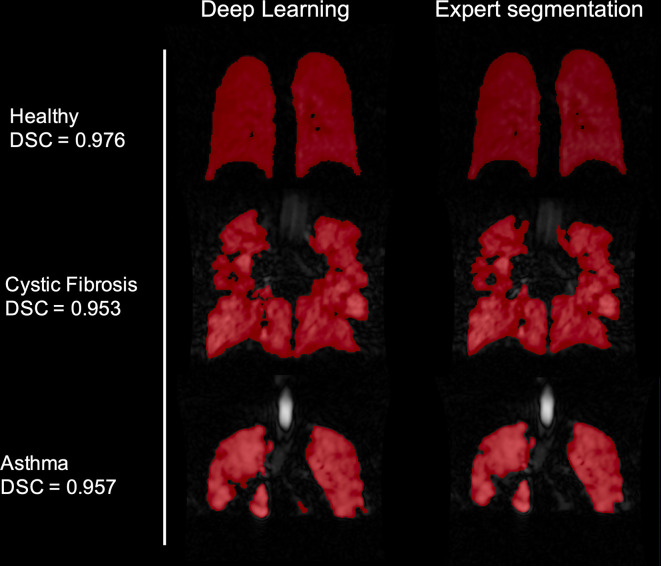
There are limited studies to date regarding pulmonary MRI segmentation, attributable perhaps to less widespread clinical use of the modality and lack of large-scale annotated pulmonary MRI data sets. However, pulmonary MRI techniques, such as contrast-enhanced lung perfusion MRI and hyperpolarised gas ventilation MRI, can provide further insights into pulmonary pathologies currently not possible with alternative techniques. 90 Quantitative biomarkers derived from hyperpolarised gas MRI, including the ventilated defect percentage, require accurate segmentation of ventilated and whole-lung volumes which can be very time consuming when performed manually. Example images of DL-based hyperpolarised gas MRI segmentations are provided in Figure 9.
Figure 9.
Example images from the authors’ own work using deep learning for hyperpolarised gas MRI segmentation. The 129Xe MR ventilation images are taken from three subjects in a testing set, a healthy volunteer, asthma patient and cystic fibrosis patient. The patient images selected are characterised by significant ventilation defects. These are compared to expert segmentations of the same image. DSC values are displayed for all images. DSC, Dice similarity coefficient.
Tustison et al 47 used CNNs to provide fast, accurate segmentations for hyperpolarised gas and proton MRI. 47 A 2D U-Net was used for hyperpolarised gas MRI segmentation whilst a 3D U-Net was used for proton MRI segmentation. They introduced a novel template-based data augmentation method to expand the limited lung imaging data. Hyperpolarised gas and proton MR images were segmented with DSC values of 0.94 ± 0.03 and 0.94 ± 0.02, respectively. Zha et al evaluated DL-based proton MRI segmentation, which yielded an average DSC of 0.965 across both lungs, outperforming conventional region growing and k-means techniques. 46
X-ray segmentation
Although the majority of segmentation studies reviewed used CT and MRI, early studies focused on X-ray segmentation. 77,79 This was due to the public availability of large-scale, annotated X-ray datasets, such as the Japanese Society of Radiological Technology (JSRT) 91 and Montgomery 92 data sets, enabling researchers to experiment with large numbers of images not previously accessible. The majority of X-ray studies reviewed used these datasets, making comparisons between methods more applicable. 32,50,51,64,78,79
Registration
Image registration is the process of transforming a moving image onto the spatial domain of a fixed image. Registration is used in numerous applications within the lung imaging field, including adaptive radiotherapy, 93 computation of functional lung metrics such as the VDP 94 and generation of surrogates of regional lung function from multi-inflation CT 95 or 1H MRI. 96
However, most image registration algorithms assume that the moving and fixed images’ topology are the same. This is not always the case in lung imaging as often functional images do not follow the same topology as structural images, especially in individuals with severe pathologies where functional lung images may show substantial heterogeneity. 97 Studies describing DL-based pulmonary registration applications are summarised in Table 3.
Table 3.
Summary of reviewed studies using deep learning for lung image registration
| Study | Modality | Disease | Public data set | Number of subjects | Dimensionality | Architecture | Preprocessing |
Percentage data split
(training*/testing) |
Performance |
|---|---|---|---|---|---|---|---|---|---|
| Eppenhof et al. (2018) 98 | 4DCT | Lung cancer | DIR-LAB, CREATIS | 17 | 3D | Modified VGG | Synthetic DVFs for data augmentation | 42 (CREATIS) / 58 (DIR-LAB) | TRE = 4.02±3.08 |
| Eppenhof & Pluim (2019) 24 | 4DCT | Lung cancer | DIR-LAB, CREATIS | 17 | 3D | Modified U-Net | Synthetic DVFs for data augmentation, Resized, Pre-computed body mask, intensity-based lung mask < −250 HU | 42 (CREATIS) / 58 (DIR-LAB) | TRE = 2.17±1.89 mm |
| Ali & Rittscher (2019) 99 | 4DCT | Lung cancer | DIR-LAB, CREATIS | 17 | 2D | Conv2Wrap (Linear and Deformable ConvNet) | 58 (DIR-LAB) / 42 (CREATIS) | DSC = 0.90 JSC = 0.84 |
|
| Sentker et al. (2018) 37 | 4DCT | Lung cancer | DIR-LAB, CREATIS | 86 | 3D | GDL-FIRE4D U-Net with VarReg | Normalisation [0,1], Cropped to same input size, Pre-computed body mask | 69/31 (DIR-LAB, CREATIS, In house) | TRE (DIR-LAB) = 2.50±1.16 mm TRE (CREATIS) = 1.74±0.57 mm |
| Fletcher and Baltas (2020) 38 | 4DCT | Lung cancer | DIR-LAB, CREATIS, Sunnybrook | 31 | 3D | U-Net one-shot learning | Pre-computed body mask, Normalisation [mean = 0, SD = 1] | LOOCV (DIR-LAB) 0/100 (CREATIS) |
TRE (DIR-LAB) = 1.83±2.35 mm TRE (CREATIS) = 1.49±1.59 mm |
| Fu et al. (2020) 25 | 4DCT | Lung cancer | DIR-LAB | 20 | 3D | LungRegNet (CourseNet, FineNet) | Vessel enhancement, Clipped at −700 HU | 5-fold CV, DIR-LAB testing | MAE (in house)=52.1±18.4 TRE (in house)=1.00±0.53 TRE (DIR-LAB) = 1.59±1.58 mm |
| Jiang et al. (2020) 26 | 4DCT | Lung cancer | DIR-LAB, SPARE | 32 | 3D | MJ-CNN | Clipped [-1000,–200 HU], Normalisation [0,0.2] | 75 (SPARE, DIR-LAB) / 25 (DIR-LAB) | TRE = 1.58±1.19 mm |
| De Vos et al.(2019) 27 | 4DCT, CT | Lung cancer | DIR-LAB, NLST | 2070 | 3D | DLIR framework ConvNet | Clipped [-1000,–200 HU], Normalisation [0,1] | 99 (NLST) / 1 (NLST, DIR-LAB) | DSC (NLST) = 0.75±0.08 HD (NLST) = 19.34±13.41 TRE (DIR-LAB) = 5.12±4.64 mm |
| Sokooti et al. (2017) 100 | CT | COPD | 19 | 3D | RegNet CNN | Synthetic DVFs for data augmentation, Initial affine registration | 63/37 (SPREAD) |
TRE = 4.39 ± 7.54 mm | |
| Sokooti et al. (2019) 101 | CT, 4DCT | Lung cancer, COPD | SPREAD, DIR-LAB | 39 | 3D | RegNet CNN (U-Net) | Synthetic DVFs for data augmentation, Initial affine registration | 54 (SPREAD, DIR-LAB COPD) / 46 (SPREAD, DIR-LAB) | TRE (DIR-LAB) = 1.86±2.12 mm |
| Blendowski & Heinrich (2019) 60 | CT | COPD | DIR-LAB | 10 | 3D | CNN | Cropped to lung region | LOOCV (DIR-LAB) |
