Abstract
A newly isolated methanogen, strain DMS1T, is the first obligately anaerobic archaeon which was directly enriched and isolated from a freshwater sediment in defined minimal medium containing dimethyl sulfide (DMS) as the sole carbon and energy source. The use of a chemostat with a continuous DMS-containing gas stream as a method of enrichment, followed by cultivation in deep agar tubes, resulted in a pure culture. Since the only substrates utilized by strain DMS1T are methanol, methylamines, methanethiol (MT), and DMS, this organism is considered an obligately methylotrophic methanogen like most other DMS-degrading methanogens. Strain DMS1T differs from all other DMS-degrading methanogens, since it was isolated from a freshwater pond and requires NaCl concentrations (0 to 0.04 M) typical of the NaCl concentrations required by freshwater microorganisms for growth. DMS was degraded effectively only in a chemostat culture in the presence of low hydrogen sulfide and MT concentrations. Addition of MT or sulfide to the chemostat significantly decreased degradation of DMS. Transient accumulation of DMS in MT-amended cultures indicated that transfer of the first methyl group during DMS degradation is a reversible process. On the basis of its low level of homology with the most closely related methanogen, Methanococcoides burtonii (94.5%), its position on the phylogenetic tree, its morphology (which is different from that of members of the genera Methanolobus, Methanococcoides, and Methanohalophilus), and its salt tolerance and optimum (which are characteristic of freshwater bacteria), we propose that strain DMS1T is a representative of a novel genus. This isolate was named Methanomethylovorans hollandica. Analysis of DMS-amended sediment slurries with a fluorescence microscope revealed the presence of methanogens which were morphologically identical to M. hollandica, as described in this study. Considering its physiological properties, M. hollandica DMS1T is probably responsible for degradation of MT and DMS in freshwater sediments in situ. Due to the reversibility of the DMS conversion, methanogens like strain DMS1T can also be involved in the formation of DMS through methylation of MT. This phenomenon, which previously has been shown to occur in sediment slurries of freshwater origin, might affect the steady-state concentrations and, consequently, the total flux of DMS and MT in these systems.
Dimethyl sulfide (DMS) has an impact on global warming and acid precipitation and on the global sulfur cycle because of its oxidation products (e.g., methanesulfonic acid and SO2), which are released into the atmosphere. For this reason transformations of volatile organic sulfur compounds have been intensively studied during the past few decades. Microbial formation and degradation of DMS and methanethiol (MT) have been shown to have a significant effect on the total flux of sulfur compounds in the atmosphere (13, 14, 21).
In freshwater sediments, formation of MT and DMS is balanced by degradation of these compounds, which results in low steady-state concentrations (18–20). In contrast to marine and estuarine systems, in which DMS originates mainly from dimethylsulfoniumpropionate, volatile organic sulfur compounds in freshwater sediments are derived mainly from methylation of sulfide and MT (7, 15, 18).
In systems with high salt contents, degradation of MT and DMS has been attributed to members of various groups of bacteria, including aerobes (e.g., thiobacilli and Methylophaga spp.) (4, 34–36) and anaerobes (anoxygenic phototrophs, sulfate reducers, and methanogens) (8, 16, 17, 23, 25, 26, 33, 39). The activity of members of these trophic groups depends on the light intensity and the availability of oxygen or alternative electron acceptors, such as sulfate and nitrate. Due to oxygen limitation in freshwater sediments, DMS and MT are degraded mainly anaerobically by means of methanogenic activity (19). Methanogenic conversion of MT and DMS in sediment slurries was first demonstrated by Zinder and Brock (40, 41). Since then, various methanogens have been isolated with DMS or MT from marine, estuarine, salt marsh, and salt lake sediments (8, 12, 16, 17, 23, 25). These methanogens belong to the genera Methanosarcina, Methanolobus, and Methanosalsus. Although methanogens have been identified as the dominant consumers of DMS and MT in sulfate-poor freshwater sediments (18–20, 40, 41), previous attempts to isolate methanogens which are able to grow on DMS or MT from such sediments were unsuccessful (33, 41). Moreover, production of methane or carbon dioxide (or [14C]methane and [14C]carbon dioxide) from MT or DMS (or [14C]MT and [14C]DMS) was not detected in pure cultures of methanogens isolated from nonsaline systems (e.g., Methanobacterium ruminantium, Methanobacterium thermautotrophicum, and Methanosarcina barkeri cultures) (28, 41).
In this paper we describe the isolation of a nonhalophilic methylotrophic methanogen, strain DMS1T, from the sediment of a eutrophic freshwater pond on the campus of the Dekkerswald Institute, Nijmegen, The Netherlands. This strain has the salt tolerance characteristic of freshwater bacteria and is able to use DMS, MT, methanol, and methylamines for growth and methanogenesis. Characteristics of strain DMS1T are discussed in relation to its ecological niche. Phylogenetic analysis revealed that this strain represents a novel genus in the family Methanosarcinaceae.
MATERIALS AND METHODS
Source of inoculum.
Samples were collected from the top layer (5 to 10 cm) of sediment (depth, 50 cm) in a eutrophic freshwater pond on the Campus of Dekkerswald Institute, Nijmegen, The Netherlands. The sediment samples were obtained by suction and placed in anaerobic bottles as described by Lomans et al. (18).
Slurry incubation.
Sediment slurries were prepared and incubated as described previously by Lomans et al. (18–20). Bromoethanesulfonic acid (BES), sodium molybdate, sodium tungstate, DMS, and MT were added from pH-neutral anaerobic stock solutions in distilled water.
Media and culture techniques.
Cultivation was carried out in 60- and 120-ml serum bottles filled with 25 and 50 ml of medium, respectively. The defined sulfide-reduced and bicarbonate-buffered medium of Widdel and Bak (37) was modified slightly and used for isolation. This medium contained (per liter) 1.00 g of NaCl, 0.25 g of Na2SO4, 0.25 g of NH4Cl, 0.20 g of KH2PO4, 0.50 g of KCl, 0.4 g of MgCl2 · 6H2O, 0.1 g of CaCl2 · 2H2O, 0.30 g of Na2S · 7H2O, and 2.50 g of NaHCO3. Sodium sulfide was sterilized separately and added to the basal medium 1 to 2 h before it was used. One milliliter of a trace element solution containing the following compounds was added per liter of medium: nitrilotriacetic acid (NTA) (13.0 g/liter), FeSO4 (0.12 g/liter), H3BO4 (0.30 g/liter), H2SeO3 (0.32 g/liter), KAl(SO4)2 · 12H2O (0.32 g/liter), CuSO4 · 5H2O (0.32 g/liter), CoCl2 · 6H2O (0.32 g/liter), NaMoO4 · 2H2O (0.032 g/liter), NiCl2 · 6H2O (0.31 g/liter), ZnSO4 · 7H2O (0.32 g/liter), and MnCl2 · 4H2O (0.32 g/liter). NTA and FeSO4 were dissolved in 600 ml of distilled water by adding NaOH. After the other components were added to the NTA-FeSO4-NaOH solution, the final pH was adjusted to 6.5 and the volume was adjusted to 1,000 ml with distilled water. The bottles were sealed with black butyl rubber stoppers, gassed with an O2-free N2-CO2 mixture (80/20, vol/vol), and sterilized (121°C, 20 min).
