ABSTRACT
Cysteine-rich knob domains can be isolated from the ultralong heavy-chain complementarity-determining region (CDR) 3, which are unique to a subset of bovine antibodies, to create antibody fragments of ~4 kDa. Advantageously, the N- and C- termini of these small binding domains are in close proximity, and we propose that this may offer a practical route to engineer extrinsic binding specificity into proteins. To test this, we transplanted knob domains into various loops of rat serum albumin, targeting sites that were distal to the interface with the neonatal Fc receptor. Using knob domains raised against the clinically validated drug target complement component C5, we produced potent inhibitors, which exhibit an extended plasma half-life in vivo via attenuated renal clearance and neonatal Fc receptor-mediated avoidance of lysosomal catabolism. The same approach was also used to modify a Camelid VHH, targeting a framework loop situated at the opposing end of the domain to the CDRs, to produce a small, single-chain bispecific antibody and a dual inhibitor of Complement C3 and C5. This study presents new protein inhibitors of the complement cascade and demonstrates a broadly applicable method to engineer target specificity within polypeptide chains, using bovine knob domains.
KEYWORDS: Bovine antibodies, complement C5, knob domain, serum albumin, bispecific, VHH
Introduction
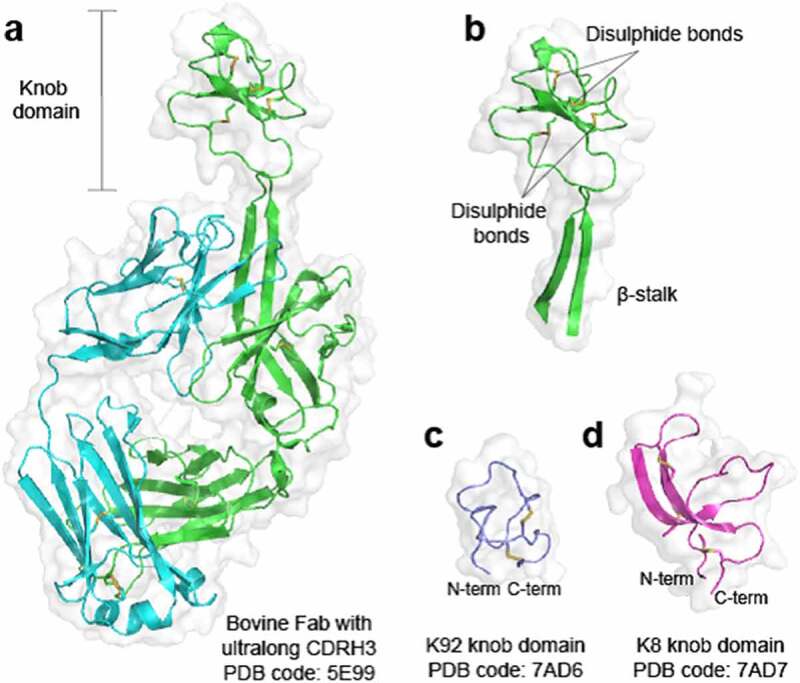
The structurally unique, disulfide-rich paratopes found within a subset of bovine immunoglobulin G (IgG) and immunoglobulin M (IgM) antibodies with ultralong heavy-chain complementarity-determining regions 3 (CDRH3) have intrigued scientists since they were first reported in the late 1990s.1 The ultralong CDRH3 exhibit a conserved structural motif where a β-ribbon “stalk” protrudes from the surface to present a disulfide-rich “knob domain”,1,2 which may be the sole point of contact with the antigen3 (Figure 1a,b). We have previously shown that knob domains can function independently of the IgG scaffold and β-stalk domain, to create small antibody fragments of some 3–6 kDa,5 which were able to finely modulate Complement component C5.6 Co-crystal structures of two knob domain peptides (Protein Data Bank (PDB) accession codes: 7AD6 and 7AD7)4 show that the N- and C- termini remain in close proximity when isolated from the bovine antibody scaffold, as they would be when attached to the β-stalk in an ultralong CDRH3 of a bovine fragment antigen-binding (Fab) region1,2,6 (Figure 1c,d).
Figure 1.
Crystal structures of bovine knob domains Panel (a) shows a bovine Fab with an ultralong CDRH32. The CDRH3 is shown in isolation in panel (b) with networks of disulfide bonds highlighted. Panels (c and d) show the K92 and K8 knob domains, respectively,4 where the N- and C- termini remain in close proximity.
The figure shows the crystal structure of a bovine Fab with an ultralong CDRH3, the disulfide-rich knob domains are highlighted.
Unusually for antibody fragments, knob domains are readily amenable to chemical synthesis and by this route we have exploited the proximity of the termini to produce head-to-tail cyclized knob domains, which may confer further resistance to exopeptidases in vivo.7 In this study, we propose that the proximity of the termini also affords opportunities for protein engineering by targeting protein loops as insertion sites. Despite a knob domain comprising at least 30 amino acids, due to its folded nature, the apparent disruption to the loop might be equivalent to a much smaller linear peptide, providing a route to insert, small, high affinity binding domains into proteins, without fusing to the terminus.
The knob domains used in this study have been raised against complement C5, which is the primary effector protein of the terminal pathway of the complement cascade. Activation of the either the classical pathway (CP), lectin pathway (LP) or alternative pathway (AP) results in cleavage of C5 into C5b, which initiates formation of the lytic terminal complement complex, and the pro-inflammatory anaphylatoxin, C5a. For cleavage of C5, two C5 convertases exist, the CP C5 convertase, C4bC2aC3b, and the AP C5 convertase, C3bBbC3b. We have previously developed knob domains that prevent C5 activation by the AP and CP (K57)4 or partially inhibit C5 activation via the AP (K92).4
As low-molecular weight therapeutic agents, knob domains display a short plasma half-life (t1/2) when administered systemically. We measured a t1/2 of 17 minutes for the unmodified K57 knob domain following administration of a 10 mg/kg intravenous (IV) dose to rats,7 which appears symptomatic of renal clearance.8 For therapeutic applications, it is critical that compound endures at the site of action, consequently various approaches to extend the t1/2 of low molecular weight proteins and peptides have been explored.8
This study exploits the proximal termini of knob domains to create C5 neutralizing constructs from serum albumin, a 66 kDa plasma protein with a circulatory t1/2 of ~19 days,9 mediated via the neonatal Fc receptor (FcRn), a salvaging partner common to IgG and albumin, which attenuates the lysosomal catabolism of both proteins.10,11
Due to its long t1/2, albumin has been harnessed for the pharmacokinetic (PK) extension of low molecular weight proteins, peptides, and small molecules. Notable Food and Drug Administration-approved examples include an interferon alpha-albumin fusion protein (albinterferon Alfa-2B, for chronic hepatitis C);12 the glucagon-like peptide-1 agonist semaglutide, which contains an albumin-binding fatty acid moiety,13 (Rybelsus®, for diabetes);14 and the albumin-binding small molecule paclitaxel (Abraxane®, for pancreatic cancer).15 When engineering albumin fusion proteins, the C-terminus has been shown to be required for FcRn binding16 and in this study we present routes to engineer antigen specificity into albumin, independently of the termini.
To highlight further opportunities for engineering with knob domains, we also generated a single-chain bispecific from a camelid VHH fragment. One of the challenges of the conventional IgG bispecific format is the requirement to efficiently pair heterologous domains, known as the chain-association issue.17 To increase the efficiency of chain pairing, Fc engineering approaches such as knobs-into-holes (KiH)18 and controlled Fab arm exchange19 have been developed to enforce heavy-chain heterodimerization. Single-chain formats are attractive as they circumvent the chain-association issue entirely, potentially affording improved manufacturability. Here, we present a single-chain-bispecific VHH that neutralizes its cognate antigen, human Complement C3,20 but also prevents activation of Complement C5, by virtue of insertion of an anti-C5 knob domain, achieving effective neutralization of the complement cascade in vitro.
The fusion proteins presented here may offer new therapeutic proteins for complement-related pathologies where myasthenia gravis, paroxysmal nocturnal hemoglobin urea and atypical hemolytic uremic syndrome are diseases of note. Importantly, the methods described here may be applied more broadly to generate single-chain multivalent or bispecific proteins.
