Abstract
Pseudomonas chlororaphis MA 342 is a potent biocontrol agent that can be used against several seed-borne diseases of cereal crops, including net blotch of barley caused by the fungus Drechslera teres. In this study, strain MA 342 was tagged with the gfp gene (encoding the green fluorescent protein) in order to study the fate of cells after seed inoculation. The gfp-tagged strain, MA 342G2, had the same biocontrol efficacy as the wild type when it was applied at high cell concentrations to seeds but was less effective at lower cell concentrations. By comparing cell counts determined by microscopy to the number of CFU, we found that the number of culturable cells was significantly lower than the total number of bacteria on seeds which were inoculated and dried for 20 h. Confocal microscopy and epifluorescence stereomicroscopy were used to determine the pattern of MA 342G2 colonization and cell aggregation on barley seeds. Immediately after inoculation of seeds, bacteria were found mainly under the seed glume, and there was no particular aggregation pattern. However, after the seeds were sown, irregularly distributed areas of bacterial aggregation were found, which reflected epiphytic colonization of glume cells. There was a trend towards bacterial aggregation near the embryo but never within the embryo. Bacterial aggregates were regularly found in the groove of each seed formed by the base of the coleoptile and the scutellum. Based on these results, we suggest that MA 342 colocalizes with the pathogen D. teres, which facilitates the action of the fungistatic compound(s) produced by this strain.
Seed-borne diseases in cereals can cause serious economic losses (19), and therefore, seeds are commonly treated with fungicides to suppress these diseases. Since fungicides may affect human health and the environment and since pathogens can develop resistance to fungicides, alternative control measures, such as biological control, are much sought after. In the last decade many microorganisms have been tested to determine their abilities to suppress seed- and soil-borne pathogens in agriculture (21). However, microorganisms that exhibit biocontrol potential in tests in vitro and/or in bioassays have often exhibited inconsistent behavior under field conditions. This has been the major impediment to large-scale use of biocontrol agents in agriculture (27).
In a recent screening for potential biocontrol agents that can be used against seed-borne net blotch of barley caused by the fungal pathogen Drechslera teres, Pseudomonas chlororaphis MA 342 was found to be a very effective and consistent biocontrol agent (14). This bacterium applied to seeds has been as effective as conventional fungicides and has exhibited consistent behavior under field conditions since 1991 in Sweden and since 1994 in more than 10 European countries (14, 17). Strain MA 342, which was patented by the Swedish company BioAgri AB (Stockholm, Sweden), has been used on a commercial scale in Sweden under the trade name Cedomon since 1998 and probably will be registered as a biocontrol agent in several other European countries in 1999.
Although MA 342 has been proven to be an effective biocontrol agent, little is known about its distribution on the plant from the seed inoculum. Such information may help clarify the means by which MA 342 protects the plant from fungal diseases. Presently, we know neither where the bacterium is located on the seed after inoculation nor to what extent MA 342 colonizes the seed and other plant parts. Although there have been studies of seed inoculation of cereal crops and subsequent colonization of the roots by other bacteria (18, 26), we are not aware of any study describing the actual pattern of colonization of biocontrol bacterial cells on cereal seeds. Cereal seeds have a complex structure, and it is not known to which parts the bacteria attach and which parts the bacteria colonize. A barley seed, for example, is surrounded by a pericarp, which is in turn surrounded by a husk (or glume) (11, 16, 19). The method used to apply bacteria to seeds may affect the distribution and pattern of colonization of the bacteria and subsequently the efficacy of the biocontrol agent. Therefore, it is important to know which parts of the seed need to be colonized for effective biocontrol to occur.
Recently, molecular techniques have been used to detect and enumerate microbes in situ on plant surfaces (2, 3, 10, 23). One promising technique relies on a marker system that uses the gfp gene, which encodes the green fluorescent protein (GFP), from Aequorea victoria (5); this technique has shown promise to monitor pseudomonads (23). GFP is a useful biomarker because it does not require any substrate or cofactor in order to fluoresce. Therefore, cells tagged with GFP can be enumerated in situ, and samples do not need to be disturbed by techniques such as fixing, washing, hybridization, or staining (22). Workers have developed GFP cassettes for chromosomal integration and expression of gfp in a variety of bacteria (4, 10, 23). These cassettes can be integrated into the chromosomes of different bacterial strains. Single cells with chromosomal integration of gfp can be identified by epifluorescence microscopy or confocal microscopy (23). For enhanced fluorescence intensity, two copies of gfp can be integrated into the chromosome (22, 24, 25). This marker system allows workers to study specific bacteria in environmental samples in situ with a minimum of sample preparation, thus avoiding possible disturbance of the natural cell colonization pattern.