After sterilization (121°C, 20 min) the medium was supplemented with 1 ml of an anaerobic sterile vitamin stock solution per liter of medium; this solution contained (per liter) 0.1 g of p-aminobenzoate, 0.1 g of riboflavin, 0.2 g of thiamine, 0.2 g of nicotinate, 0.5 g of pyridoxin, 0.1 g of pantothenate, 0.1 g of cobalamin, 0.02 g of biotin, 0.05 g of folate, and 0.05 g of lipoate.
Carbon sources were added from anaerobic sterile stock solutions that had been prepared in distilled water 1 to 2 h before inoculation. Immediately before inoculation, the medium was supplemented with small amounts of sodium dithionite obtained from a freshly prepared stock solution (final concentration, 0.08 g/liter) and with Fe(II)Cl2 (final concentration, 10 mg/liter) since this stimulated the growth of strain DMS1T significantly (see below). The bottles were incubated in the dark at 30°C.
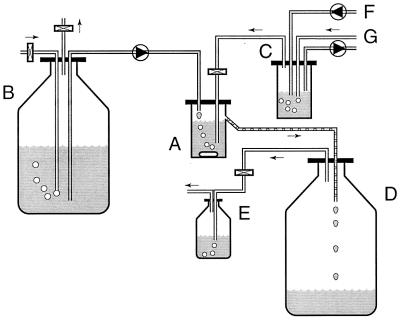
The isolate was enriched in an anaerobic chemostat that was fed with low concentrations of DMS (20 to 250 nmol per ml of headspace) in the N2-CO2 (80/20, vol/vol) gas stream that was passed through the system (Fig. 1). After enrichment, strain DMS1T was isolated by the deep agar method as described by Pfennig (27). The absence of contaminants was confirmed by growing a culture in the medium described above supplemented with yeast extract (0.5%) and Trypticase peptone (0.5%) and also by fluorescence microscopy. Stock cultures were transferred monthly into fresh medium, and cultures containing medium supplemented with glycerol (5%) were stored in glass ampoules under an N2-CO2 atmosphere (80/20, vol/vol) at −80°C.
FIG. 1.
Schematic diagram of the continuous gas flow chemostat used for enrichment and isolation of Methanomethylovorans hollandica DMS1T. Medium was pumped into the culture vessel, resulting in a dilution rate of 0.005 to 0.02 h−1. The energy and carbon source for growth was added via the gas stream bubbling through the chemostat. An oxygen-free N2-CO2 gas stream was passed through a regulation vessel. DMS from a stock solution was pumped into this vessel. The DMS was then sparged out of the water phase, and the DMS-containing gas was then passed through the culture vessel. The concentration of DMS in the incoming gas could be regulated by altering the N2-CO2 gas flow or the pump flow rate of the DMS stock solution. In this way, the dilution rate and the concentration of DMS to which the culture was exposed could be regulated independently. A, culture vessel; B, medium stock preparation; C, regulation vessel; D, waste vessel; E, gas trap bottle; F, tubing connected to a DMS stock solution; G, tubing connected to an oxygen-free N2-CO2 gas stream (0.3 atm). Shaded tubing is tubing that contained liquid. Tubing with droplets is tubing that contained both gas and liquid, whereas tubing without shading or droplets is tubing that contained gas. Rectangles represent sterile (gas) filters. Circles represent peristaltic pumps; the triangles in the circles indicate the direction of pumping. The arrows indicate the direction of the gas flow.
Determining optimal growth conditions.
Specific growth rates were determined by measuring the amount of methane formed during growth on methanol. The specific growth rate during exponential growth was analyzed by linear regression of the logarithm of the total amount of methane that accumulated versus time. When the effects of environmental parameters (pH, temperature, and salt concentration) were tested, growth rates were determined with cultures adapted to the conditions used. We transferred cultures under these conditions at least two times sequentially. In particular, in order to obtain a culture at a higher osmolarity, it was necessary to transfer a culture in several steps to media having progressively higher osmotic values.
Cell suspension experiments.
Conversion of DMS and MT was studied by using samples from chemostat cultures. After a cell suspension was harvested and placed in a 120-ml serum bottle (reduced with 0.5 ml of a solution containing 4 g of sodium dithionite per liter; final concentration, 17 mg/liter), 7- to 10-ml aliquots were dispensed into 60-ml serum bottles that had been reduced with 0.1 ml of 100 mM Ti(III) citrate (final concentration, 1 to 1.4 mM) (38). The suspensions were flushed with N2 to remove the endogenous substrates (DMS and MT) and sulfide. The cell suspensions were incubated at 32°C with shaking (100 rpm).
Analytical techniques.
Gas samples (0.5 to 1.0 ml) were removed from the incubation bottles with pressure lock syringes and were analyzed to determine their methane, MT, and DMS contents with a Hewlett-Packard model 5890 gas chromatograph equipped with a flame ionization detector and a Porapak Q column (80/100 mesh) (10). Specific determinations of sulfur compounds (H2S, MT, and DMS) in gas samples from the incubation mixtures were performed with a Packard model 438A gas chromatograph equipped with a flame photometric detector and a Carbopack B HT100 column (40/60 mesh) as described previously (3, 18).
Microscopy and photography.
Transmission and scanning electron micrographs were obtained with a Philips model 201 transmission electron microscope and a JEOL model T300 scanning electron microscope by using cells from a late-exponential-phase culture that had been fixed with glutaraldehyde (2%, wt/vol) in 50 mM sodium chloride and osmium tetroxide (1%, wt/vol) and dehydrated in absolute ethanol.
Phylogenetic analysis.
Strain DMS1T DNA was isolated by crushing a cell pellet (obtained from 20 ml of culture) in liquid N2 by using a mortar and pestle (11). The homogenate was suspended in 4 ml of TE extraction buffer (10 mM Tris-HCl, 1 mM EDTA; pH 7.5). After sodium dodecyl sulfate (SDS) (1%, wt/vol) and proteinase K (50 μg/ml) were added, the suspension was incubated for 30 min at 50°C. Then the lysate was mixed with an equal volume of cold isopropanol and centrifuged (10 min, 10,000 × g, 4°C). The resulting pellet was dissolved in TE extraction buffer and subjected to phenol-chloroform-isoamyl alcohol (25:24:1) extraction. The pellet obtained after ethanol precipitation of the water phase was dissolved in 100 μl of sterile demineralized water and stored at −20°C. The DNA was used as a template for PCR amplification of approximately 1,350- and 1,000-base segments of the 16S rRNA gene. The PCR conditions were as follows: 2.5 mM MgCl2, annealing temperature of 55°C, and 30 cycles. The PCR amplification primers used were REV007 (5′-GTTGATCCTGCCAGAGGYYA-3′), ARC1326 (5′-TGTGTGCAAGGAGCAGGGAC-3′), REV915 (5′-GTGCTCCCCCGCCAATTCCT-3′) (29), and 23S047 (5′-CCCBGGGCTTATCGCAGCTT-3′) (29). The amplification products were ligated in the pCR II vector and transformed into the Escherichia coli cells of a TA cloning kit (Invitrogen). Plasmid DNA of clones were isolated by using a FlexiPrep kit (Pharmacia P-L Biochemicals Inc.). The sequences of the cloned PCR products, which represented the original 16S rRNA sequence, were analyzed with a DNA sequencer (Applied Biosystems model 373A) by using the Taq DyeDeoxy terminator cycle sequencing method (1, 24). Besides the primers of the TA cloning kit (M13FOR and M13REV) and primers mentioned by Raskin et al. (29) (ARC915, REV344, and MC1109), the following primers were used for sequencing: REV007 (5′-GTTGATCCTGCCAGAGGYYA-3′), ARC1326 (5′-TGTGTGCAAGGAGCAGGGAC-3′), REV954 (5′-TCAAGCTAAAGACTTTACCA-3′), and REV1299 (5′-CGGTTTCCAACATAGCGCGG-3′). These primers were designed in our laboratory and were based on known sequences of other methanogens. Primer REV954 was designed on the basis of a partial sequence of the 16S rRNA gene of strain DMS1T but did not appear to be highly strain specific. The resulting sequences were assembled to produce a 1,449-base continuous DNA sequence. The deduced 16S rRNA sequence of strain DMS1T was aligned with homologous 16S rRNA sequences of closely related members of the Archaea domain by using the Pileup method (The Dutch CAOS/CAMM Center Facility, Nijmegen, The Netherlands). These 16S rRNA sequences were obtained from the GenBank/EMBL and Ribosomal Database Project databases. Distance matrix trees were constructed by using the method of Fitch and Margoliash (9) and the neighbor-joining method of Saitou and Nei (30) in the FITCH and NEIGHBOR programs of the PHYLIP (version 3.4) program package (6). Parsimony and bootstrap parsimony analyses were performed by using the DNAPARS and DNABOOT programs as implemented in the PHYLIP package.