Results
Design and expression of rat albumin-knob domain fusion proteins
As an approach to extend plasma half-life, we engineered knob domains into rat serum albumin (RSA). We noted the presence of repeating alpha-helical hairpin motifs within RSA, which are also present in serum albumins from other mammals, including humans.21 Within these sites, a poly-cysteine motif creates a stabilizing intra-hairpin disulfide and a second inter-helix disulfide, which further constrains the alpha-helix hairpin (Figure 2a).
Figure 2.
Bovine knob peptides insertion into the non-FcRn-binding loops of rat albumin.Panel (a) shows the crystal structure of the human serum albumin, in purple, in complex with FcRn, in green/cyan (PDB accession code: 4N0U)21 The corresponding sites of knob domain insertion in rat albumin, (helices 1–4; positions A83, E195, E387 and A586, respectively) are shown. Panel (b) non-reducing SDS-PAGE (1 ug load) of WT RSA (1), RSA-K57 H1 (2), RSA K92-H2 (3), RSA-K92 H3 (4) and RSA-K57 H4 (5). Panel (c) The Far UV CD spectra suggest minimal changes in secondary structure compared to the WT RSA and fusion proteins
The figure shows the crystal structure of human serum albumin with the equivalent insertion sites used in this study for rat serum albumin, highlighted. A non-reducing gel with a single band for each purified construct shows an apparent increase in molecular weight arising from insertion of the knob domains. Contiguous CD spectra for the wild type and modified albumins are also shown.
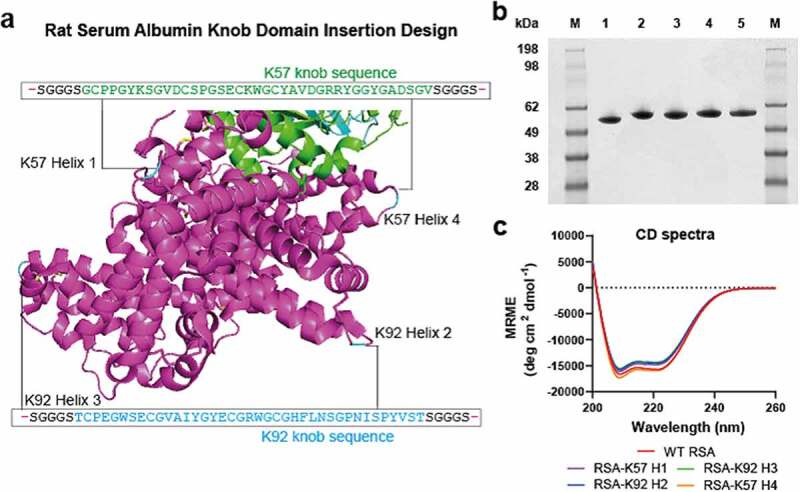
We targeted these stable loop motifs as insertion sites, at regions that were distal to the rat FcRn (rFcRn) interface, with a view to conserving this interaction. We used two knob domain peptides, K57 and K92, which bind C5 with equilibrium rate constants (KD) of 1.4 nM and <0.6 nM, respectively.5 By this approach, four fusion proteins were produced, RSA-K57 Helix 1 (insertion site: A83 [based on mature sequence]), RSA-K92 Helix 2 (insertion site: E195), RSA-K92 Helix 3 (insertion site: E387) and RSA-K57 Helix 4 (insertion site: A586), which were compared to the wild-type (WT) RSA (Figure 2a and Supplementary 2).
The albumin-fusion proteins were transiently expressed in Expi293 human embryonic kidney (HEK) cells and, following preparative nickel affinity and size exclusion chromatography, yielded 0.1–0.2 g/L. Analysis by non-reducing sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and high performance liquid chromatography-size exclusion chromatography (HPLC-SEC [Figure 2b, Supplementary 1]) detected a single species, although a shouldering of the peak was evident for RSA-K92 Helix 2. For all samples, the liquid chromatography/mass spectrometry (LC/MS) data were consistent with the predicted isotype patterns (Supplementary 5). Analysis of the far ultraviolet (UV) circular dichroism (CD) spectra indicated that the RSA-knob domain fusions were contiguous to RSA, suggesting that there was little perturbation of the secondary structure arising from the insertion of the knob domain (Figure 2c, Table 1).
Table 1.
Bestsel analysis of the CD spectra for the constructs reported in this study
| Knob-fusion proteins | ||||||||
|---|---|---|---|---|---|---|---|---|
| Secondary structure | WT RSA | RSA-K57 H1 | RSA-K92 H2 | RSA-K92 H3 | RSA-K57 H4 | hC3nb1-ScFc | hC3nb1-K57-ScFc | K57 knob |
| Helix | 40.6 | 39.9 | 39.2 | 37.1 | 43.8 | 4.4 | 2.5 | 0 |
| Antiparallel | 10.4 | 9.4 | 9.7 | 11.1 | 8.2 | 34 | 36.5 | 38.8 |
| Parallel | 4.7 | 5.3 | 5.8 | 5.7 | 4.6 | 0 | 0 | 0 |
| Turn | 13.7 | 12.4 | 11.9 | 12.4 | 11.6 | 13.7 | 14.6 | 14 |
| Others | 30.6 | 33 | 33.3 | 33.6 | 31.8 | 47.9 | 46.4 | 47.2 |
Binding and in-vivo PK of rat albumin-knob domain fusion proteins
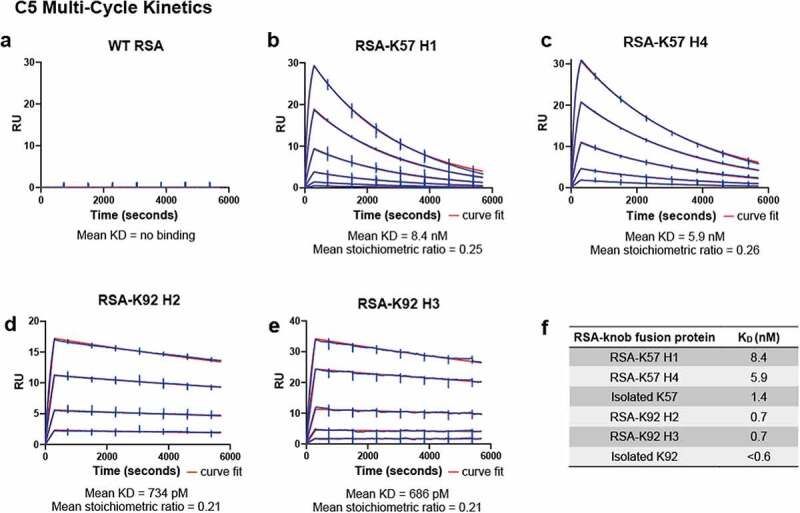
Surface plasmon resonance (SPR) was used to measure affinity for C5. Data are reported for a series of Biacore multi-cycle kinetics experiments in Table 2. We report equilibrium rate constants (KD) and individual association and dissociation rate constants (Kon and Koff) for the RSA-K57 and RSA-K92 fusions to C5. While the WT RSA did not bind C5 (Figure 3a), the four RSA-knob fusions exhibited high affinity for C5, with KD in the picomolar to low nanomolar range (Figure 3b-f). Of note, the RSA-K92 fusions were exceptionally tight binders, consistent with published data for the isolated K92 knob domain.5
Table 2.
Summary of C5 binding kinetics from biacore multi-cycle kinetics of RSA-knob domain fusion proteins. Data from n = 4 experiments
| Fusion Protein | Kon | Kon 95% CI | Koff | Koff 95% CI | KD | KD 95% CI |
|---|---|---|---|---|---|---|
| RSA-K57 H1 | 5.08E+04 | 1.31E+04 | 4.09E-04 | 6.68E-05 | 8.43E-09 | 2.23E-09 |
| RSA-K92 H2 | 7.75E+04 | 1.85E+04 | 5.53E-05 | 1.01E-05 | 7.34E-10 | 1.60E-10 |
| RSA-K92 H3 | 8.65E+04 | 5.56E+03 | 5.88E-05 | 1.19E-05 | 6.86E-10 | 1.75E-10 |
| RSA-K57 H4 | 5.23E+04 | 1.64E+04 | 2.84E-04 | 9.57E-06 | 5.91E-09 | 2.09E-09 |
| WT RSA | No Binding | - | No Binding | - | No Binding | - |
Figure 3.