TRE = 3.00 ± 0.48 mm |
| Qin et al. (2019) 102 | CT, MRI | COPD | COPDGene | 1000 | 2D | UMDIR-LaGAN | Cross-modality registration, transformation into domain invariant latent space | 90/10 (COPDGene) |
DSC = 0.967±0.03 HD = 8.257±4.43 mm MCD = 0.71±0.44 mm |
| Galib et al. (2019) 39 | CT, CBCT | Healthy, COPD, Lung cancer | DIR-LAB, VCU | 27 | 3D | CNN | Normalisation [0,1] | 37 (DIR-LAB) / 63(VCU) | AUC-ROC = 0.882±0.11 CI=68% |
| Ferrante et al. (2018) 40 | X-ray | Healthy, Lung nodule | JSRT | 247 | 2D | U-Net | Normalisation [0–1], Domain adaption Cardiac MR | 81/19 (JSRT) |
MAD ≈ 6.3 CMD ≈ 5 mm DSC ≈ 0.9 |
| Mahapatra et al. (2018) 103 | X-ray | Multiple | NIH-ChestXray14 | 420 | 2D | JRSNet (cycleGAN with U-Net) | Joint segmentation and registration | NR (SCR, NIH-ChestXray14) |
TRE = 7.75 mm |
| Stergios et al. (2018) 28 | MRI | Systemic sclerosis, healthy | 41 | 3D | CNN with transformation layer | Clipped [0, 1300], Normalisation [0,1] | 68/32 | DSC = 0. 915 ± 2.33 Euclydian error = 4.358 mm |
AUR-ROC, Area under curve-receiver operator characteristic; CMD, Contour mean distance; CNN, Convolutional neural network; COPD, Chronic obstructive pulmonary disorder; CV, Cross-validation; DLIR, Deep learning image registration; DSC, Dice similarity coefficient; HD, Hausdorff distance; HU, Hounsfield unit; JSC, Jaccard similarity coefficient; LOOCV, Leave-one-out cross-validation; MAD, Mean absolute differences; MAE, Mean absolute error; MCD, Mean contour distance; MRF, Markovian random field; TRE, Target registration error; VGG, Visual geometry group.
Eppenhof and Pluim 24 built upon previous work by Lafarge et al 98 using publicly available data sets to directly map displacement vector fields from inspiratory and expiratory CT pairs using a 3D U-Net with extensive data augmentation. Synthetic transforms were used to directly train the network as the deformation fields are known. The approach achieved fast, accurate registrations, reducing mean TRE from 8.46 to 2.17 mm. The results are further validated using landmarks from multiple observers, indicating the level of interobserver variability. Notwithstanding, only 24 images for testing and training were used, limiting the study’s generalisability. In addition, synthetic transforms do not directly represent real transforms likely found in patients.
Other approaches use a CNN to learn expressive local binary descriptors from landmarks before applying Markov random field registration. 60 This is compared to a method using handcrafted local descriptors with high self-similarity, facilitating faster computation. The results suggest that a combination of both CNN-learned descriptors and handcrafted features produce the best registration results.
In a generic registration approach, a U-Net-like architecture with a differentiable spatial transformer that can register both X-ray and MR images was used. 40 The algorithm was evaluated using the contour mean distance (CMD). CMD was approximately 5 mm on average across the testing data. Whilst this is a less accurate registration than other methods reviewed, it is more broadly applicable; the generic algorithm (in this case trained on X-ray and MR images) can learn features that are independent of modality. By fixing these weights and adding additional layers, transfer learning can then be applied to a specific modality; the additional data across modalities may lead to improved results. 104
Reconstruction
Image reconstruction is the process of generating a usable image from the raw data acquired by a scanner. CT and SPECT reconstruction fundamentally differ from MRI reconstruction and, as such, the role of DL in these applications is also different. CT and SPECT reconstruction use analytic (e.g. filtered backprojection) or iterative algorithms to produce 3D images from projections taken at multiple angles around a subject. MRI reconstruction, in contrast, produces images by transforming raw k-space data via Fourier transforms. Full details of image reconstruction methods have been described elsewhere. 105,106 Studies describing DL-based lung image reconstruction applications are summarised in Table 4.
Table 4.
Summary of reviewed studies using deep learning for lung image reconstruction
| Study | Modality | Disease | Number of patients | Dimensionality | Architecture | Preprocessing |
Percentage data split
(training*/testing) |
Performance |
|---|---|---|---|---|---|---|---|---|
| Beaudry et al. (2019) 41 | 4D cone beam CT | Lung cancer | 16 | 2D | Sino-Net (Modified U-Net) | Cropped to same input size, Sinogram Normalisation [0,1] | 88/12 | RMSE Translational = 1.67 mm (other metrics given) |
| Lee et al. (2019) 107 | CT | COPD | 60 | 2D | FCN | No sinogram used | Dataset 1: 80/20 Dataset 2: 40/60 |
Mean reduction RMSE (Dataset 1) = 65.7±15.8% Mean reduction RMSE (Dataset 2) = 59.6±5.5% |
| Ge et al. (2020) 108 | CT | Liver lesion | 5413 | 2D | ADAPTIVE-NET CNN | Convert from HU to linear attenuation coefficient | 90/10 | PSNR = 43.15±1.9 SSIM = 0.968±0.013 Normalized RMSE = 0.0071±0.002 |
| Duan et al. (2019) 42 | HP Gas MRI | COPD, nodule, PTB, healthy, asthma | 72 | 2D | C-Net and F-Net (U-Net based) | Under sampled K-space (AF = 4), Removed SNR below 6.6, Normalisation [0,1] | NR | MAE = 4.35% SSIM = 0.7558 VDP bias = 0.01±0.91% |
| Dietze et al. (2019) 109 | 99 mTc-MAA SPECT | Liver Cancer | 128 | 2D | CNN | Initial filtered back projection | 94/6 | LSF = 5.1% CNR = 12.5 |
CNN, Convolutional neural network; CNR, Contrast to noise ratio; COPD, Chronic obstructive pulmonary disorder; EIT, Electrical impedance tomography; HU, Hounsfield unit; LSF, Lung shunting fraction; MAE, Mean absolute error; PSNR, Peak signal to noise ratio; PTB, Pulmonary tuberculosis; RMSE, Root mean square error; SSIM, Structural similarity index metric; VDP, Ventilation defect percentage; VDP, Volume defect percentage; 99mTc-MAA, Technetium-99m macroaggregated albumin.