Nucleotide sequence accession number.
The deduced, almost complete sequence (1,449 bases) of the 16S rRNA gene of strain DMS1T has been deposited in the GenBank database under accession no. AF120163.
RESULTS
Enrichment and isolation of strain DMS1T.
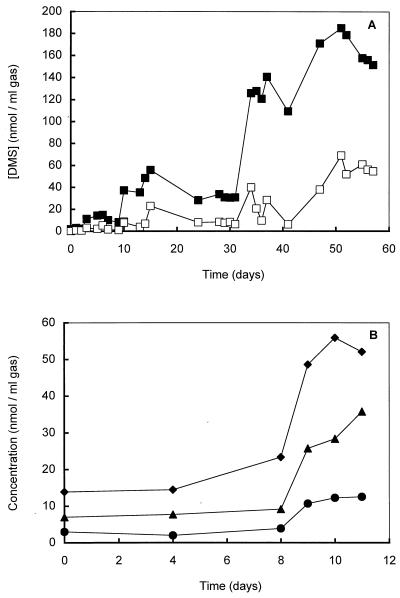
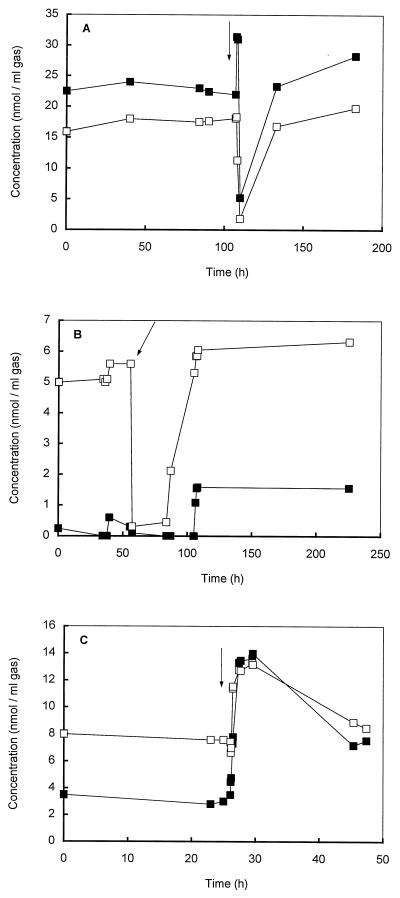
We examined various sediment samples and used the sediment slurry from a eutrophic freshwater pond (Campus of Dekkerswald Institute, Nijmegen, The Netherlands) which exhibited the highest level of DMS consumption (±7 nmol per ml of sediment slurry · h−1) (19, 20) for isolation of anaerobic DMS-consuming microorganisms. After unsuccessful enrichment on DMS in sterilized pore water amended with Trypticase peptone (final concentration, 0.02%), yeast extract (final concentration, 0.02%), or rumen fluid (final concentration, 1% [vol/vol]), mineral anaerobic medium (see above) was inoculated with 10% (vol/vol) sediment slurry. Enhancement of the DMS consumption rate in dilutions of the sediment slurries was found to be very difficult (the maximal DMS degradation rate measured was 12 nmol of DMS per ml of slurry · h−1), and transfers did not result in a DMS-degrading enrichment culture. One of the diluted sediment incubation preparations was used to inoculate a chemostat that was fed with DMS via the N2-CO2 (80/20, vol/vol) gas stream at increasing concentrations (2 to 250 nmol per ml of headspace) (Fig. 1 and 2A). The advantage of this system was that unused DMS and the products (MT and H2S) did not accumulate to inhibiting levels in the system, since they were flushed out of the system. In this way, a stable chemostat culture with a low H2S concentration could be maintained. Conversion of DMS in the chemostat was revealed by the difference between the concentration of DMS in the incoming gas and the concentration of DMS in the outgoing gas and by the presence of methane and MT in the outgoing gas (Fig. 2). Use of the chemostat (dilution rate, 0.01 h−1) resulted in an enrichment culture that had a much higher DMS-degrading capacity (±800 nmol per ml of culture fluid · h−1) and consisted of only one morphologically distinct methanogen. Attempts to isolate the DMS-degrading methanogen by using a liquid dilution series with DMS were unsuccessful. Eventually, cultivation of a single colony obtained from a dilution series in which methanol and DMS in deep agar tubes were used led to a pure culture of a DMS-degrading methanogen, which was designated strain DMS1T. A microscopic and F420 fluorescence microscopic analysis of cultures grown on medium supplemented with methanol (20 mM), yeast extract (0.5%), and Trypticase peptone (0.5%) or on the same medium without methanol revealed that the culture was free of contaminants.
FIG. 2.
(A) Time courses of the DMS concentrations in the incoming gas (■) and outgoing gas (□) in the chemostat. The chemostat was inoculated (10%) with a 10-fold-diluted sediment slurry (see text) that had been preincubated with DMS. The slurry was prepared from a eutrophic pond sediment (Campus of Dekkerswald Institute). (B) Time courses (start-up phase) of the amount of DMS consumed (▴) and the amounts of MT (●) and methane (⧫) produced by a chemostat inoculated with a pure culture of strain DMS1T.
Morphology.
