SUMMARY
The aim of this study was to characterize updated HIV subtypes in Yunnan to determine their origins and distribution within the population. RT–PCR of both the gag and env genes were sequenced from Yunnan province inhabitants newly diagnosed with HIV-1. Sequence data from 290 samples were used for statistical analysis of subtype distribution and phylogenetic tree construction. Distribution data were adjusted to account for different geographical distributions of HIV-1 subtypes in the population. Phylogenetic analysis revealed six HIV-1 subtypes in Yunnan, including eight types of unique recombination forms (URFs). The most prevalent subtypes in this province, CRF07_BC (18·9%), CRF08_BC (39·1%), CRF01_AE (22·4%), and URFs (subtype C, 5·9% and subtype B, 4·5%), were all recombinants. We found significant differences in the distribution of these HIV-1 subtypes not only geographically, but also between various ethnic groups and with respect to transmission routes. Our findings indicate a complex population of HIV-1 subtypes, URFs, and recombinant subtypes in Yunnan province. This diversity could make the prevention and control of HIV infection in Yunnan more difficult due to the possibility of virus recombination or infection by multiple subtypes.
Key words: HIV-1, recombination, subtype, Yunnan
INTRODUCTION
Yunnan province is one of the regions to suffer most from the HIV epidemic in China [1], with over 90 000 residents infected with the virus [2]. This area was the site of the initial HIV-1 outbreak in China in injecting drug users (IDUs) in 1989. From 1989 to 1993, HIV spread through Yunnan along the drug trafficking routes that exist across China from Myanmar and Laos [3]. In 1994, the HIV epidemic entered a new phase involving spread outside of IDUs through transmission by female sex workers into the general population [4]. This phase of the HIV epidemic has been characterized by the appearance of numerous HIV strains, multiple subtypes, and circulating recombinant variants with the capacity for further recombination [5–7]. Understanding the emergence and spread of new HIV strains is important in meeting the challenges of HIV prevention and treatment posed by HIV diversity.
The molecular epidemiology of HIV-1 has changed rapidly in Yunnan province. The initial HIV outbreak in 1989 was caused by HIV subtype B [8]. From 1991 to 1996, subtypes B′, B′/C recombinant, and the CRF01_AE subtype appeared successively, and the B′ subtype became predominant; by 2001, evidence suggested that new recombinant HIV strains were continually circulating in Yunnan [8]. In 2004, the population of HIV-1 subtypes was found to have further changed to C/B′ (42·9%), A/E (50%) and B′ (7·1%) [9].
Yunnan is thus the origin of the major HIV-1 subtypes now circulating in China [9, 10]. The special geographical environment, presence of multiple ethnic cultures and lifestyles, coexistence of a variety of transmission routes (drugs and sex), and the continuous migration of high-risk populations into Yunnan possibly led to a more diverse and complex population of HIV subtypes through virus recombination. Yunnan province thus plays a central role in determining the molecular characteristics of HIV throughout China. Comprehensive, systematic, and sustained HIV-1 molecular epidemiological studies in Yunnan province are therefore essential for developing AIDS prevention strategies, clinical treatments, and anti-HIV drugs and vaccines for all of China. To this end, the aim of this study was to characterize HIV subtypes in newly identified, untreated cases in Yunnan in 2006 and to determine their origins and distribution within the population.
MATERIALS AND METHODS
Subject sampling
The study population included 6443 Yunnan province inhabitants who were newly diagnosed with HIV-1 in 2006. Samples of serum or plasma were collected at VCT (Voluntary Counselling and Testing) site hospitals and sentinel surveillance sites in Yunnan. All samples were sorted, numbered according to prefecture, and selected using systematic random sampling. Of the 644 samples randomly selected, 290 (45%) were successfully amplified in both the env and gag genes. The reason for the lower amplification efficiency was either a relatively low viral load or, more likely, nucleic acid degradation due to sub-optimal storage conditions. While all HIV-positive samples were supposed to be kept at −70 °C, some had been stored at −20 °C due to lack of storage facilities at −70 °C. In fact, our usual amplification efficiency using fresh samples is about 90%.
