Abstract
In the omics era, saliva, a filtrate of blood, may serve as an alternative, noninvasive biospecimen to blood, although its use for specific metabolomic applications has not been fully evaluated. We demonstrated that the saliva metabolome may provide sensitive measures of traffic-related air pollution (TRAP) and associated biological responses via high-resolution, longitudinal metabolomics profiling. We collected 167 pairs of saliva and plasma samples from a cohort of 53 college student participants and measured corresponding indoor and outdoor concentrations of six air pollutants for the dormitories where the students lived. Grand correlation between common metabolic features in saliva and plasma was moderate to high, indicating a relatively consistent association between saliva and blood metabolites across subjects. Although saliva was less associated with TRAP compared to plasma, 25 biological pathways associated with TRAP were detected via saliva and accounted for 69% of those detected via plasma. Given the slightly higher feature reproducibility found in saliva, these findings provide some indication that the saliva metabolome offers a sensitive and practical alternative to blood for characterizing individual biological responses to environmental exposures.
Keywords: traffic-related air pollution, high-resolution metabolomics, blood metabolome, saliva metabolome, pathway analysis, metabolic perturbations
Graphical Abstract
INTRODUCTION
Saliva is a complex mixture of fluids from the salivary glands, residual food particles, and oral bacteria.1 In general, most salivary components are either excreted by the salivary glands or transported from the blood by passive diffusion and active transport.2 Correspondingly, it is possible that many components that are present in blood can be also found in saliva, and therefore, saliva is widely used as an alternative to blood for disease diagnosis and biomonitoring.3,4 Indeed, saliva specimens have increasingly been used in clinical diagnosis, given its relative stability at room temperature and ease of collection compared to blood.2 Although scientists have started utilized saliva in the omics (i.e., emerging techniques for global assessment of molecules) era for biomarker identification,5–9 blood is still the most commonly used biospecimen due to wider coverage of analyte detection and often higher concentration of analytes of interest. Thus, the use of saliva in omics applications should be interrogated carefully with a comprehensive comparison with blood. However, there are currently few evaluations of the saliva and blood metabolome (i.e., the comprehensive analysis of metabolites in saliva and blood) for use in environmental health and epidemiological applications.
The adverse effects of exposure to traffic-related air pollution (TRAP) on the respiratory and cardiovascular systems and other human organ systems have been well documented.10,11 Multiple biological mechanisms, including oxidative stress and systemic inflammation, have been proposed to play critical roles in the progression of disease associated with TRAP exposures.12 Despite growing observational evidence, less is known about the specific biological pathways and molecules underlying the TRAP toxicity.10 Notably, TRAP consists of hundreds of different organic and inorganic components with a high degree of chemical and physical heterogeneity, adding substantial difficulty in conducting traffic pollution exposure assessments.13 An additional challenge lies in the development and application of sensitive and specific TRAP biomarkers to measure internal exposures and corresponding physiologic responses, owing to the interindividual heterogeneity in pharmacokinetics and the complexity of numerous endogenous pathways that may mediate responses.12 With the lack of refined exposure characterization and biomarkers, our understanding of the molecular mechanisms underlying TRAP toxicity has been limited. To date, the field of air pollution health has focused narrowly on relationships between single toxicants and a limited set of biomarkers or biological pathways.
High-resolution metabolomics (HRM), an analytical platform that couples high resolution mass spectrometry (HRMS) with liquid or gas chromatography, has emerged as a powerful tool for estimating internal exposures to complex environmental mixtures (such as TRAP) and understanding underlying biological mechanisms. HRM measures thousands of metabolic features associated with exogenous exposure and endogenous processes.14 Ongoing investigations by our group, along with other researchers, have demonstrated the feasibility of using blood metabolomics to link external air pollution exposure to internal exposures and subsequent biological responses.15–21 Despite the growing interest in applying HRM to the study of air pollution and health, the majority of these studies were cross-sectional in design, providing a single “snapshot” of the metabolomic profile in individuals. Thus, these studies were not able to assess metabolic changes induced by TRAP exposure over time. Repeated measure designs increase statistical power for detecting significant associations given the same sample size as a single measure. Therefore, longitudinal studies with repeated biospecimen collection can be used to enhance the evidence that can be derived from studies using HRM.22,23
Most HRM applications, to date, have used blood as a primary biospecimen. Drawing blood is invasive and potentially painful and requires trained personnel, making longitudinal collection relatively costly. Our group has collected saliva samples concurrent with blood, facilitating comparisons of metabolic profiles and, potentially, TRAP-induced biological mechanisms identifiable in these biospecimens.17,24 Previously, we reported common biological pathways, including leukotriene metabolism, vitamin E metabolism, and cytochrome P450 metabolism, perturbed by the short-term exposures to TRAP in both the blood and saliva metabolome collected from the same individuals.17 These biological pathways have been documented to be predominantly associated with oxidative stress and acute inflammatory responses, which are believed to be essential mechanisms underlying TRAP-related health effects.12,25–27 Nevertheless, much less is known on whether the saliva metabolome contains the same information as the blood metabolome for use in environmental applications to detect and characterize biological responses associated with complex mixtures such as TRAP.
To address these questions, we conducted a secondary analysis based on a panel study of 53 participants exposed to varying TRAP levels, with repeated saliva and plasma measurements.17 We followed an established untargeted Metabolome-Wide Association Study (MWAS) workflow, where metabolic profiles will be analyzed without prior knowledge of their chemical identity, to examine changes in metabolic features and metabolic pathways associated with TRAP.17 To account for the correlated nature of the data from repeated measures, we employed a linear mixed-effect model (LMM) which allows to effectively partition the overall variation of the dependent variable into components of between- and within-subject changes and evaluate the effect of exposure free of the confounding from time-independent variables.28 To evaluate the use of the saliva metabolome in characterizing internal exposures to TRAP, we compared HRM profiles from saliva and blood samples collected from the same individual exposed to short-term TRAP. We tested for consistency between features present in both biospecimens and identified unique and common metabolic features and enriched biological pathways associated with TRAP in either biospecimen. We hypothesized that the saliva metabolome had the potential to discover novel biomarkers and biological pathways compared to plasma and improve our understanding of the underlying molecular mechanisms of TRAP toxicity.
METHODS
In this study, we worked with longitudinal biospecimens (blood and saliva) previously collected from the prospective Dorm Room Inhalation to Vehicle Emissions (DRIVE) study. DRIVE study was an intensive 12-week field study where we conducted traditional single and multipollutant traffic indicator measurements on the campus of the Georgia Institute of Technology (GIT) in Atlanta, Georgia.29 The campus is located adjacent to a section of highways where Interstates 75 and 85 merge (10 lanes), one of the most heavily trafficked highway arteries in the US.29 In the current analysis, we extracted metabolic features from 167 pairs of matched plasma and saliva metabolomic profiles separately and simultaneously from these individuals over four different time points, to identify metabolic features unique in saliva as well as any overlapping metabolic features that were present in both biospecimens. We then performed a comprehensive comparison on the feature quality and correlation of feature intensity (i.e., relative concentration) among the overlapping features and unique features. We also performed a metabolome-wide association analysis on unique and overlapping features to detect significant features and biological pathways associated with TRAP and evaluated saliva’s potential for discovering novel biomarkers and biological pathways compared to plasma.
Exposure Assessment.
During the DRIVE study, we conducted exposure assessment on several ubiquitous TRAP using samplers sited at varying monitoring locations.17 A detailed description of the DRIVE study design along with the sampling methods and quality control/assurance procedures can be found elsewhere.17,29 In the current analysis, we used exposure measurements conducted at two student dormitories, namely, “Near Dorm” and “Far Dorm”, located 20 m and 1.4 km away from the highway source, respectively.17 Each dormitory consisted of an indoor site and an outdoor site, which allowed us to account for indoor–outdoor exposure discrepancies. Identical sampling instrumentation was deployed inside each dorm and utilized a three-way valve to alternate sampling between indoor and outdoor air. Details with respect to site information, indoor location, inlet height, and inlet radius clearance have been previously reported.29–31 A suite of traffic-related air pollutants (TRAPs) was measured at each monitoring site continuously, including carbon monoxide (CO), nitric oxide (NO), nitrogen dioxide (NO2), nitric oxides (NOx), fine particulate matter (PM2.5), and black carbon (BC). Two identical sets of five pollutant sampling instrumentation were used to measure air pollutant concentrations at time scales from minutes to days and weeks. We used the daily average of indoor and outdoor air pollution concentrations measured during one month prior to the sampling date of blood at each dorm as the surrogate of personal exposure for the participants living in their respective dorms.
Study Panel.
We recruited a participant panel during the DRIVE field sampling campaign from GIT undergraduate students who lived in either the Near Dorm or the Far Dorm. In sum, 54 students completed the entire study protocol, including 24 and 30 students from the Near Dorm and Far Dorm, respectively. We also collected baseline information on socio-demographics and preliminary and health status using questionnaires filled by participants at the time of recruitment.
High Resolution Metabolomics.
We collected fasting venous blood every month and saliva samples every week from all 54 participants during the 12-week sampling period. In total, 175 plasma samples (average of 3.2 repeated samples per participant) and 621 2-mL vials of saliva (average of 11.5 repeated samples per participant) were collected. We conducted HRM on all paired blood and saliva samples (i.e., samples collected during the same sampling session from the same individuals) using established protocols.24,32 For the comparison of plasma and saliva, we analyzed 167 pairs of saliva and blood samples from 53 (one participant who provided saliva samples only was excluded) participants who gave consent to providing both biospecimens at the same visit in the current analysis.