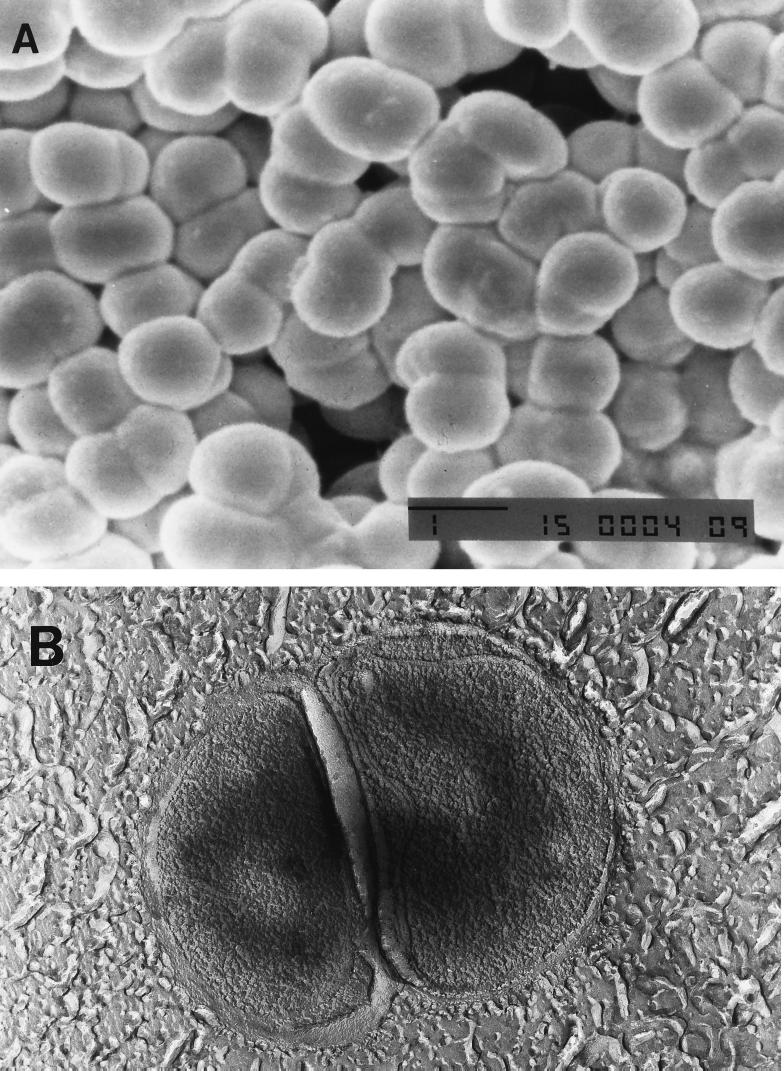
Deep agar colonies of strain DMS1T were white circular disks which reached a diameter of 2 mm in 2 weeks when they were grown on methanol. Phase-contrast microscopy and scanning and transmission electron micrographs revealed that the morphology of the cells could best be described as intermediate between the morphology of typical Methanosarcina cell clusters and the morphology of the coccoid cells characteristic of Methanolobus and Methanococcoides species (Fig. 3). The cells occurred mainly in clusters consisting of two or four cells, and the clusters formed large aggregates consisting of hundreds of cell clusters. The average diameter of individual cells was 1 to 1.5 μm. Motility was not observed. Individual cells and cell clusters or aggregates did not lyse within 15 min after SDS was added to a final concentration of 1.0 g per liter. The isolate stained gram negative.
FIG. 3.
Electron micrographs of cell clusters in a late-logarithmic-phase culture of Methanomethylovorans hollandica DMS1T grown on methanol. (A) Scanning electron micrograph clearly showing large aggregates of cell clusters consisting of two to four irregular coccoid cells. (B) Transmission electron micrograph of a freeze-etched sample. Bar, 1 μm.
Optimal growth conditions.
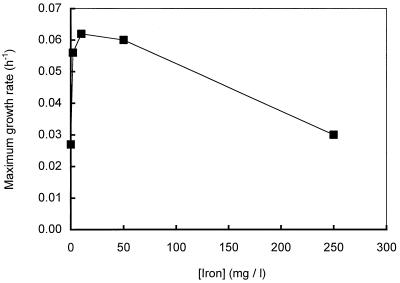
Initially, the logarithmic growth phase of cultures of strain DMS1T was short and was followed by a relatively long linear methane formation phase which indicated that no growth or slow growth occurred. To determine the optimal growth conditions, the effects of vitamins, complex nutrients, trace elements, pH, temperature, and salt concentration were tested by using cultures of strain DMS1T growing on methanol. Addition of FeCl2 to a culture significantly stimulated the growth of strain DMS1T and resulted in a short lag phase and a longer logarithmic growth phase. Since addition of other metals (NiCl2, CoCl2, and NaMoO4) or addition of larger amounts of vitamin and trace element solutions (see above) did not result in additional stimulation of the growth of strain DMS1T, only FeCl2 was added in all subsequent experiments. The FeCl2-amended cultures were characterized by large quantities of a black precipitate consisting of FeS and FeS2. To avoid unnecessary large quantities of the precipitates in cultures of strain DMS1T, we determined the minimal concentration of FeCl2 which resulted in maximal stimulation of growth (Fig. 4). Addition of FeCl2 to a final concentration of 10 mg/liter resulted in maximal stimulation of strain DMS1T growth.
FIG. 4.
Effect of adding iron (FeCl2) on the maximum growth rates of cultures of Methanomethylovorans hollandica growing on methanol.
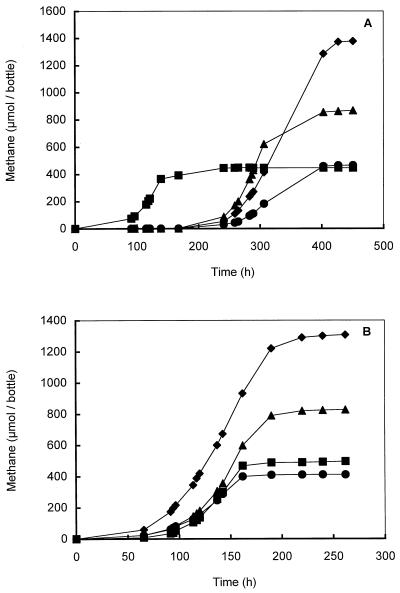
Strain DMS1T grew optimally between pH 6.5 and 7.0. The growth rate was low (<0.01 h−1) or there was no growth in cultures having pH values below 6.0 and above 8.0. The optimum temperature was about 34 to 37°C, and no growth was observed at temperatures above 40°C. Strain DMS1T exhibited optimal growth at NaCl concentrations of 0 to 40 mM. The salt tolerance range of the strain was 0 to 300 mM. At salt concentrations above 400 mM no growth was observed. The tolerance of strain DMS1T to high concentrations of sulfide was tested by growing the strain on methanol in the presence of various sulfide concentrations, and these experiments revealed that sulfide concentrations of 8 mM and higher affected the growth of strain DMS1T. Similarly, we tested the tolerance of strain DMS1T to high DMS concentrations by adding various concentrations of DMS to cultures growing on methanol. The maximal growth rate of strain DMS1T on methanol remained unaffected at DMS concentrations up to 20 mM (the highest concentration tested); however, cultures containing 10 and 20 mM DMS had dramatically longer lag phases than cultures containing 0, 2.5, and 5 mM DMS had.
Catabolic substrates.