The training data set includes internal validation data
CT/SPECT images can be reconstructed accurately using Monte-Carlo-based iterative reconstruction 110 ; however, this process is computationally expensive and time-consuming. 111 In addition, multiple studies have demonstrated the success of analytical methods such as filtered backprojection. 105 Building upon this, CNNs have been used to speed up the process of filtered backprojection to shorten reconstruction times. 109 The results suggest DL can accurately reconstruct SPECT images in under 10 sec. Furthermore, the authors compare clinical metrics, such as the lung shunting fraction (LSF), between methods in a specific time frame. DL produced an LSF of 4.7% comparable to 5.8% for Monte-Carlo methods, indicating the potential for use in clinical applications. 109
Multiple studies have employed DL for MRI reconstruction 112 but only one published study has applied it to pulmonary MRI. 42 MRI of the lungs can take upwards of 10 sec to acquire, often requiring that patients maintain inflation levels for a significant period; this can be particularly challenging for patients with severe lung pathologies. Compressed sensing can be used to reconstruct randomly undersampled k-space in conjunction with regularisation methods to produce accurate reconstructions in hyperpolarised gas MRI 113,114 and enables reduced acquisition time without significantly reducing image quality. A coarse-to-fine neural network has been proposed to yield an accurate hyperpolarised gas MRI scan with an accelerating factor of 8 (undersampled 1/8 of k-space). 42 The method can also improve inherent spatial coregistration accuracy when acquiring proton and hyperpolarised gas MRI in the same breath, 115 possibly alleviating the need for substantial post-acquisition image registration.
Tangentially related to the goal of image reconstruction, images can also be improved further using image enhancement at the post-acquisition stage. Multiple studies have shown the effectiveness of using CNNs combined with gradient regularisation and superresolution modules to enhance low-dose CT images with noise and artefacts, potentially limiting radiation exposure without degrading image quality. 116,117
Synthesis
Image synthesis, also referred to as regression, is the process of generating artificial images of unknown target images from given source images. Synthesis has been applied to a range of applications, such as generating functional or metabolic images from structural images. For example, estimating contrast-based functional images from routinely acquired non-contrast structural modalities reduces the need for additional scans, specialised equipment and administration of contrast agents. Even within traditional model-based techniques, accurate synthesis has proved challenging due to the complex mathematical functions mapping input to output images. The development of DL architectures such as GANs enables a more unsupervised approach, which lends itself to the complex problem of synthesis. 9 Studies describing DL-based lung image synthesis applications are summarised in Table 5.
Table 5.
Summary of reviewed studies using deep learning for lung image synthesis
| Study |
Modality
(original ⇒ target) |
Disease | Number of subjects | Dimensionality | Model | Preprocessing |
Percentage data split
(training*/testing) |
Performance |
|---|---|---|---|---|---|---|---|---|
| Bi et al. (2017) 118 | CT ⇒ FDG PET | Lung cancer | 50 | 2D | Multichannel-GAN (U-Net) | Manual segmentation of tumour/lymph nodes, axial slices containing tumours only | 50/50 | MAE = 4.6 PSNR = 28.06 |
| Jang et al. (2019) 119 | CT ⇒ 99 mTc-MAA SPECT perfusion | Lung cancer | 54 | 2D | Conditional GAN | Resized images, segmentation and removal of bone, soft tissue and heart | 91/9 | MS-SSIM = 0.87 γ index 2%/2mm = 97.7±1.2% |
| Zhong et al. (2019) 61 | 4DCT ⇒ CT ventilation | Lung cancer, COPD | 82 | 2D | Deep CNN | Images cropped to ROI | 10-fold CV | MSE = 7.6% γ index 5%/5mm = 80.6±1.4% SSIM = 0.880±0.035 |
| Liu et al. (2020) 43 | 4DCT ⇒ 99 mTc-Technegas SPECT ventilation | Lung cancer, oesophageal cancer | 50 | 2D | U-Net | Pre-computed lung mask, normalisation [0,1], post-processing normalisation [90th percentile] | 10-fold CV | Spearman’s ρ = 0.73±0.17 DSC = 0.73±0.09 |
| Ren et al. (2019) 29 | CT ⇒ 99 mTc-MAA SPECT perfusion | Lung cancer | 30 | 3D | U-Net | Clipped [-1000,–300 HU] for segmentation, normalisation [0,1] | 83/17 | Correlation coefficient = 0.53 ± 0.14 |
| Preiswerk et al. (2018) 120 | Ultrasound ⇒ MRI | NR | 7 | 3D | LRCN | PCA = 10 components | 66/33 (conducted in time segments) | SSE = 39.0 ± 12 |
| Olberg et al. (2018) 44 | MRI ⇒ CT | NR | 41 | NR | GAN (U-Net) | Normalisation [NR], pre-computed body mask | 90/10 | 3D γ index passing rate 99.2% Lung V20% difference = 0.11% |
CNN, Convolutional neural network; COPD, Chronic obstructive pulmonary disease; FDG, Fluorine-18‐fluorodeoxyglucose; GAN, Generative adversarial network; HU, Hounsfield unit; LRCN, Long-term recurrent convolutional network; MAE, Mean absolute error; MSE, Mean square error; MS-SSIM, Multi-scale structural similarity index metric; NR, Not reported; PCA, Principle component analysis; PSNR, Peak signal to noise ratio; ROI, Region of interest; SSE, Sum of squared error; 99mTc-MAA, Technetium-99m macroaggregated albumin.
DL has been used to generate synthetic fluorine-18‐fludeoxyglucose (FDG) PET images from CT images via a GAN. 118 The GAN’s inputs were varied to include either a CT image, label, or both CT and corresponding label; the multichannelled GANs (M-GAN) provided the most accurate synthetic PET images, demonstrating that multiple inputs increase synthesis accuracy. To explore this further, the authors also evaluate the synthetic PET images by feeding them into a network as training data. The network aims to delineate tumours by learning relationships from the training data; the data were then divided into real PET images and synthetic PET images. The trained model was then evaluated on unseen tumour detection problems. The synthetic PET-trained network produced 2.79% lower recall accuracy. This indicates that, as a whole, the synthetic PET images are closely related to the real images in terms of tumour identification. The paper posits that synthetic PET images can be used as additional training data in other DL tasks. However, it is unclear if synthetic PET images can be used in treatment planning and other clinical tasks with this level of accuracy. 118
GANs have continued to show promise in synthesis problems. 119 CT images have been used to generate SPECT images via a conditional GAN (cGAN) instead of a CNN. 29 The method used a 2D GAN with 49 patients consisting of 3054 2D images as training data; the testing data contains 5 patients. cGANs differ from the regular GAN architecture by using both the observed image and a random noise vector, mapping these to the output image instead of only the noise vector. The generator used is based on the U-Net architecture with multiple inputs. Synthetic and real SPECT images were compared using the multiscale structural similarity index measure (MS-SSIM), yielding MS-SSIM = 0.87. Further analysis used a γ index with a passing rate of 97.7±1.2% with 2%/2 mm. The authors note qualitatively that errors occur more frequently at the base of the lungs, possibly caused by the increased deformation in this region. A key limitation for synthesis methods is the errors introduced by the registration of source and target images. Consequently, it has been suggested that images that are not matched anatomically due to breathing discrepancies are excluded, 119 complicating validation for clinical adoption. 29,119
A major application of DL image synthesis is for MR-guided radiotherapy. The current paradigm in radiotherapy is to derive electron density information required for dose calculations directly from CT scans; MRI does not directly provide this information. DL has been invoked to generate pseudo-CT images for use in MR-guided stereotactic body radiotherapy using GANs, precluding the need for CT. 44
Zhong et al used a CNN to synthesise ventilation images from 4DCT scans. 61 Whilst good performance was observed, the major limitation of this study is that the target images in the training phase were CT-based surrogates of ventilation generated from aligned inspiratory and expiratory CT scans via deformable registration and computational modelling. These images are still the subject of intense validation efforts. 121 Using more direct measures of regional lung function, such as hyperpolarised gas MRI, and larger data sets are critical to the success of future work in structure-to-function DL synthesis applications.