In this paper we describe the distribution pattern on barley seeds of the bacterial biocontrol agent P. chlororaphis MA 342 chromosomally tagged with gfp.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
P. chlororaphis MA 342 was originally isolated from roots of craw berry (Empetrum nigrum L.) (14). Strain MA 342G2 was obtained by chromosomally tagging MA 342 with the gfp gene as described below. The strains were grown at 28°C on Luria-Bertani (LB) medium amended with kanamycin (75 μg/ml) when appropriate. The cultures used for seed inoculation were prepared by growing bacteria for 48 h at 20°C in 0.5× TSB liquid medium (15 g of Difco tryptic soy broth per liter) that did not contain the antibiotic.
Tagging MA 342 with gfp.
Methods and molecular tools for tagging pseudomonads with the gfp gene have been described elsewhere (22–25). MA 342 was grown in LB medium for 6 h, and cells were collected by centrifugation and washed three times in cold sterile distilled water. The final cell concentration was approximately 1010 cells/ml. To 100 μl of the concentrated cell suspension, 200 ng of purified gfp delivery plasmid DNA (pUTgfp2X) (23) was added, and the mixture was transferred to an electroporation cuvette. The electroporation conditions used were 12.5 kV/cm, 25 mF, and 200 Ω, which were provided by a Gene Pulser electroporation device (Bio-Rad Laboratories, Hercules, Calif.). After electroporation, the cell suspension was diluted in 1 ml of LB medium and incubated for 40 min at 28°C before it was plated onto LB medium plates amended with kanamycin (20 μg/ml).
Seed inoculation.
Twenty-five grams of barley seeds that were naturally infested with D. teres (Sacc.) Shoemaker was mixed with 7.5 ml of a MA 342G2 culture grown in TSB medium diluted as appropriate with 0.01 M MgSO4 in a 180-ml plastic cup, and the preparation was shaken by hand for 1 min. After mixing, the seeds were spread onto a plastic dish and air dried for 20 h before they were sown. When smaller amounts of seeds were inoculated, proportional volumes of bacterial cultures were used. Seeds from two lots were used; cultivar Golf seeds and cultivar Svani seeds were moderately and heavily infested with the fungal pathogen, respectively. The latter seeds were used so that we could compare the biocontrol efficacies of the wild-type and gfp mutant strains.
Microcosms and experimental procedures.
Sandy loam soil (pH 6.3; organic matter content, 3%) was collected from the upper 10 to 15 cm of an agricultural field in Uppsala, Sweden. The soil was air dried for 2 days (complete drying was avoided) and sifted through a 0.5-cm-mesh screen. The soil was then mixed with tap water until the water content was 12% (wt/wt) (soil matrix potential pF, 2.2). The soil was placed in a plastic box with a small opening, which permitted gas exchange, and incubated at 14°C for 16 h/day and at 8°C for 8 h/day in continuous darkness for 4 days before it was used. Tubes that were 20 cm high and 4.5 cm in diameter were filled with the moist soil, and barley seeds (cultivar Golf) were planted at a depth of 2 cm. The soil was covered with a 0.5-cm layer of sand to reduce water loss by evaporation. The tubes were placed in a growth chamber at 70% relative humidity, and two temperature-light regimes were used; in one regime the tubes were incubated at 14°C in the light for 16 h/day and at 8°C in the dark for 8 h/day, and in the other regime the tubes were incubated at 6°C in continuous darkness. For studies of bacterial aggregation as determined by microscopic visualization, seeds were sown in the soil described above at a depth of 2 cm in small pots and incubated at 6 or 14°C.
Bioassay.
The biocontrol efficacies of wild-type strain MA 342 and MA 342G2 were compared as follows. Samples of seeds (cultivar Svani) were inoculated with bacterial suspensions at different doses; we used an undiluted TSB medium culture (5 × 109 CFU/ml) and 10-fold dilutions (down to 5 × 105 CFU/ml) of a TSB medium culture that resulted in five different bacterial doses. For each dose 50 inoculated seeds were sown in pots that were 18 cm in diameter and 4 cm high. Each pot was covered with a glass lid and placed at 6°C in the dark. After 10 days, the pots were moved to a greenhouse and incubated at about 20°C. The percentages of seedlings that exhibited net blotch symptoms on the first leaf were determined after 11 days of incubation. Two pots were used per dose, and the experiment was repeated twice. Data were subjected to an analysis of variance by using a split plot model with dilutions as split plots. Nontreated seeds were used as the control.