Strain DMS1T used methanol, DMS, MT, monomethylamine, dimethylamine, and trimethylamine for growth and methanogenesis but did not use H2-CO2 or acetate. Strain DMS1T could not completely reduce MT or DMS with H2. In the presence of methanol, cultures had a lag phase of about 50 h (Fig. 5A). A prolonged lag phase that was about 250 h long was observed when methanol-grown cells were transferred into monomethylamine-, dimethylamine-, or trimethylamine-containing medium (Fig. 5A). However, after repeated transfers to media containing these substrates, the lag phase was shortened to 50 h (Fig. 5B). The maximum growth rates of strain DMS1T in batch cultures containing methanol and batch cultures containing methylamines were 0.04 to 0.06 and 0.03 to 0.05 h−1, respectively. Growth on MT and DMS (maximum growth rates, 0.005 to 0.02 h−1) occurred only in batch or chemostat cultures in which DMS was added via the N2-CO2 gas stream as described above. Degradation of DMS was characterized by the presence of methane and MT in the outgoing gas stream of the chemostat (Fig. 2B).
FIG. 5.
Growth curves for strain DMS1T grown on methanol (20 mM) (■), monomethylamine (20 mM) (●), dimethylamine (20 mM) (▴), and trimethylamine (20 mM) (⧫). (A) Cultures inoculated with a methanol-grown culture. (B) Cultures after two transfers onto medium containing the appropriate substrate.
The concentration of H2S in a chemostat culture appeared to have a dramatic impact on the conversion of DMS and MT and thus on the formation of methane. Addition of sulfide to either the gas stream or the reaction vessel itself (final concentration, 1 mM; less than 50% of the sulfide concentration in methanol-containing culture medium) instantaneously resulted in a dramatic increase in the MT concentration in the outgoing gas, and this was followed by a complete collapse of the formation of both MT and methane (Fig. 6A and B). Addition of 1 ml of a similar medium that was anaerobic but not sulfide reduced resulted in increases in both the MT concentration and methane production (Fig. 6C).
FIG. 6.
Effects of adding sulfide and adding anaerobic but not sulfide-reduced medium on DMS degradation in chemostat cultures of strain DMS1T. Sulfide was added either in the gas stream (A) or to the culture vessel (1 mM) (B). Also, 1 ml of nonreduced medium was added to the culture vessel (C). The arrows indicate when compounds were added. The concentrations of methane (□) and MT (■) in the effluent gas from the chemostat were determined.
Although accurate stoichiometric analysis of degradation of DMS by strain DMS1T in the chemostat appeared to be difficult, formation of methane, MT, and H2S (detected in the outgoing gas) (Fig. 2B) indicated that DMS was probably degraded in a manner similar to the manner described for other methanogens (with MT as an intermediate), as described by the following equation:
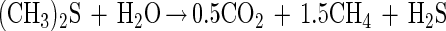 |
We were not able to estimate the stoichiometric formation of H2S because of the formation of black sulfide precipitates with iron and other metal ions present in the medium.
Cell suspension studies.
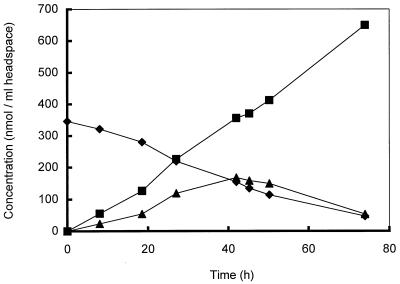
To study the conversion of DMS by strain DMS1T in more detail, samples of the culture fluid in the chemostat were frequently removed and used for cell suspension experiments performed in serum bottles. In these incubation experiments, DMS was converted to MT and methane (Fig. 7). Accumulation of MT was transient, and at lower DMS concentrations MT was converted to methane. Addition of 2-[bis(2-hydroxyethyl)amino]ethanesulfonic acid (BES) completely inhibited MT and methane production from DMS. Incubation of DMS-grown cells from the chemostat with either DMS or methanol resulted in immediate production of methane, whereas addition of DMS to methanol-grown cells did not (data not shown).
FIG. 7.
Conversion of DMS (⧫) and MT (▴) and formation of methane (■) in 9-ml cell suspension samples from the chemostat incubated in 60-ml serum bottles under an N2 atmosphere after DMS was added.
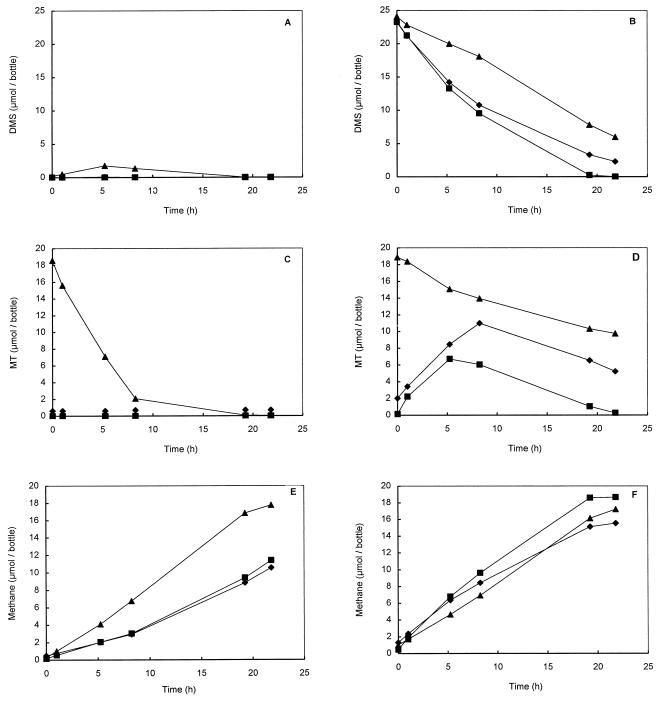
In the chemostat culture, sulfide had a dramatic impact on MT and methane formation. To determine whether this was caused by the dynamics of methyl transfer reactions or by toxic effects, DMS-degrading and methanol-degrading cell suspensions were amended with MT or sulfide. A methanol-containing cell suspension was included as a control to check for the toxic effects of MT and sulfide. The effects of MT and sulfide on DMS degradation differed dramatically from the effects on methanol degradation. Addition of MT (1.5 to 3.6 mM) to DMS-degrading cell suspensions resulted in significant decreases in DMS degradation (Fig. 8B). In contrast, addition of MT (1.5 to 3.6 mM) to the same cell suspensions amended with methanol revealed that MT at these concentrations was not toxic (Fig. 8C). Surprisingly, addition of MT to methanol-containing cell suspensions stimulated both MT degradation and methane formation (Fig. 8B and C). The MT degradation was accompanied by a strong accumulation of DMS (Fig. 8A). Apparent formation of DMS from MT and methanol has also been observed in sediment slurry preparations (20).
FIG. 8.
Conversion of DMS and MT and formation of methane by 9-ml cell suspension samples taken from the chemostat amended with methanol (A, C, and E) or DMS (B, D, and F) and incubated in 60-ml serum bottles. Symbols: ■, controls containing methanol or DMS; ▴, samples containing methanol plus MT or DMS plus MT; ⧫, samples containing methanol plus Na2S or DMS plus Na2S.
Addition of sodium sulfide (1.2 to 2.9 mM) resulted in slight decreases in DMS degradation and methane formation in DMS-containing preparations only 10 h after the addition. More MT accumulated in these preparations than in the control preparations containing DMS. Incubation of DMS-grown cells with methanol and various concentrations of Na2S revealed that like MT, sulfide was not toxic at concentrations below 4 mM.
Phylogenetic and taxonomic analysis.
