Abstract
Forty sows (PIC Camborough 1050) from a single farm were randomly selected at 112 days of gestation to evaluate if gut bacteria transverse the blood system of the sow to deposit gut microbiota into the colostrum for piglet gut inoculation via the entero-mammary pathway. Fourteen first-parity gilts and 20 third-parity sows were used for the study. At the time of farrowing, colostrum, fecal samples, and blood samples were collected to evaluate the presence of bacteria in each sample. Colostrum and blood samples were processed via centrifugation to separate the immune cell fraction. Total DNA was extracted from fecal, colostrum, and white blood cell fractions. 16S ribosomal RNA gene amplicon sequencing was conducted at the Iowa State University DNA Facility (Ames, IA) to further characterize the bacterial and archaeal taxa present within each sample. Data were analyzed using Mothur and using R v4.0.3 (R Core Team, 2020). The experimental unit was the sow. Tables were generated to demonstrate the relative abundances of bacteria and archaea present in each type of sample and also identify organisms differentially abundant between sample types. Firmicutes were the most abundant phylum in colostrum and fecal samples and Tenericutes had the greatest abundance in blood comparative to other phyla. Further evaluation of the classification of bacteria present demonstrated that a few genera of bacteria are present in all three samples. Clostridum_sensu_stricto 1 was present in high relative abundance in colostrum and moderate abundance in the feces while also being present within the blood. Other genera present in all three sample types include Ruminococcus and Mycoplasma. In conclusion, the data suggest that there are bacteria present in all three locations of the sow at the time of farrowing and that first parity sows have different microbial populations than third parity sows.
Keywords: bacteria, entero-mammary, microbiota, swine
INTRODUCTION
Without cross-placental antibody transfer, piglets are born without antibodies and are highly susceptible to environmental pathogens during the first few days of life. To provide protection, the piglet needs to receive colostrum from the sow. While it is known that a sow provides colostral antibodies and maternal mononuclear lymphocytes, little is known if the sow is able to pass viable bacteria to the piglet (Bandrick et al., 2014). In humans and cows, it has been demonstrated that dendritic cells can sample and pull viable, potentially beneficial bacteria from the maternal intestinal lumen and transfer them to the mammary gland and ultimately into the milk for microbiome establishment in the neonate (Rodriguez, 2014; Young et al., 2015). This process is called the entero-mammary pathway. While this process has the ability to establish a good microbiome environment, it also has the potential to move infectious agents such as human immunodeficiency virus, herpes, and cytomegalovirus in humans (Lamounier et al., 2004). Furthermore, this process allows for neonatal immune cell imprinting for the early establishment of maturing the immune system (Al Nabhani and Eberl, 2020). Little information has documented if the sow utilizes the entero-mammary pathway and if there are colostral microbiome differences between the first and third parity sow. Therefore, the objective of this study was to evaluate microbial populations present in the colostrum, blood, and feces of sows at the time of farrowing swine and to evaluate the related difference between first and third parity sows.
MATERIALS AND METHODS
The trial was approved by the Iowa State University Animal Care Committee (IACUC #19-330).
The study was conducted at a 2,500 head commercial sow farm located within the United States. All animal care practices were conducted by following the routine farm management procedures and Pork Quality Assurance guidelines (National Pork Board, Pork Quality Assurance, 2012).
Animals
Forty pregnant females (PIC Camborough 1050) from a single farm with no acute pathogenic infections (porcine reproductive and respiratory virus [PRRS], swine influenza [IAV-S], or porcine epidemic diarrhea virus [PEDV]) were randomly selected at the time of farrowing for enrollment into the study. The farm was identified as Mycoplasma hyopneumoniae stable positive. Fourteen first parity sows and 20 third-parity sows were used for the study. The animals all farrowed within 26 days of each other and were fed a common commercial diet manufactured at a local feed mill. Sows were allowed ad-libitum access to feed and water prior to farrowing and throughout lactation. The diet was formulated to meet or exceed nutrition requirements (NRC, 2012).
Sample Collection
The underline was disinfected with a 2% chlorohexidine solution (Patterson Veterinary Supply, Greely, CO) using sterile gauze (Bound Tree Medical, Dublin, OH). After 10 min, the second teat on the right side of the sow was swabbed using a sterile Amies charcoal swab (Starplex Scientific, Etobicoke, Ontario, Canada) to be retained for microbiological culture at a later time. The swab was immediately placed on ice until later stored in a −20 °C freezer. A sterile gloved (Kimberly Clark, Irving, TX) hand was used to express the colostrum at the time of farrowing. Sows were required to have at least two piglets farrowed and have not expelled the placenta to be eligible for the study. The initial drops of colostrum were expelled and discarded and then 20 mL of colostrum were manually expressed into a sterile 50 mL sterile conical tube (Thermo Scientific, Waltham, MA). The sample was immediately placed on ice and stored in a −20 °C freezer. Blood was collected via sterile venipuncture of the ear vein using a butterfly catheter (BD, Franklin Lakes, NJ). Ten mL of blood was collected into ethylenediaminetetraacetic acid tubes (BD, Franklin Lakes, NJ). Fecal samples were collected directly from the rectum with a gloved hand and placed into a sterile 50 mL conical tube and stored at −80 °C until further processing.
