Abstract
Diel periodicity in the expression of key genes involved in carbon and nitrogen assimilation in marine Synechococcus spp. was investigated in a natural population growing in the surface waters of a cyclonic eddy in the northeast Atlantic Ocean. Synechococcus sp. cell concentrations within the upper mixed layer showed a net increase of three- to fourfold during the course of the experiment (13 to 22 July 1991), the population undergoing approximately one synchronous division per day. Consistent with the observed temporal pattern of phycoerythrin (CpeBA) biosynthesis, comparatively little variation was found in cpeBA mRNA abundance during either of the diel cycles investigated. In marked contrast, the relative abundance of transcripts originating from the genes encoding the large subunit of ribulose bisphosphate carboxylase/oxygenase (rbcL) and glutamine synthetase (glnA) showed considerable systematic temporal variation and oscillated during the course of each diel cycle in a reciprocal rhythm. Whereas activation of rbcL transcription was clearly not light dependent, expression of glnA appeared sensitive to endogenous changes in the physiological demands for nitrogen that arise as a natural consequence of temporal periodicity in photosynthetic carbon assimilation. The data presented support the hypothesis that a degree of temporal separation may exist between the most active periods of carbon and nitrogen assimilation in natural populations of marine Synecoccoccus spp.
Marine Synechococcus spp. are among the most abundant and cosmopolitan members of the photosynthetic picoplankton: a taxonomically mixed assemblage of small (<2.0-μm diameter) prokaryotic and eukaryotic microorganisms that account for the major fraction of primary production in the world’s open oceans (40). Although genetically divergent (14, 51, 54, 66, 67), oceanic strains of Synechococcus spp. belong to a physiologically coherent group (marine cluster A) of cyanobacteria within the unicellular order Chroococcales (59). All known isolates are non-nitrogen-fixing, obligate photoautotrophs (59, 62) that produce spectroscopically distinct biliproteins (including phycoerythrin [PE]) as accessory components of their light-harvesting apparatuses (1, 35, 36). Many are also capable of flagellum-free swimming motility and, in one demonstrated case at least, display positive chemotaxis toward a variety of organic and inorganic nitrogenous compounds (64).
Following their first description in 1979 (22, 61), a reasonable (if incomplete) understanding has developed of the biological and physicochemical factors that regulate the growth and productivity of marine Synechococcus spp. in the world’s oceans (18, 19, 23, 24, 29, 46, 47, 56, 60, 65, 66). While there are undoubted exceptions to the paradigm, these organisms are generally at their most conspicuous (but not necessarily their most productive) in oligotrophic surface waters, where the limited supply of inorganic nutrients appears to exclude competition in the form of larger and potentially faster-growing eukaryotic phytoplankton (19, 23, 62).
Estimates of Synechococcus sp. growth rates in open waters are somewhat variable, but for active populations they tend to cluster around a mean of about one division per day (7, 18, 29, 56). Like many groups of marine phytoplankton, there is mounting evidence that the daily progression through the Synechococcus sp. cell cycle may be entrained in situ by the natural diel alternation in irradiance. While a number of previous studies have clearly hinted at the phenomenon (7, 8, 25, 62), the recent cytological investigation conducted by Vaulot and coworkers (56) was the first to confirm that DNA replication and cell division can become tightly synchronized in Synechococcus spp. growing in open waters. If such rhythmic behavior is a common feature of natural populations, an obvious rationale can be developed to explain several earlier reports (20, 25, 34) of diel oscillations in cell rRNA content and macromolecular composition in these organisms. Like the diel phasing of DNA replication and cytokinesis (56), temporal patterns of this type are an entirely predictable feature of synchronously dividing populations.
Diel rhythms in the synthesis and accumulation of various mRNAs in natural populations of Synechococcus spp. and other marine cyanobacteria have also been described (10, 26, 37, 38, 70), but whether these might have their origin in cell cycle-related events has not received consideration. Cell division in the marine Synechococcus sp. strain WH7803 is known to be under circadian control (50), and there is convincing preliminary evidence that expression of the rbcL gene (encoding the large subunit of the Calvin cycle enzyme, ribulose bisphosphate carboxylase/oxygenase [RubisCO]) may also be regulated in this way (37). It is an open question, however, whether rbcL transcription in Synechococcus spp. is controlled directly by as-yet-unidentified clock genes or is regulated in response to temporal oscillations in other metabolic processes that may themselves be circadian in nature.
The RubisCO genes of marine Synechococcus spp. are phylogenetically distinct from those of other cyanobacteria and may have been acquired comparatively recently by lateral gene transfer (63). In the marine Synechococcus sp. strain WH7803, both RubisCO subunits are cotranscribed with a homologue of the Synechococcus sp. strain PCC7942 ccmK gene (63), which encodes a structural component of the carboxysome, the site of CO2 fixation in cyanobacteria. RubisCO expression is enhanced considerably by light and, as in other cyanobacteria (11, 27), appears to be regulated primarily at the level of transcription. In this regard, the temporal pattern of RubisCO expression displayed by Synechococcus sp. strain WH7803 in culture closely parallels that seen in natural populations of photosynthetic picoplankton growing in subtropical oceanic surface waters (37, 38).
By analogy to the adaptive strategies adopted by other cyanobacteria growing at low concentrations of combined nitrogen, the principal route of inorganic nitrogen assimilation in marine Synechococcus spp. is thought to occur via the glutamine synthetase (GS)-glutamate synthase pathway and to be dependent upon photosynthetically generated ATP and reductant (31, 53). While the existence of alternative pathways has not been excluded experimentally, from what is known of the kinetic properties of these systems in other cyanobacteria (32) it appears unlikely they could make other than a very minor contribution to N assimilation in Synechococcus spp. at the low ambient nitrogen concentrations typical of oceanic surface waters.
The pathways of carbon and nitrogen assimilation are metabolically linked at the level of GS in cyanobacteria, and the enzyme is extensively regulated in response to a variety of environmental signals. GS is rapidly inactivated in the dark and also by ammonium ions in the well-studied model system Synechocystis sp. strain PCC6803 and several other cyanobacteria (45, 49). The nature of the reversible modification system present in Synechocystis sp. strain PCC6803 is distinct from that described for the GS of enteric bacteria, however, in that adenylylation is not involved. Nevertheless, some elements of the signal transduction system that lead to the activation or inactivation of GS do appear to be conserved between cyanobacteria and enteric bacteria. The regulatory protein PII (encoded by glnB) is present in both groups but in cyanobacteria is activated by phosphorylation rather than uridinylylation (16). As with GS, activation of PII is regulated by nitrogen availability and is dependent upon photosynthetic electron transport (17, 45).
Transcription of glnA and glnB in Synechococcus sp. strain PCC 7942 and Synechocystis sp. strain PCC 6803 is regulated by the positive transcription factor NtcA, a functional analogue of the nitrogen regulator of enteric bacteria, NtrC (17, 57). NtcA is widely distributed among cyanobacteria (including the marine strain Synechococcus sp. strain WH7803 [28]) and in the absence of ammonium binds to the promoter regions of a suite of N-regulated genes, including glnA and glnB. Transcription of glnA increases two- to threefold in cells of Synechococcus sp. strain PCC7942 switched from ammonium- to nitrate-containing growth medium and by a somewhat greater margin (5- to 10-fold) during nitrogen deprivation (13).