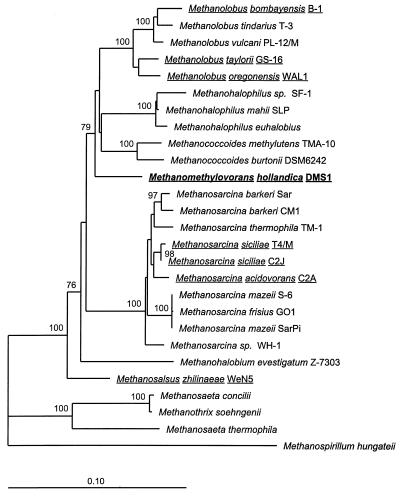
Two DNA fragments of about 1,000 and 1,350 bases long that were homologous to the rRNA gene of strain DMS1T were amplified in vitro, cloned in the pCR II vector and Escherichia coli, and partially sequenced. Chimera Check analysis of the Ribosomal Database Project data (22) revealed that the strain DMS1T 16S rRNA gene sequence was derived from a single target DNA sequence. Database searches for homologous sequences of 16S rRNA genes of other organisms revealed that the highest similarity value (94.5%) was obtained with the sequences of strain DMS1T and Methanococcoides burtonii. A phylogenetic tree based on a matrix of binary phylogenetic distances which were calculated from the alignment of archaeal 16S rRNA sequences clearly showed that strain DMS1T clusters within the family Methanosarcinaceae (Fig. 9). In addition to the 16S rRNA sequences of closely related methanogens, all known sequences of other DMS-degrading methanogens were incorporated in this tree. The topology of the tree was reevaluated by varying the positions included on the basis of the degrees of conservation and by applying alternative treeing methods (parsimony and bootstrap parsimony analysis).
FIG. 9.
Phylogenetic tree based on a distance matrix prepared from an alignment of partial 16S rRNA sequences (1,404 bases) of Methanomethylovorans hollandica and closely related methanogens. Reference sequences were obtained from the GenBank, EMBL, and Ribosomal Database Project databases. Methanospirillum hungateii was used as the outgroup. Scale bar, 10 base substitutions per 100 bases. The names of methanogens which are able to utilize DMS as a carbon source are underlined.
DISCUSSION
Isolation and physiology of strain DMS1T.
The newly isolated methanogen strain DMS1T is the first obligately anaerobic archaeon which has been directly enriched and isolated from a freshwater sediment in a defined minimal medium containing DMS as the sole carbon and energy source. In contrast to most previously described isolation procedures used for saline environments (5, 8, 12, 16, 17, 23, 32) strain DMS1T could not be obtained from dilution series of batch enrichment cultures. Using a chemostat with a continuous DMS-containing gas stream as a method of enrichment, followed by cultivation in deep agar tubes, resulted in a pure culture. The success of the continuous gas flow chemostat was probably due to maintenance of low sulfide and MT concentrations in the culture, which enhanced DMS conversion (see below). Apparently, DMS degradation by methanogens in saline environments is affected less by MT accumulation and sulfide accumulation. This may be due to the higher in situ concentrations of these compounds which the methanogens experience in these habitats (14, 16). Strain DMS1T occurred as large aggregates of cell clusters that usually consisted of two to four irregularly shaped cocci, which resembled the cocci of Methanosarcina species. Cells of strain DMS1T do not have a protein-containing cell wall, since the cells did not lyse after addition of SDS (1.0 g per liter). Like all other known DMS-degrading methanogens isolated from marine, estuarine, and salt lake sediments, strain DMS1T disproportionated DMS to methane, carbon dioxide, and sulfide with MT as an intermediate (8, 16). Strain DMS1T differs from all other DMS-degrading methanogens, since it was isolated from a freshwater pond and required NaCl concentrations (0 to 0.04 M) that are typical of the NaCl concentrations required by freshwater microorganisms for growth. Since the only substrates utilized by strain DMS1T are methanol, methylamines, MT, and DMS, this organism is considered an obligately methylotrophic methanogen like most other DMS-degrading methanogens.
Incubation experiments performed with cell suspensions obtained from chemostat cultures revealed that strain DMS1T cannot completely reduce MT or DMS with H2, as has been reported for methanol reduction by cultures of Methanosphaera stadtmanae and Methanosphaera cuniculi (2). Compared to growth on methanol (growth rate, 0.04 to 0.06 h−1) and methylamines (growth rate, 0.03 to 0.05 h−1), growth on DMS and MT was slow (growth rate, 0.005 to 0.02 h−1). These growth rates were obtained only in batch or chemostat cultures in which DMS was supplied in a continuous gas stream. Chemostat experiments revealed that accumulation of MT or sulfide significantly affected DMS degradation. This was confirmed by the inhibition of DMS degradation by MT observed in cell suspension experiments (Fig. 8). Transient accumulation of DMS in MT-containing cultures also indicated that transfer of the first methyl group during DMS degradation is a reversible process. This was confirmed by the formation of DMS in suspensions of strain DMS1T cells containing methanol plus MT. Transient accumulation of DMS in MT-containing cultures has also been reported by other authors (8). Accumulation of MT and sulfide in batch cultures makes the methyl transfer energetically less favorable. This may explain the lack of success in previous attempts to enrich DMS-degrading methanogens from freshwater sediments.
Production of the enzymes needed for degradation of methylamines has to be induced, since inoculation of methanol-grown cells of strain DMS1T into mono-, di-, or trimethylamine-containing medium resulted in long initial lag phases (Fig. 5). These lag phases were shorter after repeated transfers on these substrates. Addition of methanol to DMS-grown cells resulted in immediate formation of methane, indicating that production of the enzymes needed for methanol conversion is constitutive.
Phylogenetic and taxonomic analysis.
In its physiological properties, strain DMS1T resembles previously described methanogenic archaea belonging to the genera Methanolobus, Methanohalophilus, and Methanococcoides, the genera to which most of the other DMS- and MT-degrading methanogens belong. Like the methanogens belonging to these genera, strain DMS1T is an obligate methylotroph. In contrast to all of the members of these genera, however, strain DMS1T has the low salt tolerance typical of nonhalophilic microorganisms. Since the morphology of strain DMS1T resembles the morphology of methanogens belonging to the genera Methanolobus and Methanococcoides, as well as the genus Methanosarcina, a precise phylogenetic analysis of the 16S rRNA gene of strain DMS1T had to be performed in order to classify strain DMS1T. On the basis of its position on the phylogenetic tree, we concluded that strain DMS1T is clearly not related to the genus Methanosarcina. The results of the bootstrap analysis, as indicated by the bootstrap values on the phylogenetic tree, revealed, however, that the branching of the genera Methanolobus, Methanococcoides, and Methanohalophilus and strain DMS1T is not well defined. A comparison of the distance between two species belonging to one genus (e.g., Methanococcoides burtonii and Methanococcoides methylutens) with the distance between a species belonging to one of the three genera and strain DMS1T clearly showed that strain DMS1T does not belong to these genera. According to a combined analysis of both the 16S rRNA gene and the MCRI gene (31) of methanogens, levels of similarity of 98% or higher represent interspecies relationships, whereas levels of similarity of 90 to 95% represent intergeneric relationships.
On the basis of its low level of similarity with the most closely related methanogen, Methanococcoides burtonii (94.5%), its position on the phylogenetic tree, its morphology (which is different from the morphology of members of the genera Methanolobus, Methanococcoides, and Methanohalophilus), and its salt tolerance and optimum (which are characteristic of freshwater bacteria), we propose that strain DMS1T is a representative of a novel genus and species. This organism was named Methanomethylovorans hollandica.