The rat albumin-knob domain fusion proteins confer binding to C5 by SPR multi-cycle kinetics.Panels (a–e) show representative sensorgrams for binding to human C5, the data are shown in blue with the curves fit from a 1:1 binding model shown in red. The mean equilibrium dissociation constant (KD) values and stoichiometric values are shown (n = 4). Table f is representing the mean KDs in nM compared to the respective isolated knob peptide from our previous reports.5
shows representative surface plasmon resonance sensorgrams for the rat albumin-knob domain fusion proteins showing the presence or absence of binding to human C5 in accordance with the aims of the study. The mean KD and stoichiometric values are shown below the graphs. A table representing the mean KDs in nM compared to the respective isolated knob peptide is shown from our previous reports.
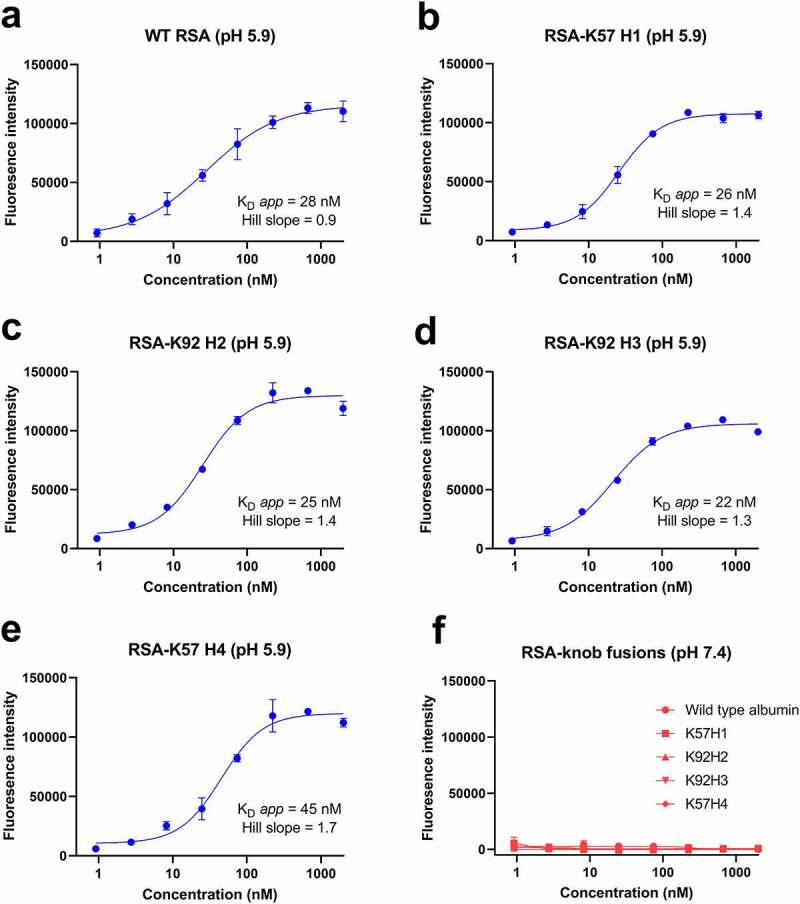
Having established binding to C5, we next sought to confirm maintenance of binding to rFcRn at pH 5.9 compared to pH 7.4, using an fluorescent-linked immunosorbent assay (FLISA) method adapted from Terje-Andersen et al.22–24 For these experiments, ELISA plates were coated with rFcRn and serial dilutions of WT RSA and RSA-knob domain fusions were incubated at either pH 5.9 or pH 7.2. We measured apparent equilibrium dissociation constants (KD app) of <45 nM for the RSA-knob domain fusions at pH 5.9, indistinguishable from WT RSA (Figure 4a-f). As expected, in a manner consistent with the pH-dependent nature of the interaction, no binding occurred under neutral pH for either the WT or modified rat albumins (Figure 4f).
Figure 4.
The rat albumin-knob fusion proteins confer pH-dependent binding to rFcRn.Panels (a–e) show binding to rFcRn for the RSA-knob fusion proteins, measured at pH 5.9 and 7.4 is shown (n = 3). The apparent KD at pH 5.9, calculated from n = 3 experiments, ranging from 22 nM to 45 nM, are shown. Panel (f) no binding to rFcRn was observed at pH 7.4.
The figure shows binding curves, illustrating that the modified rat albumins display rFcRn binding equivalent to wild-type rat albumin.
To ensure that the combination of increased molecular weight and pH-dependent binding to rFcRn extended the t1/2 of the knob domain, we measured the in vivo PK of three of the RSA fusion proteins, RSA-K92 Helix 2, RSA-K92 Helix 3 and RSA-K57 Helix 4, in Sprague Dawley rats. Following administration of a 5 mg/kg IV dose, drug concentrations were quantified by FLISA and subjected to a non-compartmental analysis (Figure 5 and Table 3). The three RSA fusions displayed t1/2 in the range of 32–54 hours, indicating that a substantial extension in t1/2 had been achieved for the knob domains. While comparative data for the K92 knob domain are unavailable, based on previously published PK data with a chemically synthesized K57 knob domain (K57chemFE),7 plasma clearance (CLp) and t1/2 were increased for RSA-K57 Helix 4, by 22-fold and 120-fold, respectively, relative to the K57 peptide (K57chemFE t1/2 = 1.6 hours/CLp = .8 mL/min/kg).
Figure 5.
Pharmacokinetics of rat albumin-knob domain fusion proteins. Quantification of compound levels in plasma was performed by FLISA, following dosing of RSA-K92 H2, RSA-K92 H3 and RSA-K57 H4 at 5 mg/kg IV in Sprague Dawley rats
The figure displays a plot of plasma compound levels versus time following intra venous dosing of RSA-K92 H2, RSA-K92 H3 and RSA-K57 H4 to Sprague Dawley rats.
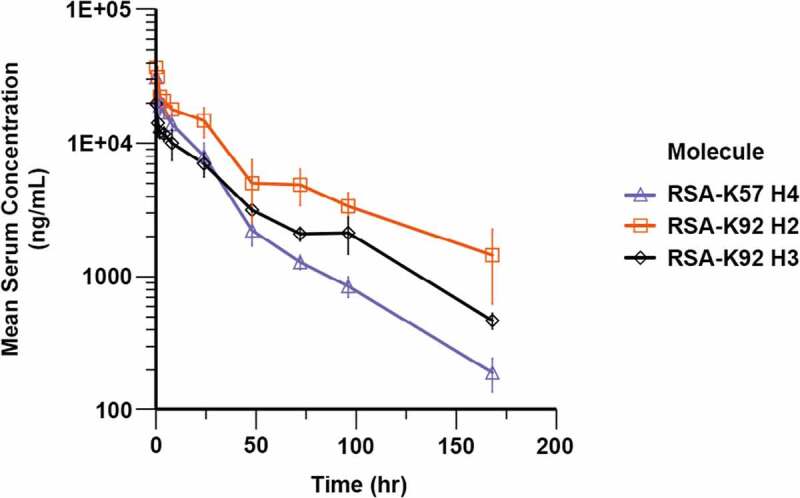
Table 3.