Sample Processing
Samples were processed and analyzed in a similar fashion as described by Young et al. (2015). In short, white blood cells (WBCs) from colostrum were separated via centrifugation and stored at −80 °C until processed for DNA extraction. WBCs from plasma were isolated as follows: the plasma tubes were centrifuged at 2,000×g for 10 min with no brake applied. The plasma layer was removed and discarded. The buffer coat layer containing the WBCs was slowly collected using a transfer pipet and placed into a microcentrifuge tube.
Microbial DNA Extraction, 16S Ribosomal RNA Gene Sequencing, and Sequence Analysis
Total DNA was extracted from fecal, colostrum, and WBC fractions (blood) using the Qiagen DNeasy Powerlyzer Powersoil Kit (Qiagen, Hilden, Germany). Two process controls were sequenced to account for known inherent DNA in the extraction kit. For all samples, mechanical lysis was accomplished using Fisher Scientific Beadmill 24, and DNA concentrations were measured using a Qubit 3 fluorometer (Invitrogen, Carlsbad, CA). DNA sequencing was conducted at Iowa State University DNA Facility (Ames, IA) using the Illumina MiSeq platform (Illumina, San Diego, CA). In brief, the DNA from each sample was amplified using Platinum Taq DNA Polymerase (Thermo Fisher Scientific) with one replicate per sample using universal 16S ribosomal RNA (rRNA) gene bacterial primers [515F (5ʹ-GTGYCAGCMGCCGCGGTAA-3ʹ) and 806R (5ʹ-GGACTACNVGGGTWTCTAAT-3ʹ)] amplifying the variable V4 region. Polymerase chain reaction (PCR) was carried out with an initial denaturation step at 94 °C for 3 min, followed by 45 s of denaturing at 94 °C, 20 s of annealing at 50 °C, and 90 s of extension at 72 °C. This process was repeated for 35 cycles, concluding with a 10-minute extension at 72 °C. The resulting PCR products were then purified with the QIAquick 96 PCR Purification Kit (Qiagen, Hilden, Germany). Barcoded amplicons were mixed at equal molar ratios and used for Illumina MiSeq paired-end sequencing with 150 bp read length and cluster generation with 10% PhiX control DNA on an Illumina MiSeq platform. Sequence analysis was conducted using Mothur V1.43.0 following the Mothur MiSeq Standard Operating Procedure (Schloss et al., 2009). Paired-end reads were merged and quality filtered using the “make.contigs” command in mothur resulting in 12,922,059 initial contigs. Subsequently, all sequences were screened using a minimum read length of 252 bp, a zero ambiguities threshold, and a maximum homopolymer length of eight bases. Possible chimeric sequences were removed using the “chimera.vsearch” command in mothur using the SILVA.gold reference database provided by the mothur website. The 7,889,668 remaining high-quality sequences were used for de novo Operational Taxonomic Units (OTUs) clustering at 99% similarity using the mothur v1.43.0 “cluster.split” command. OTUs were taxonomically classified using the SILVA SSU 132 release as a reference in mothur (Quast et al., 2013).
Data Analysis
Data were analyzed using R v4.0.3 (R Core Team, 2020). OTUs in the blood with less than 100 times the average relative abundance in blood samples compared with process controls were removed. In addition, OTUs representing less than 10 sequences and unclassified or vertebrae OTUs were excluded from the analysis. To reduce the number of spurious OTUs, all OTUs represented by less than 10 reads were deleted.
To compare community structures between sample locations and parities, Chao1, Shannon Diversity, and Simpson’s alpha-diversity indices were calculated using the estimate_richness function of the phyloseq package (v1.32.0; McMurdie and Holmes, 2013). Differences in alpha-diversity measures were detected using the Kruskal-Wallis nonparametric method, using the DunnTest function of the DescTools R package (v 0.99.39) with a Benjamini-Hochberg adjustment (Signorell et al., 2021). Using similar methods, differences in the relative abundances of specific genera of interest (Clostridium_ sensu_stricto_1, Lactobacillus, and Mycoplasma) were compared between parities (1 vs. 3+) at each sample location. The differential abundance of the top 200 OTUs was analyzed with PROC GLIMMIX of SAS (Version 9.4, SAS Inst., Cary, NC) using a negative binomial distribution offset by the total sequence count for each sample. False discovery rate P values were corrected using the MULTTEST procedure of SAS. For OTUs with a Q < 0.05, the LOG2 fold change was calculated to compare abundance between parities and sample locations.
To compare beta-diversity among groups, Bray-Curtis dissimilarity coefficients were calculated for each sample and subjected to permutational multivariate analysis of variance, and homogeneity of variance was tested using betadisper (Oksanen et al., 2015). Results were considered significant at P ≤ 0.05 and a trend at P > 0.05 and ≤ 0.10.