Sampling and enumeration of bacteria.
Seed samples were obtained at the time of sowing and 1, 3, 7, and 14 days after sowing. The seeds were carefully removed from the soil, and any seminal roots were cut off. Individual seeds were blended in 0.01 M MgSO4 for about 30 s until the glume was removed and fragmented. Blending for longer periods did not result in higher numbers of bacteria in the suspension. The number of green fluorescent cells (extracted from seeds) was determined by fluorescence microscopy. Suspensions were filtered through black cellulose nitrate filters (diameter, 47 mm; pore size, 0.45 μm). Cells in 10 to 30 microscopic fields (the field diameter was adjusted to 0.36 mm with the diaphragm) that contained at least 200 cells were counted with a Leitz Aristoplan fluorescence microscope with a ×50 objective (total magnification, ×500). The filter set included a bandpass excitation filter at 470/40 nm and a longpass emission filter (500 LP). The numbers of culturable cells were determined by dilution plating on TSA (10 g of Difco TSB and 15 g of agar/liter) amended with kanamycin (25 μg/ml). Four seeds were assayed for each sampling time. Analyses were performed by using log10-transformed data. An analysis of variance was performed for both the total number of green fluorescent cells and the number of culturable cells on seeds. The number of culturable cells and the number of green fluorescent cells extracted from seeds were compared by performing pairwise t tests. The null hypothesis of no difference was tested against the one-sided alternative hypothesis of a lower number of culturable cells than the total number of cells.
Microscopic visualization of bacteria on seeds.
Aggregations of fluorescent bacterial cells were visualized by using a fluorescence stereomicroscope (model MZ12; Leica AG, Heerbrugg, Switzerland) equipped with a double set of fluorescence filters (excitation 480/40 nm, dichroic 505LP nm, and emission 510LP nm), a high-pressure mercury vapor burner, and a ×1 planapochromatic objective. Seed samples obtained at different times after sowing were gently brushed with a paintbrush in order to remove attached soil particles and then were directly inspected and dissected under the stereomicroscope. Fujicolor Reala ISO 100 photographic film (Fuji Photo Film U.S.A., Inc.) was used for photographs. For more detailed resolution of fluorescent bacterial cells, a model LSM510 laser scanning confocal microscope (Carl Zeiss, Jena, Germany) was used. The light sources were two lasers that provided three excitation wavelengths, 488 nm (Ar) and 543 and 633 nm (HeNe). The following emission filters were used: CH1:LP650 (blue), CH2:BP505-530 (green), and CH3:BP560-615 (red). The images were the result of pseudocolor merging of the outputs of three channels. Three-dimensional rendering of the stack of images was obtained by using the software 3D for LSM510, version 1.4 (Carl Zeiss). Cryosectioning of seeds was performed with a cryostat (model CM 1800; Leica). Cryosections that were 15 to 20 μm thick were directly analyzed by confocal microscopy.
RESULTS
Tagging MA 342 with gfp and biocontrol activity.
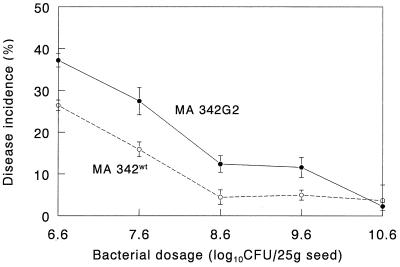
Strain MA 342G2 was selected from a group of eight gfp mutants based on its high-level green fluorescence intensity. In addition, the colony morphology and growth patterns in TSB medium (15 g/liter) and minimal medium (0.4% glucose) were similar to those of the wild type (results not shown). No white colonies appeared after repeated transfers of the tagged strain onto nonselective media. Therefore, the integration of two copies of gfp into the chromosome appeared to be stable. At lower bacterial doses the gfp mutant had a reduced ability to control seed-borne net blotch (Fig. 1) (P = 0.009, as determined by the F test). The interaction between strains and bacterial doses was significant at P = 0.0517, indicating that at a high dose the tagged and wild-type strains suppressed seed-borne net blotch to the same extent (Fig. 1).
FIG. 1.
Suppression of seed-borne net blotch in barley by P. chlororaphis MA 342 (wild type [wt]) and the gfp-tagged derivative strain MA 342G2. Data points are means based on three replicates. Bars indicate standard errors. The level of disease incidence in the control was 45.9% ± 2.2%.
