Abstract
The change of activity and abundance of Nitrosospira and Nitrospira spp. along a bulk water gradient in a nitrifying fluidized bed reactor was analyzed by a combination of microsensor measurements and fluorescence in situ hybridization. Nitrifying bacteria were immobilized in bacterial aggregates that remained in fixed positions within the reactor column due to the flow regimen. Nitrification occurred in a narrow zone of 100 to 150 μm on the surface of these aggregates, the same layer that contained an extremely dense community of nitrifying bacteria. The central part of the aggregates was inactive, and significantly fewer nitrifiers were found there. Under conditions prevailing in the reactor, i.e., when ammonium was limiting, ammonium was completely oxidized to nitrate within the active layer of the aggregates, the rates decreasing with increasing reactor height. To analyze the nitrification potential, profiles were also recorded in aggregates subjected to a short-term incubation under elevated substrate concentrations. This led to a shift in activity from ammonium to nitrite oxidation along the reactor and correlated well with the distribution of the nitrifying population. Along the whole reactor, the numbers of ammonia-oxidizing bacteria decreased, while the numbers of nitrite-oxidizing bacteria increased. Finally, volumetric reaction rates were calculated from microprofiles and related to cell numbers of nitrifying bacteria in the active shell. Therefore, it was possible for the first time to estimate the cell-specific activity of Nitrosospira spp. and hitherto-uncultured Nitrospira-like bacteria in situ.
Immobilized microorganisms are used for the purification of a variety of wastewaters in fixed-film treatment plants with systems such as trickling filters, rotating biological contactors, or fluidized beds (7). In all these systems, the organisms responsible for treatment are present in a microbial biofilm. Unlike in well-mixed activated sludge basins, sequential transformation of the sewage compounds may occur while the wastewater passes through the filter, and pronounced gradients of, e.g., oxygen, dissolved organic carbon, or ammonium can be measured in the bulk water along such a reactor (7). In comparison, changes of the underlying microbial communities and activities within the biofilm are more difficult to assess. However, these data are needed for the improvement of mathematical models used to design and dimension fixed-filter reactors.
Fluorescence in situ hybridization (FISH) with rRNA-targeted oligonucleotide probes is a reliable tool for the direct identification and quantification of bacteria in their natural environment (1, 3). Microsensors have been developed for various compounds for the determination of substrate and product gradients and activities on a micrometer scale (22, 30). The combination of both methods (2) has been shown to have great potential for direct observations of structure and function of sulfate-reducing (28) and nitrifying biofilms (32, 34). In this study, a lab-scale fluidized bed reactor (12) was used as a model system for the in situ analysis of structure and function of a whole biofilm reactor. The nitrifying community of this reactor was recently identified as Nitrosospira and Nitrospira spp. by the rRNA approach (32). Here, we present data on the changes of abundance and activity of these nitrifying populations with the bulk water gradient along the reactor. In a more ecological context, this can be regarded as an example of how environmental parameters structure microbial communities.
In an additional experiment, the microsensor measurements were repeated under an excess of substrate (ammonium or nitrite) to test the nitrification potential of the system and to evaluate the maximum specific activities of its components. This again is important for process engineers to model the system and also to estimate the competitiveness of yet-uncultured species, such as Nitrospira spp., in the environment.
(A preliminary account of part of this work appeared in the Proceedings of the 2nd International Conference on Microorganisms in Activated Sludge and Biofilms, International Association on Water Quality, Berkeley, Calif., 1997.)
MATERIALS AND METHODS
Reactor operation.
The conical 360-ml continuous-upflow reactor used as a model system has been described in detail by de Beer et al. (12). For this study, the reactor was fed with mineral medium containing 72 μM NH4+ (influent concentration), and the liquid phase was recirculated at a rate of 1.8 ml s−1 (32). The temperature was kept at 30°C. The chemical gradients that developed along the reactor are displayed in Table 1. The conical shape of the vertical reactor column creates a flow velocity gradient that stabilizes aggregates of different diameter and density at different heights in the column according to their settling velocity. Aggregate samples were taken from three different points of the reactor, labeled A1 through A3 (Table 1).
TABLE 1.
Axial gradient along the nitrifying fluidized bed reactor
| Sample | Distance from inlet (cm) | [O2] (μM) | pH | [NH4+] (μM) | [NO2−] (μM) | Aggregate diam (mm) |
|---|---|---|---|---|---|---|
| Inlet | 0 | 236 | 8.0 | 72 | 0 | — |
| A1 | 10 | 212 | 7.9 | 50 | 3 | 2.0–2.5 |
| A2 | 30 | 142 | 7.7 | 40 | 7 | 1.5–2.0 |
| A3 | 50 | 87 | 7.5 | 0 | 4 | 0.8–1.3 |
| Outlet | 80 | 59 | 7.3 | 0 | 0 | — |
Microsensor measurements.
Clark-type O2 microsensors (29) and liquid ion-exchanging membrane (LIX) microsensors for NH4+, NO2−, and NO3− (11) were prepared and calibrated as described previously. Tip diameters were <10 μm for O2, 5 μm for NH4+ and NO3−, and 15 μm for NO2− microsensors.
Aggregates were placed in a flow cell and perfused with medium, and microprofiles were recorded at steps of 25 μm from the bulk liquid into the aggregate as described by de Beer et al. (12) and Schramm et al. (32). Measurements were performed under in situ conditions, i.e., when [O2], [NH4+], [NO2−], and pH in the medium were adjusted to the values at the respective sampling points (referred to as in situ conditions [Table 1]) and in air-saturated mineral medium containing 300 μM NH4+ (referred to as incubation conditions). Profiles of aggregate samples from point A3 were also recorded in air-saturated mineral medium containing 300 μM NO2− (labeled A3nitrite). [NO3−] was always 100 μM.