Principal coordinate analysis (PCoA) was conducted for visualization of the overall variation in bacterial and archaeal communities and canonical correlation analysis was used to show the variation strictly due to sample location and parity. These plots were constructed using Bray-Curtis dissimilarity measures and the ordinate and plot_ordination functions of phyloseq (v1.32.0). All plots were constructed using ggplot2 (v3.3.3) in R 4.0.3 (Wickham, 2016).
RESULTS
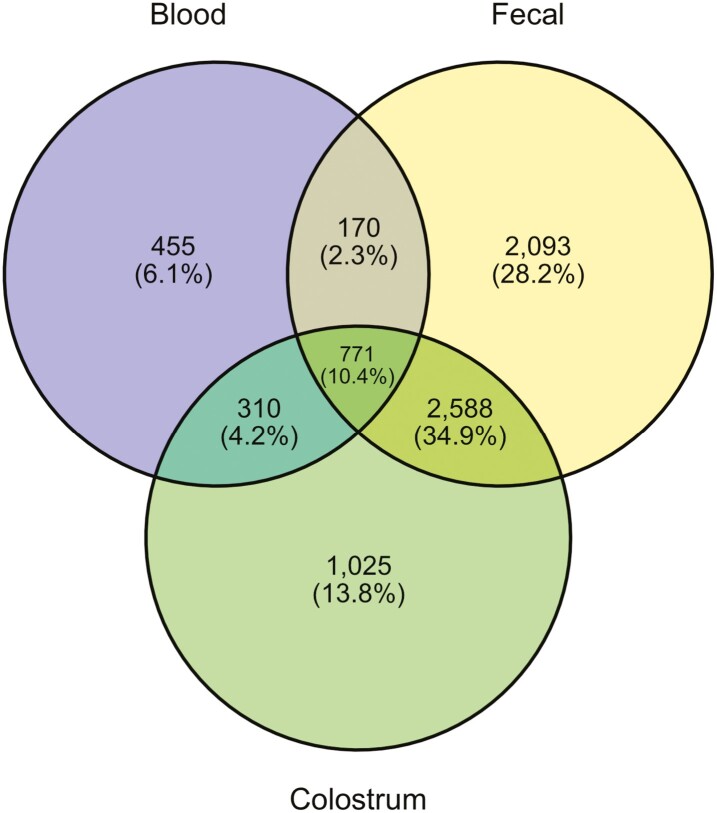
After length filtering, quality control, and chimera removal, a total of 7,085,431 16S rRNA sequences were obtained from all samples (min = 3,350; max = 188,448; median = 74,239). The mean (±SEM) number of sequences obtained from feces (n = 30), colostrum (n = 34), and blood (n = 34) was 134,168 ± 5,805, 75,566 ± 4,807, and 14,446 ± 2,018, respectively. Based on 99% similarity, 7,412 de novo OTUs were identified across all sample types, with 5,622 identified in fecal samples, 4,694 in colostral samples, and 1,706 in blood samples. The number of unique and shared OTUs between tissue types is visually represented by the Venn diagram (Figure 1). In addition, 97.0% of the reads were bacterial and 3.0% were archaeal.
Figure 1.
Venn diagram showing the number of Operational Taxonomic Units (OTUs) assigned in blood, fecal, and colostrum samples.
Comparison of the Microbiota of Samples From Different Locations
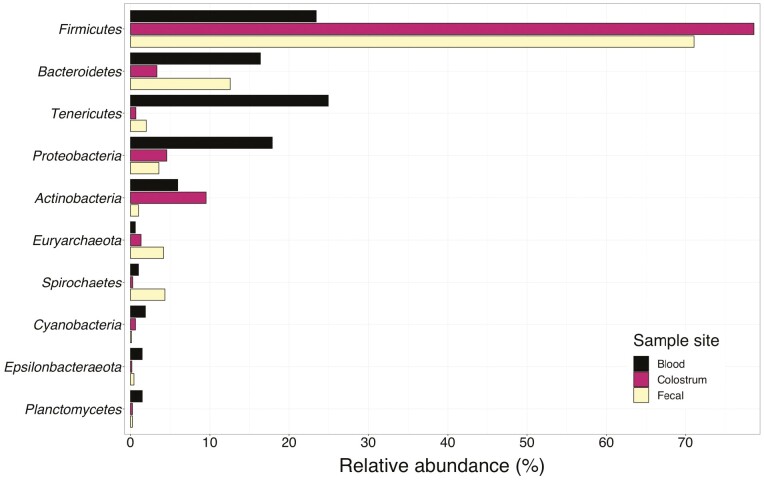
Within fecal samples, there were 26 phyla identified with Firmicutes and Bacteroidetes being the most abundant at 72% and 12% relative abundance, respectively. Highly abundant genera within fecal samples included: Lactobacillus (14%), Clostridium_sensu_stricto_1 (12%), Ruminococcaceae_UCG-005 (5%), Lachnospiraceae_unclassified (5%), and Treponema_2 (5%). There were 28 phyla identified within colostrum samples with Firmicutes and Actinobacteria being the most abundant at 81% and 9%, respectively. Highly abundant genera within colostrum samples included: Clostridium_sensu_stricto_1 (30%), Romboutsia (10%), Turicibacter (9%), Terrisporobacter (8%), and Lactobacillus (5%). There were 27 phyla identified within blood samples with Tenericutes, Firmicutes, and Proteobacteria being the most abundant, representing 40%, 18%, and 15% of all reads, respectively. Highly abundant genera within blood samples included: Mycoplasma (40%), Endozoicomonas (3%), Rikenellaceae_RC9_gut_group (2%), and Lachnospiraceae_unclassified (2%).