Each sample was treated with two volumes of acetonitrile and analyzed using liquid chromatography with high-resolution mass spectrometry (LC-HRMS) techniques (Thermo Scientific Q-Exactive HF). Each sample was randomized into blocks of 40 for blinding and analyzed in triplicate by two technical columns, hydrophilic interaction liquid chromatography (HILIC) with positive electrospray ionization (ESI) and C18 hydrophobic reversed-phase chromatography with negative ESI. The use of two columns with different degrees of hydrophilicity could enhance the coverage of metabolic feature detection via maximizing the separation performance on study samples. Two quality control pooled reference plasma samples, which included internal standard samples (NIST 1950)33 and the pooled human plasma purchased from Equitech Bio were included at the beginning and end of each analytical batch for normalization, control for background noise, and batch evaluation. The detailed rationale for inclusion of specific quality control samples could be found elsewhere.32 Briefly, the pooled human plasma samples were used for batch effect correction in the experimental phase, and the internal standard samples were used to normalize signal intensities across batches relative to a calibrated reference, which enabled interstudy or interlaboratory comparison. Additionally, in the data preprocessing phase, we evaluated the feature extraction quality based on the coefficient of variation (CV) within technical triplicates, variability across pooled human plasma, percent missing values, and signal-to-noise ratio. The variability across pooled human plasma was illustrated in Figure S3. Raw data files output from LC-HRMS were converted to .mzML files using ProteoWizard and extracted using apLCMS (v. 6.6.1)34 with modifications by xMSanalyzer (v. 2.0.6.1).35 Detected ion signals (referred to as “metabolic features”) were uniquely defined by their mass-to-charge ratio (m/z, the molecular weight of the cation divided by its charge) and retention time obtained in mass spectrometry. As study samples were run in triplicate, a CV could be obtained for each study sample by feature (for each subject, the CVm for feature m is the standard deviation divided by the arithmetic mean of the triplicates), and we selected the median of CVm across subjects as a feature-specific measure of feature m to assess the overall reliability of the corresponding feature. Only metabolic features detected in more than 15% of plasma or saliva samples with a median CV among technical replicates less than 30% were included. To reduce the possibility of false matching for identifying overlapping features, we applied different extraction strategies for unique and overlapping features, respectively (Figure 1). Specifically, to detect the overlapping features, saliva and plasma metabolomic profiles were processed together, the resulting feature table of which would only contain metabolic features present in both biospecimens (Figure 1A). The apLCMS coupled with xMSanalyzer applied a series of adaptive approaches to conduct feature detection, feature alignment, and noise removal across all plasma and saliva metabolomic profiles (i.e., spectra), which could minimize the risk of matching errors compared to the matching of m/z with an arbitrary tolerance to detect the overlapping features in saliva and plasma. To detect unique features in saliva, we processed the saliva and plasma metabolomic profiles separately and filtered out the nonunique features using a mass error threshold of 10 ppm and a retention time difference of 50 s (i.e., we excluded the metabolic features that could find a matched counterpart in plasma metabolomic profiles based on m/z and retention time; Figure 1B). In addition to the suggested tolerances in previous literature, we chose such relatively lenient tolerances regarding m/z and retention time shifts in order to minimize the possibility of false detection of unique features.35,36
Figure 1.
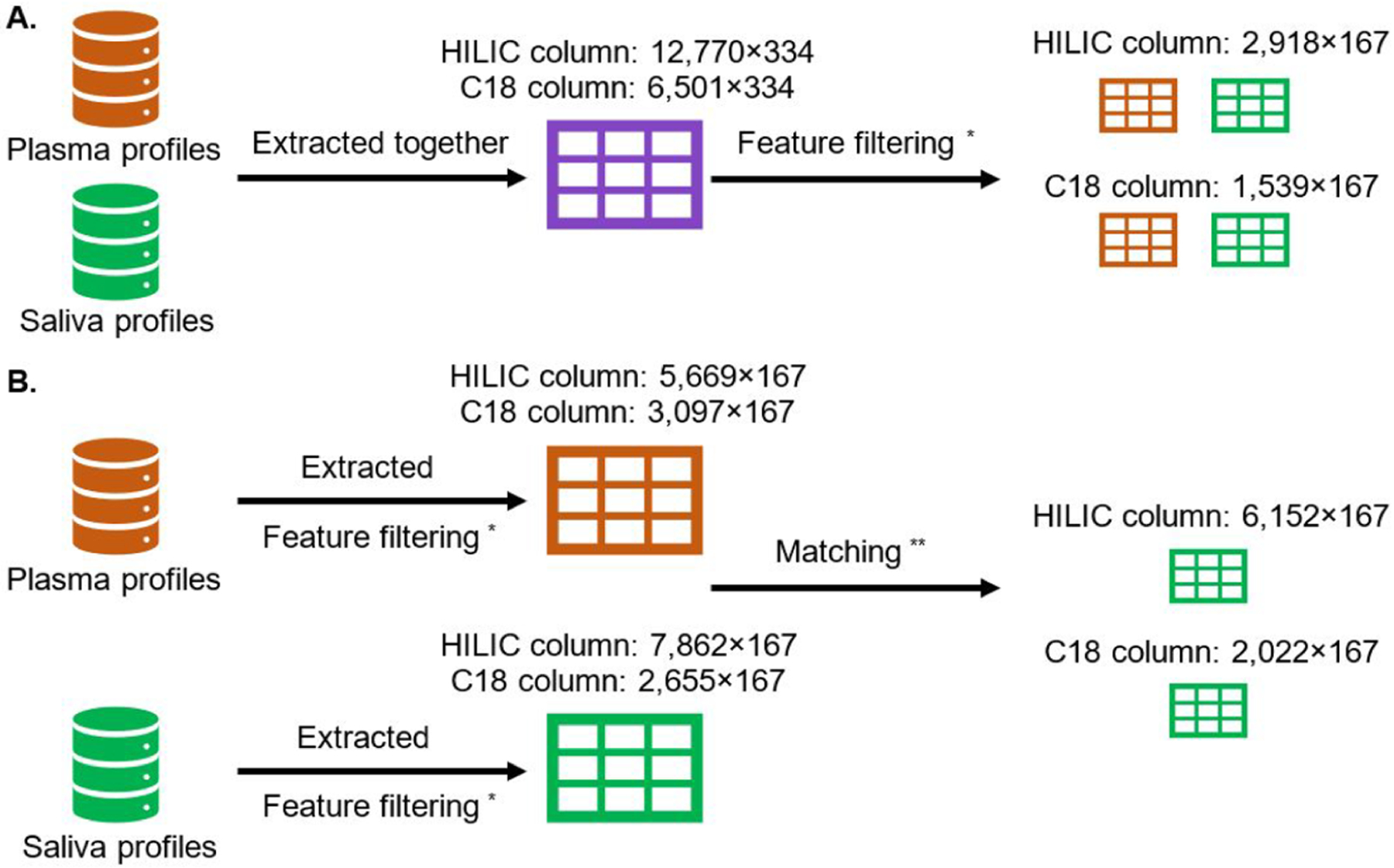
Metabolic feature extraction and data processing. We used apLCMS with xMSanalyzer to extract metabolic features from individual metabolic profiles and applied a series of statistical approaches at the feature (m/z, retention time) level across multiprofiles to match and align features and remove potential noise peaks. (A) Saliva and plasma metabolomic profiles were processed together, the resulting feature table of which only contained metabolic features present in both biospecimens [number of metabolic features × (number of plasma samples + number of saliva samples)]. (B) Saliva and plasma metabolomic profiles were processed separately, and we filtered out the nonunique features using a mass error threshold of 10 ppm and a retention time difference of 50 s. The resulting feature tables contained metabolic features unique to saliva (number of metabolic features × number of saliva samples). *Filtering criteria: present in ≥15% of both biofluids with a median coefficient of variation among technical replicates < 30%. **Matching criteria: differences in mass to charge ratio (m/z) < 10 ppm and retention time < 50 s.
Data Analysis.
As an initial metabolomic data processing step, we first averaged the intensities across triplicate samples. We compared feature quality and intensity consistency of overlapping metabolic features between plasma and saliva samples. Specifically, we assessed feature quality by two parameters: the proportion of plasma or saliva samples in which the feature was present (i.e., presence or proportion of nonmissing values) and the median CV across the triplicate for each feature, which could reflect the reproducibility of the metabolic feature. We also examined the same characteristics of the unique metabolic features extracted from the saliva samples using the strategy shown in Figure 1B. Next, we assessed correlation among the log-transformed intensities of the overlapping features detected in both plasma and saliva samples using the Pearson correlation coefficient with the assumption of a linear relationship between saliva and plasma concentrations, as most components were transported from blood to saliva by passive diffusion and active transport.2 We examined three types of correlations: (1) the subject-specific correlation, (2) the feature-specific correlation, and (3) the grand correlation. The subject-specific correlations provided a correlation coefficient for each subject across all of the overlapping features between plasma and saliva, which reflected between-subject differences in the metabolic link between plasma and saliva. For the feature-specific correlation, we examined the correlation for each overlapping feature. The feature-specific correlation described the metabolic link of each feature between plasma and saliva without considering the individual characteristics. To generate the grand correlation, we first averaged the log-transformed intensities across plasma and saliva samples, respectively, by feature (i.e., there would be N pairs of averaged intensities, and N equals the number of overlapping features). Then, the Pearson correlation coefficient was calculated based on all pairs of the averaged intensities. The grand correlation provided a general view of the metabolic link between plasma and saliva among the study population. To assess the robustness of these correlations, we also calculated the Spearman correlation coefficient (Table S1).