Future research directions
The studies reviewed show that DL has significant potential to outperform more traditional methods in a wide range of lung image analysis applications. Novel ways of using DL to synthesise more training examples 122 or combine segmentation and registration in one process 103 have been shown to enhance performance. The scope of such innovation is still in its infancy, providing an opportunity for novel technical developments.
As shown through the improved performance observed by combining traditional approaches with machine learning and DL for registration, great synergy can be achieved by combining DL and conventional image processing approaches. 60
In image synthesis, researchers have developed techniques to synthesise CT images from MRI scans of the brain 123 ; similar advancements in lung imaging would allow patients to receive less radiation exposure as well as reduce the cost and time for additional scans. Using synthesis to generate functional lung images from routinely acquired structural images would allow clinicians to understand which areas of the lungs are ventilated or perfused without the need to acquire dedicated functional scans, which often require contrast agents and specialised equipment, reducing costs and acquisition times. Such applications require further DL research in architectural development and the input of lung imaging experts. Using DL for CT enhancement to reduce radiation dose or improve compressed sensing methods in MRI has the potential to reduce scan times, improving image quality and patient compliance.
Promising results have been shown for both proton MRI and hyperpolarised gas MRI segmentation 47 ; however, further work is required to demonstrate accurate MRI segmentation in an independent multicentre validation. The importance of collaborative research to boost training data and inject heterogeneity of centre and scanner will lead to more robust and generalisable models. The paucity of published DL studies in functional lung imaging (only 12.9% of reviewed studies here) provides significant opportunities for innovations and further research in this field.
The literature on CT segmentation provides a positive picture of the success of DL methods in providing fast, accurate automatic segmentations. However, producing impressive results in a research setting is no substitute for clinical validation. Long-term clinical case studies are required with large numbers of patients before these novel developments have a real impact. The ‘black box’ nature of DL methods and the lack of explainability of generated outputs can undermine clinicians and patients’ trust, despite, or even because of, an unprecedented level of hype. Another challenge is transparency; although most software used for DL is well documented and open source, a requirement for continued use, the open-source nature also generates safety concerns relating to software edits and bugs. Developing a standardised literature consensus on validation and evaluation procedures is key to ensuring transparency. All of these challenges need to be overcome before DL can live up to its full potential.
Conclusions
We have reviewed the role of DL for several lung image analysis tasks, including segmentation, registration, reconstruction and synthesis. CT-based lung segmentation was the most prevalent application where exceptional performance has been demonstrated. However, research in other applications and modalities, including functional lung imaging, is still in its infancy. A concerted effort from the research community is required to develop the field further. Before widespread clinical adoption is achievable, challenges remain concerning validation strategies, transparency and trust.
Footnotes
Acknowledgements: This work was supported by Yorkshire Cancer Research, Weston Park Cancer Charity, National Institute of Health Research and the Medical Research Council.
Contributor Information
Joshua R Astley, Email: jastley1@sheffield.ac.uk.
Jim M Wild, Email: j.m.wild@sheffield.ac.uk.
Bilal A Tahir, Email: b.tahir@sheffield.ac.uk.
REFERENCES
- 1. Wang HD, Naghavi M, Allen C, Barber RM, Bhutta ZA, Carter A, et al. Global, regional, and national life expectancy, all-cause mortality, and cause-specific mortality for 249 causes of death, 1980-2015: a systematic analysis for the global burden of disease study 2015. Lancet 2016; 388: 1459–544. doi: 10.1016/S0140-6736(16)31012-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. GBD 2016 Disease and Injury Incidence and Prevalence Collaborators . Global, regional, and national incidence, prevalence, and years lived with disability for 328 diseases and injuries for 195 countries, 1990-2016: a systematic analysis for the global burden of disease study 2016. Lancet 2017; 390: 1211–59. doi: 10.1016/S0140-6736(17)32154-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin 2015; 65: 87–108. doi: 10.3322/caac.21262 [DOI] [PubMed] [Google Scholar]
- 4. BERNERS-LEE CM. Cybernetics and forecasting. Nature 1968; 219: 202–3. doi: 10.1038/219202b0 [DOI] [Google Scholar]
- 5. Krizhevsky A, Sutskever I, Hinton GE..editors. ImageNet classification with deep convolutional neural networks. 26th annual conference on neural information processing systems; 2012 Dec 3-6, 2012; lake Tahoe. NV United States 2012;. [Google Scholar]
- 6. Milletari F, Navab N, Ahmadi SA. V-Net: fully Convolutional neural networks for volumetric medical image segmentation. Proceedings of 2016 Fourth International Conference on 3d Vision 2016;: 565–71. [Google Scholar]
- 7. de Vos BD, Wolterink JM, de Jong PA, Leiner T, Viergever MA, Isgum I. ConvNet-Based localization of anatomical structures in 3-D medical images. IEEE Trans Med Imaging 2017; 36: 1470–81. doi: 10.1109/TMI.2017.2673121 [DOI] [PubMed] [Google Scholar]
- 8. Yang J, Veeraraghavan H, Armato SG, Farahani K, Kirby JS, Kalpathy-Kramer J, et al. Autosegmentation for thoracic radiation treatment planning: a grand challenge at AAPM 2017. Med Phys 2018; 45: 4568–81. doi: 10.1002/mp.13141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kazeminia S, Baur C, Kuijper A, van Ginneken B, Navab N, Albarqouni S. Gans for medical image analysis. arXiv preprint arXiv 2018; 180906222. [DOI] [PubMed] [Google Scholar]
- 10. Goodfellow IJ, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S..editors. Generative adversarial nets. advances in neural information processing systems. Montréal CANADA Neural information processing systems foundation 2014;. [Google Scholar]
- 11. Jiang J, YC H, Tyagi N, Zhang P, Rimner A, Mageras GS. Tumor-aware, adversarial domain adaptation from CT to MRI for lung cancer segmentation. In: editors. MICCAI 2018 Lecture notes on computer science. Granada, Spain: Springer Cham; 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Negahdar M, Beymer D, Syeda-Mahmood T. F. eds. SPIE Medical Imaging. Houston, Texas, United States: SPIE-Intl Soc Optical Eng; 2018. . [Google Scholar]
- 13. Wang X, Teng P, Lo P, Banola A, Kim G et al. eds. RAMBO 2018/BIA Lecture notes on computer science. Granada, Spain: Springer Cham; 2018. . [Google Scholar]