Enhanced glnA transcription in the absence of ammonium leads to parallel increases in GS synthesis and activity (12, 13), and a similar correlation among nitrogen regimen, glnB mRNA abundance, and PII protein levels has been reported recently for Synechocystis sp. strain PCC6803 (16, 52). Expression of glnA at the transcriptional level is not light dependent in Synechococcus sp. strain PCC 7942 (6), but like psbA (encoding the D2 protein of photosystem II) and the rbcL gene of marine Synechococcus spp. (37), there is some evidence for circadian control (30).
Establishing how temporal and spatial variability in the environment structures the molecular biological responses of natural populations of marine microorganisms is a central theme in contemporary biological oceanography. The present field-based study was designed to examine the temporal (diel) periodicity in the abundance of three different Synechococcus sp. mRNAs that encode proteins central to photosynthetic light harvesting (PE; cpeBA), carbon fixation (RubisCO; rbcL), and nitrogen assimilation (glutamine synthetase; glnA). The aims of the investigation were to assess the extent to which the three genes might be differentially expressed during the diel cycle and to relate the findings to the temporal pattern of Synechococcus sp. population growth and cell division in situ. The data reported reveal a quasireciprocal rhythm in the relative abundance of rbcL and glnA mRNAs and point to at least some degree of temporal separation between the most active phases of C and N assimilation in natural populations of Synechococcus spp.
MATERIALS AND METHODS
Observations were made during a 10-day period in July 1991 at a series of drifting stations located within a cyclonic eddy in the northeast Atlantic Ocean (Fig. 1). The study site was selected during passage of the research vessel (RRS Charles Darwin; cruise CD61) to the sea area to the south of Iceland following receipt of thermal infrared satellite images of the region on 10 July 1991. During the previous month an extensive mesoscale (∼250,000-km2) bloom of coccolithophorid algae had occurred in these subpolar waters (15, 21), but by the time the research vessel arrived on station (12 July 1991) all surface expression of the bloom had disappeared (21). Mapping of surface hydrographic and biological properties within the eddy revealed that the postbloom phytoplankton community comprised a mixed assemblage of micro- and picophytoplankton dominated by diatoms and microflagellates and by Synechococcus spp., respectively.
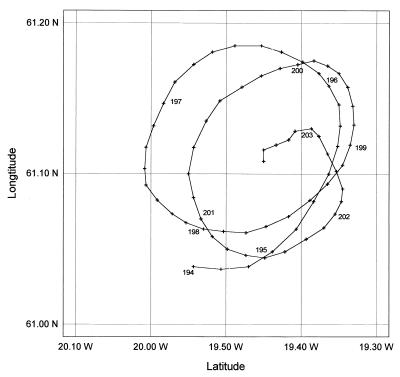
FIG. 1.
Track of the ARGOS drifter deployed on 13 July 1991 (Julian day 194). The position of the drifter at 0000 hours on each subsequent day is indicated by the day number. (Data courtesy of Bob Barrett and Robin Pingree.)
A drogued (10 m) ARGOS drifter was deployed at the center of the eddy (61°05′N, 20°W) on 13 July 1991. Thereafter, the research vessel was operated in Lagrangian mode by maintaining close station with the drifter until the morning of 22 July 1991 (for further details of the study site and its hydrographic properties, see reference 21). Plankton samples were obtained from near-surface (∼5 m) waters by using an outlet of the ship’s nontoxic seawater supply (residence time from inlet to outlet, ∼2 min) or from discrete depths by using 30-liter GO-Flo bottles (General Oceanics, Miami, Fla.) deployed from a Kevlar line. Surface nutrient concentrations were determined continuously while on station with a Technicon autoanalyzer and standard analytical procedures (5, 21).
Synechococcus sp. cell counts.
The abundance of Synechococcus sp. cells was estimated by epifluorescence microscopy (19). Seawater samples of known volume (10 to 100 ml) were prescreened through a 250-μm-mesh plankton net, and phytoplankton cells were collected by filtration on 25-mm-diameter, 0.2-μm-pore-size polycarbonate membranes (Nuclepore Corp.) overlaying a Whatman GF/F support filter. When necessary (i.e., during times of excessive ship movement), filtered samples mounted on glass slides in nonfluorescent immersion oil were stored in darkness at 4°C, but in all cases they were counted onboard ship within 24 h of collection.
Determination of PE concentrations.
Synechococcus sp. PE concentrations were determined as previously described with Synechococcus sp. strain WH7803 cells as the reference standard (69). Seawater samples (1 to 2 liters) were fractionated through 47-mm-diameter 2.0- and 0.6-μm-pore-size polycarbonate filters (Poretics Corp.) under gentle vacuum (<10 cm of Hg), and Synechococcus sp. cells retained on the 0.6-μm-pore-size filter were washed into 1.5 ml of Whatman GF/F-filtered seawater (collected from a depth of 100 m). Owing to equipment failure, it was not possible to carry out PE analyses at sea. The concentrated samples were centrifuged at 13,000 × g for 1 min, the seawater supernatant was aspirated, and the pelleted phytoplankton cells were taken up in 1 ml of 50% (vol/vol) aqueous glycerol and stored in darkness at −20°C. PE concentrations were estimated on return to the laboratory with a Perkin-Elmer LS5 spectrofluorimeter and the instrument settings and correction procedures previously reported (69).
RNA extraction and Northern analysis.
Seawater samples (10 to 20 liters) were fractionated through 90-mm-diameter, 2.0- and 0.6-μm-pore-size polycarbonate filters (Poretics Corp.) at negative vacuum pressures of 10 and 60 cm of Hg for the 2.0- and 0.6-μm-pore-size filters, respectively. Routinely, this fractionation procedure results in the retention of greater than 90% of the original Synechococcus sp. biomass on the 0.6-μm-pore-size filter whereas eukaryotic cells are either trapped by the 2.0-μm-pore-size filter or disrupted by the high vacuum pressure employed during filtration through the 0.6-μm-pore-size filter. The efficiency of the procedure was monitored by epifluorescence microscopic examination of the cell material retained on the 0.6-μm-pore-size filter. In all cases, the only intact chlorophyll-containing cells observed also exhibited PE fluorescence; i.e., only Synechococcus sp. cells were present in the 0.6- to 2.0-μm size-fractionated samples under the conditions applied.
Sample volume was determined by the amount of seawater that could be filter fractioned within 15 min of sample collection, and all nocturnal samples were processed in a darkened laboratory. Synechococcus sp. cells retained on the 0.6-μm-pore-size filter were washed with 5 ml of TEN buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA, 250 mM NaCl), resuspended in 1 ml of ice-cold extraction buffer (100 mM LiCl, 50 mM Tris-HCl [pH 7.5], 1 mM EGTA, 1% [wt/vol] sodium dodecyl sulfate [SDS]), and snap frozen prior to storage at −20°C until further processing ashore.
Total RNA was isolated by a hot-phenol extraction procedure (70). Synechococcus sp. cells were homogenized in extraction buffer at 65°C for 1 to 2 min and deproteinized with an equal volume of hot (65°C) phenol-chloroform-isoamyl alcohol (25:24:1) for 5 min. The aqueous phase was recovered by centrifugation (13,000 × g for 5 min) and reextracted with chloroform-isoamyl alcohol (24:1) at ambient temperature. Nucleic acids were precipitated from the aqueous phase with 2.5 volumes of 100% ethanol at −20°C for 24 h and recovered by centrifugation (13,000 × g for 20 min). The pelleted material was washed in 75% (vol/vol) ethanol and taken up in 100 μl of DNase buffer (100 mM sodium acetate, 10 mM MgCl2). DNA was hydrolyzed at 37°C for 30 min in the presence of 10 U of DNase (RNase free; Boehringer Mannheim). Following inactivation and removal of DNase by phenol-chloroform extraction, RNA was pelleted by ethanol precipitation and taken up in 50 μl of diethylpyrocarbonate-treated deionized water. RNA concentrations were determined by absorbance at 260 nm, and the integrity of the samples was confirmed by electrophoresis through formaldehyde agarose gels stained with ethidium bromide (3).