Chemical analysis.
[NH4+], [NO2−], and [NO3−] in the reactor bulk liquid were determined colorimetrically (Spectroquant; Merck). [O2] and pH were measured by an O2 microsensor and a pH electrode (Radiometer, Copenhagen, Denmark) which were lowered in the reactor down to the sampling points.
Calculations.
Oxygen uptake and the rates of ammonium and nitrite oxidation were determined from the O2, NH4+, and NO3− profiles as the fluxes, J, through the diffusive boundary layer (DBL). Net fluxes for O2, NH4+, NO2−, and NO3− were calculated by using Fick’s first law as follows:
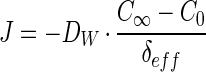 |
1 |
where DW is the molecular diffusion coefficient in water, C∞ is the bulk liquid phase concentration, C0 is the concentration at the aggregate surface, and δeff is the effective DBL thickness. The latter is defined by extrapolating the concentration gradient at the aggregate-water interface to the bulk water phase concentration. Determination of the exact thickness of δeff was done by a simple diffusion reaction model as described in detail by Ploug et al. (26). Diffusion coefficients of O2, NH4+, NO2−, and NO3− at 30°C were taken as 2.75 · 10−5, 2.25 · 10−5, 2.17 · 10−5, and 2.16 · 10−5 cm2 s−1, respectively (8, 23).
Volumetric conversion rates were calculated from the volume of the active shell and the net fluxes into whole aggregates as follows:
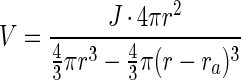 |
2 |
where 4πr2 and 4/3πr3 are the surface and the volume of the aggregate, respectively, ra is the thickness of the active shell as determined by microsensor measurements, and 4/3π(r − ra)3 is the volume of the inactive central part of the aggregate.
Oligonucleotide probes.
Previously described oligonucleotide probes specific for certain ammonia-oxidizing (25) and nitrite-oxidizing (32) bacteria were used. Their sequences and target sites are presented in Table 2. Probes were synthesized and fluorescently labeled with the hydrophilic sulfoindocyanine dyes CY3 or CY5 at the 5′ end by Interactiva Biotechnologie GmbH (Ulm, Germany).
TABLE 2.
Oligonucleotide probes applied
| Probe | Specificity | Sequence (5′ to 3′) of probe | rRNA target sitea | % FAb | [NaCl] (mM)c |
|---|---|---|---|---|---|
| NSO1225 | Ammonia-oxidizing β-Proteobacteria | CGCCATTGTATTACGTGTGA | 16S, 1225–1244 | 35 | 80 |
| NSV443 | Nitrosospira spp. | CCGTGACCGTTTCGTTCCG | 16S, 444–462 | 30 | 112 |
| NSR826 | Nitrospira spp. | GTAACCCGCCGACACTTA | 16S, 826–843 | 20 | 225 |
| NSR1156 | Nitrospira spp. | CCCGTTCTCCTGGGCAGT | 16S, 1156–1173 | 30 | 112 |
| NSR447 | Nitrospira spp. | GGTTTCCCGTTCCATCTT | 16S, 447–464 | 30 | 112 |
Escherichia coli numbering (9).
Formamide in the hybridization buffer.
Sodium chloride in the washing buffer.
Sample preparation and in situ hybridization.
Aggregates were fixed in paraformaldehyde and cut on a cryomicrotome, and the individual cross-sections (thickness, 14 μm) were immobilized on microscopic slides. This whole procedure has been described in detail by Schramm et al. (32). In situ hybridization of fixed and dehydrated aggregate sections was carried out at 46°C in an isotonically equilibrated humidity chamber according to the protocol of Amann et al. (4). Stringent hybridization conditions for the different oligonucleotide probes were adjusted by using the formamide and sodium chloride concentrations listed in Table 2 in the hybridization and washing buffers, respectively (24). Double hybridizations with two probes that require different stringencies (e.g., NSR826 plus NSR1156) were done as subsequent hybridizations, starting with the probe of higher thermal stability.
Confocal microscopy and image analysis.
Digital images of aggregates after hybridization were taken by confocal laser scanning microscopy (CLSM) on a model LSM510 (Carl Zeiss, Jena, Germany) equipped with two HeNe lasers (543 and 633 nm). We applied optical sections 1.5 and 0.7 μm thick for Nitrosospira and Nitrospira spp., respectively. As these are approximately the mean cell diameters of the two populations, the optical sections were assumed to contain single-cell layers. For each probe, cell numbers per unit of volume were derived from randomly chosen optical sections by using the area function of the standard software delivered with the instrument. A threshold value was set manually to exclude empty spaces and background fluorescence from the record, and the remaining cell area was quantified relative to the total aggregate area. Threshold levels were calibrated separately for each probe and sample by determining mean values for the area of at least 300 single cells, counting cells within a defined area, and adjusting the threshold value to match the calculated total cell area. The same calibrations were used to calculate cell numbers from the cell area values. On the assumption that there was only one layer of cells in an optical section, these numbers were regarded as cell numbers per unit of volume, where the volume was the total measured area multiplied by the thickness of the optical section. Nitrifying bacteria in the active shell were enumerated separately from those in the central part of the aggregates.
Statistical evaluation.