- 14. Sousa P, Galdran A, Costa P, Campilho A. eds. 2019 IEEE 16th International Symposium on Biomedical Imaging. Venice, Italy: Institute of Electrical and Electronics Engineers (IEEE); 2019. . [Google Scholar]
- 15. Javaid U, Dasnoy D, Lee J. A. eds. ACIVS 2018 Lecture notes on computer science. Granada, Spain: Springer Cham; 2018. . [Google Scholar]
- 16. Hofmanninger J, Prayer F, Pan J, Rohrich S, Prosch H, Langs G. Automatic lung segmentation in routine imaging is a data diversity problem, not a methodology problem. ArXiv preprint 2020; 11767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jiang J, Hu Y-C, Tyagi N, Zhang P, Rimner A, Deasy JO, et al. Cross-Modality (CT-MRI) prior augmented deep learning for robust lung tumor segmentation from small Mr datasets. Med Phys 2019; 46: 4392–404. doi: 10.1002/mp.13695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tahmasebi N, Boulanger P, Noga M, Punithakumar K. A fully Convolutional deep neural network for lung tumor boundary tracking in MRI. Annu Int Conf IEEE Eng Med Biol Soc 2018; 2018: 5906–9. doi: 10.1109/EMBC.2018.8513607 [DOI] [PubMed] [Google Scholar]
- 19. Zhong Z, Kim Y, Plichta K, Allen BG, Zhou L, Buatti J, et al. Simultaneous cosegmentation of tumors in PET-CT images using deep fully convolutional networks. Med Phys 2019; 46: 619–33. doi: 10.1002/mp.13331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhou B, Crawford R, Dogdas B, Goldmacher G, Chen A et al. eds. 2019 IEEE Winter Conference on Applications of Computer Vision; 2019. . [Google Scholar]
- 21. Park J, Yun J, Kim N, Park B, Cho Y, Park HJ, et al. Fully automated lung lobe segmentation in volumetric chest CT with 3D U-Net: validation with intra- and Extra-Datasets. J Digit Imaging 2020; 33: 221–30. doi: 10.1007/s10278-019-00223-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gerard SE, Patton TJ, Christensen GE, Bayouth JE, Reinhardt JM. FissureNet: a deep learning approach for pulmonary fissure detection in CT images. IEEE Trans Med Imaging 2019; 38: 156–66. doi: 10.1109/TMI.2018.2858202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yun J, Park J, Yu D, Yi J, Lee M, Park HJ, et al. Improvement of fully automated airway segmentation on volumetric computed tomographic images using a 2.5 dimensional convolutional neural net. Med Image Anal 2019; 51: 13–20. doi: 10.1016/j.media.2018.10.006 [DOI] [PubMed] [Google Scholar]
- 24. Eppenhof KAJ, Pluim JPW. Pulmonary CT registration through supervised learning with Convolutional neural networks. IEEE Trans Med Imaging 2019; 38: 1097–105. doi: 10.1109/TMI.2018.2878316 [DOI] [PubMed] [Google Scholar]
- 25. Fu Y, Lei Y, Wang T, Higgins K, Bradley JD, Curran WJ, et al. LungRegNet: an unsupervised deformable image registration method for 4D-CT lung. Med Phys 2020; 47: 1763–74. doi: 10.1002/mp.14065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jiang Z, Yin F-F, Ge Y, Ren L. A multi-scale framework with unsupervised joint training of convolutional neural networks for pulmonary deformable image registration. Phys Med Biol 2020; 65: 015011. doi: 10.1088/1361-6560/ab5da0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. de Vos BD, Berendsen FF, Viergever MA, Sokooti H, Staring M, Išgum I. A deep learning framework for unsupervised affine and deformable image registration. Med Image Anal 2019; 52: 128–43. doi: 10.1016/j.media.2018.11.010 [DOI] [PubMed] [Google Scholar]
- 28. Stergios C, Mihir S, Maria V, Guillaume C, Marie-Pierre R, Stavroula M. Linear and Deformable Image Registration with 3D Convolutional Neural Networks. In: Image Analysis for Moving Organ, Breast, and Thoracic Images. Granada, Spain: Springer International Publishing; 2018. [Google Scholar]
- 29. Ren G, WY H, Qin J, Cai J. Deriving lung perfusion directly from CT image using deep Convolutional neural network: a preliminary study. Artificial Intelligence in Radiation Therapy 2019;: 102–9. [Google Scholar]
- 30. Liu Y, Fu W, Selvakumaran V, Phelan M, Segars W. P et al. eds. In Medical Imaging 2019: Imaging Informatics for Healthcare, Research, and Applications. San Diego, California, United States: SPIE-Intl Soc Optical Eng; 2019. . [Google Scholar]
- 31. Akila Agnes S, Anitha J, Dinesh Peter J. Automatic lung segmentation in low-dose chest CT scans using convolutional deep and wide network (CDWN. Neural Computing and Applications 2018;. [Google Scholar]
- 32. Novikov AA, Lenis D, Major D, Hladuvka J, Wimmer M, Buhler K. Fully Convolutional architectures for multiclass segmentation in chest radiographs. IEEE Trans Med Imaging 2018; 37: 1865–76. doi: 10.1109/TMI.2018.2806086 [DOI] [PubMed] [Google Scholar]
- 33. Gaál G, Maga B, Lukács A. Attention U-Net based Adversarial architectures for chest X-ray lung segmentation. arXiv preprint arXiv 2020; 200310304. [Google Scholar]
- 34. Hatamizadeh A, Hoogi A, Sengupta D, Lu W, Wilcox B, Rubin D. Deep active lesion segmentation. Machine Learning in Medical Imaging 2019;: 98–105. [Google Scholar]
- 35. Sandkühler R, Jud C, Bauman G, Willers C, Pusterla O, Nyilas S. Weakly supervised learning strategy for lung defect segmentation. Machine Learning in Medical Imaging 2019;: 541–8. [Google Scholar]
- 36. Rajchl M, Lee MCH, Oktay O, Kamnitsas K, Passerat-Palmbach J, Bai W, et al. DeepCut: object segmentation from bounding box annotations using Convolutional neural networks. IEEE Trans Med Imaging 2017; 36: 674–83. doi: 10.1109/TMI.2016.2621185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sentker T, Madesta F, Werner R. eds. MICCAI 2018 Lecture notes on computer science. Granada, Spain: Springer Cham; 2018. . [Google Scholar]
- 38. Fechter T, Baltas D. One-Shot learning for deformable medical image registration and periodic motion tracking. IEEE Trans Med Imaging 2020; 39: 2506–17. doi: 10.1109/TMI.2020.2972616 [DOI] [PubMed] [Google Scholar]
- 39. Galib SM, Lee HK, Guy CL, Riblett MJ, Hugo GD. A fast and scalable method for quality assurance of deformable image registration on lung CT scans using convolutional neural networks. Med Phys 2020; 47: 99–109. doi: 10.1002/mp.13890 [DOI] [PubMed] [Google Scholar]
- 40. Ferrante E, Oktay O, Glocker B, Milone D. H. eds. MLMI 2018 Lecture Notes in Computer Science. Granada, Spain: Springer Cham; 2018. . [Google Scholar]
- 41. Beaudry J, Esquinas P, Shieh C. -C. eds. Proc SPIE Medical Imaging 2019. San Diego, California, United States: SPIE-Intl Soc Optical Eng; 2019. . [Google Scholar]
- 42. Duan C, Deng H, Xiao S, Xie J, Li H, Sun X, et al. Fast and accurate reconstruction of human lung gas MRI with deep learning. Magn Reson Med 2019; 82: 2273–85. doi: 10.1002/mrm.27889 [DOI] [PubMed] [Google Scholar]
- 43. Liu Z, Miao J, Huang P, Wang W, Wang X, Zhai Y, et al. A deep learning method for producing ventilation images from 4DCT: first comparison with technegas SPECT ventilation. Med Phys 2020; 47: 1249–57. doi: 10.1002/mp.14004 [DOI] [PubMed] [Google Scholar]