Northern dot blots were performed as described previously (70) following heat denaturation of samples (1 to 2 μg of RNA) in 200 μl of 5× SSC (1× SSC is 0.15 M NaCl, 15 mM Na citrate, pH 7.5)–10 mM EDTA at 65°C for 15 min. Following transfer under gentle vacuum (<4 cm of Hg) to positively charged nylon membranes (Boehringer Mannheim), the blots were air dried and the RNA was immobilized by UV irradiation at 305 nm.
Dot blots were hybridized at high stringency with double-stranded DNA probes derived from the oceanic cyanobacterium Synechococcus sp. strain WH8103 (68, 71). The probes employed were (i) a 699-bp BamHI-BglII fragment of pRBGL1.4, which includes the 5′ region and the first 630 nucleotides of rbcL; (ii) an EcoRI-SalI fragment of pPE1.3, which includes 198 bp of upstream sequence, the coding region of cpeB, and the 5′ end of cpeA of the class 1 PE operon; and (iii) the EcoRI insert of pGlnA1.1, which includes most of the coding region of glnA plus 350 bp of upstream sequence. Probe DNA was isolated in low-melting-point agarose following restriction digestion of plasmid DNA, and the recovered fragments were purified with glass milk. DNA was labelled with digoxigenin-dUTP (Boehringer Mannheim) by random priming according to the manufacturer’s recommendations.
Membranes were prehybridized for 4 h at 55°C in a solution containing 50% (vol/vol) formamide, 5× SSC, 0.1% (wt/vol) SDS, 1% (wt/vol) blocking reagent (Boehringer Mannheim), and 5% (wt/vol) dextran sulfate. Probe DNA (25-ng/ml final concentration) and sheared salmon sperm DNA (100-μg/ml final concentration) were denatured at 100°C for 10 min and added to fresh solution prior to overnight hybridization at 55°C.
Northern dot blots were stringency washed twice in 2× SSC–0.1% (wt/vol) SDS at ambient temperature for 5 min and twice again at 65°C in 0.2× SSC–0.1% (wt/vol) SDS for 30 min. Hybrids were detected by immunochemistry with alkaline phosphatase-conjugated anti-digoxigenin and the chemiluminescent substrate AMPPD (Tropix Inc.) in conjunction with Amersham Hybond enhanced chemiluminescence film (70). The resulting luminographs were quantified by densitometry with a GDS 8000 gel documentation system (UVP Ltd.) and the Gel Works 1D Advanced software package.
RESULTS
Filter fractionation of Synechococcus spp.
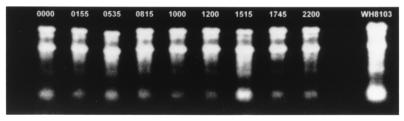
An essential requirement of several of the analyses carried out during the present study was the development of a rapid isolation procedure to concentrate Synechococcus sp. cells from bulk phytoplankton samples. The differential-fractionation technique employed was a minor refinement of an earlier protocol (69) that had been field tested during previous cruises in the northeast Atlantic Ocean (RRS Discovery cruise D189 and RRS Charles Darwin cruise CD47) and also by others for phytoplankton communities sampled off the North Carolina coast (25). In confirmation of the absence of eukaryotic algae from the fractionated samples prepared for the Northern analyses, only 16S and 23S rRNA bands were detected on denaturing RNA gels (Fig. 2) plus an additional band of intermediate molecular mass which is a diagnostic degradation product of the 23S rRNA subunit found in cyanobacterial RNA samples extracted in buffers containing low concentrations of Mg2+ ions (25, 48).
FIG. 2.
Ethidium bromide-stained formaldehyde agarose gel of filter-fractionated RNA samples collected on 20 July 1991 (diel 2). The lane on the far right contains 5 μg of RNA isolated from the marine Synechococcus sp. strain WH8103.
Temporal variability in Synechococcus sp. abundance and PE content.
Within the eddy, Synechococcus sp. cell concentrations were elevated (∼2-fold) in comparison to those of surrounding waters and showed a net increase of 3- to 4-fold between the first and last days on station (Table 1). During this period, the sea surface temperature increased by ∼1°C, leading to the development of some weak secondary structure in the upper 10 m of the water column toward the end of the study (Table 1 and Figure 3, top). Nitrate concentrations within the upper mixed layer were within the range of 0.6 to 2.1 μM, whereas ammonium concentrations increased with depth from the surface (<0.1 μM) to reach 0.15 to 0.2 μM near the base of the mixed layer (5).
TABLE 1.
Temporal variability in Synechococcus sp. cell numbers within the surface mixed layer of a cyclonic eddy in the northeast Atlantic during July 1991
| Date (July) | SSTb (°C) | Salinity (‰) | 108 Cells liter−1 |
|---|---|---|---|
| 12 | 12.34 | 35.18 | 0.76 |
| 16 | 12.39 | 35.17 | 1.32 |
| 19 | 13.03 | 35.18 | 2.9 |
| 20 | 13.29 | 35.18 | 3.76 |
| 22 | 13.29 | 35.18 | 2.72 |
| 12a | 11.72 | 35.12 | 0.43 |
| 23a | 12.47 | 35.11 | 0.66 |
Samples obtained from surrounding waters immediately adjacent to the eddy.
SST, sea surface temperature.
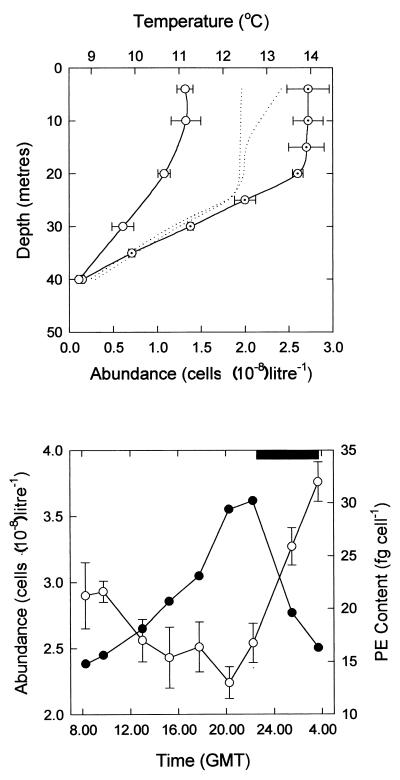
FIG. 3.
(Top) Vertical distribution of temperature (dotted lines) and Synechococcus sp. cells (± standard error [SE]) on 16 July (open circles) and 21 July (dotted circles) 1991. (Bottom) Diel variation in Synechococcus sp. cell counts (open circles; ±SE) and PE content (solid circles) on 19 to 20 July 1991 (diel 2). The solid rectangle at the top of the figure indicates the duration of the nocturnal period. GMT, Greenwich mean time.
Depth profiles of Synechococcus sp. cell numbers revealed that the population was homogeneously distributed throughout the surface mixed layer, with very few cells occurring at depths below the seasonal thermocline at approximately 25 m (Fig. 3, top). The increase in cell concentrations seen in surface waters, therefore, was indicative of similar changes in the size of the Synechococcus sp. population that occurred throughout the mixed layer. As suggested by the increase in Synechococcus sp. cell concentrations during the period on station, the contribution of the picoplankton size class (0.2 to 2.0 μm) to water column integrated primary production (0 to 40 m) increased from 15 to 17% at the start of the experiment to 24 to 29% between 18 and 22 July 1991 (5).