Cell numbers of ammonia- and nitrite-oxidizing bacteria in the active shell as well as fluxes of oxygen, ammonium, and nitrate were subjected to statistical analysis with the software package STATISTICA 4.5 (Stat-Soft Inc., Tulsa, Okla.). Data were tested for even distribution by applying Shapiro-Wilk’s W test. Because this test showed uneven distribution of all data, nonparametric statistics were used for further evaluation. We tested for differences in mean values of cell numbers and fluxes from all three samples (A1, A2, and A3) by applying a Kruskal-Wallis analysis of variance. Finally, the data were tested for differences in mean values between individual groups (A1 and A2, A1 and A3, and A2 and A3) by using the Kolmogorov-Smirnov test and the Mann-Whitney U test.
RESULTS
Microgradients.
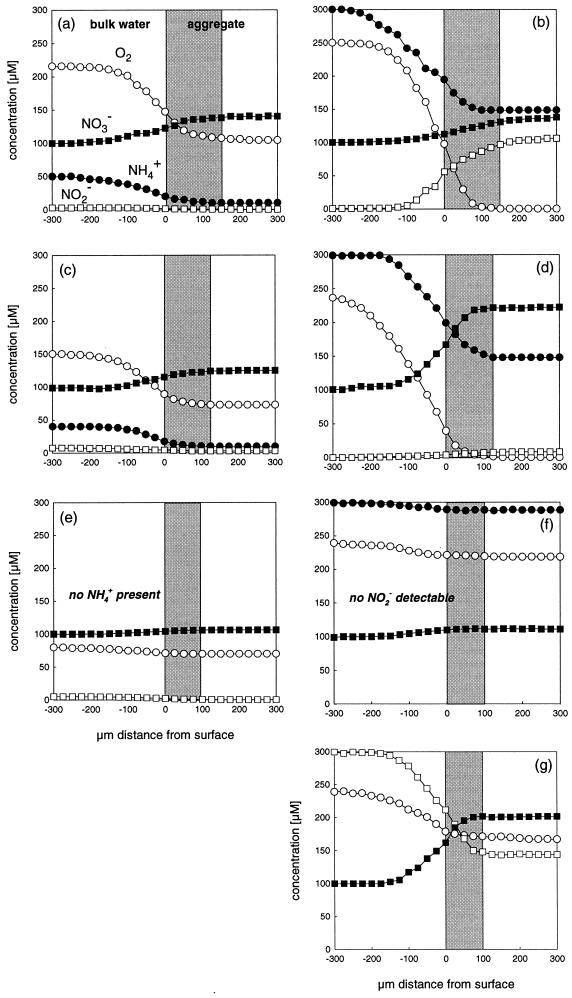
For practical reasons, three to seven profiles (Table 3) of O2, NH4+, NO2−, and NO3− were measured, each in a separate aggregate, under in situ conditions and incubation conditions for all three sampling sites. Examples of these profiles are shown in Fig. 1. The effective DBL ranged from 70 to 225 μm, depending on aggregate size, surface structure, and solute type (data not shown). Oxygen consumption and ammonium and nitrite oxidation were always restricted to a shell consisting of the outer 75 to 200 μm of the aggregate, while the central part of the aggregates appeared to be inactive. The mean thickness of the nitrifying zone as defined by ammonium, nitrite, and nitrate profiles decreased with increasing distance of the aggregate sampling site from the inlet, from about 150 μm (sample A1) to 125 μm (A2), to 100 μm (A3).
TABLE 3.
Fluxes through the aggregate-bulk liquid interface calculated from microprofiles
| Conditions and sample | Mean ± SD flux (nmol · mm−2 · h−1)a (no. of profiles)
|
|||
|---|---|---|---|---|
| Oxygen | Ammonium | Nitrite | Nitrate | |
| In situb | ||||
| A1 | −4.67 ± 1.02 (5) | −1.83 ± 0.86 (3) | −0.16 ± 0.02 (3) | 2.06 ± 0.11 (4) |
| A2 | −3.62 ± 0.96 (4) | −1.30 ± 0.43 (4) | −0.16 ± 0.05 (3) | 1.46 ± 0.14 (3) |
| A3 | −0.44 ± 0.02 (3) | Not detectable | −0.23 ± 0.07 (3) | 0.62 ± 0.01 (3) |
| Incubationc | ||||
| A1 | −13.70 ± 5.50 (7) | −6.69 ± 3.84 (3) | 3.18 ± 0.92 (3) | 4.03 ± 0.52 (3) |
| A2 | −9.78 ± 2.49 (4) | −4.60 ± 3.29 (4) | 0.34 ± 0.04 (3) | 3.85 ± 3.15 (5) |
| A3ammonium | −1.51 ± 0.01 (3) | −0.62 (1) | 0.00 (2) | 0.62 (2) |
| A3nitrite | −4.13 ± 2.51 (3) | No NH4+ present | −6.09 ± 1.02 (3) | 5.60 ± 2.47 (3) |
95% confidence limit.
In situ conditions were as measured in the reactor (Table 1).
For samples A1, A2, and A3ammonium, incubation conditions were 300 μM ammonium (pH 8.0) air saturation; for sample A3nitrite, incubation conditions were 300 μM nitrite (pH 8.0) air saturation.
FIG. 1.
Microprofiles of oxygen (○), ammonium (●), nitrite (□), and nitrate (■) in nitrifying aggregates from different sampling points in the fluidized bed reactor near inlet A1 (a, b), middle A2 (c, d), and near outlet A3 (e to g). (a, c, e) Measurements under in situ conditions (Table 1); (b, d, f) profiles in 300 μM ammonium (pH 8.0) at air saturation; (g) profiles in 300 μM nitrite (pH 8.0) at air saturation.
Under in situ conditions, oxygen was only partially consumed within the nitrifying zone. In contrast, ammonium and nitrite present in the bulk water phase were almost completely converted to nitrate within the aggregate, since less than 10 μM ammonium was found in the central part of A1 and A2 aggregates (Fig. 1a, c, and e).