Summary of pharmacokinetic data
| RSA-K57 H4 |
RSA-K92 H2 |
RSA-K92 H3 |
||||
|---|---|---|---|---|---|---|
| Estimate | C. of V. % | Estimate | C. of V. % | Estimate | C. of V. % | |
| 1V1 (ml.kg) | 614 | 6.1 | 371 | 14.0 | 774 | 14.0 |
| 1V2 (ml/kg) | 295 | 16.8 | 440 | 48.1 | 567 | 20.4 |
| 25CL (ml/hr/kg) | 28.2 | 4.5 | 11.4 | 12.5 | 25.7 | 4.7 |
| 25Q (ml/hr/kg) | 8.0 | 27.6 | 7.3 | 42.1 | 280.3 | 77.4 |
| 1T ½ (hrs) | 34 | 8.5 | 54 | 25.0 | 44 | 4.6 |
V1 = Central compartment volume, V2 = peripheral compartment volume, CL = clearance from central compartment, Q = inter-compartmental clearance
25Calculated using 2-compartmental population PK model
Functional characterization of rat albumin-knob domain fusion proteins
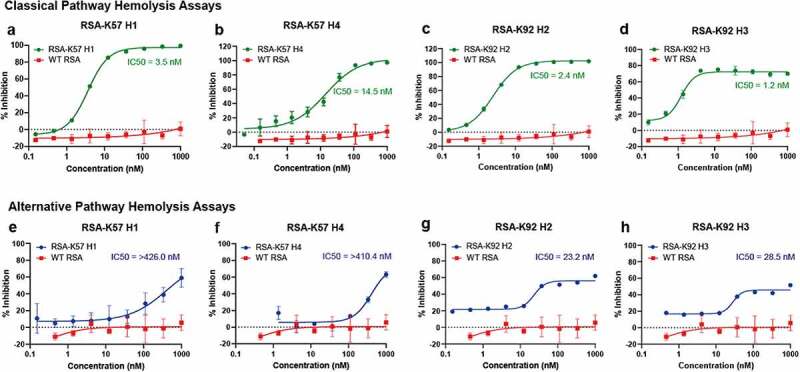
Having engineered C5 binding albumins, we next tested their inhibitory properties using in vitro assays of complement activation. Previous studies have shown that K92 is a potent partial inhibitor of the AP that has no effect on the CP or LP and that, in contrast, K57 is a potent inhibitor of terminal pathway activation arising from both the AP and CP.4 We tested the RSA-knob fusions in hemolysis assays, which measure the ability of serum complement components to lyse erythrocytes. To assay inhibition of the CP, sheep erythrocytes were sensitized with an anti-sheep red blood cell stroma antibody and the serum concentration was fixed < 1% (v/v). For the AP, unsensitized rabbit erythrocytes were assayed in serum concentration of 2.5% (v/v) with chelating agents to remove Ca2+ ions, thereby precluding contribution from the CP and LP. The engineering of K57 into RSA achieves complete inhibition of the CP (Figure 6a-d) and AP (Figure 6e-h). Interestingly, while the RSA-K92 fusion proteins show partial inhibition in the AP assay (Figure 6g), in a manner consistent with earlier studies with the isolated K92 knob domain,4 fusion to albumin achieves 100% inhibition in the CP for all the RSA fusions (Figure 6a-c) other than RSA-K92 H3 (Figure 6d) where it reaches ~80% inhibition. This may arise if the additional bulk of the RSA sterically displaces the CP C5 convertase, whereas the AP C5 convertase remains modulated via an allosteric mechanism. These data highlight the extent of the mechanistic differences between the AP and CP C5 convertase, suggesting that they each interact with C5 via distinct interfaces.
Figure 6.
The rat albumin-knob fusion proteins show inhibition of the classical and alternative pathway Unlike WT RSA, the engineered RSA-knob fusion proteins inhibit terminal pathway activation via the CP (panels a–d) and the AP (panels e–h). The mean IC50 values (n = 3) are shown on the respective graphs.
This figure shows inhibition of the classical and alternative pathway by the C5 binding rat albumin-knob domain fusion proteins that were engineered in this study.
Engineering of a bispecific single-chain VHH
We next applied this approach to generate a small, single-domain bispecific antibody, using the K57 knob domain for transplantation into an acceptor VHH scaffold. We targeted a non-binding framework loop of a VHH (Kabat #H41P), at the opposing end to the CDRs, with the aim of retaining antigen recognition via the canonical CDR loops (Figure 7a and Supplementary 4). It has been suggested previously that the framework loops of VHHs are highly conserved, owing to their importance for solubility and protein folding26 and, to our knowledge, they are not reported as sites for the insertion of non-native polypeptides.
Figure 7.
Bovine knob peptides insertion into the non-binding loops of the hC3nb1 anti-C3b VHH Panel (a) shows the crystal structure of the VHH, hC3nb1, in purple (PDB accession code: 6EHG20) in complex with C3b, in cyan/green. The site of insertion of the K57 knob domain is shown. Panel (b) the purified material is shown by non-reducing SDS-PAGE (1 µg load), lane 1 is hC3nb1-ScFc and lane 2 is hC3nb1-K57-ScFc. Panel (c) Far UV CD reveals changes in secondary structure arising from insertion of the knob domain
The figure shows the crystal structure of camelid VHH, hC3nb1, with the insertion sites used in this study highlighted. A non-reducing gel with a single band for each purified construct shows an apparent increase in molecular weight in the main band arising from insertion of the knob domains. Non-contiguous CD spectra for hC3nb1 and hC3nb1-K57 are shown, alongside the random coil spectra which was measured for the isolated K57 knob domain.
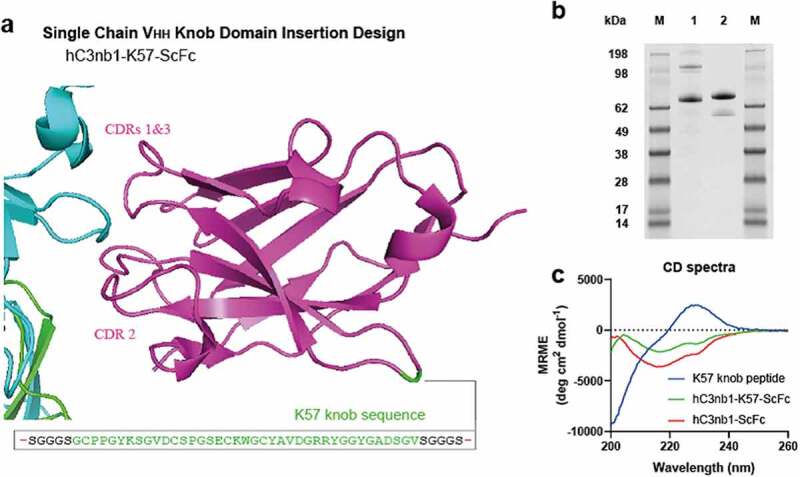
We selected a VHH that had been raised against Complement Component C3, for which an antigen bound co-crystal structure and both binding and functional data are published.20 This VHH antibody, known as hC3nb1, was derived through llama immunization with C3b and has been shown specifically to bind multiple forms of C3 (including C3, C3b, iC3b and C3(H2O), based on C3-methylamine as a mimic).20
We opted to use a poly-histidine, single-chain Fc (ScFc) tag to aid expression and purification of the VHH-knob fusion protein. After nickel affinity and gel filtration chromatography, a yield of approximately 29 mg of purified hC3nb1-K57-ScFc protein per liter was obtained, relative to 35 mg of purified protein per liter for hC3nb1-ScFc. The protein was analyzed by non-reducing SDS-PAGE (Figure 7b) and SEC-UHPLC, which confirmed that the sample was a single species (Supplementary 3), and LC/MS data were consistent with the predicted isotype pattern (Supplementary 5).
Comparative analysis by far UV CD revealed that hC3nb1-K57-ScFc showed changes in the spectra, relative to hC3nb1-ScFc, due to the introduction of the K57 knob domain. The K57 knob domain alone produced a spectrum indicative of an unordered/random coil structure. (Figure 7c). Secondary structure analysis of these spectra using Bestsel,27 indicates a modest 1.5% increase in disordered content arising from insertion of K57 into hC3nb1-ScFc (Table 1).
Binding and functional characterization of a bispecific single-chain VHH
We used SPR to measure binding of the engineered VHH-knob domain fusion protein, hC3nb1-K57-ScFc, to the cognate hC3nb1 antigen, C3. Based on an average of n = 3 experiments, the mean KD of hC3nb1 for C3 was 1.59 nM, while for the hC3nb1-K57-ScFc fusion a mean KD of 3.70 nM was determined, indicating a modest ~2-fold reduction in affinity upon incorporation of the knob domain (Table 5 and Figure 8a-c). Consistent with earlier studies, no interaction between the isolated K57 knob domain and C3 was detected.5
Table 5.
Summary of C3 binding kinetics from biacore multi-cycle kinetics of VHH knob domain fusion protein. Data from n = 3 experiments
| Fusion Protein | Kon | Kon 95% CI | Koff | Koff 95% CI | KD | KD 95% CI |
|---|---|---|---|---|---|---|
| hC3nb1-ScFc | 7.55E+04 | 2.49E+03 | 1.20E-04 | 1.24E-06 | 1.59E-09 | 5.94E-11 |
| hC3nb1-K57-ScFc | 4.63E+04 | 1.43E+03 | 1.71E-04 | 9.64E-06 | 3.70E-09 | 2.87E-10 |
Figure 8.