Growth of the bacterial population in the spermosphere.
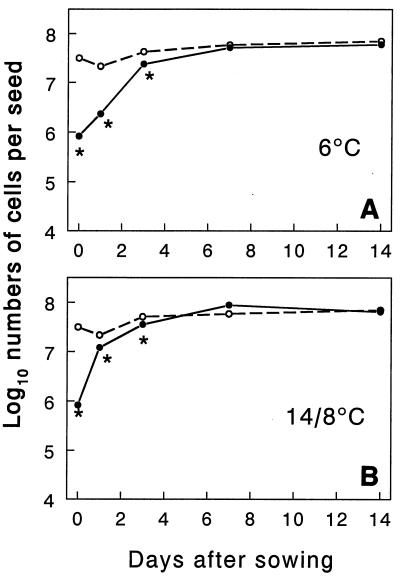
At the time of inoculation the GFP fluorescent cells were culturable, since total counts and CFU values were similar (7 × 109 cells or CFU/ml of TSB medium). However, after 20 h of drying, less than 10% of the gfp-tagged cells extracted from seeds were culturable (Fig. 2). After inoculated seeds were sown, the number of culturable cells increased, and the number of cells reached a value similar to the number of gfp-tagged cells counted by microscopy between 3 and 7 days. The number of gfp-tagged cells also increased, and the initial population about doubled after 7 days (P = 0.0001, as determined by the F test). The number of culturable cells increased faster when seeds were incubated with a cycle consisting of 16 h of light at 14°C and 8 h of darkness at 8°C than when seeds were incubated at 6°C in continuous darkness, as shown by the significant interaction between time and temperature (P = 0.0128, as determined by the F test). The populations of gfp fluorescent cells and culturable cells remained stable after day 7 (Fig. 2).
FIG. 2.
Numbers of green fluorescent cells (○) and culturable cells (●) of bacterial strain MA 342G2 on barley seeds. Seeds were mixed with a bacterial suspension, dried for 20 h, sown and incubated at 6°C in continuous darkness (A) or with cycles consisting of 16 h of light at 14°C and 8 h of darkness at 8°C (B). Data are means based on four seeds. The asterisks indicate significantly lower numbers of culturable cells than green fluorescent cells (P < 0.05, as determined by a t test). The pooled standard deviations were 0.18 for the number of green fluorescent cells, 0.20 for the number of culturable cells, and 0.11 for the number of green fluorescent cells minus the number of culturable cells.
In situ microscopic visualization of bacteria.
The high-level fluorescence intensity of the gfp-tagged cells resulted in good visualization of single bacteria on the seeds, as determined by microscopy. After inoculation and drying of the seeds, green fluorescent bacteria were scattered mainly on the inner side of the seed glume. At this time the bacterial cells were only loosely aggregated, and consequently it was very difficult to assess whether they were located intra- or intercellularly (data not shown). After sowing, more aggregation of bacteria was observed on seeds at the time that the roots and shoots began to elongate (Fig. 3 through 5). Single bacteria were visible on seeds, as determined by confocal microscopy (results not shown), but the weak clustering of cells did not permit macroscopic visualization by epifluorescence stereomicroscopy. In contrast, little or no colonization of the roots was observed, and no colonization of the leaves was observed.