Because nitrification was obviously substrate limited under reactor conditions, we also measured microprofiles while aggregates were incubated in 300 μM ammonium, sufficient to saturate ammonia oxidation, under air saturation to obtain some information about the nitrification potential (Fig. 1b, d, and f). In A1 and A2 aggregates, oxygen was now depleted within the nitrifying zone, while ammonium was consumed to concentrations down to 100 to 150 μM within the aggregate. Nitrite accumulated to significant concentrations (∼100 μM) in A1 aggregates but only to negligible levels in A2. Almost no change in the concentration of oxygen, ammonium, or nitrate was detected in A3 aggregates, showing very low ammonium oxidation potential. Therefore, a third set of microprofiles was measured in these aggregates, supplying 300 μM nitrite under air saturation (Fig. 1g). Oxygen and nitrite were consumed but not depleted within the active shell, resulting in the enhanced production of nitrate.
Rate calculations.
Net fluxes of oxygen, ammonium, nitrite, and nitrate through the aggregate-bulk liquid interface were calculated from the microprofiles and are summarized in Table 3. In general, the fluxes measured under incubation conditions exceeded the in situ fluxes. The fluxes of oxygen and ammonium and those of nitrate under in situ conditions were significantly higher for samples A1 than for samples A3 (P < 0.001). In contrast, no significant difference of fluxes was found between samples A1 and A2 or between all nitrate fluxes under incubation conditions. The ratio of the fluxes for NH4+-O2-NO3− was close to 1:2:1 for all samples when all species were present. When ammonium was absent (i.e., in samples A3in situ and A3nitrite), the ratios of the fluxes of NO2−-O2-NO3− were 1:1.9:2.7 and 1:0.7:0.92, respectively.
Volumetric conversion rates of oxygen and ammonium were calculated from the net fluxes into the active layer of the aggregates, whereas the rates of nitrite oxidation were calculated from the net fluxes out of this active layer (Table 4). Again, the rates were higher under incubation conditions than in situ, and no significant difference was found in conversion rates between A1 and A2 aggregates. However, volumetric rates were significantly higher in samples A1 than in samples A3 (P < 0.002), with one exception; nitrite oxidation was higher in A3nitrite than in A1 under incubation conditions, indicating high nitrite oxidation potential at the top of the reactor.
TABLE 4.
Volumetric conversion rates calculated from microprofiles
| Conditions and sample | Mean ± SD volumetric rate (nmol · mm−3 · h−1)
|
||
|---|---|---|---|
| Respiration | Ammonium oxidation | Nitrite oxidation | |
| In situ | |||
| A1 | 35.82 ± 7.82 | 14.01 ± 6.65 | 15.77 ± 0.87 |
| A2 | 33.40 ± 8.86 | 11.98 ± 3.99 | 13.44 ± 1.27 |
| A3 | 5.44 ± 0.22 | No NH4+ present | 7.65 ± 0.15 |
| Incubation | |||
| A1 | 105.03 ± 42.16 | 51.29 ± 29.43 | 30.90 ± 3.97 |
| A2 | 90.15 ± 22.95 | 42.45 ± 30.38 | 35.46 ± 29.07 |
| A3ammonium | 18.51 ± 0.23 | 7.61 | 7.65 |
| A3nitrite | 50.75 ± 30.84 | No NH4+ present | 68.86 ± 30.32 |
In situ detection, quantification, and specific reaction rates of nitrifying bacteria.
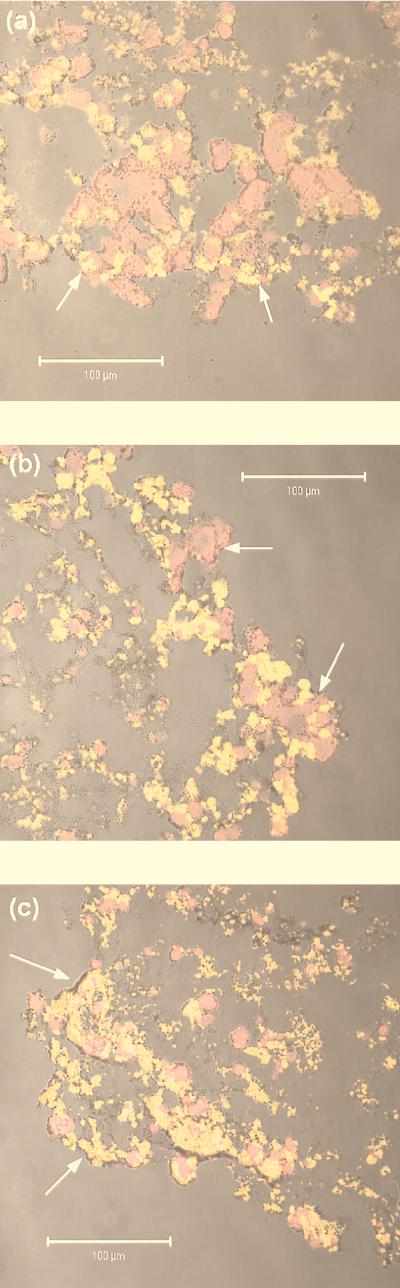
The principal composition of the nitrifying community of the reactor had been resolved previously as Nitrosospira and Nitrospira spp. by the rRNA approach (32). Here, the stratification of these populations within the aggregates as well as along the reactor column is reported (Table 5 and Fig. 2). The relative close match of cell numbers for probes NSO1225 and NSV443, targeting all ammonia-oxidizing β-Proteobacteria and all known members of the genus Nitrosospira, respectively, confirmed our observation that Nitrosospira spp. represent the vast majority, if not all, of the ammonia-oxidizing bacteria in the system. The numbers of ammonia oxidizers were significantly higher at the bottom (A1 [Fig. 2a]) than at the top (A3 [Fig. 2c]) of the reactor (P < 0.001), whereas no significant difference was evident between A1 and A2 or between A2 and A3. Many fewer cells were detected in the center of all aggregates than in the outer shell. Moreover, ammonia oxidizers formed substantially larger cell clusters in the nitrifying zone than in the inner part of the aggregate (Fig. 2). Concerning the stratification within a single aggregate, the same observation is also true for nitrite-oxidizing bacteria of the genus Nitrospira as detected by a combination of probes NSR826 and NSR1156. The combination of these probes targets all known Nitrospira-like sequences from freshwater habitats. In contrast, the cell volume of Nitrospira spp. was significantly higher at the top of the reactor than at the bottom (P > 0.0001) and equaled in aggregate A2 the cell volume of Nitrospira spp. (cf. Fig. 2). However, due to their much smaller cell size, numbers of nitrite-oxidizing bacteria exceeded those of ammonia-oxidizing bacteria by more than 1 order of magnitude. Interestingly, a distinct, less-abundant population of Nitrospira spp., as detected by probe NSR447, was restricted almost exclusively to the active shell of the aggregates.