We then used an untargeted MWAS workflow to identify changes in metabolic features and metabolic pathways associated with TRAP among the overlapping plasma and saliva features, along with alterations associated with the unique saliva features, respectively. LMM were used to assess associations between the metabolic feature intensity (i.e., relative concentration) and levels of TRAPs with subject-specific intercepts. The weekly average outdoor or indoor levels of the six single-species air pollutants (i.e., BC, CO, NO, NO2, NOx, and PM2.5) were used as the independent variables in LMM as
| (1) |
where Yij referred to the log-transformed intensity of a specific metabolic feature for subject i at sampling visit j. μ and θi referred to the fixed-effect intercept and the subject-specific random effect, respectively. Pollutantij referred to the weekly average outdoor or indoor level of the traffic related pollutant at the dorm location for subject i at sampling visit j. β1 referred to the coefficient for the air pollutant, indicating the change in log-transformed feature intensity per unit increase in pollution. Dorm location (i.e., “Near Dorm” or “Far Dorm”) was controlled to account for potential confounding resulting from nontraffic-pollutant-related factors that were associated with the dorm location. Age (continuous), gender (binary), body mass index (continuous), and race (categorical) were included in the LMM to adjust for the potential confounding. We also controlled for Moving Daysij, the total number of days between the date of sampling visit j and the date that subject i moved into the dorm, and Timepointij, the time point order (i.e., month number for plasma or saliva) when the sample was collected from subject i at sampling visit j.
The numbers of significant metabolic features [i, p < 0.05; ii, adjusted p < 0.05 using Benjamini-Hochberg (BH) procedure] associated with corresponding TRAPs were summarized by biospecimen and feature type. Results after BH correction were presented using Manhattan plots in the Supporting Information, which plot the retention time of each metabolic feature on the x axis against the −log 10(raw p values) for β1 from eq 1 on the y axis (Figure S1). All statistical analyses were performed in R (v.4.0.3).
Metabolic Pathway Enrichment Analysis and Metabolite Annotation.
Pathway enrichment analysis can identify biological pathways that are statistically significantly enriched by metabolites based on a known biological network, which is a key step in MWAS to help researchers obtain functional interpretation of the high-throughput data. To predict molecular mechanisms and biological functions associated with significant features, we conducted pathway enrichment on metabolic features meeting raw p < 0.05 using mummichog (v.2.0.6), a bioinformatics platform that infers and categorizes functional biological activity directly from mass spectrometry output.37 Pathway enrichment analyses were conducted separately for 72 sets of significant features from each of the linear mixed models: (six air pollutants (CO, NO, NO2, NOx, PM2.5, and BC) × two environmental settings (indoor and outdoor) × two types of chromatography column (HILIC and C18) × three categories of metabolic features (the overlapping features in saliva, the overlapping features in plasma, and the unique features in saliva). A p value for each pathway was generated by penalizing pathways with fewer significant metabolic features and assigning greater significance to pathways with more significant features using a gamma distribution.37 We classified pathways with p < 0.05 for at least two of the TRAPs models by the same chromatography column, and with at least three significant features from the experimental data matched with pathway metabolites. We presented the final results in a metabolic-pathway–TRAPs heat map, with each cell of the heat map representing a statistical association between each of the metabolic pathways and each of the corresponding indoor/outdoor TRAPs. We also generated Venn diagrams manually to visualize the metabolic pathways by biospecimens. Pathway analysis was conducted in Python (v.2.7.16). Then, we confirmed the identities of significant metabolic features by comparison of m/z, retention time, and ion dissociation patterns to authentic chemical reference standards analyzed in our lab using the identical method and instrument parameters via tandem mass spectrometry. To reduce the possibility of false positive findings, each of the annotated features was inspected for spectrum peak quality and purity by manual examination of their respective extracted ion chromatographs (EICs, the spectral plots of each metabolic signal observed at a chosen m/z value of mass spectra recorded as a function of retention time38) created via xMSanalyzer.
RESULTS
Fifty-three participants provided 167 pairs of plasma and saliva samples during four time points, spaced approximately 21–35 days apart from each other. Participants living in “Near Dorm” or “Far Dorm” had generally similar demographic characteristics according to the baseline information collected from each participant at the commencement of sampling (Table 1). The detailed indoor and outdoor TRAP concentrations are summarized in Table S2.
Table 1.
Traffic Pollutant Levels and Baseline Demographic Data of the Study Participants
| variable | overall | Near Dorm (N = 24) | Far Dorm (N = 29) |
|---|---|---|---|
| traffic pollutant levelsa | |||
| BC (μg/m3), mean (SD) | 0.91 (0.92) | 1.04 (0.63) | |
| CO (ppb), mean (SD) | 391 (101) | 246 (110) | |
| NO (ppb), mean (SD) | 22.8 (8.5) | 17.6 (8.0) | |
| NO2 (ppb), mean (SD) | 23.2 (7.8) | 23.3 (1.8) | |
| NOx (ppb), mean (SD) | 46.0 (16.1) | 40.9 (7.8) | |
| PM2.5 (μg/m3), mean (SD) | 13.0 (0.9) | 14.2 (1.7) | |
| demographic characteristics | |||
| age (SD) | 19.3 (0.8) | 19.2 (0.9) | 19.4 (0.8) |
| BMI (SD) | 23.1 (3.1) | 22.5 (3.2) | 23.6 (3.1) |
| gender, n (%) | |||
| female | 24 (45) | 12 (50) | 12 (41) |
| male | 29 (55) | 12 (50) | 17 (59) |
| race, n (%) | |||
| African American | 3 (6) | 1 (4) | 2 (7) |
| Asian | 17 (32) | 7 (29) | 10 (34) |
| Mexican | 1 (2) | 0 (0) | 1 (3) |
| White | 32 (60) | 16 (67) | 16 (56) |
| academic year, n (%) | |||
| freshman | 30 (57) | 16 (67) | 14 (48) |
| sophomore | 7 (13) | 4 (17) | 3 (10) |
| junior | 1 (2) | 1 (4) | 0 (0) |
| senior | 15 (28) | 3 (12) | 12 (41) |
| days in dorm prior to baseline, mean (SD) | 69 (119) | 87 (161) | 55 (67) |
Weekly average outdoor level at each site across the 4 weeks.
We detected 12 770 overlapping features from plasma and saliva samples in the HILIC column and 6501 in the C18 column, respectively. After feature data quality filtering, 2918 and 1539 features remained. In contrast, we obtained 6152 and 2022 metabolic features unique to saliva in the HILIC and C18 columns, respectively. As shown in Table 2, for the overlapping features detected by the HILIC column, the median of presence (i.e., the proportion of plasma or saliva samples in which a feature was present) in plasma was slightly higher than that in saliva (100.0% vs 96.1%), while no difference was observed for the C18 column (100.0% vs 100.0%). As for the unique saliva features, the median of presence was substantially lower than that of the overlapping features (Table 2). According to the median CVs, most features were comparably reproducible in either plasma or saliva. The grand correlation was 0.56 (p < 0.001) and 0.89 (p < 0.001) for the HILIC and C18 columns, respectively. The subject-specific correlations had a median of 0.64 (range: 0.15 to 0.97) and 0.91 (range: 0.40 to 0.99) for the HILIC and C18 columns, respectively (Figure 2A). All of these subject-specific Pearson correlation coefficients were significantly different from 0 after multiple comparison corrections using the Benjamini–Hochberg (BH) procedure. On the other hand, the feature-specific correlations were primarily weak (Figure 2B), indicating substantial between-subject variability in the formation of saliva. We observed a similar pattern of these correlation estimates using Spearman correlation in the sensitivity analysis.
Table 2.
Distributions of the Presence and the Median Coefficient of Variance (CV) for Features among the Corresponding Samples
| overlapping | ||||
|---|---|---|---|---|
| propertya | plasma | saliva | unique saliva | |
| HILIC column | presenceb | 100.0 (92.8, 100.0) | 96.1 (72.5, 100.0) | 32.9 (21.6, 57.5) |
| Median CVc | 13.0 (7.4, 20.7) | 11.0 (5.6, 18.9) | 16.2 (10.5, 22.4) | |
| C18 column | presenceb | 100.0 (96.1, 100.0) | 100.0 (87.1, 100.0) | 42.3 (23.2, 88.7) |
| Median CVc | 14.5 (8.7, 21.3) | 10.3 (5.7, 17.5) | 17.1 (11.5, 23.1) | |
All results were displayed as median (first quartile, third quartile) due to the skewed distribution of feature properties.
The proportion of plasma or saliva samples in which a specific feature was present.
The median of the coefficient of variance for each feature across the triplicates from one biological sample.
Figure 2.