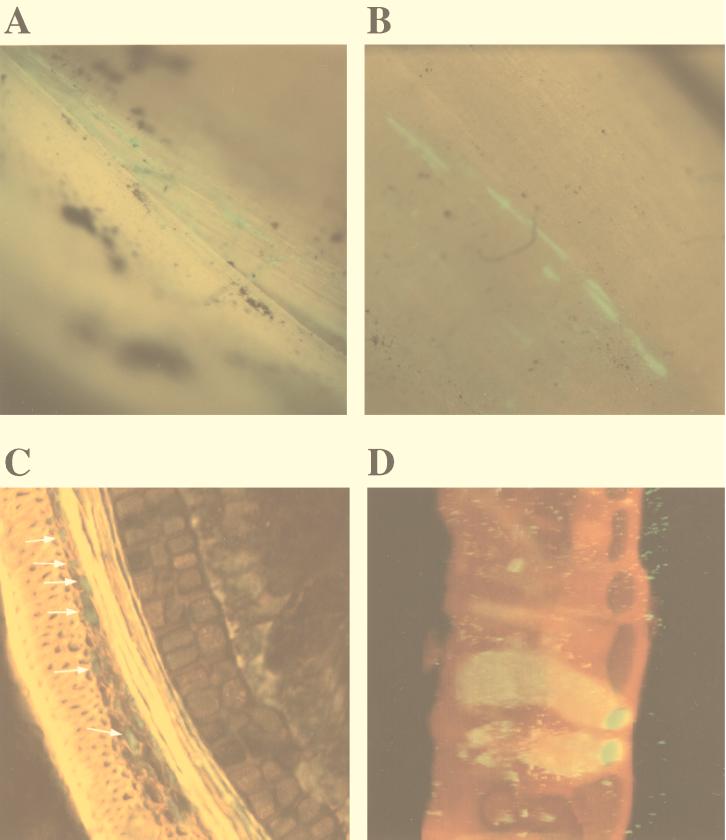
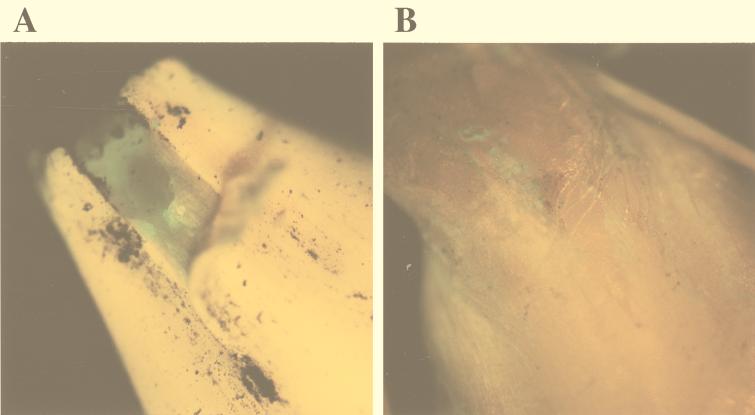
FIG. 3.
Epifluorescence stereomicroscope images (A and B) and confocal microscope images (C and D) of the external layers of a barley seed inoculated with MA 342G2 after incubation for 1 to 2 days in soil at 14°C. (A and B) Streaks of fluorescent bacteria at the edges of the glume (A) and on the external side of the glume (B). Magnification range, ×10 to ×40. (C) Confocal microscope image of a 15-μm-thick transverse section of a germinating seed. Green fluorescent bacteria are visible in the parenchymatous layer of the glume. (D) Confocal microscope projection of a stack of images of spots corresponding to the spots in panel B (on a different seed). Magnification, ×400. Scan zoom, 1.7.
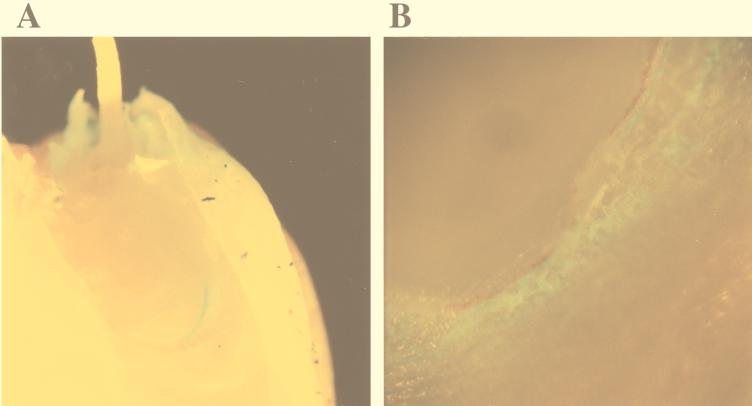
FIG. 5.
Photographs of the area at the base of the coleoptile of a barley embryo, obtained with the epifluorescence stereomicroscope. (A and B) The emerging shoot was removed, and the green fluorescent spots around and under it, corresponding to fluorescent bacteria, were exposed. (C) The emerging shoot was lifted in order to reveal the heavy localized colonization by bacteria surrounding its base. Magnification range, ×10 to ×40.
One day after seeds were sown, we observed green fluorescent streaks that were irregularly scattered on the seed glume external surface, as determined by epifluorescence stereomicroscopy (Fig. 3A and B). Often these green fluorescent streaks appeared at the edges of the glume (Fig. 3A). Inspection of the seed surface by confocal microscopy revealed that the long ribbonlike green fluorescing spots corresponded to intracellular colonization of cells of the seed glume (Fig. 3D). Glume epidermal cells were found to be packed with green fluorescing MA 342G2 cells (Fig. 3D). Transverse sections of the glume, analyzed by confocal microscopy, revealed that MA 342G2 cells also colonized the parenchymatous layers of the glume (Fig. 3C, arrows). However, in the numerous seed cross-sections carefully analyzed in this study, green fluorescent bacteria were never observed in or below the aleuronic cell layer of the seed.