TABLE 5.
Quantification of nitrifying bacteria by FISH
| Sample | No. (± SD) of cells (107 · mm−3) detected by probe
|
|||
|---|---|---|---|---|
| NSO1225 | NSV443 | NSR826/NSR1156 | NSR447 | |
| A1 | ||||
| Shella | 5.55 ± 2.75 | 4.30 ± 0.70 | 62.1 ± 17.9 | 1.71 ± 0.90 |
| Centerb | 1.80 ± 0.62 | 1.35 ± 0.13 | 12.3 ± 11.6 | 0.06 ± 0.02 |
| A2 | ||||
| Shell | 4.39 ± 2.22 | 4.80 ± 2.56 | 78.5 ± 25.4 | 1.75 ± 0.90 |
| Center | 1.92 ± 0.59 | 1.87 ± 0.54 | 31.3 ± 19.0 | 0.07 ± 0.02 |
| A3 | ||||
| Shell | 2.74 ± 0.38 | 2.24 ± 1.17 | 93.4 ± 55.0 | 3.9 ± 0.71 |
| Center | 0.97 ± 0.57 | 1.14 ± 0.21 | 42.1 ± 27.3 | 0.14 ± 0.12 |
Number of cells within the active, nitrifying shell of the aggregates.
Number of cells within the inactive central part of the aggregates.
FIG. 2.
CLSM images of part of aggregate cross-sections after FISH with CY5-labelled probe NSV443, specific for Nitrosospira spp. (in red), and with CY3-labelled probes NSR826 plus NSR1156, specific for Nitrospira spp. (in yellow). For each picture, two confocal images and the respective phase-contrast image were combined. Aggregates from A1 (a), A2 (b), and A3 (c) are shown. The aggregate surface is indicated by arrows. Bar = 100 μm.
Cell numbers were used to calculate specific oxidation rates of ammonium and nitrite per cell from the volumetric ammonium and nitrite oxidation rates (Table 6). Generally, the specific reaction rates of Nitrosospira spp. were 1 order of magnitude higher than those of Nitrospira spp., and rates were higher under incubation conditions than in situ. However, no significant change in the specific rates per cell was observed along the reactor column. The only exception was the ammonium oxidation rate of Nitrosospira spp. for aggregate A3, which was considerably lower than the rates for aggregates A1 and A2 when incubated with 300 μM ammonium.
TABLE 6.
Specific conversion rates of nitrifying bacteria in the active shell
| In situ conditions
|
Incubation conditions
|
||||
|---|---|---|---|---|---|
| Sample | Specific conversion rate (10−4 pmol · cell−1 · h−1)
|
Sample | Specific conversion rate (10−4 pmol · cell−1 · h−1)
|
||
| AORa | NORb | AOR | NOR | ||
| A1 | 2.5 ± 2.4 | 0.2 ± 0.1 | A1 | 9.2 ± 9.9 | 0.5 ± 0.2 |
| A2 | 2.7 ± 2.3 | 0.2 ± 0.1 | A2 | 9.7 ± 11.8 | 0.5 ± 0.5 |
| A3 | 0 | 0.1 ± 0.05 | A3ammonium | 2.8 ± 0.4 | 0.1 ± 0.05 |
| A3nitrite | 0 | 0.7 ± 0.8 | |||
Ammonium oxidation rate (± SD) per NSO1225-positive cell.
Nitrite oxidation rate (± SD) per NSR826/NSR1156-positive cell.
DISCUSSION
Accuracy of the calculations.
The nitrifying aggregates investigated in this study were rather heterogeneous regarding their irregular surface structures and the patchy distribution of nitrifying bacteria within the active shell (Fig. 2). Consequently, the measured profiles showed some variability, and the standard deviations of both calculated conversion rates and FISH quantification were rather high. The ratio of the fluxes for NH4+-O2-NO3−, however, was close to the expected stoichiometric ratio of 1:2:1, thus lending support to the mean values obtained. In contrast, the flux ratio in sample A3 (in situ conditions) clearly indicates that not enough profiles have been measured to reliably calculate average fluxes of nitrite and/or nitrate. A small source of uncertainty, again due to the irregular shape of the aggregates, was the determination of the radius required to calculate volumetric conversion rates. However, the error introduced by a deviation of ±100 μm amounted to only about 2% and therefore can be disregarded.