Synechococcus sp. cell division was phased to the brief nighttime period that is characteristic of northern latitudes during the summer months, the population undergoing approximately one division, and hence one round of the cell cycle, per day (Fig. 3, bottom). Consistent with the observed diel pattern of growth and cell division, the PE content of Synechococcus sp. cells increased throughout the daylight hours but was reduced by about half following the late-evening/nocturnal separation of daughter cells (Fig. 3, bottom).
Although the numbers of Synechococcus sp. cells approximately doubled between evening and nighttime samples, the net generation time of the population was on the order of 4 to 5 days (Table 1). Since the study was conducted within the same water mass and the depth of the mixed layer did not change significantly during the period on station (Fig. 3, top; see also ref. 5), this discrepancy (and the net losses in Synechococcus sp. cell numbers observed during the daylight hours [Fig. 3, bottom]) suggests that the population was being actively grazed.
Diel variability in Synechococcus mRNA abundance.
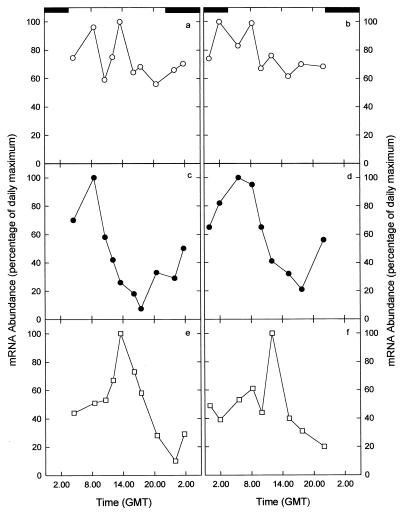
Changes in the relative abundance of three Synechococcus sp. mRNAs were measured over the course of two diel cycles (17 to 18 and 19 to 20 July 1991) toward the end of the period on station. Comparatively little temporal variability in cpeBA mRNA levels was observed; transcript levels oscillated somewhat (dynamic range, ∼2), but no consistent evidence of a distinct temporal pattern was evident during either diel cycle (Fig. 4a and b). By contrast, marked temporal variability in rbcL and glnA expression occurred and resulted in about an order of magnitude difference in mRNA levels between the respective daily maxima and minima in the abundance of both transcripts (Fig. 4c to f).
FIG. 4.
Diel variation in the abundance of cpeBA mRNA (a and b), rbcL mRNA (c and d), and glnA mRNA (e and f) at the northeast-Atlantic Lagrangian station 17 to 18 July (left-hand panels) and 19 to 20 July (right-hand panels) 1991. The solid rectangles at the tops of the upper panels indicate the nocturnal periods. GMT, Greenwich mean time.
RubisCO mRNA increased in abundance during the short nighttime period to reach a maximum by midmorning (∼0800). Transcript levels declined steadily thereafter before increasing once more from late afternoon (∼1800) to dusk. While the abundance of glutamine synthetase transcripts varied over approximately the same dynamic range, the temporal periodicity in glnA mRNA was distinct from that observed for rbcL. The peak in rbcL mRNA abundance was observed about 4 h postdawn, whereas during both diel cycles the highest abundance of glnA transcripts occurred at or around local midday (∼1300 Greenwich mean time). The abundance of glnA transcripts showed only small increases during the first few hours of the diel cycle before increasing sharply (∼2-fold) to the daily maximum. Thereafter, glnA mRNA levels declined throughout the afternoon and evening to reach a minimum that was coincident with the initiation of cell division around dusk.
DISCUSSION
Diel patterns of Synechococcus sp. cell division and PE synthesis.
The entrainment of Synechococcus sp. cell division to a particular phase of the diel cycle (Fig. 3b) has been documented previously for both coastal and open waters (7, 8, 42, 56, 62). In agreement with the observations reported here, these earlier studies indicate that oceanic populations most frequently undergo division during the late evening or night. It is intriguing, therefore, that cell division in rapidly growing coastal communities is initiated somewhat earlier in the diel cycle (8, 62). While such contrasting behavior may point to fundamental differences in cell cycle control between coastal and oceanic Synechococcus spp., the few seasonal records available provide some evidence that the peak in the frequency of dividing cells is shifted to a later phase in the diel cycle in coastal waters during the autumn and winter months (8, 62). Perhaps under these circumstances, in situ growth rate (one or fewer divisions per day) and environmental constraints on the diel phasing of the cell cycle more closely mirror those typical of Synechococcus sp. populations growing in open waters under presumably less favorable physicochemical conditions. Such a contention is certainly supported by the observation that phosphate additions to surface seawater samples from the Mediterranean Sea advanced subsequent progression through the Synechococcus sp. cell cycle by several hours (56).
Two distinct temporal patterns of DNA synthesis have been described for Synechococcus spp. growing in laboratory culture under alternating light-dark cycles (2, 4), but neither pattern is consistent with the development of true cell cycle synchrony. In both coastal and oceanic isolates a significant proportion of cells enter the light cycle in G2 having already completed progression through S during the latter half of the previous photoperiod. In the natural populations studied by Vaulot and colleagues, by contrast, all Synechococcus sp. cells entered the photoperiod in G1 having progressed through G2/M during the previous evening and night (56). Similar significant differences between the cell cycle behavior of laboratory cultures and that of natural populations have also been reported for the related photosynthetic prokaryote Prochlorococcus marinus. Progression through the cell cycle is highly synchronized to the diel periodicity in irradiance in natural populations of P. marinus but, as with Synechococcus spp., this behavior is not reproduced under standard laboratory conditions (55, 56).
The apparent discrepancy between laboratory models of Synechococcus sp. cell division and field observations remains unexplained but might be reconciled by examining the quantum requirement for passage through the light-dependent cell cycle checkpoints in G1 and G2 originally proposed by Chisolm and coworkers (2, 4). If the quantum requirement for progression through G1 into S phase is significantly higher than that required for progression through G2/M, then under natural illumination (in which, as Vaulot et al. [56] point out, incident irradiance shows a sinusoidal temporal distribution) cells may become stalled in G1 during the last few hours of fading daylight, whereas the dark block in G2 might still be passed. Quite clearly, such a scenario could lead to the rapid establishment of cell cycle synchrony under natural illumination because, irrespective of when division took place during the latter half the previous diel cycle, all cells should enter the next diurnal period in G1. It is not an absolute requirement of this model to propose the existence of two distinct light control circuits to regulate the G1 and G2 checkpoints in order to promote cell cycle synchrony under natural illumination. Newly born cells in G1 have approximately half the pigment complement and, presumably, half the light absorption capacity of G2 cells that are just about to undergo division (Fig. 3, bottom).
In addition to the diel periodicity in DNA content that results from cell cycle synchronization (56), the entrainment of Synechococcus sp. cell division to the natural photoperiod predicts a number of other cell cycle-related effects on the daily pattern of macromolecular synthesis. The most obvious of these is the exponential increase in stable RNA (rRNA and tRNA) and protein content that must accompany progression through the cell cycle. Bulk measurements of biosynthetic processes performed on natural populations are unlikely to resolve such temporal changes in cell composition, however, unless they are corrected for (or are independent of) loss terms such as grazing and advection as well as the influence of other members of the planktonic community. Nevertheless, where specific estimates of Synechococcus sp. cell composition have been attempted with natural populations the expected temporal pattern of macromolecular synthesis has emerged (20, 25).