- 44. Olberg S, Zhang H, Green OL, Mazur TR, Yang D, Hugo GD, et al. Deep Learning-Based pseudo CT reconstruction for Mr Only-Guided radiation therapy of lung SBRT. Int J Radiat Oncol Biol Phys 2018; 102: e309–10. doi: 10.1016/j.ijrobp.2018.07.969 [DOI] [Google Scholar]
- 45. Xu J, Liu H. Segmentation of Pulmonary CT Image by Using Convolutional Neural Network Based on Membership Function. In: editors. 2017 IEEE International Conference on Computational Science and Engineering (CSE) and IEEE International Conference on Embedded and Ubiquitous Computing (EUC. Guangzhou, China: Institute of Electrical and Electronics Engineers Inc; 2017. [Google Scholar]
- 46. Zha W, Fain SB, Schiebler ML, Evans MD, Nagle SK, Liu F. Deep convolutional neural networks with. J Magn Reson Imaging 2019; 50: 1169–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tustison NJ, Avants BB, Lin Z, Feng X, Cullen N, Mata JF, et al. Convolutional neural networks with template-based data augmentation for functional lung image quantification. Acad Radiol 2019; 26: 412–23. doi: 10.1016/j.acra.2018.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Soans R. E, Shackleford J. A. eds. SPIE Medical Imaging. Houston, Texas, United States: SPIE-Intl Soc Optical Eng; 2018. . [Google Scholar]
- 49. Zhu J, Zhang J, Qiu B, Liu Y, Liu X, Chen L. Comparison of the automatic segmentation of multiple organs at risk in CT images of lung cancer between deep convolutional neural network-based and atlas-based techniques. Acta Oncol 2019; 58: 257–64. doi: 10.1080/0284186X.2018.1529421 [DOI] [PubMed] [Google Scholar]
- 50. Hooda R, Mittal A, Sofat S. An efficient variant of Fully-Convolutional network for Segmenting lung fields from chest radiographs. Wireless Personal Communications 2018; 101: 1559–79. doi: 10.1007/s11277-018-5777-3 [DOI] [Google Scholar]
- 51. Mittal A, Hooda R, Sofat S. LF-SegNet: a fully Convolutional Encoder–Decoder network for Segmenting lung fields from chest radiographs. Wireless Personal Communications 2018; 101: 511–29. doi: 10.1007/s11277-018-5702-9 [DOI] [Google Scholar]
- 52. Zhao X, Li L, Lu W, Tan S. Tumor co-segmentation in PET/CT using multi-modality fully convolutional neural network. Phys Med Biol 2018; 64: 015011. doi: 10.1088/1361-6560/aaf44b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Moriya T, Roth H. R, Nakamura S, Oda H, Nagara K et al. eds. Proc SPIE 10578, Medical Imaging 2018: Biomedical Applications in Molecular, Structural, and Functional Imaging. Houston, Texas, USA: SPIE-Intl Soc Optical Eng; 2018. . [Google Scholar]
- 54. Kalinovsky A, Liauchuk V, Tarasau A. Lesion detection in CT images using deep learning semantic segmentation technique. ISPRS - International Archives of the Photogrammetry, Remote Sensing and Spatial Information Sciences 2017; XLII-2/W4: 13–17. doi: 10.5194/isprs-archives-XLII-2-W4-13-2017 [DOI] [Google Scholar]
- 55. Anthimopoulos M, Christodoulidis S, Ebner L, Geiser T, Christe A, Mougiakakou S. Semantic segmentation of pathological lung tissue with dilated fully Convolutional networks. IEEE J Biomed Health Inform 2019; 23: 714–22. doi: 10.1109/JBHI.2018.2818620 [DOI] [PubMed] [Google Scholar]
- 56. Gao M, Xu Z, Lu L, Harrison A. P, Summers R. M et al. eds. MLMI 2016 Lecture Notes in Computer Science. Athens, Greece: Springer Cham; 2016. . [Google Scholar]
- 57. Wang C, Hayashi Y, Oda M, Itoh H, Kitasaka T, Frangi AF. Tubular structure segmentation using spatial fully connected network with radial distance loss for 3D medical images. Medical Image Computing and Computer Assisted Intervention – MICCAI 2019;: 348–56. [Google Scholar]
- 58. Garcia-Uceda Juarez A, Selvan R, Saghir Z. De Bruijne M. a joint 3D UNet-Graph neural network-based method for airway segmentation from chest CTS. Machine Learning in Medical Imaging 2019;: 583–91. [Google Scholar]
- 59. Juarez AGU, Tiddens H, de Bruijne M. Automatic airway segmentation in chest CT using convolutional neural networks. Granada, Spain: Springer Cham; 2018. pp. 16–20. [Google Scholar]
- 60. Blendowski M, Heinrich MP. Combining MRF-based deformable registration and deep binary 3D-CNN descriptors for large lung motion estimation in COPD patients. Int J Comput Assist Radiol Surg 2019; 14: 43–52. doi: 10.1007/s11548-018-1888-2 [DOI] [PubMed] [Google Scholar]
- 61. Zhong Y, Vinogradskiy Y, Chen L, Myziuk N, Castillo R, Castillo E, et al. Technical note: deriving ventilation imaging from 4DCT by deep convolutional neural network. Med Phys 2019; 46: 2323–9. doi: 10.1002/mp.13421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ioffe S, Szegedy C..editors. Batch normalization: accelerating deep network training by reducing internal covariate shift. Proceedings of the 32nd International Conference on International Conference on Machine Learning 2015;. [Google Scholar]
- 63. Huang L, Yang D, Lang B, Deng J. eds. Decorrelated batch normalization In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. Salt Lake City: UT: IEEE; 2018. . [Google Scholar]
- 64. Dai W, Dong N, Wang Z, Liang X, Zhang H et al. eds. DLMIA 2018 Lecture Notes in Computer Science. Granada, Spain: Springer Cham; 2018. . [Google Scholar]
- 65. Bluemke DA, Moy L, Bredella MA, Ertl-Wagner BB, Fowler KJ, Goh VJ, et al. Assessing Radiology Research on Artificial Intelligence: A Brief Guide for Authors, Reviewers, and Readers-From the Radiology Editorial Board. Radiology 2020; 294: 487–9. doi: 10.1148/radiol.2019192515 [DOI] [PubMed] [Google Scholar]
- 66. Moher D, Liberati A, Tetzlaff J, Altman DG, Altman D, Antes G. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. American College of Physicians 2009;: 264–9. [Google Scholar]
- 67. Pehrson LM, Nielsen MB, Lauridsen CA. Automatic pulmonary nodule detection applying deep learning or machine learning algorithms to the LIDC-IDRI database: a systematic review. Mdpi Ag 2019;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lobo P, Guruprasad S. Classification and Segmentation Techniques for Detection of Lung Cancer from CT Images. In: editors. International Conference on Inventive Research in Computing Applications (ICIRCA. Coimbatore: Institute of Electrical and Electronics Engineers Inc; 2018. [Google Scholar]
- 69. Chassagnon G, Vakalopoulou M, Paragios N, Revel M-P. Artificial intelligence applications for thoracic imaging. Eur J Radiol 2020; 123: 108774. doi: 10.1016/j.ejrad.2019.108774 [DOI] [PubMed] [Google Scholar]
- 70. Dong X, Lei Y, Wang T, Thomas M, Tang L, Curran WJ, et al. Automatic multiorgan segmentation in thorax CT images using U-net-GAN. Med Phys 2019; 46: 2157–68. doi: 10.1002/mp.13458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lustberg T, van Soest J, Gooding M, Peressutti D, Aljabar P, van der Stoep J, et al. Clinical evaluation of atlas and deep learning based automatic contouring for lung cancer. Radiother Oncol 2018; 126: 312–7. doi: 10.1016/j.radonc.2017.11.012 [DOI] [PubMed] [Google Scholar]
- 72. Soliman A, Shaffie A, Ghazal M, Gimel'Farb G, Keynton R, El-Baz A..editors. A novel CNN segmentation framework based on using new shape and appearance features. 2018 25th IEEE International Conference on Image Processing (ICIP); 2017 Oct 7-10, 2018; Athens, Greece: IEEE Computer Society. [Google Scholar]