Methods
HIV RNA was extracted (Viral RNA Extraction kit, Qiagen, Germany) and the gag and env genes were amplified using nested PCR [One Step RNA PCR kit (AMV), TaKaRa, Shiga, Japan]. PCR primer sequences were as follows [11, 12]: First round gag gene: upstream, GAG-L (5′-TCGACGCAGGACTCGGCTTGC-3′); downstream GAG-E2 (5′-TCCAACAGCCCTTTTTCCTAGG-3′). Second round gag gene: upstream, GUX (5′-AGGAGAGAGATGGGTGCGAGAGCGTC-3′); downstream, GDX (5′-GGCTAGTTCCTCCTACTCCCTGACAT-3′). First round env gene: upstream, 44F (5′-ACAGTRCARTGYACACATGG-3′); downstream, 35R (5′-CACTTCTCCAATTGTCCITCA-3′). Second round env gene: upstream, 33FM (5′-TGTAAAACGACGGCCAGTCTGTTIAATGGCAGICTAGC-3′); downstream, 48RM (5′-CAGGAAACAGCTATGACCRATGGGAGGRGYATACAT-3′). The sequencing primers were M13F (5′-5TGTAAAACGACGGCCAGT-3′) and M13R (5′-CAGGAAACAGCTATGACC-3′).
The RT–PCR amplification conditions for the gag region were as follows. Round 1:50 °C for 30 min, 94 °C for 2 min, 55 °C for 1 min, 72 °C for 2 min, 1 cycle; 94 °C for 30 s, 55 °C for 45 s, 72 °C for 1 min 30 s, 30 cycles; 72 °C for 10 min, 4 °C maintenance. Round 2:94 °C for 2 min, 55 °C for 1 min, 72 °C for 90 s, 1 cycle; 94 °C for 30 s, 55 °C for 45 s, 72 °C for 1 min 30 s, 30 cycles; 72 °C for 10 min, 4 °C maintenance. RT–PCR amplification conditions for the env region were as follows. Round 1:50 °C for 30 s, 94 °C for 2 min, 50 °C for 1 min, 72 °C for 4 min, 1 cycle; 94 °C for 30 s, 50 °C for 30 s, 72 °C for 2 min, 35 cycles; 72 °C for 10 min, 4 °C maintenance. Round 2:95 °C for 2 min; 95 °C for 15 s, 55 °C for 30 s, 72 °C for 75 s, 5 cycles; 95 °C for 15 s, 62 °C for 30 s, 72 °C for 1 min, 25 cycles; 72 °C for 10 min, 4 °C maintenance. The amplified PCR products were separated using gel electrophoresis and were then extracted and purified from the gel for sequencing.
Products were sent to Biomed Co. (China) for sequencing on an ABI 371 automated DNA sequencer (Applied Biosystems, USA).
SeqScanner 4.7 software (Applied Biosystems, USA) was used to clean up the obtained sequence data, and the ambiguous bases at both ends were spliced. Contig Express (Invitrogen, USA) was used for sequence assembly and editing; proofreading was performed using BioEdit software (Ibis Biosciences Inc., USA). Data sequences were compared to subtype reference strain sequences downloaded from the Los Alamos HIV database website [13]. MEGA 4.1 software (Biodesign Institute, Arizona State University, USA) was used for phylogenetic analysis of sequences longer than 800 bp in the gag region and 380 bp in the env region. The Neighbour-Joining (NJ) phylogenetic tree was constructed by repeating the bootstrap calculations 1000 times. If the obtained sequence formed a stable and reliable phylogenetic cluster (bootstrap values >70%) with the corresponding standard reference sequence, it was considered to be of the subtype represented by that reference strain. Sequences of shorter length were submitted to the HIV database for comparison to determine the subtype. Recombination analysis was performed for some sequences using Simplot 3.5 software [14], and the possibility of fragment recombination was investigated.
Statistical analysis
Age was presented as mean with standard deviation (s.d.); other categorical variables were presented as count with percentage. To decrease the influence of sampling error, the estimated proportions of HIV-1 infection routes and subtypes were corrected according to the geographical distribution in the population (Supplementary Fig. S1) as follows: a total of 6443 newly diagnosed HIV-1-infected patients who had not yet received anti-HIV therapy were residing in all 16 cities of Yunnan province in 2006. For sampling, 2% of these patients were sampled from those cities with more than 1000 newly diagnosed HIV-1 cases in 2006, 3% sampling for cities with 500–999 identified HIV-1 cases, 5% sampling for cities with 250–499 cases, 10% sampling for cities with 100–249 cases, 20% sampling for cities with 50–99 cases, and 50% sampling for cities with fewer than 50 cases. The total number of cases sampled should not be fewer than 250.