As a further case in point, the cell content of the biliprotein PE was measured directly during the present study and found to increase in a quasilogarithmic fashion throughout the daylight hours (Fig. 3, bottom). Assuming that the Synechococcus sp. population within the eddy was in balanced growth (a not-unreasonable assumption given the comparative lack of variability in the environment), this suggests a more or less constant rate of synthesis (relative to the general protein pool) during all phases of the cell cycle.
Perhaps as a consequence of the comparatively short duration of the nocturnal period, the decline in cell PE content after dusk could be largely explained by the effect of dilution through cell division rather than by a nighttime decrease in the net rate of synthesis. Periodicity in energy supply need not in fact lead to very large variations in the rate of protein synthesis, at least not in cyanobacteria grown in light-dark cycles with comparatively long light periods (41). At lower latitudes, however, the nocturnal rate of PE synthesis in marine Synechococcus spp. does appear to be somewhat lower even after allowance for dilution effects following cell division (68). There is also some evidence that the rate of Synechococcus sp. ribosome synthesis declines during the rather longer hours of darkness experienced at more southerly locations during the summer months (25).
Temporal variation in gene expression.
Consistent with the diel pattern of PE synthesis, the relative abundance of Synechococcus sp. cpeBA mRNA showed comparatively little temporal variability when normalized, as in the present case, to total RNA. Since cell ribosome content must also be a temporal variable in synchronously dividing populations, the comparative lack of variability in the normalized abundance of cpeBA mRNA suggests that either the PE operon was expressed constitutively or the cpeBA message is unusually stable. In fact, the half-life of cpeBA mRNA in the marine cyanobacterium Synechococcus sp. strain WH7803 is of the same order (10 to 15 min) as those determined for several other genes (9, 28, 68), and there is no a priori reason to believe that the natural population should differ markedly in this regard.
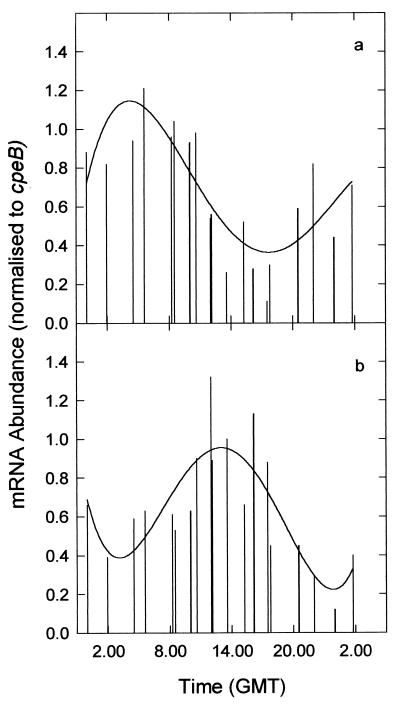
The pronounced diel variability and periodicity in Synechococcus sp. rbcL and glnA mRNAs, by contrast, is much more consistent with the operation of specific controls at the transcriptional and/or posttranscriptional levels. Although some of the temporal variability in mRNA abundance may reflect global changes in transcription rates as the population progressed through the cell cycle, such effects are perhaps as likely to influence the overall abundance of transcripts from the PE operon as they are to influence the diel pattern of RubisCO or GS expression. It is significant, therefore, that the temporal periodicities in Synechococcus sp. rbcL and glnA mRNAs remain markedly conservative features of the natural population when each is normalized to cpeBA mRNA (Fig. 5). Irrespective of whether the PE operon was truly constitutively expressed or not, it is clearly apparent that rbcL and glnA were differentially regulated during the diel cycle, both with respect to each other and with respect to cpeBA.
FIG. 5.
Histogram showing temporal variation in the abundance of Synechococcus sp. rbcL (a) and glnA (b) transcripts. Data from both diel studies are included and are normalized to the diel variability in cpeBA mRNA abundance. Both data sets are fitted with fourth-order polynomial regression curves (r = 0.675 [a] and 0.644 [b]). GMT, Greenwich mean time.
The diel variability in rbcL mRNA abundance observed in this subpolar Synechococcus sp. population differed somewhat from that shown previously for picoplankton-dominated surface waters from coastal and offshore sites in the Gulf of Mexico (38, 39). Whereas these earlier studies reported a midday maximum in transcript abundance, the temporal pattern of RubisCO expression revealed here is more reminiscent of the diel rhythm in rbcL mRNA found by these same investigators in a more detailed recent study (37). However, unlike the northeast Atlantic Synechococcus sp. population, in which rbcL mRNA increased from late afternoon to reach near-maximal levels by daybreak, there was no evidence of a comparable nighttime increase in transcript abundance in the subtropical populations studied by Pichard et al. (37). Nevertheless, the periodicity (i.e., the temporal evolution of maxima and minima) in rbcL mRNA observed in the present investigation was very similar to that found in the studies of Paul and coworkers described above and by Watson and Tabita (63) and Pichard et al. (37) for laboratory cultures of Synechococcus sp. strain WH7803 and P. marinus, respectively.
The rbcL gene of Synechococcus sp. strain WH8103 is 97% identical at the nucleotide level and 99% at the amino acid level (GenBank accession no. AF148536 [68]) to the homologue present in strain WH7803. Neither sequence is particularly closely related to the rbcL gene probe derived from the freshwater cyanobacterium Synechococcus sp. strain PCC6301 used by Pichard and coworkers (37–39) to examine gene expression in subtropical picoplankton. It is of interest, therefore, that similar diel rhythms in rbcL expression have been observed for both sequence types even though the target organisms recognized by each probe are likely to be somewhat different.
In marked contrast to the temporal pattern of rbcL expression, maxima in glnA transcript levels occurred some 5 to 6 hours later in the diel cycle (i.e., at the midpoint of the daylight period) and coincided with the approach of the daily minimum in rbcL mRNA abundance. Whether similar periodic differences also occurred in the diel patterns of RubisCO and glutamine synthetase translation and activity was not determined, but the differential accumulation of their respective mRNAs is certainly consistent with such a temporal separation. For RubisCO at least, shifts in rbcL transcription have been shown to be closely linked to temporal variations in the rate of C fixation (37), but no comparable field investigations have been attempted to examine the diel pattern of nitrogen assimilation in these organisms.
Diurnal periodicity in photosynthetic potential may be a rather general feature of natural populations of Synechococcus spp., since very similar rhythmic behavior (characterized by a midday peak in CO2 fixation capacity) to that found by Pichard et al. (37) has been noted by others under widely different environmental conditions (43, 44). Not least because variation in the intracellular C/N ratio is of such key importance in regulating the primary N-assimilatory enzyme, glutamine synthetase (12, 32, 33, 53), temporal periodicity of this kind in C fixation must also play some role in determining the pattern of N assimilation in natural populations if, as would appear to be the case, diel rhythms in RubisCO expression are the norm rather than the exception in Synechococcus spp. (37–39, 63).
In cyanobacteria, as in the well-studied prokaryotic models Escherichia coli and Bacillus subtilis, expression of glutamine synthetase and other nitrogen-assimilatory enzymes is regulated by signal transduction systems sensitive to the balance of C and N entering intermediary metabolism (12, 13, 53, 57). Transcription of glnA is enhanced under physiological conditions in which the ratio between the relative rates of C and N assimilation increases to the point where intracellular metabolites at the amino level of reduction become depleted, i.e., during periods of N starvation or, conversely, as a result of increases in the prevailing rate of C fixation. The combined effects of energy limitation and the catabolism of C reserves during the nocturnal phase of the diel cycle predicts that natural populations of Synechococcus spp. are likely to enter the diurnal period in a C-depleted state. Under such circumstances, it is perhaps not too surprising that transcription of the RubisCO operon (and hence C fixation potential) should be transiently up-regulated in anticipation of, and during, the first few hours of daylight.