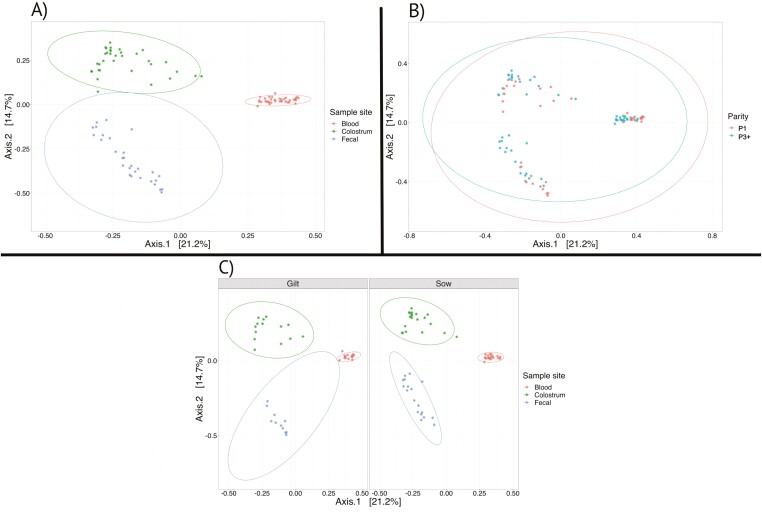
Differences in overall microbial community structure between the three sampling sites have been visualized in PCoA plots (Figure 2). The PCoA plot revealed clear separation and clustering between the three sampling sites and suggests that the communities present in the fecal and colostral samples are more similar to each other than to those found in the blood (WBCs) samples.
Figure 2.
Principal coordinates analysis (PCoA) based on Bray-Curtis distances was conducted for visualization of overall beta diversity of the microbial communities between sampling sites (A), among parities (B), and between sampling sites separated by parity (C).
When the diversity indices were compared, fecal samples showed greater microbial diversity (Shannon, P < 0.001), species richness (Chao1, P < 0.001), and microbial community evenness (Simpson, P < 0.001) compared with colostrum samples. Compared with blood samples, colostrum showed greater diversity (P < 0.001) and species richness (P < 0.001); however, community evenness did not differ between blood and colostrum samples (P = 0.92). In addition, fecal samples showed greater community evenness (P < 0.001) compared with blood samples.
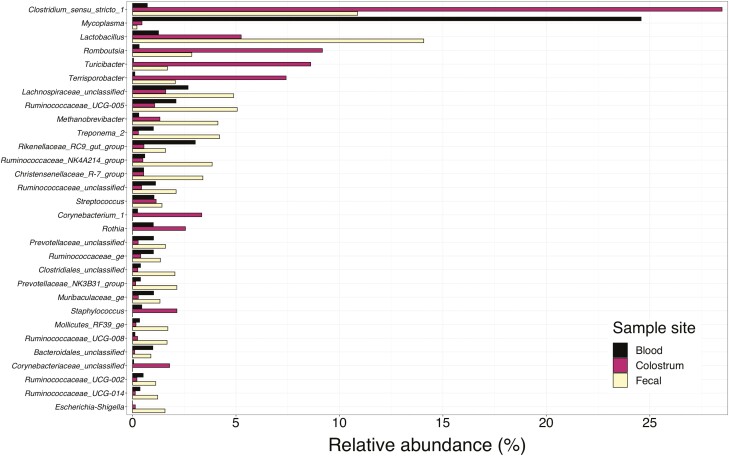
A total of 11 phyla were present in all three sample locations in at least one animal with Firmicutes, Bacteroidetes, and Tenericutes having the greatest incidence (Figure 3). Further evaluation of the classification of bacteria present demonstrated that multiple genera of bacteria are present in all three sample types (Figure 4). However, the primary interest of this study is to investigate the overlap between these communities by identifying OTUs and taxa present in all three sampling sites, particularly within samples from the same animal (Table 1). Thirty of the sampled animals had at least one OTU that was present in all three of their sampling sites. Of these animals, 18 were third parity sows (90% of the third parity sows sampled) and 12 were first parity sows (86% of the first parity sows sampled). Clostridum_sensu_stricto_1 OTU 12 was present in high abundance in colostrum and moderate abundance in the feces while also being present within the blood. Other organisms present in all three sample types at a lesser abundance than those mentioned previously included Ruminococcaceae_UCG-005 and Mycoplasma.