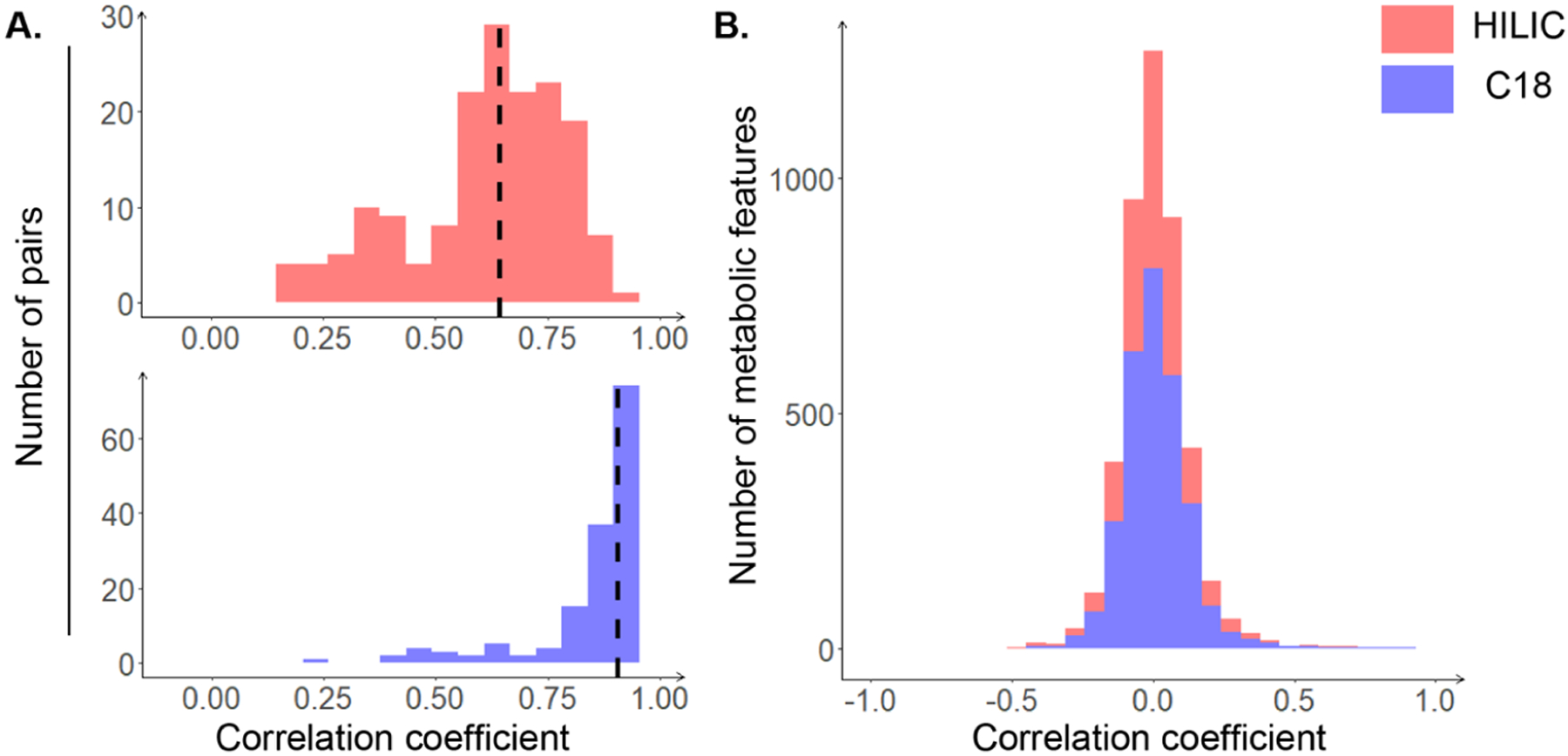
Histogram of Pearson correlation coefficients. (A) Subject-specific correlations: correlations for each pair of plasma and saliva samples across the metabolic features present in both biospecimens. The median correlation coefficient is marked as the dashed line. (B) Feature-specific correlations: correlations for each metabolic feature across all of the participants. The results of HILIC and C18 columns are colored in pink and purple, respectively.
We conducted and analyzed 72 sets of MWAS models (12 indoor/outdoor TRAPs among metabolic features present in both biospecimens and among features unique to saliva, with each analyzed using two chromatography columns). In general, for the overlapping features, we detected more features significantly associated with TRAPs in plasma than saliva; there were more significant features associated with TRAPs among the unique features of saliva compared to the number of significant features among the overlapping features of saliva (Table 3).
Table 3.
Number of Significant Metabolic Features [p < 0.05 (FDRB–H < 0.05 in Parentheses)] for Each Set of Metabolomics-Wide Association Study (MWAS) Models by the HILIC and C18 Columnsa
| BC | CO | NO2 | NO | NOx | PM2.5 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| column | biomatrixb | in | out | in | out | in | out | in | out | in | out | in | out |
| HILIC | plasma | 549 (7)c | 517 (32) | 761 (257) | 803 (314) | 529 (223) | 619 (250) | 409 (0) | 308 (0) | 538 (85) | 505 (0) | 515 (204) | 444 (163) |
| saliva | 128 (0) | 83 (0) | 133 (0) | 130 (0) | 123 (0) | 130 (0) | 126 (0) | 213 (0) | 181 (0) | 185 (0) | 145 (0) | 124 (0) | |
| saliva (unique) | 341 (4) | 299 (2) | 391 (0) | 390 (1) | 410 (6) | 411 (6) | 365 (1) | 393 (2) | 426 (2) | 425 (4) | 416 (4) | 367 (3) | |
| C18 | plasma | 151 (2) | 122 (0) | 253 (21) | 277 (24) | 160 (12) | 202 (15) | 128 (0) | 118 (1) | 141 (14) | 128 (13) | 159 (14) | 123 (12) |
| saliva | 60 (0) | 54 (0) | 73 (0) | 71 (0) | 67 (0) | 77 (0) | 65 (0) | 68 (0) | 83 (0) | 85 (0) | 79 (0) | 49 (0) | |
| saliva (unique) | 102 (1) | 92 (3) | 116 (0) | 118 (3) | 111 (1) | 113 (1) | 113 (1) | 118 (1) | 119 (0) | 123 (1) | 113 (0) | 102 (2) | |
Acronym: BC, black carbon; CO, carbon monoxide; NO2, nitrogen dioxide; NO, nitric oxide; NOx, nitrogen oxide; PM2.5, fine particulate matter; in, indoor; out, outdoor.
The metabolic features were classified into three groups: the overlapping features in plasma, the overlapping features in saliva, and the unique features in saliva.
The number of significant metabolic features after multiple comparison corrections is shown in parentheses.
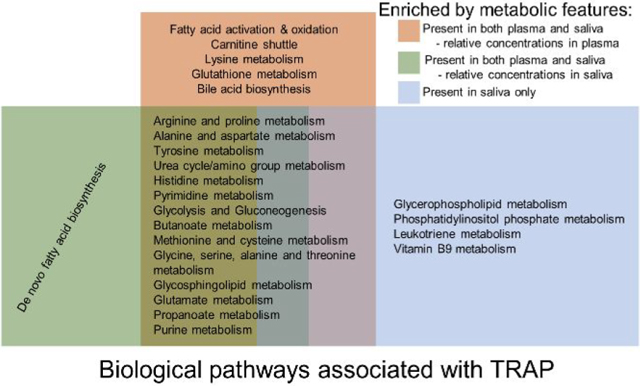
Using mummichog, we further investigated which biological pathways were enriched by the significant metabolic features associated with at least two TRAPs and compared pathways identified in either plasma or saliva. Twenty-six biological pathways were associated with at least two TRAPs based on the overlapping features detected from plasma, including biological pathways primarily involved in fatty acid metabolism and amino acid metabolism, such as fatty acid activation and oxidation, arginine and proline metabolism, and alanine and aspartate metabolism, while 16 were based on the overlapping features detected from saliva, such as pyrimidine metabolism, glycolysis and gluconeogenesis, and butanoate metabolism (Figures 3 and 4). All detected biological pathways were closely related to fatty acid metabolism and amino acid metabolism. Fifteen perturbed biological pathways were shared by saliva and plasma, which mainly belonged to amino acid metabolism (Figures 3 and 4).
Figure 3.
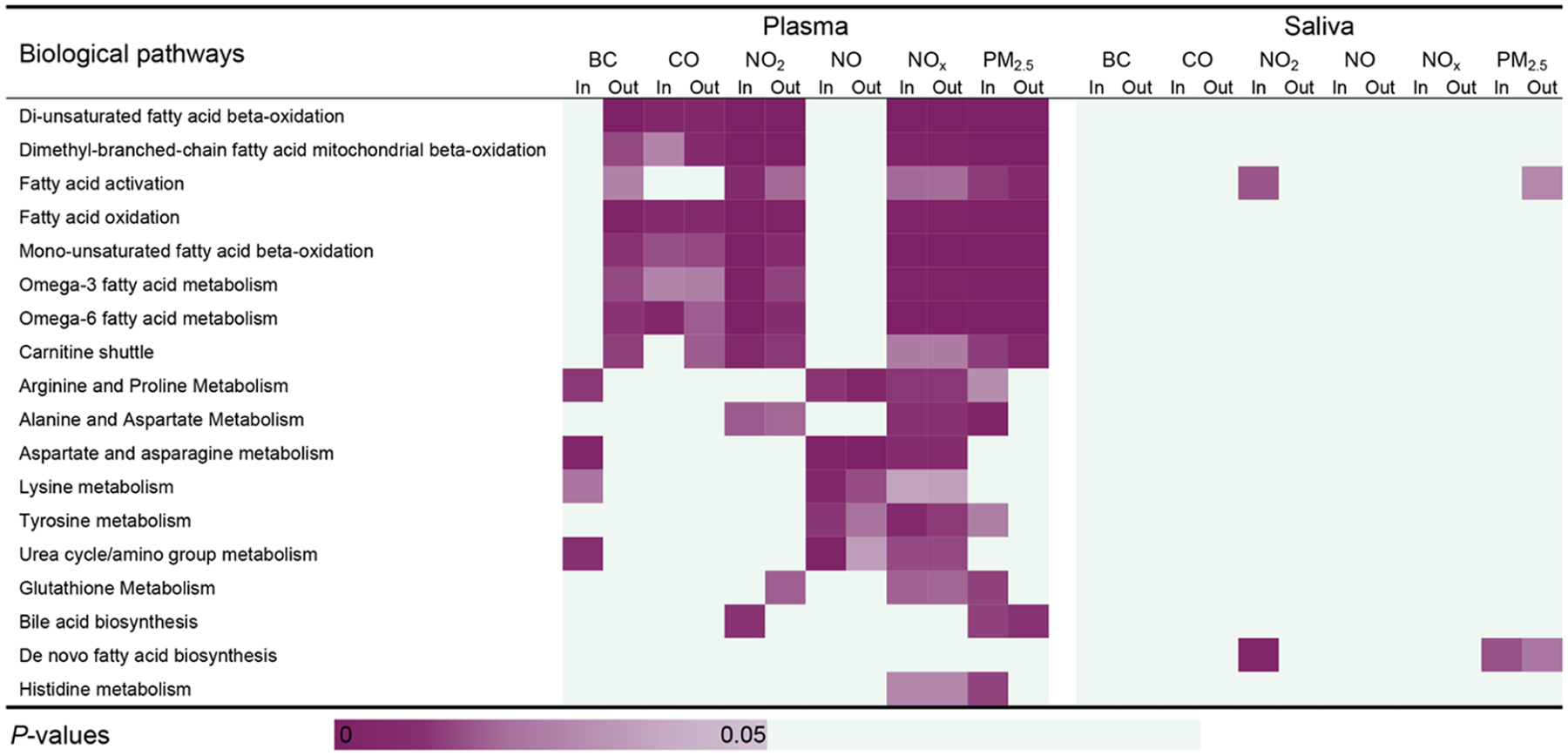
Metabolic pathways enriched by the metabolic features present in both plasma and saliva by the HILIC column. The cells are shaded according to strength of association (i.e., p value) between each of the metabolic pathways and significant features that were associated with each indoor/outdoor single traffic pollutant indicator. Pathways are ordered according to the total number of the significant pathway-traffic pollutant pairs (p < 0.05) in the HILIC column. Each pathway is enriched by at least three significant annotated overlapping features by either plasma or saliva.