While specific feedback controls of this kind are not necessarily excluded, they are insufficient to explain why the diel rhythm in rbcL mRNA abundance is also maintained in Synechococcus spp. held in constant light (37), i.e., under experimental conditions that might be expected to eliminate the temporal oscillations in carbon reserves and cell C/N ratio experienced by the population in situ. This type of rhythmic behavior clearly suggests that some element of circadian control may be involved in the regulation of the Synechococcus sp. RubisCO operon (37). It does not follow, however, that circadian control need invoke clock-regulated trans-acting elements that interact with the rbcL promoter directly (30). From an organizational standpoint at least, one might argue that coordinating RubisCO expression with the cell cycle probably offers a more parsimonious molecular solution to address the same problem (i.e., one of intermittent energy supply) if, as in Synechococcus sp. strain WH7803, cell division is itself controlled by a circadian oscillator (50).
Whether rbcL is regulated directly or indirectly by a circadian rhythm, the fact remains that there is a dramatic increase in the capacity for C fixation in natural populations of Synechococcus spp. as the solar altitude increases toward midday (37, 38, 43, 44). Not least because this accelerating supply of newly synthesized carbon skeletons should rapidly deplete the available pool of intracellular nitrogen, it is the likely imbalance in the cell C/N ratio that arises from the temporal periodicity in Synechococcus sp. C fixation rates that also provides the most conservative interpretation of the diel pattern of glnA mRNA abundance. The rhythm in glnA mRNA abundance observed was clearly distinct from that for rbcL and is at least consistent with the notion that a shift up in glutamine synthetase synthesis and activity may occur during the latter half of the diurnal period in natural populations of Synechococcus spp. as a direct result of the enhanced rates of C assimilation somewhat earlier in the diel cycle.
An element of temporal periodicity in N-assimilation rates is a not entirely unexpected feature of synchronously growing populations of microorganisms like Synechococcus spp., since the bulk of the cell’s macromolecules are produced during the latter half of the cell cycle. The growth of the individual, like that of the population in general, is an exponential function, and hence the cell’s demand for nutrients (including nitrogen) must also increase in a quasilogarithmic manner as the cell cycle progresses. This feature of the daily pattern of macromolecule biosynthesis in natural populations of Synechococcus spp. has been noted by others (34) and is further illustrated by examination of the diel pattern of PE production reported here (Fig. 3, bottom). Very little change in cell PE content was observed during the hours up to midday compared with the near doubling in concentrations that occurred thereafter, prior to the nocturnal separation of daughter cells.
A number of features emerge from the present study that may have wider implications for our understanding of the factors that regulate carbon and nitrogen assimilation in natural populations of marine Synechococcus spp. Endogenous changes in physiological demands that arise as an inevitable consequence of the daily progression through the cell cycle are, perhaps, as important in determining the periodic abundance of specific mRNAs in these organisms as are changes in exogenous environmental variables, such as the diel alternation in irradiance. The differential expression of Synechococcus sp. rbcL and glnA mRNAs reported here clearly suggests that these key C- and N-assimilatory enzymes may be maximally expressed at different periods during the cell cycle in a manner analogous to the reciprocal control of RubisCO and nitrogenase gene expression found in the diazotrophic cyanobacterium Synechococcus sp. strain RF-1 (11). Whether, like that of RubisCO (37), the activity of glutamine synthetase in marine Synechococcus spp. also shows some diel variability requires further investigation, since the occurrence of temporal changes in nitrogen assimilation capacity is likely to modify our general overview of (i) niche specialization among marine phytoplankton and (ii) the specific role that these picoplanktonic cyanobacteria may play in the nutrient dynamics of the surface oceans.
ACKNOWLEDGMENTS
This work was supported by research grants from the Natural Environment Research Council (NERC), the Joint Environment Programme of National Power and Powergen, and the University of Stirling.
I acknowledge access to ship time during the NERC Biogeochemical Ocean Flux Study and the encouragement and support of R. P. Harris, M. Whitfield, and others at Plymouth Marine Laboratory and the Marine Biological Association.
REFERENCES
- 1.Alberte R S, Wood A M, Kursar T A, Guillard R R L. Novel phycoerythrins in marine Synechococcus spp.: characterisation, and evolutionary and ecological implications. Plant Physiol. 1984;75:732–739. doi: 10.1104/pp.75.3.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armbrust E V, Bowen J D, Olson R J, Chisolm S W. Effect of light on the cell cycle of a marine Synechococcus strain. Appl Environ Microbiol. 1989;55:425–432. doi: 10.1128/aem.55.2.425-432.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Siedman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1994. [Google Scholar]
- 4.Binder B J, Chisolm S W. Cell cycle regulation in marine Synechococcus sp. strains. Appl Environ Microbiol. 1995;61:708–717. doi: 10.1128/aem.61.2.708-717.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyd P, Pomroy A, Bury S, Savidge G, Joint I. Micro-algal carbon and nitrogen uptake in post-coccolithophore bloom conditions in the northeast Atlantic, July 1991. Deep-Sea Res. 1997;44:1497–1517. [Google Scholar]
- 6.Bustos S A, Schaefer M R, Golden S S. Different and rapid responses of four cyanobacterial psbA transcripts to changes in light intensity. J Bacteriol. 1991;172:1998–2004. doi: 10.1128/jb.172.4.1998-2004.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campbell L, Carpenter E J. Diel patterns of cell division in marine Synechococcus spp. (cyanobacteria): the use of the frequency of dividing cells technique to measure growth rate. Mar Ecol Prog Ser. 1986;32:139–148. [Google Scholar]
- 8.Carpenter E J, Campbell L. Diel patterns of cell division and growth rates of Synechococcus spp. in Long Island Sound. Mar Ecol Prog Ser. 1988;47:179–183. [Google Scholar]
- 9.Carr N G, Mann N H. The oceanic cyanobacterial picoplankton. In: Bryant D A, editor. The molecular biology of cyanobacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. pp. 27–48. [Google Scholar]