Figure 3.
Average relative abundance of the top 10 microbial phyla present in blood, colostrum, and fecal samples of lactating sows.
Figure 4.
Average relative abundance of the top 30 genera present in blood, colostrum, and fecal samples of lactating sows.
Table 1.
Operational Taxonomic Units (OTUs) present in all sample sites (feces, colostrum, and blood) in either first or third parity sows
| OTU | Phylum | Order | Family | Genus | Paritya | |
|---|---|---|---|---|---|---|
| 1 | 3+ | |||||
| 12 | Firmicutes | Clostridiales | Clostridiaceae_1 | Clostridium_sensu_stricto_1 | 2 | 13 |
| 9 | Tenericutes | Mycoplasmatales | Mycoplasmataceae | Mycoplasma | 8 | 6 |
| 17 | Firmicutes | Clostridiales | Ruminococcaceae | Ruminococcaceae_NK4A214_group | 3 | 7 |
| 46 | Firmicutes | Clostridiales | Clostridiaceae_1 | Clostridium_sensu_stricto_1 | 4 | 6 |
| 16 | Firmicutes | Clostridiales | Lachnospiraceae | Lachnospiraceae_AC2044_group | 5 | 5 |
| 28 | Firmicutes | Selenomonadales | Veillonellaceae | Megasphaera | 4 | 6 |
| 20 | Firmicutes | Clostridiales | Ruminococcaceae | Ruminococcaceae_UCG-005 | 3 | 6 |
| 62 | Firmicutes | Clostridiales | Ruminococcaceae | Ruminococcaceae_NK4A214_group | 4 | 4 |
| 21 | Firmicutes | Clostridiales | Lachnospiraceae | Lachnospiraceae_unclassified | 1 | 6 |
| 115 | Firmicutes | Clostridiales | Lachnospiraceae | Lachnospiraceae_unclassified | 3 | 3 |
| 82 | Firmicutes | Erysipelotrichales | Erysipelotrichaceae | Solobacterium | 1 | 5 |
| 81 | Firmicutes | Clostridiales | Ruminococcaceae | Ruminococcaceae_UCG-005 | 2 | 4 |
| 77 | Firmicutes | Clostridiales | Ruminococcaceae | Ruminococcaceae_UCG-005 | 2 | 4 |
| 61 | Bacteroidetes | Bacteroidales | Muribaculaceae | Muribaculaceae_ge | 3 | 3 |
| 70 | Firmicutes | Erysipelotrichales | Erysipelotrichaceae | Erysipelotrichaceae_unclassified | 3 | 2 |
| 139 | Bacteroidetes | Bacteroidales | Prevotellaceae | Prevotellaceae_UCG-003 | 2 | 3 |
| 38 | Bacteroidetes | Bacteroidales | Rikenellaceae | dgA-11_gut_group | 1 | 4 |
| 49 | Firmicutes | Clostridiales | Ruminococcaceae | Ruminococcaceae_unclassified | 2 | 3 |
| 43 | Firmicutes | Clostridiales | Clostridiales_unclassified | Clostridiales_unclassified | 2 | 3 |
| 25 | Tenericutes | Mycoplasmatales | Mycoplasmataceae | Mycoplasma | 3 | 2 |
| 78 | Bacteroidetes | Bacteroidales | Prevotellaceae | Prevotellaceae_NK3B31_group | 1 | 4 |
| 51 | Firmicutes | Lactobacillales | Aerococcaceae | Aerococcus | 0 | 4 |
| 957 | Firmicutes | Clostridiales | Peptostreptococcaceae | Romboutsia | 1 | 3 |
| 84 | Bacteroidetes | Bacteroidales | Rikenellaceae | Rikenellaceae_RC9_gut_group | 1 | 3 |
| 59 | Actinobacteria | Corynebacteriales | Corynebacteriaceae | Corynebacterium_1 | 1 | 3 |
| 136 | Bacteroidetes | Bacteroidales | Rikenellaceae | Rikenellaceae_RC9_gut_group | 2 | 2 |
| 63 | Proteobacteria | Oceanospirillales | Endozoicomonadaceae | Endozoicomonas | 1 | 3 |
| 58 | Spirochaetes | Spirochaetales | Spirochaetaceae | Treponema_2 | 1 | 3 |
| 68 | Firmicutes | Clostridiales | Ruminococcaceae | Ruminococcaceae_UCG-008 | 0 | 4 |
| 89 | Firmicutes | Clostridia_unclassified | Clostridia_unclassified | Clostridia_unclassified | 2 | 2 |
| 19 | Actinobacteria | Corynebacteriales | Corynebacteriaceae | Corynebacterium_1 | 2 | 2 |
| 42 | Firmicutes | Clostridiales | Lachnospiraceae | Coprococcus_3 | 2 | 2 |
| 223 | Firmicutes | Clostridiales | Ruminococcaceae | Ruminococcaceae_UCG-009 | 0 | 4 |
| 45 | Firmicutes | Clostridiales | Ruminococcaceae | Ruminococcaceae_ge | 2 | 2 |
| 34 | Firmicutes | Clostridiales | Christensenellaceae | Christensenellaceae_R-7_group | 0 | 4 |
Count of the number of animals which had OTU present in all three sample sites (feces, colostrum, and blood)
Parity by Location Analysis
When comparing the diversity measures between parities, there was no evidence for a difference in the microbial diversity (Shannon, P = 0.35), species richness (Chao1, P = 0.14), or community evenness (Simpson, P = 0.67) in fecal samples of first and third parity sows. Within colostrum samples, first parity sows showed greater microbial diversity (P = 0.03) and evenness (P = 0.05); however, there was no evidence for an effect of parity on species richness (P = 0.26). Within blood samples, third parity sows showed greater diversity (P = 0.01) and evenness (P = 0.02), and no difference in species richness (P = 0.31) compared with first parity sows. Additionally, there is no clustering by parity apparent when observing beta diversity of microbial communities revealed by PCoA (Figure 2) plots, suggesting that parity has no significant impact on microbial community structure.