After the glume was peeled off the germinating seed, green fluorescent patches were observed both on the inner side of the glume and on the outer side of the pericarp, and there was a trend toward a higher level of colonization on the embryo half of the seed (Fig. 4). During root elongation, green fluorescent spots were always visible on the pericarp cell layers surrounding the growing embryo (Fig. 4B). At a high magnification (during confocal microscopy) we did not observe any intracellular aggregation of bacteria in the pericarp layers around the embryo. When we peeled off the pericarp layer further (Fig. 4B), we found that there were no green fluorescent cells on the naked embryo surface underneath (results not shown).
FIG. 4.
Photographs of the embryo areas of germinating barley seeds obtained with the epifluorescence stereomicroscope. (A) Green fluorescent areas corresponding to bacterial cells near the embryo under the glume. (B) Green fluorescent spots corresponding to bacterial cells near the point of root emergence observed after the glume was peeled off. Magnification range, ×10 to ×40.
The patterns of seed colonization by MA 342G2 described above revealed a certain variability, but heavy colonization of the groove between the coleoptile and scutellum was always observed in the approximately 100 seeds examined. Figure 5 shows epifluorescence stereomicroscope images of seeds taken when the shoot was 2 to 6 mm long. When the shoot was excised from the embryo, strong aggregation of MA 342G2 cells was visible, and these bacteria formed a semicircle of strong green fluorescence (Fig. 5A). This region of heavy colonization between the coleoptile and the scutellum was also visible when the shoot was gently lifted (Fig. 5C). Emergence of the shoot, as well as strong bacterial aggregation, occurred later at 6°C than at 14°C.
DISCUSSION
Tagging bacterial strain MA 342 with the gfp gene enabled us to both microscopically count green fluorescent cells and localize the specific green fluorescing bacteria on barley seeds in situ. In addition, the number of culturable cells on a selective medium could be determined, and the number of nonculturable green fluorescent cells could be assessed by microscopy. After inoculated barley seeds were dried for 1 day, more than 90% of the green fluorescent cells extracted from the seeds were nonculturable. It is not clear if these nonculturable cells were viable. The increase in the number of culturable cells after sowing (Fig. 1) may have been due to multiplication of culturable cells present on the seeds and/or resuscitation of nonculturable cells. We have previously found that gfp-tagged bacteria remain fluorescent even during long-term starvation, when they are no longer metabolically active (23). However, it is not known if dead cells retain the GFP, which could be a limitation to the use of gfp as a marker to estimate the total sizes of viable cell populations.
Each of the eight gfp-tagged transformants had a biocontrol effect similar to that of the wild-type strain MA 342 when seeds were treated with an undiluted TSB medium culture, but each organism exhibited reduced activity when seeds were treated with 10-fold dilutions of the culture. The reduced ability to suppress seed-borne net blotch at lower cell densities compared to the ability of the wild-type strain was more likely due to a metabolic burden imposed by GFP and kanamycin resistance expression enhanced by the stress conditions than to insertion of the transposon in a gene essential for biological control. Negative effects on the environmental fitness of genetically modified microorganisms containing marker gene integrations in the chromosome inserted at nonessential sites have been observed in other pseudomonads as well (8).
The high GFP fluorescence intensity of the mutant used (MA 342G2) was useful for microscopic studies of bacterial aggregation on seeds. In this study we macroscopically visualized fluorescent bacterial aggregates on seeds by fluorescence stereomicroscopy for the first time (Fig. 3A and B and Fig. 4 and 5). This method allowed us to describe the colonization pattern on a much larger scale than the scale allowed by scanning electron microscopy or confocal microscopy.
Bacterial colonization of plant surfaces, such as roots and leaves, appears to occur in the form of aggregates or microcolonies (3, 6, 7, 12). Although for biocontrol purposes bacteria are often applied to seeds, there have not been many studies on bacterial colonization patterns of seeds. Bacterial colonization of the outer surfaces of sugar beet and cotton seeds has been studied by using scanning electron microscopy, conventional microscopy, and confocal microscopy with conventional staining techniques (9, 15). These studies showed that bacteria occurred in microcolonies or aggregates in grooves or cracks on the outer seed surface. We are not aware of any study which has revealed colonization of different cell layers of the seed coat, pericarp, or other seed envelopments, such as the glume in the case of barley seeds. Since the seed-enveloping parts often consist of dead tissue (e.g., the pericarp of sugar beet seeds [9]), we expect that colonization of these cell layers may occur.