- 10.Chen Y-B, Dominic B, Mellon M, Zehr J P. Circadian rhythm of nitrogenase gene expression in the diazotrophic filamentous nonheterocystous cyanobacterium Trichodesmium sp. strain IMS 101. J Bacteriol. 1998;180:3598–3605. doi: 10.1128/jb.180.14.3598-3605.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chow T-J, Tabita F R. Reciprocal light-dark transcriptional control of nif and rbc expression and light-dependent posttranslational control of nitrogenase activity in Synechococcus sp. strain RF-1. J Bacteriol. 1994;176:6281–6285. doi: 10.1128/jb.176.20.6281-6285.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen-Kupiec R, Gurevitz M, Zilberstein A. Expression of glnA in the cyanobacterium Synechococcus sp. strain PCC 7942 is initiated from a single nif-like promoter under various nitrogen conditions. J Bacteriol. 1993;175:7727–7731. doi: 10.1128/jb.175.23.7727-7731.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cohen-Kupiec R, Zilberstein A, Gurevitz M. Characterization of cis elements that regulate the expression of glnA in Synechococcus sp. strain PCC 7942. J Bacteriol. 1995;177:2222–2226. doi: 10.1128/jb.177.8.2222-2226.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Douglas S E, Carr N G. Examination of genetic relatedness of marine Synechococcus spp. using restriction fragment length polymorphisms. Appl Environ Microbiol. 1988;54:3071–3078. doi: 10.1128/aem.54.12.3071-3078.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fernandez E, Boyd P, Holligan P M, Harbour D S. Production of organic and inorganic carbon within a large scale coccolithophore bloom in the north Atlantic Ocean. Mar Ecol Prog Ser. 1993;97:271–285. [Google Scholar]
- 16.Forchhammer K, Tandeau de Marsac N. Functional analysis of the phosphoprotein PII (glnB gene product) in the cyanobacterium Synechococcus sp. strain PCC 7942. J Bacteriol. 1995;177:2033–2040. doi: 10.1128/jb.177.8.2033-2040.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia-Dominguez M, Florencio F J. Nitrogen availability and electron transport control expression of glnB gene (encoding PII protein) in the cyanobacterium Synechocystis sp. PCC 6803. Plant Mol Biol. 1997;35:723–734. doi: 10.1023/a:1005846626187. [DOI] [PubMed] [Google Scholar]
- 18.Glover H E, Prezelin B B, Campbell L, Wyman M, Garside C G. A nitrate dependent Synechococcus bloom in surface Sargasso Seawater. Nature (London) 1988;331:161–163. [Google Scholar]
- 19.Glover H E, Prezelin B B, Campbell L, Wyman M. Pico- and ultraplankton Sargasso Sea communities: variability and comparative distributions. Mar Ecol Prog Ser. 1988;49:127–139. [Google Scholar]
- 20.Glover H E, Smith A E. Diel patterns of carbon incorporation into biochemical constituents of Synechococcus spp. and larger algae in the Northwest Atlantic Ocean. Mar Biol. 1988;97:259–267. [Google Scholar]
- 21.Harris R P, Boyd P, Harbour D S, Head R N, Pingree R D, Pomroy A J. Physical, chemical and biological features of a cyclonic eddy in the region of 61°10′N 19°50′W in the North Atlantic. Deep-Sea Res Part I. 1997;44:1815–1838. [Google Scholar]
- 22.Johnson P W, Sieburth J M. Chroococcoid cyanobacteria in the sea: a ubiquitous and diverse biomass. Limnol Oceanogr. 1979;24:928–935. [Google Scholar]
- 23.Joint I R. Physiological ecology of picoplankton in various ocean provinces. Can Bull Fish Aquat Sci. 1986;214:287–309. [Google Scholar]
- 24.Kana T M, Glibert P M. Effect of irradiances up to 2000 μE m−2 s−1 on marine Synechococcus WH7803. II. Photosynthetic responses and mechanisms. Deep-Sea Res. 1987;34:497–516. [Google Scholar]
- 25.Kramer J G, Singleton F L. Measurement of rRNA variations in natural communities of microorganisms on the Southeastern U.S. continental shelf. Appl Environ Microbiol. 1993;59:2430–2436. doi: 10.1128/aem.59.8.2430-2436.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kramer J G, Wyman M, Zehr J P, Capone D G. Diel variability in transcription of the structural gene for glutamine synthetase (glnA) in natural populations of the marine diazotrophic cyanobacterium Trichodesmium thiebautii. FEMS Microbiol Ecol. 1996;21:187–196. [Google Scholar]
- 27.Li L-H, Tabita F R. Transcription control of ribulose bisphosphate carboxylase/oxygenase activase and adjacent genes in Anabaena species. J Bacteriol. 1994;176:6697–6706. doi: 10.1128/jb.176.21.6697-6706.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lindell D, Padan E, Post A F. Regulation of ntcA expression and nitrite uptake in the marine Synechococcus sp. strain WH7803. J Bacteriol. 1998;180:1878–1886. doi: 10.1128/jb.180.7.1878-1886.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu H, Campbell L, Landry M R. Growth and mortality rates in Prochlorococcus and Synechococcus measured with a selective inhibitor technique. Mar Ecol Prog Ser. 1995;116:277–287. [Google Scholar]
- 30.Liu Y, Tsinoremas N F, Hirschie Johnson C, Lebedeva N V, Golden S S, Ishiura M, Kondo T. Circadian orchestration of gene expression in cyanobacteria. Genes Dev. 1995;9:1469–1478. doi: 10.1101/gad.9.12.1469. [DOI] [PubMed] [Google Scholar]
- 31.Meeks J C, Wolk C P, Lockau W, Schilling N, Shaffer P W, Chien W-S. Pathways of assimilation of [13N]-N2 and 13NH4+ by cyanobacteria with and without heterocysts. J Bacteriol. 1978;134:125–130. doi: 10.1128/jb.134.1.125-130.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Merida A, Flores E, Florencio F J. Regulation of Anabaena sp. strain PCC 7120 glutamine synthetase activity in a Synechocystis sp. strain PCC 6803 derivative strain bearing the Anabaena glnA gene and a mutated host glnA gene. J Bacteriol. 1992;174:650–654. doi: 10.1128/jb.174.2.650-654.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Merida A, Leurentop L, Candau P, Florencio F. Purification and properties of glutamine synthetase from the cyanobacteria Synechocystis sp. strain PCC 6803 and Calothrix sp. strain PCC 7601. J Bacteriol. 1990;172:4732–4735. doi: 10.1128/jb.172.8.4732-4735.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olson R J, Chisolm S W, Zettler E R, Armbrust E V. Pigments, size, and distribution of Synechococcus in the North Atlantic and Pacific Oceans. Limnol Oceanogr. 1990;35:45–58. [Google Scholar]
- 35.Ong L J, Glazer A N. Structural studies of phycobiliproteins in unicellular marine cyanobacteria. In: Stevens S E, Bryant D A, editors. Light-energy transduction in photosynthesis: higher plant and bacterial models. New York, N.Y: American Society of Plant Physiologists; 1988. [Google Scholar]
- 36.Ong L J, Glazer A N, Waterbury J B. An unusual phycoerythrin from a marine cyanobacterium. Science. 1984;224:80–83. doi: 10.1126/science.224.4644.80. [DOI] [PubMed] [Google Scholar]
- 37.Pichard S L, Campbell L, Kang J B, Tabita F R, Paul J H. Regulation of ribulose bisphosphate carboxylase gene expression in natural phytoplankton communities. I. Diel rhythms. Mar Ecol Prog Ser. 1996;139:257–265. [Google Scholar]
- 38.Pichard S L, Frischer M E, Paul J H. Ribulose bisphosphate carboxylase gene expression in subtropical marine phytoplankton populations. Mar Ecol Prog Ser. 1993;101:55–65. [Google Scholar]
- 39.Pichard S L, Paul J H. Detection of gene expression in genetically engineered microorganisms and natural phytoplankton populations in the marine environment by mRNA analysis. Appl Environ Microbiol. 1991;51:1721–1727. doi: 10.1128/aem.57.6.1721-1727.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Platt T, Li W K W. Photosynthetic picoplankton. Can Bull Fish Aquat Sci. 1986;214:583. [Google Scholar]
- 41.Post A F, Loogman J G, Mur L R. Photosynthesis, carbon flows and growth of Oscillatoria agardhii Gomont in environments with a periodic light supply. J Gen Microbiol. 1986;132:2129–2136. [Google Scholar]