The VHH- knob domain fusion protein confers binding and function in hemolysis assays.Panels (a–b) show representative sensorgrams for hC3nb1-ScFc and hC3nb1-K57-ScFc binding to human C3 by SPR multi-cycle kinetics, the data are shown in green. Panels (d–e) show representative sensorgrams for hC3nb1-ScFc and hC3nb1-K57-ScFc binding to human C5 by SPR multi-cycle kinetics, the data are shown in blue. All data shows the curves fit from a 1:1 binding model shown in red. The mean KD and stoichiometric values (n = 3) are shown below the graphs. Tables c and f are representing the mean KDs in nM compared to the respective isolated knob peptide from our previous reports.5 Panel (g) shows the hC3nb1-ScFc (orange) inhibits the CP,20 however, the addition of a C5 inhibiting domain in hC3nb1-K57-ScFc (green) markedly increased the potency of CP antagonism. Panel (h) shows the hC3nb1-ScFc (orange) inhibits the AP, as previously reported,20 but the addition of a C5 inhibiting domain in hC3nb1-K57-ScFc (blue) markedly increased the potency of AP antagonism. The mean IC50 values (n = 3) are shown on the respective graphs
This figure shows representative surface plasmon resonance sensorgrams for the VHH-knob fusion protein showing the presence or absence of binding to human C5 and C3 in accordance with the aims of the study. The mean KD and stoichiometric values are shown below the graphs. Tables representing the mean KDs in nM compared to the respective isolated knob peptide are shown from our previous reports. Increased inhibition of the classical and alternative pathway are also shown in this figure by the addition of the C5 binding domain.
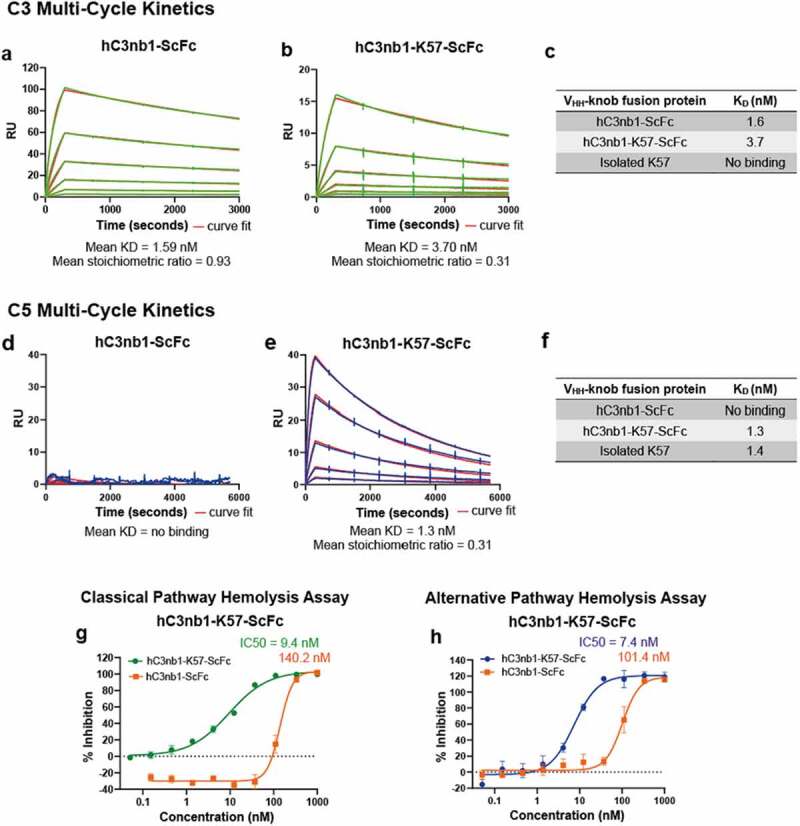
Having established that C3 binding was retained following insertion of K57, we again used SPR to measure binding to C5. In these experiments, hC3nb1-K57-ScFc bound C5 with a mean KD of 1.52 nM (Table 4 and Figure 8d-f). As expected, there was no measurable interaction between the hC3nb1-ScFc and human C5 (Figure 8d).
Table 4.
Summary of C5 binding kinetics from biacore multi-cycle kinetics of VHH knob domain fusion protein. Data from n = 4 experiments
| Fusion Protein | Kon | Kon 95% CI | Koff | Koff 95% CI | KD | KD 95% CI |
|---|---|---|---|---|---|---|
| hC3nb1-K57-ScFc | 4.45E+05 | 2.29E+05 | 3.36E-04 | 5.65E-06 | 1.31E-09 | 1.377E-09 |
| hC3nb1-ScFc | No Binding | - | No Binding | - | No Binding | - |
Having shown individual binding to C5 and C3, we wished to determine if our hC3nb1-K57-ScFc could mediate the formation of a ternary complex by binding both C5 and C3 simultaneously. Biolayer interferometry (BLI) experiments were performed using both antigens as analytes, as described by Krah et al.28 Immobilized hC3nb1-ScFc and hC3nb1-K57-ScFc were subject to binding cycles of C3 and C5 (Figure 9a-b), and while both showed an association with C3, hC3nb1-K57-ScFc was able to subsequently associate with C5, indicating ternary complex formation (Figure 9b).
Figure 9.
Bilayer interferometry confirms simultaneous binding to C3 and C5 by the VHH-knob domain fusion protein.Panel (a) shows hC3nb1-ScFc binding to C3 with no subsequent binding following addition of C5. Panel (b) the hC3nb1-K57-ScFc knob fusion shows binding to C3 and C5 indicating formation of a ternary complex.
The figure shows binding data generated by bilayer interferometry. The addition of K57 into hC3nb1 permits formation of a ternary complex with C3 and C5, in contrast to hC3nb1 which only forms a binary complex with C3.
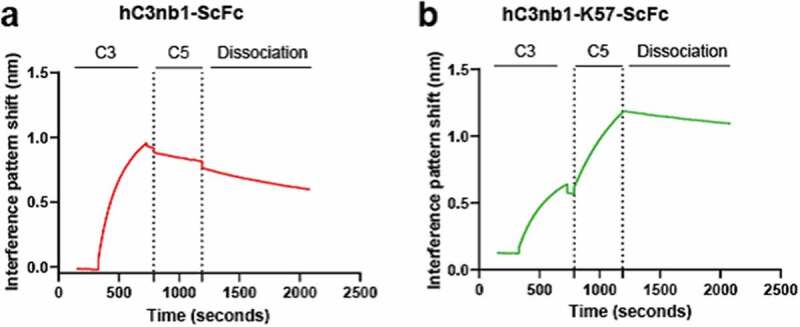
Finally, we tested our anti-C3/C5 VHH bispecific in hemolysis assays, where both hC3nb1-ScFc and hC3nb1-K57-ScFc were potent inhibitors of the CP (Figure 8g) and AP (Figure 8h). The hC3nb1-K57-ScFc bispecific exhibits dual inhibition of C3 and C5, improving the potency compared to the single-targeted VHH, hC3nb1-ScFc. In the AP assay the addition of the C5-targeting K57 knob domain increases the potency of AP antagonism from ~100 nM to 7.4 nM. (Figure 8h). Correspondingly, a 15-fold increase in potency was observed in the CP hemolysis assay. This improved in vitro potency may equate to a lower dose in vivo.
Discussion
This study presents new biological inhibitors of the complement cascade and a new method by which small autonomous paratopes can be grafted into proteins to introduce target specificity. We show that small, immune-derived knob domain peptides can be inserted into various protein loops to provide a second, non-canonical paratope, to rapidly engineer target specificity into single polypeptide chains.
We have shown that knob domains can be fused into rat serum albumin, a protein that can extend plasma half-life by virtue of a pH-dependent interaction with FcRn. The fusion of the knob domains, K57 and 92 into albumin showed minimal effect on the secondary structure compared to rat albumin and retained pH-dependent interaction with rFcRn. These proteins effectively increase the molecular weight of knob domains above the renal threshold, extending the in vivo PK of the knob domains.
A virtue of the single-chain VHH bispecific format described here is that the two antibody entities are expressed as a single polypeptide chain, neatly evading the requirement to efficiently pair heterologous chains. These 18 kDa bispecific antibodies may be useful for immunotherapy, where their small size may aid penetration of solid tissues.
We noted that insertion of the K57 knob domain into the hC3nb1 VHH scaffold slightly decreases the dissociation constant for its canonical antigen, C3. Although the binding of the VHH to its native target reduced slightly, good affinity for C3 was retained and we therefore conclude the VHH can accommodate the insertion of a knob domain.