The high GFP fluorescence intensity of the MA 342 mutant allowed us to visualize by confocal microscopy the bacteria in deeper cell layers of the glume, which presumably consist of dead cells, with a minimum of sample preparation, which avoided changes in the spatial distribution of the bacteria which might otherwise have occurred during sample preparation. We found that P. chlororaphis MA 342G2 colonizes barley seeds during germination and that there is a strong aggregation pattern only at specific sites on the seed (Fig. 3 through 5). The strong aggregation of bacteria near the embryo (Fig. 5) may be due to release of high concentrations of nutrients at this site, where there is actively growing tissue. Many bacteria were present in the glume, where the fungal pathogen D. teres probably was also present (1, 20). The colonization pattern of MA 342 thus appears to be similar to the colonization pattern of the pathogen; both microorganisms are present on sites around the embryo and in the glume. We hypothesize that a close physical relationship between the pathogen and the biocontrol organism is required for biocontrol. This hypothesis is supported by the fact that MA 342 is not effective against the seed-borne pathogen Ustilago nuda, which is known to be present mainly in the seed embryo (13).
The main mechanism by which MA 342 suppresses disease is probably via antibiosis as strain MA 342 produces an antifungal compound, 2,3-deepoxy-2,3-didehydrorhizoxin, and the biocontrol abilities of mutants which have lost the ability to produce this compound are substantially reduced (13). We hypothesize that protection of barley seedlings from fungal infection occurs mainly by suppression of fungal growth by 2,3-deepoxy-2,3-didehydrorhizoxin in the glume and that this suppression is favored by colocalization of MA 342 and the pathogen.
ACKNOWLEDGMENTS
This work was supported by the Carl Tryggers Foundation, by the Swedish Foundation for Strategic Research, by the Swedish Council for Engineering Science (J.K.J. and R.T.), and by the Swedish Foundation for Environmental Research.
REFERENCES
- 1.Agarwal V K, Sinclair J B. Principles of seed pathology. Boca Raton, Fla: CRC Press; 1997. [Google Scholar]
- 2.Amann R, Kühl M. In situ methods for assessment of microorganisms and their activities. Curr Opin Biotechnol. 1998;1:352–358. doi: 10.1016/s1369-5274(98)80041-6. [DOI] [PubMed] [Google Scholar]
- 3.Assmus B, Hutzler P, Kirchhof G, Amann R, Lawrence J K, Hartmann A. In situ localization of Azospirillium brasilense in the rhizosphere of wheat with fluorescently labelled rRNA-targeted oligonucleotide probes and scanning confocal laser microscopy. Appl Environ Microbiol. 1995;61:1013–1019. doi: 10.1128/aem.61.3.1013-1019.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bloemberg G V, O’Toole G A, Lugtenberg B-J J, Kolter R. Green fluorescent protein as a marker for Pseudomonas spp. Appl Environ Microbiol. 1997;63:4543–4551. doi: 10.1128/aem.63.11.4543-4551.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chalfie M, Tu Y, Euskirchen G, Ward W W, Prasher D C. Green fluorescent protein as a marker for gene-expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 6.Chin A W, De-Priester W, Van-Der-Bij A, Lugtenberg B-J J. Description of the colonization of a gnotobiotic tomato rhizosphere by Pseudomonas fluorescens biocontrol strain WCS365, using scanning electron microscopy. Mol Plant Microbe Interact. 1997;10:79–86. [Google Scholar]
- 7.Dandurand L M, Schotzko D J, Knudsen G R. Spatial patterns of rhizoplane populations of Pseudomonas fluorescens. Appl Environ Microbiol. 1997;63:3211–3217. doi: 10.1128/aem.63.8.3211-3217.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Leij F A A M, Thomas C E, Bailey M J, Whipps J M, Lynch J M. Effect of insertion site and metabolic load on the environmental fitness of a genetically modified Pseudomonas fluorescens isolate. Appl Environ Microbiol. 1998;64:2634–2638. doi: 10.1128/aem.64.7.2634-2638.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fukui R, Poinar E, Bauer I P H, Schroth M N, Hendson M, Wang X L, Hancock J G. Spatial colonization patterns and interaction of bacteria on inoculated sugar beet seeds. Phytopathology. 1994;84:1338–1345. [Google Scholar]