- 73. Zhou X, Takayama R, Wang S, Hara T, Fujita H. Deep learning of the sectional appearances of 3D CT images for anatomical structure segmentation based on an FCN voting method. Med Phys 2017; 44: 5221–33. doi: 10.1002/mp.12480 [DOI] [PubMed] [Google Scholar]
- 74. Gerard S. E, Herrmann J, Kaczka D. W, Reinhardt J. M. eds. RAMBO 2018, BIA 2018, TIA 2018 Lecture Notes in Computer Science. Granada, Spain: Springer Cham; 2018. . [Google Scholar]
- 75. Hu Q, de F Souza LF, Holanda GB, Alves SSA, Dos S Silva FH, Han T, de FSLF HGB, Dos SSFH HT, et al. An effective approach for CT lung segmentation using mask region-based convolutional neural networks. Artif Intell Med 2020; 103: 101792. doi: 10.1016/j.artmed.2020.101792 [DOI] [PubMed] [Google Scholar]
- 76. Xu M, Qi S, Yue Y, Teng Y, Xu L, Yao Y, et al. Segmentation of lung parenchyma in CT images using CNN trained with the clustering algorithm generated dataset. Biomed Eng Online 2019; 18: 2. doi: 10.1186/s12938-018-0619-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Hwang S, Park S. Accurate lung segmentation via Network-Wise training of Convolutional networks. Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support 2017; 10553: 92–9. [Google Scholar]
- 78. Souza JC, Bandeira Diniz JO, Ferreira JL, França da Silva GL, Corrêa Silva A, de Paiva AC. An automatic method for lung segmentation and reconstruction in chest X-ray using deep neural networks. Comput Methods Programs Biomed 2019; 177: 285–96. doi: 10.1016/j.cmpb.2019.06.005 [DOI] [PubMed] [Google Scholar]
- 79. Wang C. eds. SCIA 2017, Part II, LNCS 10270 Lecture notes on computer science. Tromsø, Norway: Springer Cham; 2017. . [Google Scholar]
- 80. Chen W, Wei H, Peng S, Sun J, Qiao X, Liu B. Hsn: hybrid segmentation network for small cell lung cancer segmentation. IEEE Access 2019; 7: 75591–603. doi: 10.1109/ACCESS.2019.2921434 [DOI] [Google Scholar]
- 81. Imran A-A-Z, Hatamizadeh A, Ananth SP, Ding X, Tajbakhsh N, Terzopoulos D. Fast and automatic segmentation of pulmonary lobes from chest CT using a progressive dense V-network. Comput Methods Biomech Biomed Engin 2020; 8: 509–18. doi: 10.1080/21681163.2019.1672210 [DOI] [Google Scholar]
- 82. Vakalopoulou M, Chassagnon G, Bus N, Marini R, Zacharaki E. I et al. eds. MICCAI 2018, LNCS 11073 Lecture notes on computer science. Granada, Spain: Springer Cham; 2018. . [Google Scholar]
- 83. Park B, Park H, Lee SM, Seo JB, Kim N. Lung segmentation on HRCT and volumetric CT for diffuse interstitial lung disease using deep Convolutional neural networks. J Digit Imaging 2019; 32: 1019–26. doi: 10.1007/s10278-019-00254-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Suzuki Y, Yamagata K, Yanagawa M, Kido S, Tomiyama N. Weak supervision in convolutional neural network for semantic segmentation of diffuse lung diseases using partially annotated dataset: SPIE. 2020;.
- 85. Wang G, Li W, Zuluaga MA, Pratt R, Patel PA, Aertsen M, et al. Interactive medical image segmentation using deep learning with Image-Specific fine tuning. IEEE Trans Med Imaging 2018; 37: 1562–73. doi: 10.1109/TMI.2018.2791721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Edmunds D, Sharp G, Winey B. Automatic diaphragm segmentation for real-time lung tumor tracking on cone-beam CT projections: a convolutional neural network approach. Biomed Phys Eng Express 2019; 5: 035005. doi: 10.1088/2057-1976/ab0734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Taha AA, Hanbury A. Metrics for evaluating 3D medical image segmentation: analysis, selection, and tool. BMC Med Imaging 2015; 15: 29. doi: 10.1186/s12880-015-0068-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Mukesh M, Benson R, Jena R, Hoole A, Roques T, Scrase C, et al. Interobserver variation in clinical target volume and organs at risk segmentation in post-parotidectomy radiotherapy: can segmentation protocols help? Br J Radiol 2012; 85: e530–6. doi: 10.1259/bjr/66693547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Chlebus G, Meine H, Thoduka S, Abolmaali N, van Ginneken B, Hahn HK, et al. Reducing inter-observer variability and interaction time of Mr liver volumetry by combining automatic CNN-based liver segmentation and manual corrections. PLoS One 2019; 14: e0217228. doi: 10.1371/journal.pone.0217228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Woodhouse N, Wild JM, Paley MNJ, Fichele S, Said Z, Swift AJ, et al. Combined helium-3/proton magnetic resonance imaging measurement of ventilated lung volumes in smokers compared to never-smokers. J Magn Reson Imaging 2005; 21: 365–9. doi: 10.1002/jmri.20290 [DOI] [PubMed] [Google Scholar]
- 91. Shiraishi J, Katsuragawa S, Ikezoe J, Matsumoto T, Kobayashi T, Komatsu K, et al. Development of a digital image database for chest radiographs with and without a lung nodule: receiver operating characteristic analysis of radiologists' detection of pulmonary nodules. AJR Am J Roentgenol 2000; 174: 71–4. doi: 10.2214/ajr.174.1.1740071 [DOI] [PubMed] [Google Scholar]
- 92. Jaeger S, Karargyris A, Candemir S, Folio L, Siegelman J, Callaghan F, et al. Automatic tuberculosis screening using chest radiographs. IEEE Trans Med Imaging 2014; 33: 233–45. doi: 10.1109/TMI.2013.2284099 [DOI] [PubMed] [Google Scholar]
- 93. Moro J, Ribes S, Caselles O, Parent L. Evaluation of two registration techniques applied to lung adaptive radiotherapy. Physica Medica 2013; 29: e7. doi: 10.1016/j.ejmp.2013.08.025 [DOI] [Google Scholar]
- 94. Hughes PJC, Horn FC, Collier GJ, Biancardi A, Marshall H, Wild JM. Spatial fuzzy c-means thresholding for semiautomated calculation of percentage lung ventilated volume from hyperpolarized gas and 1 H MRI. J Magn Reson Imaging 2018; 47: 640–6. doi: 10.1002/jmri.25804 [DOI] [PubMed] [Google Scholar]
- 95. Tahir BA, Hughes PJC, Robinson SD, Marshall H, Stewart NJ, Norquay G, et al. Spatial comparison of CT-based surrogates of lung ventilation with hyperpolarized Helium-3 and xenon-129 gas MRI in patients undergoing radiation therapy. Int J Radiat Oncol Biol Phys 2018; 102: 1276–86. doi: 10.1016/j.ijrobp.2018.04.077 [DOI] [PubMed] [Google Scholar]
- 96. Bauman G, Puderbach M, Deimling M, Jellus V, Chefd'hotel C, Dinkel J, et al. Non-contrast-enhanced perfusion and ventilation assessment of the human lung by means of Fourier decomposition in proton MRI. Magn Reson Med 2009; 62: 656–64. doi: 10.1002/mrm.22031 [DOI] [PubMed] [Google Scholar]
- 97. Tahir BA, Van Holsbeke C, Ireland RH, Swift AJ, Horn FC, Marshall H, et al. Comparison of CT-based lobar ventilation with 3He MR imaging ventilation measurements. Radiology 2016; 278: 585–92. doi: 10.1148/radiol.2015142278 [DOI] [PubMed] [Google Scholar]
- 98. In Lafarge M. W, Moeskops P, Veta M, Pluim J. P. W, Eppenhof K. A. J et al. eds. SPIE Medical Imaging. Houston, Texas, United States: SPIE-Intl Soc Optical Eng; 2018. . [Google Scholar]
- 99. Ali S, Rittscher J. Conv2Warp: an unsupervised deformable image registration with continuous convolution and Warping machine learning in medical imaging. 2019;: 489–97.