Figure 4.
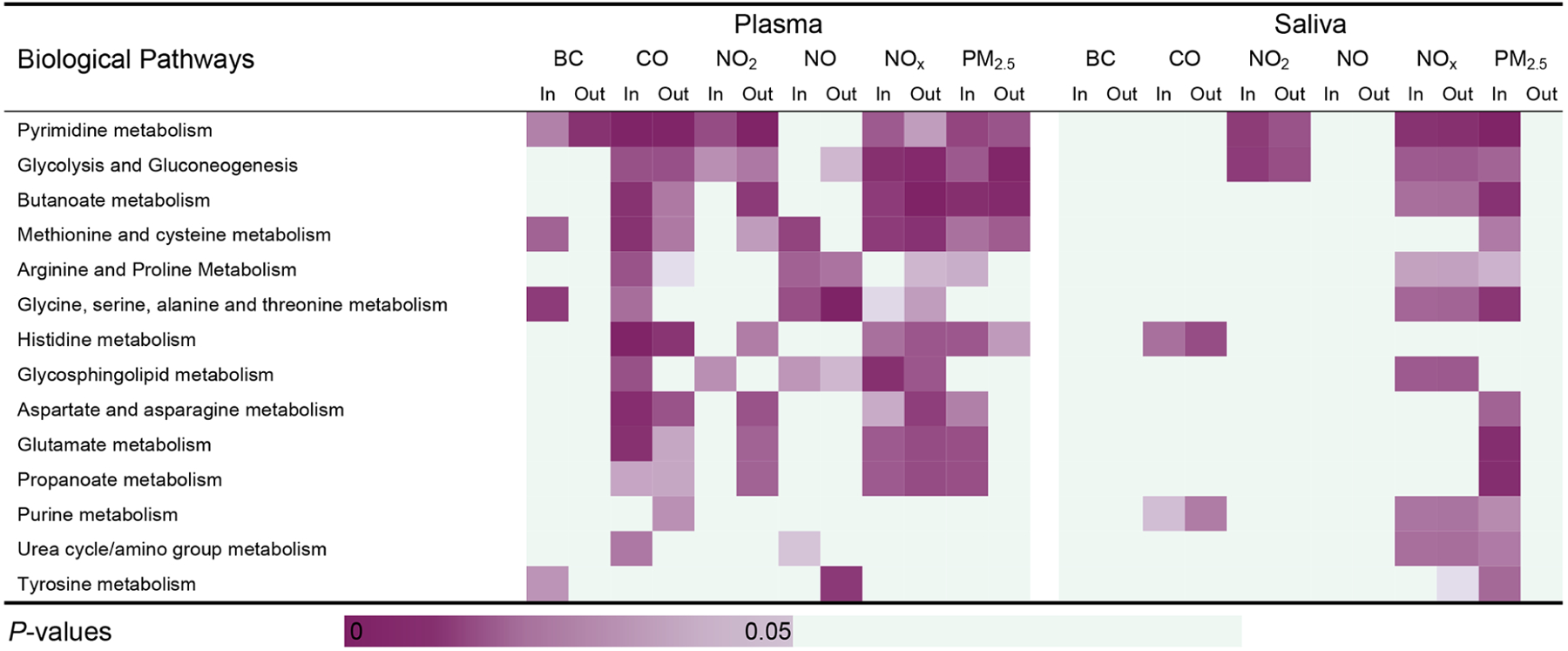
Metabolic pathways enriched by the metabolic features present in both plasma and saliva by the C18 column. The cells are shaded according to strength of association (i.e., p value) between each of the metabolic pathways and significant features that were associated with each indoor/outdoor single traffic pollutant indicator. Pathways are ordered according to the total number of the significant pathway-traffic pollutant pairs (p < 0.05) in the C18 column. Each pathway is enriched by at least three significant annotated overlapping features by either plasma or saliva.
We also conducted a pathway analysis on the metabolic features unique to saliva (Figure 5). In total, 16 pathways were detected, among which seven were also found in the overlapping features of saliva, including pyrimidine metabolism; purine metabolism; glycine, serine, alanine, and threonine metabolism; aspartate and asparagine metabolism; glutamate metabolism; tyrosine metabolism; and urea cycle/amino group metabolism, while 10 of them were found in the overlapping features of plasma including several biological pathways involved in lipid metabolism. In other words, more than the half of the biological pathways enriched by significant metabolic features unique to saliva were not able to be detected via the overlapping features in saliva. Six biological pathways were uniquely detected by the metabolic features unique to saliva compared to the overlapping features in saliva and plasma. In total, we found 25 biological pathways significantly associated at least two TRAPs via saliva, to which the overlapping and unique metabolic features contributed equally. Eighteen out of 26 (~69%) biological pathways in plasma associated with air pollutants were also found among the 25 pathways in saliva when considering the overlapping and unique features together.
Figure 5.
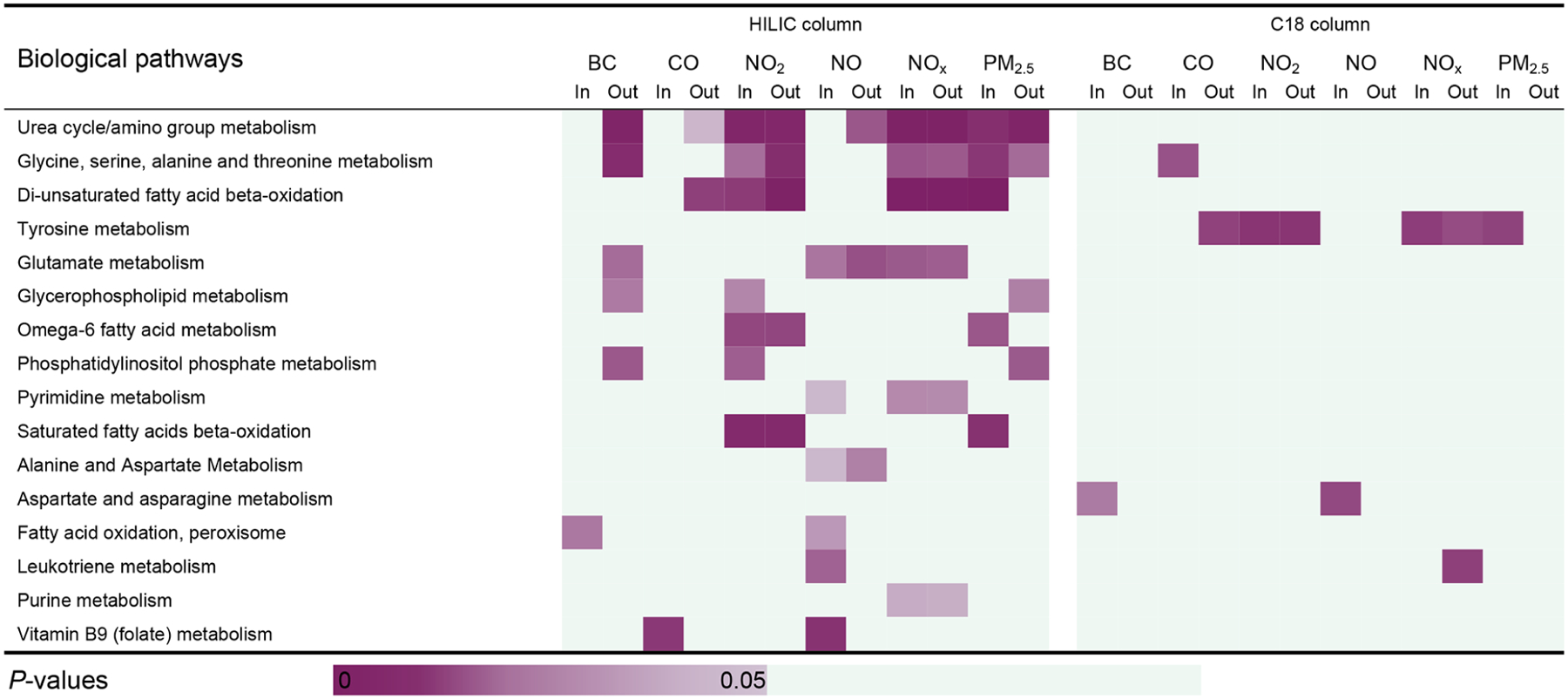
Metabolic pathways enriched by the metabolic features unique to saliva. The cells are shaded according to strength of association (i.e., p value) between each of metabolic pathways and significant features that were associated with each indoor/outdoor single traffic pollutant indicator. Pathways are ordered according to the total number of the significant pathway-traffic pollutant pairs (p < 0.05) in both columns. Each pathway is enriched by at least 3 significant annotated unique features.