Ecological niche of Methanomethylovorans hollandica.
Like the concentrations of MT and DMS in marine, estuarine, and salt lake sediments, the concentrations of MT and DMS in freshwater sediments are low due to the balance between the formation and degradation of these compounds (18). Degradation of MT and DMS occurs anaerobically due to the steep oxygen gradient at the water column-sediment interface, which results in anaerobic conditions in the sediment (19). Various inhibition studies performed with the specific inhibitors BES and sodium tungstate revealed that most (about 95%) of the endogenously produced MT in freshwater sediments is converted by methanogenic archaea (20, 40, 41). Microscopic analysis of DMS-containing sediment slurries with a fluorescence microscope revealed the presence of methanogens which were morphologically identical to Methanomethylovorans hollandica, the organism described in this study. The enrichment conditions used resembled the in situ conditions and therefore made it plausible that Methanomethylovorans hollandica DMS1T is the most important DMS and MT consumer in its natural freshwater environment. This conclusion is supported by the physiological properties of the organism (substrate use, low growth rate, and iron requirement). Whether Methanomethylovorans hollandica is also an important utilizer of methanol and methylamines in situ remains to be investigated. The obligately methylotrophic archaea are able to form a stable community along with the acetoclastic and hydrogenotrophic methanogens.
Due to the reversibility of the DMS conversion, methanogens like strain DMS1T can also be involved in the formation of DMS through methylation of MT. This phenomenon, which previously has been shown to occur in sediment slurries with a freshwater origin (19), might affect the steady-state concentration and consequently the total flux of DMS (and MT) in these systems. Moreover, stimulation of the production of MT and DMS by high concentrations of sulfide (18) also appears to inhibit degradation of these compounds. Therefore, higher steady-state concentrations and fluxes of MT and DMS in sulfide-rich freshwater systems are conceivable.
Description of Methanomethylovorans hollandica gen. nov., sp. nov.
Methanomethylovorans hollandica (Me.tha.no.me.thy.lo′vo.rans. M. L. n. methanum, methane; M. L. n. methylum, methyl; L. adj. vorans, devouring; M. L. n. Methanomethylovorans, methane producing, methyl group consuming; hol.lan′di.ca. L. adj. hollandica, from The Netherlands [Holland], referring to the origin of the type strain). Cells are irregular, nonmotile, coccoid (diameter, 1 to 1.5 μm), and gram negative. Cells normally occur in clusters consisting of two to four cells which form large aggregates. Cells are not sensitive to lysis by 1.0 g of SDS per liter. Trimethylamine, dimethylamine, monomethylamine, methanol, DMS, and MT are catabolic substrates, but H2-CO2 and acetate are not. Growth is most rapid in the presence of 0 to 0.04 M NaCl, and no growth occurs at NaCl concentrations higher than 0.4 M. Optimal growth occurs at pH 6.5 to 7.0, and no growth occurs at pH values lower than 6.0 and higher than 8.0. Growth is most rapid at 34 to 37°C, and very slow or no growth occurs at temperatures below 12°C and above 40°C. Type strain DMS1 was isolated from a slurry prepared from a eutrophic pond sediment (Campus of Dekkerswald Institute, Nijmegen, The Netherlands). Strain DMS1T has been deposited in the culture collection of the Deutsche Sammlung von Mikroorganismen (Braunschweig, Germany).
ACKNOWLEDGMENTS
We thank Wim Willems of the Technical Service Department, University of Nijmegen, for constructing the chemostat headplate, Ron Hochstenbach and Wander Sprenger for discussions related to the 16S rRNA sequence analysis, and Erik van Wesel for assistance with electron microscopy.
This work was supported by The Netherlands Organization for the Advancement of Pure Research (NWO) as part of the program “Verstoring van Aardsystemen.”
REFERENCES
- 1.Applied Biosystems, Inc. Taq DyeDeoxy terminator cycle sequencing kit user bulletin no. 901497, revision E. Foster City, Calif: Applied Biosystems, Inc.; 1992. [Google Scholar]
- 2.Biavati B, Vasta M, Ferry J G. Isolation and characterization of “Methanosphaera cuniculi” sp. nov. Appl Environ Microbiol. 1988;54:768–771. doi: 10.1128/aem.54.3.768-771.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Derikx P J L, Op den Camp H J M, van der Drift C, van Griensven L J L D, Vogels G D. Odorous sulfur compounds emitted during production of compost used as a substrate in mushroom cultivation. Appl Environ Microbiol. 1990;56:176–180. doi: 10.1128/aem.56.1.176-180.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Zwart J M M, Nelisse P N, Kuenen J G. Isolation and characterization of Methylophaga sulfidovorans sp. nov.: an obligate methylotrophic aerobic, DMS oxidizing bacterium from a microbial mat. FEMS Microbiol Ecol. 1996;20:261–271. [Google Scholar]
- 5.Elberson M A, Sowers K R. Isolation of an aceticlastic strain of Methanosarcina siciliae from marine canyon sediments and emendation of the species description for Methanosarcina siciliae. Int J Syst Bacteriol. 1997;47:1258–1261. doi: 10.1099/00207713-47-4-1258. [DOI] [PubMed] [Google Scholar]
- 6.Felsenstein J. Numerical methods of inferring evolutionary trees. Q Rev Biol. 1982;57:379–404. [Google Scholar]
- 7.Finster K, King G M, Bak F. Formation of methyl mercaptan and dimethyl sulfide from methoxylated aromatic compounds in anoxic marine and freshwater sediments. FEMS Microbiol Ecol. 1990;74:295–302. [Google Scholar]
- 8.Finster K, Tanimoto Y, Bak F. Fermentation of methanethiol and dimethylsulfide by a newly isolated methanogenic bacterium. Arch Microbiol. 1992;157:425–430. [Google Scholar]
- 9.Fitch W M, Margoliash E. Construction of phylogenetic trees: a method based on mutation distances as estimated by cytochrome c sequences is of general applicability. Science. 1967;155:279–284. doi: 10.1126/science.155.3760.279. [DOI] [PubMed] [Google Scholar]
- 10.Hutten T J, de Jong M H, Peeters B P H, van der Drift C, Vogels G D. Coenzyme M derivatives and their effects on methane formation from carbon dioxide and methanol by cell extracts of Methanosarcina barkeri. J Bacteriol. 1981;145:27–34. doi: 10.1128/jb.145.1.27-34.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jarrell K F, Faguy D, Hebert A M, Kalmokoff M L. A general method of isolating high molecular weight DNA from methanogenic archaea (archaebacteria) Can J Microbiol. 1992;38:65–68. doi: 10.1139/m92-010. [DOI] [PubMed] [Google Scholar]
- 12.Kadam P C, Ranade D R, Mandelco L, Boone D R. Isolation and characterization of Methanolobus bombayenis sp. nov., a methylotrophic methanogen that requires high concentrations of divalent cations. Int J Syst Bacteriol. 1994;44:603–607. [Google Scholar]