A potentially surprising feature of this study was that, once inserted into larger protein carriers, knob domains against C5 exhibit different functional behavior and, in the case of the RSA-K92 proteins, appeared functionally competitive. This switch in compound mechanism is possibly a consequence of the larger constructs sterically displacing the activating enzyme complexes. Our previous study highlighted the capacity of knob domains to allosterically modulate C5,4 but where an orthosteric mechanism is desired the additional bulk of a fusion construct may be beneficial. Notably, both RSA-K92 constructs were allosteric inhibitors of the C5 activation by the AP convertase, but fully competitive, and probably orthosteric, inhibitors of the CP. This shows that the enzymatic machinery of the CP and AP C5 convertase have distinct interfaces, which may be partially, but not wholly, overlapping.
Approaches that use knob domains as non-canonical paratopes could readily be applied to other immune or nonimmune proteins, as a pliable ‘plug and play’ platform to introduce improved affinity or multi-valency. Insertions of multiple knob domains could also be trialed to harness avidity or to create polyvalent single-chain constructs. We propose that this simple engineering method is highly adaptable and allows the rapid engineering of multi-valent protein constructs.
Materials and methods
Protein expression and purification
The constructs were designed, with custom synthesis and cloning performed by ATUM into a mammalian expression vector. Expi293F HEK cells were transfected using ExpiFectamine (Invitrogen [Cat # A14525]), as per the manufacturer’s instructions. Cells were cultured for 6 days at 37°C. The cultures were centrifuged, and the supernatants were filtered using 0.22 μm sterifilters. The supernatants were purified on Nickel-Sepharose Excel capture resin columns (GE Healthcare [Cat # 17371206]) using an Akta pure (GE Healthcare). The column was equilibrated with 10 column volumes (CV) of phosphate-buffered saline (PBS). The cell supernatants were loaded at 1.0 mL/minute, the captured protein was washed with 10x CV high salt buffer (0.5 M NaCl, PBS, pH 7.3), then with 10x CV of Buffer A (0.5 M NaCl, 0.025 M Imidazole, PBS, pH 7.3), followed by elution with 10x CV with Buffer B (0.5 M NaCl, 0.25 M Imidazole, PBS, pH 7.3). Between each sample, the column was washed with 0.1 M NaOH, followed by re-equilibration into PBS. The eluant from the nickel purification was then subject to preparative size exclusion chromatography (HiLoad 26/600 SuperDex 200 pg) into PBS. The samples were analyzed by non-reducing SDS-PAGE and stored at −80°C.
SEC
The SEC for the RSA-knob domain fusions was performed on an Agilent 1200 series HPLC instrument using a Phenomex, Yarra™ 3 µm LC column (300 x 7.8 mm) heated to 30°C. Ten µL of a 1 mg/ml sample was injected and run for 27 minutes at a flow rate of 1 mL/minute using PBS as the mobile phase, measuring UV at 254 and 280 nm resulted in a signal eluting at approximately 7.5 minutes. The percentage monomer was obtained in ChemStation (version B.04.03) by integrating the area between 5.5 and 9 minutes and the area of the largest signal.
The VHH-knob domain fusion protein performed on a Waters Acquity UPLC system. The column used was an ACQUITY UPLC Protein BEH SEC column (200 Å, 1.7 µm, 4.6 mm X 150 mm). The column was equilibrated in 0.2 M PBS before 1 µl of sample was loaded onto the column at a flow rate of 0.35 mL/minute. This was followed by 0.2 M PBS at the same flow rate for 10 minutes (~1.6 CV). Data was analyzed using processing methods on Empower 3 software (Waters).
Circular dichroism
The proteins were buffer exchanged into 100 mM phosphate, pH 7.0, 100 mM NaF using 0.5-mL Zeba desalting columns (ThermoFisher Scientific) to 0.3 mg/mL. The CD spectra were acquired using a 1 mm pathlength cell. The scan range was 200–260 nm with a step size of 0.5 nm and an averaging time of 1s. Scans were performed in triplicate. An air and buffer blank were measured and manually subtracted from the spectra with the mean residue molar ellipticity calculated and adjusted for concentrations.
Protein QC by LC/MS
LC/MS was performed on the intact protein samples diluted with PBS to 0.1 mg/mL and reduced with 20 mM tris(2-carboxyethyl) phosphine in 150 mM ammonium acetate at 37°C for 60 minutes. Data were acquired using a Waters ACQUITY UPLC System connected to a Waters Xevo G2 Q-ToF mass spectrometer operated with MassLynx Software. LC conditions were as follows: BioResolveT RP mAb Polyphenyl, 450 Å, 2.7 µm column held at 80°C with a flow rate of 0.6 mL/minute. The mobile phase buffers were: water/0.02% trifluoroacetic acid (TFA)/0.08% formic acid (solvent A) and 95% acetonitrile/5% water/0.02% TFA/0.08% formic acid (solvent B). A reverse phase gradient was run from 5% to 50% solvent B over 8.80 minutes with a 95% solvent B wash and re-equilibration. UV data were acquired at 280 nm. MS conditions were as follows: Ion mode: ESI positive ion, resolution mode, mass range: 400–5000 m/z and external calibration with NaI.
Surface plasmon resonance: multi-cycle kinetics, C5
Multi-cycle kinetics experiments were performed using Biacore 8 K and 8 K+ instruments (GE Healthcare). Following normalization of a Biacore sensor chip CM5, human C5 protein (purified from serum, as described)29 was amine coupled as follows: flow cells 1 and 2 were activated using a 1:2 molar ratio of 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide and N-hydroxysuccinimide (flow rate 10 µL/minute; contact time, 30s). A 5 µg/mL solution of human C5 in 10 mM sodium acetate at pH 4.5 was immobilized in flow cell two only and, finally, both flow cells were blocked with 1 M ethanolamine-HCl, pH 8 (flow rate 10 µL/minute; contact time, 420 s). This typically resulted in immobilization of 230–730 response units (RU). To derive kinetics, five point, three-fold serial dilutions of analyte (range of 100–0.4 nM) were prepared in HBS-EP+ buffer. For each injection, a flow rate of 40 µL/minute, a contact time of 300 s and dissociation time of 5400 s was used. After each injection, regeneration of the surface was performed with sequential injections of 2 M MgCl2 (flow rate 30 µL/min; contact time 30s). The data was fitted with the reference surface subtracted using a Biacore evaluation software 1:1 binding model to determine the binding kinetics.
Rat FcRn binding FLISA
Black ELISA plates (Nunc [Cat# P8741]) were coated overnight at 4°C with 100 µL/well of 5 µg/mL rat FcRn (Acro Biosystems [Cat # FCM-R5287]) solution in PBS. The rFcRn solution was removed and the plates were blocked with 200 µL/well of PBS, 10% [v/v] Sea Block (Thermo [Cat# 37527]), at room temperature (RT), for a minimum of 1 hour. The plates were then washed three times with Wash Buffer (100 mM sodium phosphate [pH 5.9], 0.05% [v/v] Tween 20). The rat albumin-knob fusion proteins were serially diluted in Assay Buffer (100 mM sodium phosphate [pH 5.9] with 10% [v/v] Sea Block), at pH 5.9 or pH 7.2, plated at 100 µL/well and incubated at RT for 1 hour. The plates were then washed at the appropriate pH, as previously described, and a 1:1000 dilution of an anti-rat albumin monoclonal FITC labeled antibody (Life Technologies [Cat# PA1-86695]), prepared in Assay Buffer (pH 5.9 or 7.2), was plated at 100 µL/well and incubated on the assay plate for 45 minutes. Finally, the plate was washed at the appropriate pH, 100 µL/well of Assay Buffer was added and the plate was read on a Pherastar FX plate reader (Exc: 495 nm/Em: 520 nm). Data were analyzed in GraphPad Prism software, using a 4-parameter logistic model.
Pharmacokinetics
The plasma PK was studied for RSA-K92 Helix 2, RSA-K92 Helix 3 and RSA-K57 Helix 4 in male Sprague–Dawley rats. Compounds were administered intravenously via the tail vein. Blood samples were taken at 5 minutes and at 1, 2, 4, 8, 24, 72 and 96 hours. The blood was collected into Li heparin tubes and spun to prepare plasma samples for a bioanalysis. PK properties were analyzed using Phoenix 64 v.8.3.3.33 (Certara). Initial evaluation to determine half-life was conducted using non-compartmental analysis (NCA). NCA was also used to provide initial estimates for two-compartmental population pharmacokinetic model analysis. PK estimations of central and peripheral volume and clearance were conducted using all data from rats in the dosing groups.