We further matched the significant metabolic features (FDRB–H < 0.05) to a list of in-house authentic reference standards based on m/z using a tolerance of 10 ppm. We confirmed the chemical identities of one metabolic feature, cytosine, which negatively associated with indoor and outdoor NO2 and indoor PM2.5 concentrations (Figure S2).
DISCUSSION
We compared metabolomic profiles concurrently expressed in saliva and blood in order to assess the feasibility of using saliva as an alternative, less invasive, and easier to collect TRAP biospecimen. Broadly, we observed fewer associations between saliva and TRAP as compared to plasma. No overlapping features in saliva were significantly associated with any TRAPs after multiple comparison correction, while hundreds of significant features were found in plasma. For instance, 204 and 163 overlapping features in plasma were associated with the indoor and outdoor exposure to PM2.5, respectively.
Regarding comparative plasma and saliva, there were several key findings from this analysis. First, the overlapping features were more prevalent among the biospecimens from all participants than the unique features of saliva according to the presence, indicating a highly various composition of unique molecules in saliva. The resulting high presence of overlapping metabolic features in both plasma and saliva had slightly lower median feature-specific CVs (HILIC, 11.0%; C18, 10.3%) than plasma (HILIC, 13.0%; C18, 14.5%), suggesting that features detected in the saliva metabolome might be more reproducible than those found in the blood metabolome. Third, in the current study, the ratios between the numbers of the overlapping and unique features of saliva were about 1:2 and 3:4 in the HILIC and C18 columns, respectively. In a previous proteome study conducted by Yan et al., about 27% (i.e., 1:3) of the whole-saliva proteins were found in plasma.39,40 The ratios of our study were both higher than that reported by Yan et al., potentially due to the relatively high feature coverage of HRM.
A number of unique features of saliva were found associated with TRAPs. From the pathway analyses, the biological pathways enriched by both the overlapping features of plasma and saliva belonged primarily to amino acid metabolism, which suggested that active protein metabolism took place in both plasma and saliva. The biological pathways enriched by the overlapping features of plasma covered the majority of the pathways enriched by those of saliva. In other words, the overlapping features of saliva did not provide additional information on the biological responses of the human body to TRAP. On the other hand, the unique features of saliva were enriched in six biological pathways that were not detected through the overlapping saliva and plasma features, including glycerophospholipid metabolism, phosphatidylinositol phosphate metabolism, saturated fatty acids beta-oxidation, fatty acid oxidation in peroxisome, leukotriene metabolism, and folate metabolism. This finding indicated that the saliva metabolome could provide additional information on internal exposures and health responses associated with TRAP exposures compared to the blood metabolome, though further validation is warranted. Of specific note was the enrichment in saliva of leukotriene metabolism, a biological pathway actively involved in asthmatic and allergic reactions and prolonged inflammatory reactions,41 which was consistently associated with multiple indoor and outdoor TRAPs for both plasma and saliva in our previous analysis and in other independent studies.16–18,42,43 When considering the overlapping and unique features as a whole, saliva was capable of detecting 69% of biological pathways enriched by the overlapping features in plasma.
We also compared the metabolic features in saliva and plasma in terms of their intensity consistency. The feature-specific correlation between saliva and plasma varied substantially, with correlation strength ranging from weak to strong, while the strengths of the subject-specific correlations across all features were moderate to strong. The results were broadly consistent with those reported previously.4,39,44–47 Williamson et al. conducted a comparison of levels of 27 cytokines between plasma and saliva among 50 healthy college students, and the relationships varied by cytokine with the highest correlation strength of 0.34.4 The evidence on the significance of the correlation between interlukin-6 (IL-6) levels in plasma with those in saliva remained inconsistent,44,45 and no correlation has been reported for the levels of C-reactive protein and soluble IL-6 receptor.44,46 Poll et al. found that the concentration of salivary cortisol was strongly correlated with total serum cortisol,47 and Loo et al. revealed that the abundances of the immunoglobulins present in saliva and plasma were highly correlated.39,40 In sum, the feature-specific correlation between saliva and plasma could vary by different chemicals and metabolites, partly due to the variation in oral microbiota.48 Conversely, the high subject-specific correlations we observed in our study provided an initial indication of the potential of using the saliva metabolome as a surrogate for plasma. Although subject-specific correlations had a slightly wider range in both the HILIC and C18 columns, which indicated the interindividual heterogeneity in pharmacokinetics, most subjects had a moderate to high correlation in the overlapping features between saliva and plasma, suggesting that those common features present in saliva might be capable of serving as surrogates to their counterparts in plasma. The finding reaffirmed the validity of using saliva for disease diagnosis and biomonitoring and the feasibility of saliva metabolome as an alternative to blood.
There are some limitations to this study. First, given the nature of the panel study, the sample size of the participants was relatively small, and the findings may be unduly influenced by individual participants. To address this, we conducted repeated biomonitoring over four time points for each participant, which reduces impacts associated with intra-individual variation to some degree and enhances the statistical power. Second, we employed outdoor and indoor measures as surrogates for individual exposures and did not incorporate the individual daily mobility into exposure assessment, which may have led to the introduction of measurement error that might either under- or overestimate true TRAP exposures. The effect estimates of air pollution should be viewed cautiously with this caveat. Despite the potential measurement error, continuous GPS data collected from part of the participants (N = 43) over the course of 11 sampling weeks indicated a clear pattern that participants spent a majority of their time inside or near their respective dorms.29,49 In addition to the stationary monitoring used in the current analysis, 43 participants conducted personal exposure measurements, and the personal exposure levels of specific air pollutants (e.g., black carbon and nitrogen dioxide) were found to be moderately and significantly associated with measurements both outside and inside of the dorms.29,49 Thus, the error due to daily mobility might not be substantial. In addition, MWAS using the untargeted technique is at a relatively higher risk of false discovery compared to conventional targeted analysis. To reduce the potential high risk of false discovery in untargeted metabolomics, we employed a suite of approaches in both the experimental and the statistical analyses phases, including adding pooled human plasma and internal standard samples, correcting batch effect, using distinct strategies to detect the overlapping and unique features, adjusting for potential confounders, conducting multiple testing corrections via the BH procedure, and reporting biological pathways only if they are enriched with more than two significant features and associated with at least two air pollutant measures. Despite all of these approaches used, the biological interpretation of the untargeted metabolomics still hinged on the ability to accurately identify metabolites. Fourth, due to the uncertainties in metabolite annotation and arbitrary classification of biological pathways, mummichog is inherent to increased risk of false-positive discoveries. Thus, the pathway enrichment results need to be interpreted with caution, and further studies are warranted to validate these findings. Finally, we used the R packages apLCMS coupled with xMSanalyzer to preprocess raw metabolomics profiles, which used a model-based tolerance level search and kernel density-based iterative splitting to avoid forcing hard cutoffs of m/z for feature detection.34 In this process, the m/z of the resulting metabolic features was learned from the input profiles to ensure high sensitivity. As a result, the feature tables extracted from the metabolomic profiles in saliva and plasma, respectively, might not be directly comparable due to the slight difference in m/z. We had to use two extraction strategies in the current analysis to detect the unique and overlapping features.
In spite of these limitations, we believe our study had several notable strengths. First, the study participants were recruited from two student dormitories that had significantly different levels of exposures to TRAP. Although the enrollment was not random, the demographic characteristics of the participants from both dorms were similar. Second, all of the biological samples were collected in a fasting state during the same time of the day, which minimized the impact of acute dietary intake on oral microbiota and metabolic profiles.48,50 Third, we used the plasma metabolome as the reference for the saliva metabolome, which allowed us to evaluate the diagnostic power of the saliva metabolome in response to TRAP exposures. To identify the overlapping features, we performed the extraction process simultaneously on saliva and plasma metabolomic profiles, which guaranteed that resulting metabolic features were matched based on m/z and retention time stringently across all metabolomic profiles. This strategy was less jeopardized by false matching compared to matching the metabolic features after the extraction process. We calculated three types of intensity correlation for the overlapping features, and different correlations captured distinct aspects of the variation in the metabolic link between plasma and saliva. Moreover, we separated the overlapping and unique features in saliva, which allowed us to target the sources of alterations in metabolism. In other words, the changes in unique features were more sensitive to the exposure compared to the overlapping features in saliva. In addition, we matched the significant metabolic features (FDRB–H < 0.05) to a list of in-house authentic reference standards and confirmed the chemical identity of cytosine with level 1 confidence.51
Our study presents findings from among the first environmental exposure analyses examining metabolomic concordance in both salivary and plasma samples collected from a panel of healthy adults. We specifically examined whether saliva has the potential to serve as an alternative matrix to plasma for this application. Although the quality of features and the correlation of abundance were comparable between saliva and plasma, saliva was less sensitive than plasma for the detection of metabolic perturbations associated with TRAP exposures as defined by metabolic features present in both biospecimens. However, the metabolic pathways detected by saliva were comparable to those detected by plasma. Due to its easy, noninvasive, and low-cost collection, saliva may be more suitable than blood for use in large-scale environmental metabolomics applications. Future large-scale studies are needed to validate the feasibility of saliva metabolomics to identify the alterations in biological pathways associated with the exposure to complex environmental pollutants like TRAP.