- 10.Gage D J, Bobo T, Long S R. Use of green fluorescent protein to visualize early events of symbiosis between Rhizobium meliloti and alfalfa (Medicago sativa) J Bacteriol. 1996;178:7159–7166. doi: 10.1128/jb.178.24.7159-7166.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gasser G. Mikroskopische Untersuchung pflanzlicher Lebensmittel. Stuttgart, Germany: Gustav Fischer Verlag; 1973. [Google Scholar]
- 12.Hansen M, Kragelund L, Nybroe O, Sørensen J. Early colonization of barley roots by Pseudomonas fluorescens studied by immunofluorescence technique and confocal laser scanning microscopy. FEMS Microbiol Ecol. 1997;23:353–360. [Google Scholar]
- 13.Hökeberg M. Seed bacterization for control of fungal seed-borne diseases in cereals. Ph.D. thesis. Uppsala, Sweden: Swedish University of Agricultural Sciences; 1998. [Google Scholar]
- 14.Hökeberg M, Gerhardson B, Johnsson L. Biological control of cereal seed-borne diseases by seed bacterization with greenhouse-selected bacteria. Eur J Plant Pathol. 1997;103:25–33. [Google Scholar]
- 15.Hood M A, van Dijk K V, Nelson E B. Factors affecting attachment of Enterobacter cloacae to germinating cotton seed. Microb Ecol. 1998;36:101–110. doi: 10.1007/s002489900097. [DOI] [PubMed] [Google Scholar]
- 16.Hoseney R C. Principles of cereal science and technology. St. Paul, Minn: American Association of Cereal Chemists; 1992. [Google Scholar]
- 17.Johnsson L, Hökeberg M, Gerhardson B. Performance of the Pseudomonas chlororaphis biocontrol agent MA 342 against cereal seed-borne diseases in fields experiments. Eur J Plant Pathol. 1998;104:701–711. [Google Scholar]
- 18.Kragelund L, Nybroe O. Competition between Pseudomonas fluorescens Ag1 and Alcaligenes eutrophus JMP134(pHP4) during colonization of barley roots. FEMS Microbiol Ecol. 1996;20:41–51. [Google Scholar]
- 19.Neergaard P. Seed pathology. London, United Kingdom: MacMillan Press Ltd.; 1977. [Google Scholar]
- 20.Singh S. Biotic factors affecting net-blotch (Helmintosporium teres) epidemiology. Indian Phytopathol. 1962;15:195–202. [Google Scholar]
- 21.Slininger P J, VanCauwenberge J E, Shea-Wilbur M A, Bothast R J. Impact of liquid culture physiology, environment, and metabolites on biocontrol agent qualities, Pseudomonas fluorescens 2-79 versus Take-All. In: Boland G J, Kuykendall L D, editors. Plant-microbe interactions and biological control. New York, N.Y: Marcel Dekker, Inc.; 1998. pp. 329–353. [Google Scholar]
- 22.Tombolini R, Jansson J K. Monitoring of GFP-tagged bacterial cells. In: LaRossa R A, editor. Methods in molecular biology. Bioluminescence methods and protocols. Totowa, N.J: Humana Press Inc.; 1998. pp. 285–298. [DOI] [PubMed] [Google Scholar]
- 23.Tombolini R, Unge A, Davey M E, de Bruijn F J, Jansson J K. Flow cytometric and microscopic analysis of GFP-tagged Pseudomonas fluorescens bacteria. FEMS Microbiol Ecol. 1997;22:17–28. [Google Scholar]
- 24.Unge A, Tomblini R, Davey M E, de Bruijn F J, Jansson J K. GFP as a marker gene. In: Akkermans A D, van Elsas J D, de Bruijn F J, editors. Molecular microbial ecology manual. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998. pp. 1–16. [Google Scholar]
- 25.Unge A, Tombolini R, Müller A, Jansson J K. Optimization of GFP as a marker for detection of bacteria in environmental samples. In: Hastings J W, Kricka L J, Stanley P E, editors. Bioluminescence and chemiluminescence: molecular reporting with photons. Sussex, United Kingdom: Wiley & Sons; 1997. pp. 391–394. [Google Scholar]
- 26.Weller D M. Colonization of wheat roots by a fluorescent pseudomonad suppressive to Take-All. Phytopathology. 1983;73:1548–1553. [Google Scholar]
- 27.Weller D M, Thomashow L S, Cook R J. Biological control of soil-borne pathogens of wheat: benefits, risks and current challenges. In: Hokkanen H M T, Lynch J M, editors. Biological control: benefits and risks. Cambridge, United Kingdom: Cambridge University Press; 1995. pp. 149–160. [Google Scholar]