- 13.Kiene R P, Bates T S. Biological removal of dimethyl sulfide from sea water. Nature. 1990;345:702–705. [Google Scholar]
- 14.Kiene R P, Service S K. Decomposition of dissolved DMSP and DMS in estuarine waters: dependence on temperature and substrate concentration. Mar Ecol Prog Ser. 1991;76:1–11. [Google Scholar]
- 15.Kiene R P, Hines M E. Microbial formation of dimethyl sulfide in anoxic Sphagnum peat. Appl Environ Microbiol. 1995;61:2720–2726. doi: 10.1128/aem.61.7.2720-2726.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kiene R P, Oremland R S, Catena A, Miller L G, Capone D G. Metabolism of reduced methylated sulfur compounds in anaerobic sediments and by a pure culture of an estuarine methanogen. Appl Environ Microbiol. 1986;52:1037–1045. doi: 10.1128/aem.52.5.1037-1045.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Y, Boone D R, Choy C. Methanohalophilus oregonense sp. nov., a methylotrophic methanogen from an alkaline, saline aquifer. Int J Syst Bacteriol. 1990;40:111–116. [Google Scholar]
- 18.Lomans B P, Smolders A, Intven L, Pol A, Op den Camp H J M, van der Drift C. Formation of dimethyl sulfide and methanethiol in anoxic freshwater sediments. Appl Environ Microbiol. 1997;63:4741–4747. doi: 10.1128/aem.63.12.4741-4747.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lomans B P, Op den Camp H J M, Pol A, Vogels G D. Anaerobic versus aerobic degradation of dimethyl sulfide and methanethiol in anoxic freshwater sediments. Appl Environ Microbiol. 1999;65:438–443. doi: 10.1128/aem.65.2.438-443.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lomans B P, Op den Camp H J M, Pol A, Vogels G D. The role of methanogens and other bacteria in the degradation of dimethyl sulfide and methanethiol in anoxic freshwater sediments. Appl Environ Microbiol. 1999;65:2116–2136. doi: 10.1128/aem.65.5.2116-2121.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lovelock J E, Maggs R J, Rasmussen R A. Atmospheric dimethyl sulfide and the natural sulfur cycle. Nature. 1972;237:452–453. [Google Scholar]
- 22.Maidak B L, Cole J R, Parker C T, Jr, Garrity G M, Larsen N, Li B, Lilburn T G, McCaughey M J, Olsen G J, Overbeek R, Pramanik S, Schmidt T M, Tiedje J M, Woese C R. A new version of the RDP (Ribosomal Database Project) Nucleic Acids Res. 1999;27:171–173. doi: 10.1093/nar/27.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mathrani I M, Boone D R, Mah R A, Fox G E, Lau P P. Methanohalophilus zhilinae sp. nov., an alkaliphilic, halophilic, methylotrophic methanogen. Int J Syst Bacteriol. 1988;38:139–142. doi: 10.1099/00207713-38-2-139. [DOI] [PubMed] [Google Scholar]
- 24.McBride L J, Koepf S M, Gibbs R A, Salser W, Mayrand P E, Hunkapiller M W, Kronick M N. Automated DNA sequencing methods involving polymerase chain reaction. Clin Chem. 1989;35:2196–2201. [PubMed] [Google Scholar]
- 25.Ni S, Boone D R. Isolation and characterization of a dimethyl sulfide-degrading methanogen, Methanolobus siciliae HI350, from an oil well, characterization of M. siciliae T4/MT, and emendation of M. siciliae. Int J Syst Bacteriol. 1991;41:410–416. doi: 10.1099/00207713-41-3-410. [DOI] [PubMed] [Google Scholar]
- 26.Oremland R S, Zehr J P. Formation of methane and carbon dioxide from dimethylselenide in anoxic sediments and by a methanogenic bacterium. Appl Environ Microbiol. 1986;52:1031–1036. doi: 10.1128/aem.52.5.1031-1036.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pfennig N. Rhodocyclus purpureus gen. nov. and sp. nov., a ring-shaped, vitamin B12-requiring member of the family Rhodospirillaceae. Int J Syst Bacteriol. 1978;28:283–288. [Google Scholar]
- 28.Rajagopal B S, Daniels L. Investigations of mercaptans, organic sulfides, and inorganic sulfur sources for the growth of methanogenic bacteria. Curr Microbiol. 1986;14:137–144. [Google Scholar]
- 29.Raskin L, Stromley J M, Rittmann B E, Stahl D A. Group-specific 16S rRNA hybridization probes to describe natural communities of methanogens. Appl Environ Microbiol. 1994;60:1232–1240. doi: 10.1128/aem.60.4.1232-1240.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 31.Springer E, Sachs M S, Woese C R, Boone D R. Partial gene sequences for the A subunit of methyl-coenzyme M reductase (mcrI) as a phylogenetic tool for the family Methanosarcinaceae. Int J Syst Bacteriol. 1995;45:554–559. doi: 10.1099/00207713-45-3-554. [DOI] [PubMed] [Google Scholar]
- 32.Stetter K O. Genus II. Methanolobus König and Stetter 1983, 439. In: Staley J T, Bryant M P, Pfennig N, Holt J G, editors. Bergey’s manual of systematic bacteriology. Vol. 3. Baltimore, Md: The Williams & Wilkins Co.; 1989. pp. 2205–2207. [Google Scholar]
- 33.Tanimoto Y, Bak F. Anaerobic degradation of methyl mercaptan and dimethyl sulfide by newly isolated sulfate-reducing bacteria. Appl Environ Microbiol. 1994;60:2450–2455. doi: 10.1128/aem.60.7.2450-2455.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Visscher P T, Quist P, van Gemerden H. Methylated sulfur compounds in microbial mats: in situ concentrations and metabolism by a colorless sulfur bacterium. Appl Environ Microbiol. 1991;57:1758–1763. doi: 10.1128/aem.57.6.1758-1763.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Visscher P T, Taylor B F. A new mechanism for the aerobic catabolism of dimethyl sulfide. Appl Environ Microbiol. 1993;59:3784–3789. doi: 10.1128/aem.59.11.3784-3789.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Visscher P T, Taylor B F, Kiene R P. Microbial consumption of dimethyl sulfide and methanethiol in coastal marine sediments. FEMS Microbiol Ecol. 1995;18:145–154. [Google Scholar]
- 37.Widdel F, Bak F. Gram-negative mesophilic sulfate-reducing bacteria. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K H, editors. The prokaryotes. 2nd ed. IV. New York, N.Y: Springer-Verlag; 1992. pp. 3352–3378. [Google Scholar]
- 38.Zehnder A J B, Wuhrmann K. Titanium(III) citrate as a non-toxic oxidation-reduction buffering system for the culture of obligate anaerobes. Science. 1976;194:1165–1166. doi: 10.1126/science.793008. [DOI] [PubMed] [Google Scholar]
- 39.Zeyer J, Eicher P, Wakeham S G, Schwarzenbach R P. Oxidation of dimethyl sulfide to dimethylsulfoxide by phototrophic purple bacteria. Appl Environ Microbiol. 1987;53:2026–2032. doi: 10.1128/aem.53.9.2026-2032.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zinder S H, Brock T D. Methane, carbon dioxide, and hydrogen sulfide production from the terminal methiol group of methionine by anaerobic lake sediments. Appl Environ Microbiol. 1978;35:344–352. doi: 10.1128/aem.35.2.344-352.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zinder S H, Brock T D. Production of methane and carbon dioxide from methane thiol and dimethyl sulfide by anaerobic lake sediments. Nature. 1978;273:226–228. doi: 10.1128/aem.35.2.344-352.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]