Supplementary Material
ACKNOWLEDGMENTS
Support for this project was provided through a contract with the Health Effects Institute (RFA #4942-RFA13-1/14-3). We acknowledge C. Cornwell, K. Parada, S. Shim, K. Johnson, and E. Yang for their assistances in conducting the field study. We want to thank Dr. R. Weber, Dr. V. Verma, and Ms. D. Gao for their measurements of oxidative potential of fine particulate matter via DTT assay. The study used the instrumentation assembled for field studies conducted as part of the Southeastern Center for Air Pollution and Epidemiology (SCAPE), which was funded by U.S. Environmental Protection Agency STAR grant R834799. The information in this document may not necessarily reflect the views of the Agency, and no official endorsement should be inferred. We also acknowledge the tremendous support from the HERCULES exposome research center, supported by the National Institute of Environmental Health Sciences of the National Institutes of Health (P30ES019776). The study is also supported by the National Institute of Health (NIH) research grant (R21ES032117). We would like to thank Ms. V. Tran for conducting metabolomics profiling on biosamples. Specific thanks go to the administrators at Georgia Tech for the permission of conducting this study on campus and in their residence hall facilities.
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.est.2c00064.
Spearman correlation coefficients among the log-transformed intensities of the overlapping features detected in both plasma and saliva samples (Table S1); mean indoor and outdoor levels of traffic-related air pollution at Near Dorm and Far Dorm during the study period (Table S2); Manhattan plots of associations between changes in log-transformed intensities of the overlapping features with traffic-related air pollutants using the HILIC column with positive ion mode (Figure S1a); Manhattan plots of associations between changes in log-transformed intensities of the overlapping features with traffic-related air pollutants using the C18 column with negative ion mode (Figure S1b); Manhattan plots of associations between changes in log-transformed intensities of the unique features detected in saliva samples with traffic-related air pollutants (Figure S1c); number of overlapping metabolic features that were both significantly (raw p < 0.05) associated with traffic-related air pollutants in plasma and saliva (Table S3); chemical identity of the metabolic feature that was unique to saliva and significantly associated with TRAPs (FDRB–H < 0.05) in the DRIVE study (Figure S2); the distribution of feature intensities in each pooled quality control sample for all batches running for plasma and saliva samples (Figure S3) (PDF)
Complete contact information is available at: https://pubs.acs.org/10.1021/acs.est.2c00064
The authors declare no competing financial interest.
Contributor Information
Zhenjiang Li, Gangarosa Department of Environmental Health, Rollins School of Public Health, Emory University, Atlanta, Georgia 30322, United States.
Jeremy A. Sarnat, Gangarosa Department of Environmental Health, Rollins School of Public Health, Emory University, Atlanta, Georgia 30322, United States
Ken H. Liu, Clinical Biomarkers Laboratory, Department of Medicine, School of Medicine, Emory University, Atlanta, Georgia 30322, United States
Robert B. Hood, Department of Epidemiology, Rollins School of Public Health, Emory University, Atlanta, Georgia 30322, United States
Che-Jung Chang, Gangarosa Department of Environmental Health, Rollins School of Public Health, Emory University, Atlanta, Georgia 30322, United States.
Xin Hu, Clinical Biomarkers Laboratory, Department of Medicine, School of Medicine, Emory University, Atlanta, Georgia 30322, United States.
ViLinh Tran, Clinical Biomarkers Laboratory, Department of Medicine, School of Medicine, Emory University, Atlanta, Georgia 30322, United States.
Roby Greenwald, Department of Population Health Sciences, School of Public Health, Georgia State University, Atlanta, Georgia 30302, United States.
Howard H. Chang, Gangarosa Department of Environmental Health, Rollins School of Public Health, Emory University, Atlanta, Georgia 30322, United States; Department of Biostatistics and Bioinformatics, Rollins School of Public Health, Emory University, Atlanta, Georgia 30322, United States.
Armistead Russell, School of Civil and Environmental Engineering, Georgia Institute of Technology, Atlanta, Georgia 30332, United States.
Tianwei Yu, School of Data Science, The Chinese University of Hong Kong, Shenzhen, Guangdong 518172, China.
Dean P. Jones, Clinical Biomarkers Laboratory, Department of Medicine, School of Medicine, Emory University, Atlanta, Georgia 30322, United States
Donghai Liang, Gangarosa Department of Environmental Health, Rollins School of Public Health, Emory University, Atlanta, Georgia 30322, United States.
REFERENCES
- (1).Azevedo LR; De Lima AAS; Machado MAN; Gregio AMT; de Almeida PDV Saliva composition and functions: a comprehensive review. J. Contemp Dent Pract 2008, 9 (3), 72–80. [PubMed] [Google Scholar]
- (2).Kaufman E; Lamster IB The diagnostic applications of saliva–a review. Crit Rev. Oral Biol. Med 2002, 13 (2), 197–212. [DOI] [PubMed] [Google Scholar]
- (3).Pfaffe T; Cooper-White J; Beyerlein P; Kostner K; Punyadeera C Diagnostic potential of saliva: current state and future applications. Clin Chem. 2011, 57 (5), 675–687. [DOI] [PubMed] [Google Scholar]
- (4).Williamson S; Munro C; Pickler R; Grap MJ; Elswick RK Jr. Comparison of biomarkers in blood and saliva in healthy adults. Nurs Res. Pract 2012, 2012, 246178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Abraham JE; Maranian MJ; Spiteri I; Russell R; Ingle S; Luccarini C; Earl HM; Pharoah PP; Dunning AM; Caldas CJ Saliva samples are a viable alternative to blood samples as a source of DNA for high throughput genotyping. Bmg 2012, 5 (1), 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Li Y; St. John MA; Zhou X; Kim Y; Sinha U; Jordan RC; Eisele D; Abemayor E; Elashoff D; Park NH; Wong DT Salivary transcriptome diagnostics for oral cancer detection. Clin. Cancer Res 2004, 10 (24), 8442–8450. [DOI] [PubMed] [Google Scholar]
- (7).Rushing A; Sommer EC; Zhao S; Po’e EK; Barkin SLJ Salivary epigenetic biomarkers as predictors of emerging childhood obesity. Bmg 2020, 21 (1), 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Zhang A; Sun H; Wang X Saliva metabolomics opens door to biomarker discovery, disease diagnosis, and treatment. Appl. Biochem. Biotechnol 2012, 168 (6), 1718–1727. [DOI] [PubMed] [Google Scholar]
- (9).Cuevas-Cordoba B; Santiago-Garcia J Saliva: a fluid of study for OMICS. OMICS. 2014, 18 (2), 87–97. [DOI] [PubMed] [Google Scholar]
- (10).Traffic-related air pollution: a critical review of the literature on emissions, exposure, and health effects. In A Special Report of the Institute’s Panel on the Health Effects of Traffic-Related Air Pollution; Health Effects Institute, 2010. [Google Scholar]
- (11).Fox M; Koehler K; Johnson N Established and emerging effects of traffic-related air pollution. In. Traffic-Related Air Pollution; Elsevier, 2020; pp 207–228. [Google Scholar]
- (12).Miller MR; Raftis JB Evidence from toxicological and mechanistic studies. In: Traffic-Related Air Pollution; Elsevier, 2020; pp 229–279. [Google Scholar]
- (13).Beevers SD; Williams ML Traffic-related air pollution and exposure assessment. In Traffic-Related Air Pollution; Elsevier, 2020; pp 137–162. [Google Scholar]
- (14).Jones DP; Park Y; Ziegler TR Nutritional metabolomics: progress in addressing complexity in diet and health. Annu. Rev. Nutr 2012, 32, 183–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Li Z; Liang D; Ye D; Chang HH; Ziegler TR; Jones DP; Ebelt ST Application of high-resolution metabolomics to identify biological pathways perturbed by traffic-related air pollution. Environ. Res 2021, 193, 110506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Liang D; Ladva CN; Golan R; Yu T; Walker DI; Sarnat SE; Greenwald R; Uppal K; Tran V; Jones DP; Russell AG; Sarnat JA Perturbations of the arginine metabolome following exposures to traffic-related air pollution in a panel of commuters with and without asthma. Environ. Int 2019, 127, 503–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Liang D; Moutinho JL; Golan R; Yu T; Ladva CN; Niedzwiecki M; Walker DI; Sarnat SE; Chang HH; Greenwald R; Jones DP; Russell AG; Sarnat JA Use of high-resolution metabolomics for the identification of metabolic signals associated with traffic-related air pollution. Environment international. 2018, 120, 145–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Jeong A; Fiorito G; Keski-Rahkonen P; Imboden M; Kiss A; Robinot N; Gmuender H; Vlaanderen J; Vermeulen R; Kyrtopoulos S; Herceg Z; Ghantous A; Lovison G; Galassi C; Ranzi A; Krogh V; Grioni S; Agnoli C; Sacerdote C; Mostafavi N; Naccarati A; Scalbert A; Vineis P; Probst-Hensch N Perturbation of metabolic pathways mediates the association of air pollutants with asthma and cardiovascular diseases. Environ. Int 2018, 119, 334–345. [DOI] [PubMed] [Google Scholar]