For the FISH-based quantification of nitrifying populations, the threshold set point for the area measurement was critical. Thus, special effort was taken in its calibration, and occasionally the results from image analysis were compared with conventional counts of hybridized cells in defined areas of an optical slice. Since the results from both procedures never differed more than 10%, we concluded that enumeration of nitrifying bacteria by our image analysis procedure was accurate enough for our purposes. However, this was possible only because of the morphological homogeneity of the respective nitrifying populations. Another source of uncertainty was the extrapolation of these values to cell numbers per unit of volume. Biofilm samples have been reported to shrink, especially in the z direction during immobilization on microscopic slides and during dehydration (33). This would lead to underestimation of the total aggregate volume and hence to the overestimation of cell numbers per unit of volume. A comparison of the thickness of sections after hybridization with the cryosection thickness revealed a shrinkage of about 15% for all samples. Therefore, the numbers of nitrifiers per unit of volume might have been overestimated by 15%, while the specific rates per cell might have been underestimated by 15%, but both parameters were overestimated and underestimated to the same extent for all samples. For all the reasons discussed above, we are aware that the absolute numbers reported in this study might be only best estimates, especially those for the specific conversion rates per cell. Nevertheless, we are convinced that our data reliably describe the trends in the investigated system and are within the correct order of magnitude.
Analysis under in situ conditions.
The decrease of ammonium in the bulk liquid phase with reactor height (Table 1) most likely leads to decreasing numbers of ammonia-oxidizing bacteria in the active shell of the aggregates from A1 to A3 (Table 5 and Fig. 2) and to a decreasing thickness of this shell (Fig. 1a, c, and e). However, no significant change in cell numbers, volumetric ammonium oxidation rate and, consequently, the specific ammonium oxidation rate per cell of Nitrosospira spp. was detectable from A1 to A2 (Tables 4 to 6). The estimated value of approximately 0.25 fmol · cell−1 · h−1 is comparable to the specific rates reported by Wagner et al. for Nitrosococcus mobilis in activated sludge (20, 35) but is 1 to 2 orders of magnitude below the rates reported for Nitrosospira spp. and other ammonia oxidizers from pure-culture studies (6, 27). This was probably due to the apparent ammonium limitation under in situ conditions. In aggregates from the top of the reactor (A3) and in the central parts of all aggregates, ammonium was virtually absent or probably below Km as discussed previously (32), and no ammonium oxidation activity was detected (Fig. 1). Nevertheless, ammonia-oxidizing bacteria were detectable by FISH, although in significantly lower numbers than in the active zones (Fig. 2 and Table 5). This again demonstrates the capacity of ammonia-oxidizing bacteria to maintain their ribosomes, even under conditions not conducive to activity and growth (34, 35).
Nitrite oxidation was almost completely coupled to nitrite production by ammonia oxidizers. Consequently, nitrite oxidation rates decreased from A1 to A3 (Table 4), despite an increase in the number of Nitrospira spp. found in the active shell from A1 to A3 (Fig. 2 and Table 5). Specific nitrite oxidation rates per cell were always extremely low (maximum, 0.02 fmol · cell−1 · h−1). Pure culture data are currently not available for Nitrospira spp., but the values reported for Nitrobacter spp. are about 200 to 2,000 times higher (27). Again, this difference might result from unfavorable in situ conditions and/or from the differing physiological properties of these distantly related (13) nitrite oxidizers.
Nitrification activity under elevated substrate concentrations.
To test the nitrifying capacity of the system, additional measurements were performed under air saturation in medium containing 300 μM ammonium (A1, A2, and A3) or 300 μM nitrite (A3). The volumetric respiration rates in A1 and A2 are high (∼100 nmol · mm−3 · h−1) compared to the values of 2 to 40 nmol · mm−3 · h−1 reported from sediments (31), activated-sludge flocs (34a), and heterotrophic aggregates (26) or biofilms (21). They are similar, however, to rates determined earlier under the same conditions in the same system (12), in a nitrifying trickling filter biofilm (34), or in a hypersaline microbial mat (19). It should be noted that such high rates are possible only if bacteria, and hence their activities, are extremely concentrated, as is shown for the nitrifying shell in this study (Fig. 2).
The specific ammonium oxidation rates per cell under incubation conditions (Table 6) are approximately the same in A1 and A2, and both are higher than the rates under in situ conditions. Although oxygen limited, we assume these values to be close to the maximum specific activity, since higher oxygen levels previously have been shown to inhibit nitrification in the same system (12). Still, the specific activity is well below the rates reported for pure cultures (see above). Interestingly, ammonia oxidizers in A3, i.e., the population subjected to starvation in situ, developed significantly less activity, even when supplied with enough substrate (Tables 4 and 6, A3ammonium). This finding leads to the hypothesis that Nitrosospira spp. adapt to starvation by entering a dormant or inactive state that is not coupled to the reduction of the cellular ribosome level, as proven by FISH. Whether this is due to decreased activity or concentration of ammonia monooxygenase or to some other unknown mechanisms might be addressed by the application of mRNA-targeted probes or ammonia monooxygenase-targeted antibodies in the future.
Cell-specific nitrite oxidation rates increased for all sampling points when aggregates were released from nitrite limitation (Table 6). However, whether the maximum nitrite oxidation activity was reached during incubation is questionable. Ammonia oxidizers are thought to possess lower Km values for oxygen than nitrite oxidizers (14, 27). Therefore, the nitrite accumulation detected in A1 and A2 (Fig. 1b and d) might indicate that ammonia oxidizers have out-competed nitrite oxidizers for oxygen. On the other hand, Km values are available only for Nitrosomonas spp. and Nitrobacter spp., and nitrite accumulation was less pronounced in A2, although the oxygen concentration within the active layer was even lower than in A1. Furthermore, in A3, when neither oxygen nor nitrite was limiting, the specific nitrite oxidation rates were not significantly higher. For these reasons, we assume that the specific rates were at least close to the maximum activity. As mentioned for the ammonia oxidizers, these activities are much lower (100 to 900 times) than those described for Nitrobacter. The recent detection of Nitrospira-like sequences and cells in various environments (10, 13, 16, 20) and the absence of Nitrobacter spp. in similar habitats (15, 33, 36) might therefore indicate other competitive advantages of Nitrospira spp., e.g., higher substrate affinity for oxygen and/or nitrite, better adaptation to starvation, or better resistance to toxic shocks.