- 100. Sokooti H, de Vos B, Berendsen F, Lelieveldt B. P. F, Išgum I et al. eds. MICCAI 2017 Lecture Notes in Computer Science. Quebec City, QC, Canada: Springer Cham; 2017. . [Google Scholar]
- 101. Sokooti H, De Vos B, Berendsen F, Ghafoorian M, Yousefi S, Lelieveldt B. 3D Convolutional neural networks image registration based on efficient supervised learning from artificial deformations. 2019;.
- 102. Qin C, Shi B, Liao R, Mansi T, Rueckert D et al. eds. Information Processing in Medical Imaging. Cham: Springer International Publishing; 2018. . [Google Scholar]
- 103. Mahapatra D, Ge Z, Sedai S, Chakravorty R. eds. MLMI 2018 Lecture notes on computer science. Spain: Springer Cham; 2018. . [Google Scholar]
- 104. Tajbakhsh N, Shin JY, Gurudu SR, Hurst RT, Kendall CB, Gotway MB, et al. Convolutional neural networks for medical image analysis: full training or fine tuning? IEEE Trans Med Imaging 2016; 35: 1299–312. doi: 10.1109/TMI.2016.2535302 [DOI] [PubMed] [Google Scholar]
- 105. Willemink MJ, Noël PB. The evolution of image reconstruction for CT-from filtered back projection to artificial intelligence. Eur Radiol 2019; 29: 2185–95. doi: 10.1007/s00330-018-5810-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Ye JC. Compressed sensing MRI: a review from signal processing perspective. BMC Biomed Eng 2019; 1: 8. doi: 10.1186/s42490-019-0006-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Lee SM, Lee JG, Lee G, Choe J, Do KH, Kim N, et al. Ct image conversion among different reconstruction kernels without a Sinogram by using a Convolutional neural network. Korean J Radiol 2019; 20: 295–303. doi: 10.3348/kjr.2018.0249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Ge Y, Su T, Zhu J, Deng X, Zhang Q, Chen J, et al. ADAPTIVE-NET: deep computed tomography reconstruction network with analytical domain transformation knowledge. Quant Imaging Med Surg 2020; 10: 415–27. doi: 10.21037/qims.2019.12.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Dietze MMA, Branderhorst W, Kunnen B, Viergever MA, de Jong HWAM. Accelerated SPECT image reconstruction with FBP and an image enhancement convolutional neural network. EJNMMI Phys 2019; 6: 14. doi: 10.1186/s40658-019-0252-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Norberg P, Bake B, Jacobsson L, Carlsson GA, Gustafsson A. Evaluation of reconstruction techniques for lung single photon emission tomography: a Monte Carlo study. Nucl Med Commun 2007; 28: 929–36. doi: 10.1097/MNM.0b013e3282f1acac [DOI] [PubMed] [Google Scholar]
- 111. El Bitar Z, Lazaro D, Coello C, Breton V, Hill D, Buvat I. Fully 3D Monte Carlo image reconstruction in SPECT using functional regions. Fully 3D Monte Carlo image reconstruction in SPECT using functional regions, Nuclear Instruments and Methods in Physics Research Section A: Accelerators, Spectrometers, Detectors and Associated Equipment 2006;. [Google Scholar]
- 112. Hammernik K, Klatzer T, Kobler E, Recht MP, Sodickson DK, Pock T, et al. Learning a variational network for reconstruction of accelerated MRI data. Magn Reson Med 2018; 79: 3055–71. doi: 10.1002/mrm.26977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Ajraoui S, Lee KJ, Deppe MH, Parnell SR, Parra-Robles J, Wild JM. Compressed sensing in hyperpolarized 3He Lung MRI. Magn Reson Med 2010; 63: 1059–69. doi: 10.1002/mrm.22302 [DOI] [PubMed] [Google Scholar]
- 114. Sheikh K, Coxson HO, Parraga G. This is what COPD looks like. Respirology 2016; 21: 224–36. [DOI] [PubMed] [Google Scholar]
- 115. Wild JM, Ajraoui S, Deppe MH, Parnell SR, Marshall H, Parra-Robles J, et al. Synchronous acquisition of hyperpolarised 3He and 1H MR images of the lungs - maximising mutual anatomical and functional information. NMR Biomed 2011; 24: 130–4. doi: 10.1002/nbm.1565 [DOI] [PubMed] [Google Scholar]
- 116. Gou S, Liu W, Jiao C, Liu H, Gu Y, Zhang X, et al. Gradient regularized convolutional neural networks for low-dose CT image enhancement. Phys Med Biol 2019; 64: 165017. doi: 10.1088/1361-6560/ab325e [DOI] [PubMed] [Google Scholar]
- 117. Umehara K, Ota J, Ishida T. Application of super-resolution Convolutional neural network for enhancing image resolution in chest CT. J Digit Imaging 2018; 31: 441–50. doi: 10.1007/s10278-017-0033-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Bi L, Kim J, Kumar A, Feng D, Fulham M et al. eds. Synthesis of positron emission tomography (PET) images via multi-channel generative adversarial networks (GANs). In: RAMBO 2017 Lecture Notes in Computer Science. Québec City, QC, Canada: Springer Cham; , ; 2017. 2017 Sept 14. [Google Scholar]
- 119. Jang B-S, Chang JH, Park AJ, Wu H-G, Jang Bum†Sup, Wu Hong†Gyun. Generation of virtual lung single-photon emission computed tomography/CT fusion images for functional avoidance radiotherapy planning using machine learning algorithms. J Med Imaging Radiat Oncol 2019; 63: 229–35. doi: 10.1111/1754-9485.12868 [DOI] [PubMed] [Google Scholar]
- 120. Preiswerk F, Cheng C. C, Luo J, Madore B. eds. Synthesizing dynamic MRI using long-term recurrent convolutional networks. In: MLMI 2018, LNCS 11046 Lecture notes on computer science; , . Spain: Springer Cham; , ; 2018. Granada 2018 Sept 16-20. [Google Scholar]
- 121. Kipritidis J, Tahir BA, Cazoulat G, Hofman MS, Siva S, Callahan J, et al. The vampire challenge: a multi-institutional validation study of CT ventilation imaging. Med Phys 2019; 46: 1198–217. doi: 10.1002/mp.13346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Salehinejad H, Colak E, Dowdell T, Barfett J, Valaee S. Synthesizing chest X-ray pathology for training deep Convolutional neural networks. IEEE Trans Med Imaging 2019; 38: 1197–206. doi: 10.1109/TMI.2018.2881415 [DOI] [PubMed] [Google Scholar]
- 123. Nie D, Cao X, Gao Y, Wang L, Shen D et al. eds. Estimating CT image from MRI data using 3D fully convolutional networks. In: DLMIA 2016, LABELS 2016 Lecture Notes in Computer Science , . Greece: Springer Cham; 2016. Oct 21, 2016; Athens. [DOI] [PMC free article] [PubMed] [Google Scholar]