Bioanalysis by FLISA
For bioanalysis, black ELISA plates (Nunc [Cat# P8741]) were coated overnight at 2–8 C with a 2 µg/mL solution of human C5 protein in carbonate bicarbonate buffer. The plates were washed four times with Wash Buffer (PBS, 0.05% Tween 20) and blocked with a 10% (v/v) solution of Sea Block (Thermo [Cat# 37527]) in PBS for a minimum of two hours at RT. The plate was washed, and samples, standards and assay controls diluted in Assay Buffer (PBS, 10% [v/v] Sea Block, 0.05% Tween 20) were added at 100 µL/well and incubated for an hour at RT with shaking. After another round of washing, a 1:1000 dilution of FITC conjugated anti-rat serum albumin antibody (Life Technologies [Cat# PA1-86695]) was added at 100 µL/well and incubated for an hour at RT with shaking. Finally, the plate was washed, 100 µL/well of Assay Buffer was added and the plate was read on a Pherastar FX plate reader (Exc: 495 nm/Em: 520 nm). Data was analyzed in GraphPad Prism software, with sample concentrations interpolated from a standard curve fitted to 4-parameter logistic model.
Surface plasmon resonance: multi-cycle kinetics, C3
hC3nb1-ScFc and hC3nb1-K57-ScFc at 5 µg/mL was immobilized on a Biacore sensor chip CM5 as above, typically resulting in immobilizations in the range of 110–270 RU. A 5-point, 3-fold serial dilution of C3 protein (Complement Technologies, Inc. [Cat # A113]) was prepared in HBS-EP+ buffer. For each injection, a flow rate of 40 µL/minute, contact time 300 s and dissociation time 2700 s was used. Between injections, the surface was regenerated with sequential injections of 10 mM glycine-HCl pH 1.5 with a flow rate of 30 µL/minute and contact time 30s. The data was fitted with the reference surface subtracted using a Biacore evaluation software 1:1 binding model.
Complement hemolysis assay
Sheep and rabbit erythrocytes were used for the CP and AP complement assays, respectively. Sheep erythrocytes (TCS Bioscience [Cat# SB069]) were washed, and centrifuged at 800 g, in DGVB++ buffer (0.1% gelatin, 5 mM Veronal buffer, 145 mM NaCl, 2.5% glucose (w/v), 0.15 mM calcium chloride, 1 mM magnesium chloride, pH 7.3) until the supernatant appeared colorless. The erythrocytes were then sensitized with an anti-sheep erythrocyte amboceptor (Sigma [Cat# S1389]) for 30 minutes at 37°C. For the AP assay, rabbit erythrocytes (TCS Bioscience [Cat# RB053AP]) were washed with Mg++EGTA (2.5 mM veronal buffer [pH 7.3] containing 70 mM of NaCl, 140 mM of glucose, 0.1% gelatin, 7 mM of MgCl2, and 10 mM of EGTA) until the supernatant appeared clear.
For the CP, a 0.75% human serum solution was prepared in DGVB++ buffer and, for the AP, a 2.5% human serum solution was prepared in Mg+ EGTA. Ten point, three-fold serial dilutions were prepared in the respective serum solutions in a low bind plate (Costar [Cat# 10274972]), to give a range final assay range of 3 µM-0.15 nM. To 90 µL of the serial dilutions, 10 µL of the rabbit or sheep erythrocytes added and incubated for 30 minutes at 37°C at 650 rpm. Finally, 50 µL of buffer was added to each well and the plates were centrifuged at 800 g for 1 minute. 80 µL of supernatant was transferred to a 96-well flat-bottom plate (Nunc [Cat# 44–2404-21]) and absorbance was measured at 405 nm. A serum solution positive control and a 1 µM OmCI negative control were used to calculate percentage inhibition.
Biolayer interferometry
The ternary complexing experiments were performed on an Octet system (ForteBio, Pall Life Science). Anti-human Fc biosensors (AHC) were pre-wet in kinetics buffer (KB; PBS, 0.1% Tween 20, 1% BSA). The hC3nb1-ScFc and hC3nb1-K57-ScFc proteins were immobilized at 5 ug/mL on the AHC sensors for 30s, followed by sensor rinsing in kinetics buffer for 180 s. A 50 nM solution of human Complement C3 (Complement Technologies, Inc [Cat # A113]) was allowed to associate with the sensor for 400 s. After a baseline in kinetics buffer for 60s, a 400 s incubation with 50 nM Complement C5 protein was performed and dissociation measured for 900 s in kinetics buffer. For each sample, a baseline control was measured where the captured fusion protein was incubated without antigen. This control was subtracted from the binding curves and the evaluation was processed using the ForteBio data analysis software 10.0 with Savitzky-Golay filtering.
Supplementary Material
Funding Statement
The author(s) reported there is no funding associated with the work featured in this article.
Abbreviations
AP Alternative Pathway BLI Biolayer interferometry CD Circular Dichroism CDRH3 Heavy Chain Complementarity-Determining Region 3 CLp Plasma ClearanceCP Classical Pathway EDC 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide Fab Fragment Antigen-Binding Fc fragment crystallization FcRn neonatal Fc receptor FLISA Fluorescent-Linked Immunosorbent Assay ELISA Enzyme-Linked Immunosorbent Assay HEK Human Embryonic Kidney IgG Immunoglobulin G IgM Immunoglobulin MIV intravenous KD Equilibrium Rate Constant kDa Kilodaltons Koff Dissociation Rate ConstantKon Association Rate Constant LC Liquid ChromatographyMS Mass SpectrometryPBS Phosphate-Buffered Saline PDB Protein Data BankPKPharmacokineticsRSA Rat Serum Albumin RU Response Unit ScFc Single-chain Fc SDS-PAGE sulfate-polyacrylamide gel electrophoresis SEC Size Exclusion ChromatographySPR Surface Plasmon Resonance TMB Tetramethylbenzidine UPLC Ultra-High Performance Liquid Chromatography UV Ultraviolet WT Wild Type
Disclosure statement
All authors, apart from J.v.d.E, are current or previous employees of UCB Biopharma UK and may hold shares and/or stock options. A.M and A.D.G.L are inventors on patent applications relating to bovine knob domain peptides.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/19420862.2022.2076295
Data availability statement
The authors confirm that the data supporting the findings of this study are available within the article [and/or] its supplementary materials.