In principle, substrate affinities (expressed as Km values) can be estimated from ammonium and nitrite microprofiles and cell numbers by Michaelis-Menten kinetics. For this approach, cell-specific conversion rates were calculated for each data point by using the second derivative of the profiles. Relating these values to the respective substrate concentration in a Lineweaver-Burk plot, Km values as low as 40 μM ammonium (pH 7.8) and 10 μM nitrite were obtained for Nitrosospira and Nitrospira spp., respectively. This is 1 to 2 orders of magnitude lower than the Km values for Nitrosomonas europaea and most other Nitrosomonas strains (18, 27) and most Nitrobacter spp. (17, 27) found in the literature. In an ecological context, Nitrosospira and Nitrospira spp. thus could indeed be regarded as typical K strategists, with high substrate affinities and low maximum activity (or growth rate), compared to the r strategists N. europaea and Nitrobacter spp. (5, 32). However, again it must be emphasized that these data are subject to many uncertainties. First, the sample was highly heterogeneous, as discussed above; second, whether maximum reaction rates could be reached under the conditions applied is not sure, and third, the oligonucleotide probes used in this study are not specific at the species level, leaving open the possibility of phylogenetic and physiological diversity in Nitrospira spp., as discussed below. Therefore, the present data should be considered best-possible estimates, correct to within an order of magnitude.
It is tempting to speculate about the small fraction of Nitrospira spp. that was detected exclusively in the active shell of the aggregates by probe NSR447. Are they physiologically different from the main population? This which, as long as it has not been proven on pure cultures, certainly cannot be decided on the basis of this study. There are currently two dozen Nitrospira-like sequences available, probably representing at least five distinct species, should not be presumed to have identical physiologies.
In conclusion, the combination of microsensor measurements and FISH allowed a detailed analysis of the in situ structure and function of the nitrifying bioreactor on a microscale. Measurements under elevated substrate concentrations were used to extract valuable information about the in situ activity of hitherto-uncultured nitrifying bacteria.
ACKNOWLEDGMENTS
We thank G. Eickert, A. Eggers, and V. Hübner for constructing oxygen microsensors. Michael Wagner, Jens Harder, and Arzhang Khalili are acknowledged for fruitful discussions. The help of Jakob Pernthaler with statistical analysis is appreciated.
This work was supported by a grant of the Körber Foundation to R.A. and by the Max Planck Society.
REFERENCES
- 1.Amann R, Glöckner F O, Neef A. Modern methods in subsurface microbiology: in situ identification of microorganisms with nucleic acid probes. FEMS Microbiol Rev. 1997;20:191–200. [Google Scholar]
- 2.Amann R, Kühl M. In situ methods for assessment of microorganisms and their activities. Curr Opin Microbiol. 1998;1:352–358. doi: 10.1016/s1369-5274(98)80041-6. [DOI] [PubMed] [Google Scholar]
- 3.Amann R I, Ludwig W, Schleifer K-H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amann R I, Krumholz L, Stahl D A. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990;172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andrews J H, Harris R F. r- and K-selection and microbial ecology. Adv Microb Ecol. 1986;9:99–147. [Google Scholar]
- 6.Belser L W. Population ecology of nitrifying bacteria. Annu Rev Microbiol. 1979;33:309–333. doi: 10.1146/annurev.mi.33.100179.001521. [DOI] [PubMed] [Google Scholar]
- 7.Bishop P L, Kinner N E. Aerobic fixed-film processes. In: Schönborn W, editor. Biotechnology, vol. 8. Microbial degradations. 1st ed. Weinheim, Germany: VCH; 1986. pp. 113–176. [Google Scholar]
- 8.Broecker W S, Peng T-H. Gas exchange rates between air and sea. Tellus. 1974;26:21–35. [Google Scholar]
- 9.Brosius J, Dull T J, Sleeter D D, Noller H F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981;148:107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- 10.Burrel P C, Keller J, Blackall L L. Microbiology of a nitrite-oxidizing bioreactor. Appl Environ Microbiol. 1998;64:1878–1883. doi: 10.1128/aem.64.5.1878-1883.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Beer D, Schramm A, Santegoeds C M, Kühl M. A nitrite microsensor for profiling environmental biofilms. Appl Environ Microbiol. 1997;63:973–977. doi: 10.1128/aem.63.3.973-977.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Beer D, van den Heuvel J C, Ottengraf S P P. Microelectrode measurements of the activity distribution in nitrifying bacterial aggregates. Appl Environ Microbiol. 1993;59:573–579. doi: 10.1128/aem.59.2.573-579.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ehrich S, Behrens D, Lebedeva E, Ludwig W, Bock E. A new obligately chemolithoautotrophic, nitrite-oxidizing bacterium, Nitrospira moscoviensis sp. nov. and its phylogenetic relationship. Arch Microbiol. 1995;164:16–23. doi: 10.1007/BF02568729. [DOI] [PubMed] [Google Scholar]