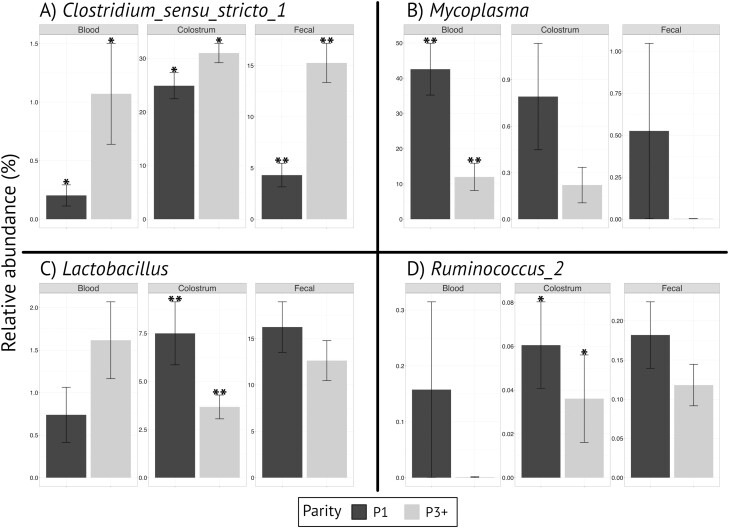
Four genera/taxa of interest with high relative abundance among sample sites and age of the sow were further evaluated (Figure 5). In comparison between first and third parity sows, Clostridium_sensu_stricto_1 was observed in greater abundance in the fecal samples (P < 0.001) and tended to be greater in the colostrum (P = 0.06) and blood (P = 0.06) of third parity sows compared with first parity sows. Lactobacillus was observed in greater abundance in the colostrum (P = 0.04) of first parity sows compared with third parity sows but was not different in the fecal (P = 0.24) or blood (P = 0.55) samples. Mycoplasma abundance was greater in the blood (P < 0.001) of first parity sows compared with third parity sows but was not different in the fecal (P = 0.25) or colostrum (P = 0.24) samples. Ruminococcus_2 abundance was not different (P > 0.10) in blood or colostrum but was higher (P < 0.05) in fecal samples from first parity sows compared with third parity sows.
Figure 5.
Average relative abundance by parity of Clostridium_sensu_stricto_1 (A), Mycoplasma (B), Lactobacillus (C), and Ruminococcus_2 (D) in blood, colostrum, and fecal samples. **Within a panel indicates difference at P ≤ 0.05. *Within a panel indicates difference at 0.05 < P ≤ 0.10.
DISCUSSION
The purpose of this study was to evaluate a possible maternal microbiota transfer from the gut of the sow to the colostrum. The predominant phylum present in feces and colostrum was Firmicutes, while Tenericutes and Firmicutes had the greatest abundance in the blood samples. Firmicutes can be further classified into highly abundant genera including that of Lactobacillus, Bacillus, and Clostridium and Tenericutes will include genera that of Mycoplasma.
The farm had been Mycoplasma hyopneumonia positive-stable for years. Replacement gilts in the gilt developer were received as naiive to this organism and had seroconverted to a positive state upon arrival to the farm. As all gilts were exposed within a very short time in the gilt developer unit prior to breeding, disease status was not used as a blocking factor. While the presence of Mycoplasma hyopneuomiae does not impact the primary objective of the study, which was to determine if the entero-mammary pathway exists, the presenence of the organism does add information to the study for further work in which disease status and parity may create microbial differences.