References
- 1.Stanfield RL, Wilson IA, Smider VV, Smider VV.. Conservation and diversity in the ultralong third heavy-chain complementarity-determining region of bovine antibodies. Science Immunology. 2016;1(1):1–12. doi: 10.1126/sciimmunol.aaf7962. PMID: 27574710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang F, Ekiert DC, Ahmad I, Yu W, Zhang Y, Bazirgan O, Torkamani A, Raudsepp T, Mwangi W, Criscitiello MF, et al. Reshaping antibody diversity. Cell. 2013;153(6):1379–93. doi: 10.1016/j.cell.2013.04.049. PMID: 23746848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stanfield RL, Berndsen ZT, Huang R, Sok D, Warner G, Torres JL, Burton DR, Ward AB, Wilson IA, Smider VV, et al. Structural basis of broad HIV neutralization by a vaccine-induced cow antibody. Sci Adv. 2020;6(22):eaba0468. doi: 10.1126/sciadv.aba0468. PMID: 32518821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Macpherson A, Laabei M, Ahdash Z, Graewert MA, Birtley JR, Schulze ME, Crennell S, Robinson SA, Holmes B, Oleinikovas V, et al. The allosteric modulation of complement C5 by knob domain peptides. eLife. 2021;10:e63586. doi: 10.7554/eLife.63586. PMID: 33570492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Macpherson A, Scott-Tucker A, Spiliotopoulos A, Simpson C, Staniforth J, Hold A, Snowden J, Manning L, van den Elsen J, Lawson ADG.. Isolation of antigen-specific, disulphide-rich knob domain peptides from bovine antibodies. PLoS Biology. 2020;18(9):e3000821. doi: 10.1371/journal.pbio.3000821. PMID: 32886672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dong J, Finn JA, Larsen PA, Smith TPL, Crowe JE Jr. Structural diversity of ultralong CDRH3s in seven bovine antibody heavy chains. Front Immunol. 2019;10:558. doi: 10.3389/fimmu.2019.00558. PMID: 30967877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Macpherson A, Birtley JR, Broadbridge RJ, Brady K, Schulze MED, Tang Y, Joyce C, Saunders K, Bogle G, Horton J, et al. The chemical synthesis of knob domain antibody fragments. ACS Chem Biol. 2021;16(9):1757–69. doi: 10.1021/acschembio.1c00472. PMID: 34406751. [DOI] [PubMed] [Google Scholar]
- 8.Markovsky E, Baabur-Cohen H, Eldar-Boock A, Omer L, Tiram G, Ferber S, Ofek P, Polyak D, Scomparin A, Satchi-Fainaro R.. Administration, distribution, metabolism and elimination of polymer therapeutics. Journal of Controlled Release: Official Journal of the Controlled Release Society. 2012;161(2):446–60. doi: 10.1016/j.jconrel.2011.12.021. PMID: 22286005 [DOI] [PubMed] [Google Scholar]
- 9.Peters T. 2 - The albumin molecule: its structure and chemical properties. In: Peters T, editor. All About Albumin. San Diego: Academic Press; 1995. p. 9–II. [Google Scholar]
- 10.Chaudhury C, Mehnaz S, Robinson JM, Hayton WL, Pearl DK, Roopenian DC, Anderson CL. The major histocompatibility complex-related Fc receptor for IgG (FcRn) binds albumin and prolongs its lifespan. J Exp Med. 2003;197(3):315–22. doi: 10.1084/jem.20021829. PMID: 12566415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Waldmann TA, Strober W. Metabolism of immunoglobulins. Prog Allergy. 1969;13:1–110. doi: 10.1159/000385919. PMID: 4186070. [DOI] [PubMed] [Google Scholar]
- 12.Colvin RA, Tanwandee T, Piratvisuth T, Thongsawat S, Hui AJ, Zhang H, Ren H, Chen P-J, Chuang W-L, Sobhonslidsuk A, et al. Randomized, controlled pharmacokinetic and pharmacodynamic evaluation of albinterferon in patients with chronic hepatitis B infection. Journal of Gastroenterology and Hepatology. 2015;30(1):184–91. doi: 10.1111/jgh.12671. PMID: 24995515. [DOI] [PubMed] [Google Scholar]
- 13.Lau J, Bloch P, Schaffer L, Pettersson I, Spetzler J, Kofoed J, Madsen K, Knudsen LB, McGuire J, Steensgaard DB, et al. Discovery of the once-weekly glucagon-like peptide-1 (GLP-1) analogue semaglutide. J Med Chem. 2015;58(18):7370–80. doi: 10.1021/acs.jmedchem.5b00726. PMID: 26308095. [DOI] [PubMed] [Google Scholar]
- 14.Lewis AL, McEntee N, Holland J, Patel A. Development and approval of rybelsus (oral semaglutide): ushering in a new era in peptide delivery. Drug Deliv Transl Res. 2021;12(1):1–6. doi: 10.1007/s13346-021-01000-w. PMID: 34024013 [DOI] [PubMed] [Google Scholar]
- 15.Huang X, Wang C, Ma T, Huang Z, Zhou H, Xu L, Zhang R, Zhao J, Zhang Y, Huang Z, et al. The efficacy of combined cisplatin and nanoparticle albumin-bound paclitaxel in a stage iv pancreatic squamous cell carcinoma patient with a somatic BRCA2 mutation: a case report. Front Oncol. 2021;11:585983. doi: 10.3389/fonc.2021.585983. PMID: 33996534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nilsen J, Trabjerg E, Grevys A, Azevedo C, Brennan SO, Stensland M, Wilson J, Sand KMK, Bern M, Dalhus B, et al. An intact C-terminal end of albumin is required for its long half-life in humans. Communications Biology. 2020;3(1):181. doi: 10.1038/s42003-020-0903-7. PMID: 32313072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Labrijn AF, Janmaat ML, Reichert JM, Parren P. Bispecific antibodies: a mechanistic review of the pipeline. Nat Rev Drug Discov. 2019;18(8):585–608. doi: 10.1038/s41573-019-0028-1. PMID: 31175342 [DOI] [PubMed] [Google Scholar]
- 18.Xu Y, Lee J, Tran C, Heibeck TH, Wang WD, Yang J, Stafford RL, Steiner AR, Sato AK, Hallam TJ, et al. Production of bispecific antibodies in “knobs-into-holes” using a cell-free expression system. mAbs. 2015;7(1):231–42. doi: 10.4161/19420862.2015.989013. PMID: 25427258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Labrijn AF, Meesters JI, de Goeij BECG, van den Bremer ETJ, Neijssen J, van Kampen MD, Strumane K, Verploegen S, Kundu A, Gramer MJ, et al. Efficient generation of stable bispecific IgG1 by controlled Fab-arm exchange. Nat Protoc. 2013;110(13):5145–50. doi: 10.1073/pnas.1220145110. PMID: 23479652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jensen RK, Pihl R, Gadeberg TAF, Jensen JK, Andersen KR, Thiel S, Laursen NS, Andersen GR. A potent complement factor C3-specific nanobody inhibiting multiple functions in the alternative pathway of human and murine complement. The Journal of Biological Chemistry. 2018;293(17):6269–81. doi: 10.1074/jbc.RA117.001179. PMID: 29497000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oganesyan V, Damschroder MM, Cook KE, Li Q, Gao C, Wu H, Dall’Acqua WF. Structural insights into neonatal Fc receptor-based recycling mechanisms. The Journal of Biological Chemistry. 2014;289(11):7812–24. doi: 10.1074/jbc.M113.537563. PMID: 24469444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sand KM, Dalhus B, Christianson GJ, Bern M, Foss S, Cameron J, Sleep D, Bjørås M, Roopenian DC, Sandlie I, et al. Dissection of the neonatal Fc receptor (FcRn)-albumin interface using mutagenesis and anti-FcRn albumin-blocking antibodies. The Journal of Biological Chemistry. 2014;289(24):17228–39. doi: 10.1074/jbc.M113.522565. PMID: 24764301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andersen JT, Dalhus B, Cameron J, Daba MB, Plumridge A, Evans L, Brennan SO, Gunnarsen KS, Bjørås M, Sleep D, et al. Structure-based mutagenesis reveals the albumin-binding site of the neonatal Fc receptor. Nat Commun. 2012;3(1):610. doi: 10.1038/ncomms1607. PMID: 22215085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andersen JT, Daba MB, Berntzen G, Michaelsen TE, Sandlie I. Cross-species binding analyses of mouse and human neonatal Fc receptor show dramatic differences in immunoglobulin G and albumin binding. The Journal of Biological Chemistry. 2010;285(7):4826–36. doi: 10.1074/jbc.M109.081828. PMID: 20018855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berens SJ, Wylie DE, Lopez OJ. Use of a single VH family and long CDR3s in the variable region of cattle Ig heavy chains. Int Immunol. 1997;9(1):189–99. doi: 10.1093/intimm/9.1.189. PMID: 9043960 [DOI] [PubMed] [Google Scholar]
- 26.Mitchell LS, Colwell LJ. Comparative analysis of nanobody sequence and structure data. Proteins. 2018;86(7):697–706. doi: 10.1002/prot.25497. PMID: 29569425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Micsonai A, Wien F, Bulyaki E, Kun J, Moussong E, Lee YH, Goto Y, Refregiers M, Kardos J. BeStSel: a web server for accurate protein secondary structure prediction and fold recognition from the circular dichroism spectra. Nucleic Acids Res. 2018;46(W1):315–22. doi: 10.1093/nar/gky497. PMID: 29893907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krah S, Schröter C, Eller C, Rhiel L, Rasche N, Beck J, Sellmann C, Günther R, Toleikis L, Hock B, et al. Generation of human bispecific common light chain antibodies by combining animal immunization and yeast display. Protein Engineering, Design and Selection. 2017;30:291–301. doi: 10.1093/protein/gzw077. PMID: 28062646. [DOI] [PubMed] [Google Scholar]
- 29.Macpherson A, Liu X, Dedi N, Kennedy J, Carrington B, Durrant O, Heywood S, van den Elsen J, Lawson ADG. The rational design of affinity-attenuated OmCI for the purification of complement C5. The Journal of Biological Chemistry. 2018;293(36):14112–21. doi: 10.1074/jbc.RA118.004043. PMID: 30030376 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article [and/or] its supplementary materials.