- 14.Focht D D, Verstraete W. Biochemical ecology of nitrification and denitrification. Adv Microb Ecol. 1977;1:135–214. [Google Scholar]
- 15.Hovanec T A, DeLong E F. Comparative analysis of nitrifying bacteria associated with freshwater and marine aquaria. Appl Environ Microbiol. 1996;62:2888–2896. doi: 10.1128/aem.62.8.2888-2896.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hovanec T A, Taylor L T, Blakis A, DeLong E F. Nitrospira-like bacteria associated with nitrite oxidation in freshwater aquaria. Appl Environ Microbiol. 1998;64:258–264. doi: 10.1128/aem.64.1.258-264.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hunik J H, Meijer H J G, Tramper J. Kinetics of Nitrobacter agilis at extreme substrate, product and salt concentrations. Appl Microbiol Biotech. 1993;40:442–448. [Google Scholar]
- 18.Hunik J H, Meijer H J G, Tramper J. Kinetics of Nitrosomonas europaea at extreme substrate, product and salt concentrations. Appl Microbiol Biotech. 1992;37:802–807. [Google Scholar]
- 19.Jørgensen B B, Des Marais D J. The diffusive boundary layer of sediments: oxygen microgradients over a microbial mat. Limnol Oceanogr. 1990;35:1343–1355. doi: 10.4319/lo.1990.35.6.1343. [DOI] [PubMed] [Google Scholar]
- 20.Juretschko S, Timmermann G, Schmidt M, Schleifer K-H, Pommerening-Röser A, Koops H-P, Wagner M. Combined molecular and conventional analysis of nitrifying bacterial diversity in activated sludge: Nitrosococcus mobilis and Nitrospira-like bacteria as dominant populations. Appl Environ Microbiol. 1998;64:3042–3051. doi: 10.1128/aem.64.8.3042-3051.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kühl M, Jørgensen B B. Microsensor measurement of sulfate reduction and sulfide oxidation in compact microbial communities of aerobic biofilms. Appl Environ Microbiol. 1992;58:1164–1174. doi: 10.1128/aem.58.4.1164-1174.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kühl, M., and N. P. Revsbech. Microsensors for the study of interfacial biogeochemical processes. In B. P. Boudreau and B. B. Jørgensen (ed.), The benthic boundary layer, in press. Oxford University Press, Oxford, England.
- 23.Li Y H, Gregory S. Diffusion of ions in sea water and in deep-sea sediments. Geochim Cosm Acta. 1974;38:703–714. [Google Scholar]
- 24.Manz W, Amann R, Ludwig W, Wagner M, Schleifer K-H. Phylogenetic oligodeoxynucleotide probes for the major subclasses of proteobacteria: problems and solutions. Syst Appl Microbiol. 1992;15:593–600. [Google Scholar]
- 25.Mobarry B K, Wagner M, Urbain V, Rittmann B E, Stahl D A. Phylogenetic probes for analyzing abundance and spatial organization of nitrifying bacteria. Appl Environ Microbiol. 1996;62:2156–2162. doi: 10.1128/aem.62.6.2156-2162.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ploug H, Kühl M, Buchholz-Cleven B, Jørgensen B B. Anoxic aggregates—an ephemeral phenomenon in the pelagic environment? Aquat Microb Ecol. 1997;13:285–294. [Google Scholar]
- 27.Prosser J I. Autotrophic nitrification in bacteria. Adv Microb Physiol. 1989;30:125–181. doi: 10.1016/s0065-2911(08)60112-5. [DOI] [PubMed] [Google Scholar]
- 28.Ramsing N B, Kühl M, Jørgensen B B. Distribution of sulfate-reducing bacteria, O2, and H2S in photosynthetic biofilms determined by oligonucleotide probes and microelectrodes. Appl Environ Microbiol. 1993;59:3840–3849. doi: 10.1128/aem.59.11.3840-3849.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Revsbech N P. An oxygen microelectrode with a guard cathode. Limnol Oceanogr. 1989;34:474–478. [Google Scholar]
- 30.Revsbech N P, Jørgensen B B. Microelectrodes: their use in microbial ecology. In: Marshall K C, editor. Advances in microbial ecology. Vol. 9. New York, N.Y: Plenum; 1986. pp. 293–352. [Google Scholar]
- 31.Revsbech N P, Madsen B, Jørgensen B B. Oxygen production and consumption in sediments determined at high spatial resolution by computer simulation of oxygen microelectrode data. Limnol Oceanogr. 1986;31:293–304. [Google Scholar]
- 32.Schramm A, de Beer D, Wagner M, Amann R. Identification and activity in situ of Nitrosospira and Nitrospira spp. as dominant populations in a nitrifying fluidized bed reactor. Appl Environ Microbiol. 1998;64:3480–3485. doi: 10.1128/aem.64.9.3480-3485.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schramm A, Larsen L H, Revsbech N P, Amann R I. Structure and function of a nitrifying biofilm as determined by microelectrodes and fluorescent oligonucleotide probes. Water Sci Tech. 1997;36:263–270. [Google Scholar]
- 34.Schramm A, Larsen L H, Revsbech N P, Ramsing N B, Amann R, Schleifer K-H. Structure and function of a nitrifying biofilm as determined by in situ hybridization and the use of microelectrodes. Appl Environ Microbiol. 1996;62:4641–4647. doi: 10.1128/aem.62.12.4641-4647.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34a.Schramm, A., et al. Unpublished data.
- 35.Wagner M, Rath G, Amann R, Koops H-P, Schleifer K-H. In situ identification of ammonia-oxidizing bacteria. Syst Appl Microbiol. 1995;18:251–264. [Google Scholar]
- 36.Wagner M, Rath G, Koops H-P, Flood J, Amann R. In situ analysis of nitrifying bacteria in sewage treatment plants. Water Sci Tech. 1996;34:237–244. [Google Scholar]