Evaluation of the microbiota in colostrum, feces, and blood between first and third parity sows demonstrated that multiple genera were present across the populations. A decrease in fecal Clostridium_sensu_stricto-1 in intra-uterine growth retardation pigs results in an increase of tumor necrosis factor, interleukin 6, and interferon-gamma as well as potential to reduce harvest weights (Huang et al., 2019; Jiang et al., 2019). However, if Clostridium_sensu_stricto-1 overgrows in the ileum of chickens after a Clostridium perfringens exposure, there is an increase in necrotic enteritis, which means that the abundance of this genus needs to be tightly regulated (Yang et al., 2019). This regulation likely is tied to other organisms within the colostrum. For example, the first parity sows had a higher relative abundance of Lactobacillus in the colostrum comparative to the third parity sows. While many Lactobacillus strains are known to have positive impacts on gut health, certain Lactobacillus can be pathogenic (Harty et al., 1994).
There are some general limitations of 16S rRNA gene-base amplicon sequencing studies. One limitation is the limited phylogenetic resolution of 16S rRNA gene amplicon sequencing due to its comparatively short read lengths and the inherent limitations of the taxonomic resolution of the 16S rRNA gene for same bacterial genera/groups which can result in an incomplete profile of the microorganisms across all samples. In addition, as this sequencing was DNA-based, it should be noted that (at least some of) the sequencing reads obtained could derive from dead microorganisms rather than live microorganisms (Poretsky et al., 2014; Muhamad Rizal et al., 2020). In addition, low microbial biomass samples, such as blood, represent another challenge for amplicon sequencing studies. Neonatal programming begins in-utero and continues through lactation as the sow passes many organisms to the piglet through the placenta, vaginal tract, and milk (Perez-Munoz et al., 2017).
The increase in Mycoplasma in the blood samples from the first parity population comparative to the third parity population is likely due to the herd introducing Mycoplasma negative first parity sows into the facility and the first parity sows were seroconverting at the time of parturition. Further evaluation of the samples indicated a tendency for significantly higher levels of Ruminococcus_2 in the fecal samples of first parity sows compared with third parity sows, but not colostrum or blood between first and third parity sows. Surendran Nair et al. (2019) demonstrated that piglets having higher fecal levels of Ruminococcus_2 had reduced lung lesion scores after a M. hyopneumoniae exposure. Ruminococcus is known to produce short-chain fatty acids, such as acetate, propionate, and butyrate. Within the last 10 years, research has demonstrated that short-chain fatty acids can modulate inflammation within the respiratory system (Trompette et al., 2012). Since the first parity sows in this study had a known M. hyopneumoniae exposure and had a significantly higher abundance of Mycoplasma in the blood compared with the third parity sows, the increase in fecal Ruminococcus_2 in the colostrum of the first parity gilt is possibly involved in controlling the pathogenicity of the disease through short-chain fatty acid production. A recent study analyzing the entero-mammary pathway in cows also found a high abundance of Mycoplasma in WBC (Young et al., 2015). More importantly, the study by (Young et al., 2015) provided the first evidence for the existence of the entero-mammary pathway in cows using amplicon sequencing. In conclusion, these data indicate that the entero-mammary pathway may exist in swine. Although the results from our study do not prove the hypothesis that translocation of gastrointestinal tract bacteria into the mammary gland does occur in the sows, they support the existence of an endogenous entero-mammary pathway for some bacteria during lactation in the sow. However, as the results from our study are based on amplicon sequencing, future studies will be needed to experimentally demonstrate the translocation of bacteria through the entero-mammary pathway.
Acknowledgments
The authors would like to acknowledge the support of farm staff that allowed the sample collection process and their assistance in helping us complete the project. This study was funded in part by the Iowa Pork Productions Association and the National Pork Board #19-213 IPPA.
Contributor Information
Laura L Greiner, Department of Animal Science, Iowa State University, Ames, IA 50011, USA.
Dalton C Humphrey, Department of Animal Science, Iowa State University, Ames, IA 50011, USA.
Shayla N Holland, Department of Animal Science, Iowa State University, Ames, IA 50011, USA.
C J Anderson, Department of Animal Science, Iowa State University, Ames, IA 50011, USA.
Stephan Schmitz-Esser, Department of Animal Science, Iowa State University, Ames, IA 50011, USA.
Conflict of Interest statement
None decleared.