- 42.Prezelin B B, Glover H E, Campbell L. Effects of light intensity and nutrient availability on diel patterns of cell metabolism and growth in populations of Synechococcus spp. Mar Biol. 1987;95:469–480. [Google Scholar]
- 43.Prezelin B B, Putt M, Glover H E. Diurnal patterns in photosynthetic capacity and depth-dependent photosynthesis-irradiance relationships in Synechococcus spp. and larger phytoplankton in three water masses in the Northwest Atlantic Ocean. Mar Biol. 1986;91:205–217. [Google Scholar]
- 44.Putt M, Prezelin B B. Observations of diel patterns of photosynthesis in cyanobacteria and nanoplankton in the Santa Barbara Channel during ‘el Nino’. J Plankton Res. 1985;7:779–790. [Google Scholar]
- 45.Reyes J C, Florencio F J. Electron transport controls transcription of the glutamine synthetase gene (glnA) from the cyanobacterium Synechocystis sp. PCC6803. Plant Mol Biol. 1995;27:789–799. doi: 10.1007/BF00020231. [DOI] [PubMed] [Google Scholar]
- 46.Scanlan D J, Mann N H, Carr N G. The response of the picoplanktonic marine cyanobacterium Synechococcus species WH7803 to phosphate starvation involves a protein homologous to the periplasmic phosphate-binding protein of Escherichia coli. Mol Microbiol. 1993;10:181–191. doi: 10.1111/j.1365-2958.1993.tb00914.x. [DOI] [PubMed] [Google Scholar]
- 47.Scanlan D J, Silman N J, Donald K M, Wilson W H, Carr N G, Joint I, Mann N H. An immunological approach to detect phosphate stress in populations and single cells of photosynthetic picoplankton. Appl Environ Microbiol. 1997;63:2411–2420. doi: 10.1128/aem.63.6.2411-2420.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Siebens H C, Trench R K. Aspects of the relation between Cyanophora paradoxa (Korschikoff) and its endosymbiotic cyanelles Cyanocyta korschnikoffiana (Hall and Claus). III. Characterization of ribosomal ribonucleic acids. Proc R Soc Lond B. 1978;202:463–472. [Google Scholar]
- 49.Silman N J, Carr N G, Mann N H. ADP-ribosylation of glutamine synthetase in the cyanobacterium Synechocystis sp. strain PCC 6803. J Bacteriol. 1995;177:3527–3533. doi: 10.1128/jb.177.12.3527-3533.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sweeney B M, Borgese M B. A circadian rhythm in cell division in a prokaryote, the cyanobacterium, Synechococcus WH7803. J Phycol. 1989;25:183–186. [Google Scholar]
- 51.Toledo G, Palenik B. Synechococcus diversity in the California Current as seen by RNA polymerase (rpoC1) gene sequences of isolated strains. Appl Environ Microbiol. 1997;63:4298–4303. doi: 10.1128/aem.63.11.4298-4303.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tsinoremas N F, Castets A M, Harrison M A, Allen J F, Tandeau de Marsac N. Photosynthetic electron transport controls nitrogen assimilation in cyanobacteria by means of posttranslational modification of the glnB gene product. Proc Natl Acad Sci USA. 1991;88:4565–4569. doi: 10.1073/pnas.88.11.4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tumer N K, Robinson S J, Haselkorn R. Different promoters for the Anabaena glutamine synthetase gene during growth using molecular or fixed nitrogen. Nature (London) 1983;306:337–342. [Google Scholar]
- 54.Urbach E, Scanlan D J, Distel D L, Waterbury J B, Chisolm S W. Rapid diversification of marine picophytoplankton with dissimilar light-harvesting structures inferred from sequences of Prochlorococcus and Synechococcus (Cyanobacteria) J Mol Evol. 1998;46:188–201. doi: 10.1007/pl00006294. [DOI] [PubMed] [Google Scholar]
- 55.Vaulot D, Marie D, Olson R J, Chisolm S W. Growth of Prochlorococcus, a photosynthetic prokaryote, in the equatorial Pacific Ocean. Science. 1995;268:1480–1482. doi: 10.1126/science.268.5216.1480. [DOI] [PubMed] [Google Scholar]
- 56.Vaulot D, LeBot N, Marie D, Fukai E. Effect of phosphorus on the Synechococcus cell cycle in surface Mediterranean waters during summer. Appl Environ Microbiol. 1996;62:2527–2533. doi: 10.1128/aem.62.7.2527-2533.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vega-Palas M, Flores E, Herrero A. NtcA, a global nitrogen regulator from the cyanobacterium Synechococcus that belongs to the Crp family of bacterial regulators. Mol Microbiol. 1992;6:1853–1859. doi: 10.1111/j.1365-2958.1992.tb01357.x. [DOI] [PubMed] [Google Scholar]
- 58.Wagner S J, Thomas S P, Kaufman R I, Nixon B T, Stevens S E., Jr The glnA gene of the cyanobacterium Agmenellum quadruplicatum PR-6 is nonessential for ammonium assimilation. J Bacteriol. 1993;175:604–612. doi: 10.1128/jb.175.3.604-612.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Waterbury J B, Rippka R. Order Chroococcales. In: Krieg N R, Holt J B, editors. Bergey’s manual of systematic bacteriology. Baltimore, Md: The Williams & Wilkins Co.; 1989. pp. 1728–1746. [Google Scholar]
- 60.Waterbury J B, Valois F W. Resistance to co-occurring phages enables marine Synechococcus communities to coexist with cyanophages abundant in seawater. Appl Environ Microbiol. 1993;59:3393–3399. doi: 10.1128/aem.59.10.3393-3399.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Waterbury J B, Watson S W, Guillard R R L, Brand L E. Widespread occurrence of a unicellular marine planktonic cyanobacterium. Nature (London) 1979;277:293–294. [Google Scholar]
- 62.Waterbury J B, Watson S W, Valois F W, Franks D G. Biological and ecological characterization of the marine unicellular cyanobacterium, Synechococcus. Can J Fish Aquat Sci. 1986;214:71–120. [Google Scholar]
- 63.Watson G M F, Tabita F R. Regulation, unique gene organization, and unusual structure of carbon fixation genes from a marine phycoerythrin-containing cyanobacterium. Plant Mol Biol. 1996;32:1103–1115. doi: 10.1007/BF00041394. [DOI] [PubMed] [Google Scholar]
- 64.Willey J M, Waterbury J B. Chemotaxis toward nitrogenous compounds by swimming strains of marine Synechococcus spp. Appl Environ Microbiol. 1989;55:1888–1894. doi: 10.1128/aem.55.8.1888-1894.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wilson W H, Joint I R, Carr N G, Mann N H. Isolation and molecular characterization of five marine cyanophages propagated on Synechococcus sp. strain WH7803. Appl Environ Microbiol. 1993;59:3736–3743. doi: 10.1128/aem.59.11.3736-3743.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wood A M. Adaptation of the photosynthetic apparatus of marine ultraphytoplankton to natural light fields. Nature (London) 1985;316:253–255. [Google Scholar]
- 67.Wood A M, Townsend D. DNA polymorphisms within the WH7803 serogroup of marine Synechococcus spp. (cyanobacteria) J Phycol. 1990;26:567–585. [Google Scholar]
- 68.Wyman, M. Unpublished observations.
- 69.Wyman M. An in vivo method for the estimation of phycoerythrin concentrations in marine cyanobacteria (Synechococcus spp.) Limnol Oceanogr. 1992;37:1300–1306. [Google Scholar]
- 70.Wyman M, Zehr J P, Capone D G. Temporal variability in nitrogenase gene expression in natural populations of the marine cyanobacterium Trichodesmium thiebautii. Appl Environ Microbiol. 1996;62:1073–1075. doi: 10.1128/aem.62.3.1073-1075.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wyman M, Davies J T, Weston K, Crawford D G, Purdie D A. Ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO) gene expression and photosynthetic activity in nutrient-enriched mesocosm experiments. Estuarine Coastal Shelf Sci. 1998;46:23–33. [Google Scholar]