- (19).van Veldhoven K; Kiss A; Keski-Rahkonen P; Robinot N; Scalbert A; Cullinan P; Chung KF; Collins P; Sinharay R; Barratt BM; Nieuwenhuijsen M; Rodoreda AA; Carrasco-Turigas G; Vlaanderen J; Vermeulen R; Portengen L; Kyrtopoulos SA; Ponzi E; Chadeau-Hyam M; Vineis P Impact of short-term traffic-related air pollution on the metabolome - Results from two metabolome-wide experimental studies. Environ. Int 2019, 123, 124–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Walker DI; Lane KJ; Liu K; Uppal K; Patton AP; Durant JL; Jones DP; Brugge D; Pennell KD Metabolomic assessment of exposure to near-highway ultrafine particles. J. Expo Sci. Environ. Epidemiol 2019, 29 (4), 469–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Xia Y; Niu Y; Cai J; Lin Z; Liu C; Li H; Chen C; Song W; Zhao Z; Chen R; Kan H Effects of Personal Short-Term Exposure to Ambient Ozone on Blood Pressure and Vascular Endothelial Function: A Mechanistic Study Based on DNA Methylation and Metabolomics. Environ. Sci. Technol 2018, 52 (21), 12774–12782. [DOI] [PubMed] [Google Scholar]
- (22).Sampson JN; Boca SM; Shu XO; Stolzenberg-Solomon RZ; Matthews CE; Hsing AW; Tan YT; Ji BT; Chow WH; Cai Q; Liu DK; Yang G; Xiang YB; Zheng W; Sinha R; Cross AJ; Moore SC Metabolomics in epidemiology: sources of variability in metabolite measurements and implications. Cancer Epidemiol Biomarkers Prev. 2013, 22 (4), 631–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Rappaport SM Implications of the exposome for exposure science. J. Expo Sci. Environ. Epidemiol 2011, 21 (1), 5–9. [DOI] [PubMed] [Google Scholar]
- (24).Ladva CN; Golan R; Greenwald R; Yu T; Sarnat SE; Flanders WD; Uppal K; Walker DI; Tran V; Liang D; Jones DP; Sarnat JA Metabolomic profiles of plasma, exhaled breath condensate, and saliva are correlated with potential for air toxics detection. J. Breath Res 2018, 12 (1), 016008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Singh U; Devaraj S; Jialal I Vitamin E, oxidative stress, and inflammation. Annu. Rev. Nutr 2005, 25, 151–174. [DOI] [PubMed] [Google Scholar]
- (26).Henderson WR Jr. The role of leukotrienes in inflammation. Ann. Int. Med 1994, 121 (9), 684–697. [DOI] [PubMed] [Google Scholar]
- (27).Gonzalez FJ Role of cytochromes P450 in chemical toxicity and oxidative stress: studies with CYP2E1. Mutat. Res 2005, 569 (1–2), 101–110. [DOI] [PubMed] [Google Scholar]
- (28).Gałecki A; Burzykowski T Linear mixed-effects model. In Linear Mixed-Effects Models Using R.; Springer, 2013; pp 245–273. [Google Scholar]
- (29).Sarnat JA; Russell A; Liang D; Moutinho JL; Golan R; Weber RJ; Gao D; Sarnat SE; Chang HH; Greenwald R; Yu T Developing Multipollutant Exposure Indicators of Traffic Pollution: The Dorm Room Inhalation to Vehicle Emissions (DRIVE) Study. Res. Rep. Health Eff Inst 2018, 196, 3–75. [PMC free article] [PubMed] [Google Scholar]
- (30).Moutinho JL; Liang DH; Golan R; Ebelt ST; Weber R; Sarnat JA; Russell AG Evaluating a multipollutant metric for use in characterizing traffic-related air pollution exposures within near-road environments. Environmental Research 2020, 184, 109389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Moutinho JL; Liang D; Golan R; Sarnat SE; Weber R; Sarnat JA; Russell AG Near-road Vehicle Emissions Air Quality Monitoring for Exposure Modeling. Atmos. Environ 2020, 224, 117318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Go YM; Walker DI; Liang Y; Uppal K; Soltow QA; Tran V; Strobel F; Quyyumi AA; Ziegler TR; Pennell KD; Miller GW; Jones DP Reference Standardization for Mass Spectrometry and High-resolution Metabolomics Applications to Exposome Research. Toxicol. Sci 2015, 148 (2), 531–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Simon-Manso Y; Lowenthal MS; Kilpatrick LE; Sampson ML; Telu KH; Rudnick PA; Mallard WG; Bearden DW; Schock TB; Tchekhovskoi DV; Blonder N; Yan X; Liang Y; Zheng Y; Wallace WE; Neta P; Phinney KW; Remaley AT; Stein SE Metabolite profiling of a NIST Standard Reference Material for human plasma (SRM 1950): GC-MS, LC-MS, NMR and clinical laboratory analyses, libraries, and web-based resources. Anal. Chem 2013, 85 (24), 11725–11731. [DOI] [PubMed] [Google Scholar]
- (34).Yu T; Park Y; Johnson JM; Jones DP apLCMS–adaptive processing of high-resolution LC/MS data. Bioinformatics. 2009, 25 (15), 1930–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Uppal K; Soltow QA; Strobel FH; Pittard WS; Gernert KM; Yu T; Jones DP xMSanalyzer: automated pipeline for improved feature detection and downstream analysis of large-scale, non-targeted metabolomics data. BMC Bioinformatics. 2013, 14, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Li Y; Li L Retention time shift analysis and correction in chemical isotope labeling liquid chromatography/mass spectrometry for metabolome analysis. Rapid Commun. Mass Spectrom 2020, 34, No. e8643. [DOI] [PubMed] [Google Scholar]
- (37).Li S; Park Y; Duraisingham S; Strobel FH; Khan N; Soltow QA; Jones DP; Pulendran B Predicting network activity from high throughput metabolomics. PLoS Comput. Biol 2013, 9 (7), No. e1003123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Smoluch M; Piechura K Basic Definitions. In Smoluch M, Grasso G, Suder P, Silberring J, Eds.; Mass Spectrometry: An Applied Approach; Wiley, 2019; pp 9–12. [Google Scholar]
- (39).Loo JA; Yan W; Ramachandran P; Wong DT Comparative human salivary and plasma proteomes. J. Dent Res 2010, 89 (10), 1016–023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Yan W; Apweiler R; Balgley BM; Boontheung P; Bundy JL; Cargile BJ; Cole S; Fang X; Gonzalez-Begne M; Griffin TJ; Hagen F; Hu S; Wolinsky LE; Lee CS; Malamud D; Melvin JE; Menon R; Mueller M; Qiao R; Rhodus NL; Sevinsky JR; States D; Stephenson JL; Than S; Yates JR; Yu W; Xie H; Xie Y; Omenn GS; Loo JA; Wong DT Systematic comparison of the human saliva and plasma proteomes. Proteomics Clin Appl. 2009, 3 (1), 116–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Salmon JA; Higgs GAJ Prostaglandins and leukotrienes as inflammatory mediators. Bmb. 1987, 43 (2), 285–296. [DOI] [PubMed] [Google Scholar]
- (42).Yan Q; Liew Z; Uppal K; Cui X; Ling C; Heck JE; von Ehrenstein OS; Wu J; Walker DI; Jones DP; Ritz B Maternal serum metabolome and traffic-related air pollution exposure in pregnancy. Environ. Int 2019, 130, 104872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Ladva CN; Golan R; Liang D; Greenwald R; Walker DI; Uppal K; Raysoni AU; Tran V; Yu T; Flanders WD; Miller GW; Jones DP; Sarnat JA Particulate metal exposures induce plasma metabolome changes in a commuter panel study. PLoS One. 2018, 13 (9), No. e0203468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Fernandez-Botran R; Miller JJ; Burns VE; Newton TL Correlations among inflammatory markers in plasma, saliva and oral mucosal transudate in post-menopausal women with past intimate partner violence. Brain Behav Immun. 2011, 25 (2), 314–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Minetto M; Rainoldi A; Gazzoni M; Terzolo M; Borrione P; Termine A; Saba L; Dovio A; Angeli A; Paccotti P Differential responses of serum and salivary interleukin-6 to acute strenuous exercise. Eur. J. Appl. Physiol 2005, 93 (5–6), 679–686. [DOI] [PubMed] [Google Scholar]
- (46).Kopanczyk R; Opris DC; Lickliter J; Bridges EG; Nazar AM; Bridges KG C-reactive protein levels in blood and saliva show no correlation in young, healthy adults. FASEB J. 2010, 24, No. lb409–lb409. [Google Scholar]
- (47).Poll EM; Kreitschmann-Andermahr I; Langejuergen Y; Stanzel S; Gilsbach JM; Gressner A; Yagmur E Saliva collection method affects predictability of serum cortisol. Clin. Chim. Acta 2007, 382 (1–2), 15–19. [DOI] [PubMed] [Google Scholar]
- (48).Gardner A; Parkes HG; So PW; Carpenter GH Determining bacterial and host contributions to the human salivary metabolome. J. Oral Microbiol 2019, 11 (1), 1617014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Liang D; Golan R; Moutinho JL; Chang HH; Greenwald R; Sarnat SE; Russell AG; Sarnat JA Errors associated with the use of roadside monitoring in the estimation of acute traffic pollutant-related health effects. Environ. Res 2018, 165, 210–219. [DOI] [PubMed] [Google Scholar]
- (50).De Filippis F; Vannini L; La Storia A; Laghi L; Piombino P; Stellato G; Serrazanetti DI; Gozzi G; Turroni S; Ferrocino I; Lazzi C; Di Cagno R; Gobbetti M; Ercolini D The same microbiota and a potentially discriminant metabolome in the saliva of omnivore, ovo-lacto-vegetarian and Vegan individuals. PLoS One. 2014, 9 (11), No. e112373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Schrimpe-Rutledge AC; Codreanu SG; Sherrod SD; McLean JA Untargeted Metabolomics Strategies-Challenges and Emerging Directions. J. Am. Soc. Mass Spectrom 2016, 27 (12), 1897–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