LITERATURE CITED
- Al Nabhani, Z., and Eberl G.. . 2020. Imprinting of the immune system by the microbiota early in life. Mucosal Immunol. 13:183–189. doi: 10.1038/s41385-020-0257-y. [DOI] [PubMed] [Google Scholar]
- Bandrick, M., Ariza-Neito C., Baidoo S. K., and Molitor T. W.. . 2014. Colostral antibody-mediated and cell-mediated immunity contributes to innate and antigen-specific immunity in pigs. Dev. Comp. Immunol. 43:114–120. doi: 10.1016/j.dci.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harty, D. W., Oakey H. J., Patrikakis M., Hume E. B., and Knox K. W.. . 1994. Pathogenic potential of lactobacilli. Int. J. Food Microbiol. 24:179–189. doi: 10.1016/0168-1605(94)90117-1. [DOI] [PubMed] [Google Scholar]
- Huang, S., Li N., Liu C., Li T., Wang W., Jiang L., Li Z., Han D., Tao S., and Wang J.. . 2019. Characteristics of the gut microbiota colonization, inflammatory profile, and plasma metabolome in intrauterine growth restricted piglets during the first 12 hours after birth. J. Microbiol. 57:748–758. doi:. [DOI] [PubMed] [Google Scholar]
- Jiang, L., Feng C., Tao S., Li N., Zuo B., Han D., and Wang J.. . 2019. Maternal imprinting of the neonatal microbiota colonization in intrauterine growth restricted piglets: a review. J. Animal Sci. Biotechnol. 10:88. doi: 10.1186/s40104-019-0397-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamounier, J. A., Moulin Z. S., and Xavier C. C.. . 2004. Recommendations for breastfeeding during maternal infections. J. Pediatr. (Rio J) . 80:S181–S188. doi: 10.2223/1252. [DOI] [PubMed] [Google Scholar]
- McMurdie, P. J., and Holmes S.. . 2013. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhamad Rizal, N. S., Neoh H-M., Ramli R., A/LK Periyasamy P. R., Hanafiah A., Abdul Samat M. N., Tan T. L., Wong K. K., Nathan S., Chieng S., Saw S. H., and Khor B. Y.. . 2020. Advantages and limitations of 16S rRNA next-generation sequencing for pathogen identification in the diagnostic microbiology laboratory: Perspectives from a middle-income country. Diagnostics. 10(10):816. doi: 10.3390/diagnostics10100816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NRC. 2012. Nutrient requirements of swine. 11th rev. edn. Washington, DC: The National Academies Press. doi: 10.17226/13298. [DOI] [Google Scholar]
- Oksanen, J., Blanchet F., Kindt R., Legendre P., Minchin P., O’Hara B., Simpson G., Solymos P., Stevens H., and Wagner H.. . 2015. Vegan: community ecology package. R package version 2.2-1. [Google Scholar]
- Perez-Muñoz, M. E., Arrieta M. C., Ramer-Tait A. E., and Walter J.. . 2017. A critical assessment of the “sterile womb” and “in utero colonization” hypotheses: Implications for research on the pioneer infant microbiome. Microbiome. 5:48. doi: 10.1186/s40168-017-0268-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poretsky, R., Rodriguez-R L. M., Luo C., Tsementzi D., and Konstantinidis K. T.. . 2014. Strengths and limitations of 16S rRNA gene amplicon sequencing in revealing temporal microbial community dynamics. PLoS One. 9:e93827. doi: 10.1371/journal.pone.0093827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pork Quality Assurance. 2012. Des Moines, IA: National Pork Board. [Google Scholar]
- Quast, C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., Peplies J., and Glöckner F.. . 2013. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez, J. M. 2014. The origin of human milk bacteria: Is there a bacterial etero-mammary pathway during late pregnancy and lactation. Adv. Nutr. 5:779–784. doi: 10.3945/an.114.007229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss, P. D., Westcott S., Ryabin T., Hall J., Hartmann M., Hollister E., Lesniewski R., Oakley B., Parks D., Robinson C., . et al. 2009. Introducing mother; open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signorell, A., Aho K., Alfons A., Anderegg N., Aragon T., Arachchige C., Arppe A., Baddeley A., Barton K., Bolker B., . et al. 2021. DescTools: tools for descriptive statistics. R package version 0.99.42 [accessed 20 October 2021]. Available from https://cran.r-project.org/package=DescTools. [Google Scholar]
- Surendran Nair, M., Eucker T., Martinson B., Neubauer A., Victoria J., Nicholson B., and Pieters M.. . 2019. Influence of pig gut microbiota on Mycoplasma hyopneumoniae susceptibility. Vet. Res. 50:86. doi: 10.1186/s13567-019-0701-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trompette, A., Yadava K., Gollwitzer E., Sichelstiel A., Nicod L., and Marsland B.. . 2012. Short-chain fatty acids are potent modulators of allergic airway inflammation. Eur. Respir. J. 40:P2362. [Google Scholar]
- Wickham, H. 2016. ggplot2: elegant graphics for data analysis. New York: Springer-Verlag. [Google Scholar]
- Yang, W. Y., Lee Y., Lu H., Chou C. H., and Wang C.. . 2019. Analysis of contributory gut microbiota and lauric acid against necrotic enteritis in Clostridium perfringens and Eimeria side-by-side challenge model. PLoS One. 14(5):e0205784. doi: 10.1371/journal.pone.0205784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, W., Hine B. C., Wallace O. A. M., Callaghan M., and Bibiloni R.. . 2015. Transfer of intestinal bacterial components to mammary secretions in the cow. PeerJ. 3:e888. doi: 10.7717/peerj.888. [DOI] [PMC free article] [PubMed] [Google Scholar]