To calculate the distribution of newly diagnosed HIV-1 subtypes throughout Yunnan province in 2006, we first calculated the percentage of each HIV-1 subtype in each city based on 290 samples (total number), then estimated the total number of patients with that HIV-1 subtype in each city by multiplying this percentage by the number of newly identified HIV-1 infections in that city. Finally, the distribution of newly diagnosed HIV-1 subtypes in 2006 throughout the whole of Yunnan province was calculated as follows:
where i = 1 − 16, representing city i; Ei is the estimated number of HIV-1 subtypes in each city i; Ei = p×Ni, where pi is the percentage of HIV-1 subtype in city i based on 290 samples and Ni is the total number of HIV-1 patients in city i newly diagnosed in 2006.
The difference in subjects' age between various HIV-1 subtypes was tested using the one-way ANOVA test. The associations between categorical variables were tested using Fisher's exact test. Multiple comparisons of the proportion of HIV-1 subtypes in each of the two groups were performed using the Z test with Bonferroni corrections. All statistical analyses were two-sided and performed using SPSS v. 15.0 software (SPSS Inc., USA). P < 0·05 was considered statistically significant.
RESULTS
Characteristics of HIV-1 patients
General characteristics of the 290 HIV-1 patients are given in Table 1. The patients had a mean age of 32·3 years, and over 60% of the HIV-1 patients were male. The majority race was Han (69·0%), followed by Dai (5·86%), and Hui (5·52%); all other races were <5% of the total population. Over 60% of the patients were married.
Table 1.
Characteristics of HIV-1 patients*
| n (%) | |
|---|---|
| Age† (years) | |
| 1–18 | 11 (3·8) |
| 19–25 | 65 (22·4) |
| 26–35 | 121 (41·7) |
| 36–45 | 61 (21·0) |
| ⩾ 46 | 32 (11·0) |
| Gender | |
| Male | 178 (61·4) |
| Female | 112 (38·6) |
| Ethnic group | |
| Han | 200 (69·0) |
| Dai | 17 (5·9) |
| Hui | 16 (5·5) |
| Yi | 14 (4·8) |
| Bai | 11 (3·8) |
| Zhuang | 8 (2·8) |
| Jingpo | 7 (2·4) |
| Hani | 5 (1·7) |
| Other | 12 (4·1) |
| Marital status | |
| Married | 179 (61·7) |
| Unmarried | 89 (30·7) |
| Divorced/widowed | 18 (6·2) |
| Unknown | 4 (1·4) |
| Education level | |
| Illiterate | 17 (5·9) |
| Elementary school | 71 (24·5) |
| Junior high school | 114 (39·3) |
| Senior high school | 57 (19·7) |
| College or above | 17 (5·9) |
| Unknown | 14 (4·8) |
N = 290.
Mean age, 32·3 ± 11·0 years.
Distribution of HIV-1 subtypes
There were six HIV-1 subtypes, including eight unique recombination forms (URFs), represented in the 290 samples, as described in Table 2. After adjustment for geographical distribution, the corrected percentages showed that the top three HIV-1 subtypes in Yunnan were CRF08_BC (39·1%), CRF01_AE (18·9%), and CRF07_BC (22·4%). URFs were the fourth most prevalent HIV-1 subtype, contributing 9·2% of the population, including BC, 4·3%; 01_C, 1·5%; 08_01, 0·7%; C_01, 0·7%; 01_BC, 0·4%; B01, 0·9%; BC_01, 0·4%; 07_01, 0·4%. Subtypes B and C were present in 5·9% and 4·5% of the population, respectively.
Table 2.
Distribution of HIV-1 subtypes and transmission routes (N = 20)
| Count (%) | Corrected value (%)* | |
|---|---|---|
| HIV-1 subtype | ||
| CRF08_BC | 129 (44·5) | 39·1 |
| CRF07_BC | 61 (21·0) | 18·9 |
| CRF01_AE | 57 (19·7) | 22·4 |
| B | 11 (3·8) | 5·9 |
| C | 8 (2·8) | 4·5 |
| URFs | 24 (8·3) | 9·2 |
| BC | 9 (3·1) | 4·3 |
| 01_C | 5 (1·7) | 1·5 |
| 08_01 | 3 (1·0) | 0·7 |
| C_01 | 2 (0·7) | 0·7 |
| 01_BC | 2 (0·7) | 0·4 |
| B01 | 1 (0·3) | 0·9 |
| BC_01 | 1 (0·3) | 0·4 |
| 07_01 | 1 (0·3) | 0·4 |
| HIV-1 transmission route | ||
| Sexual transmission | 166 (57·2) | 54·8 |
| Injecting drug use | 100 (34·5) | 38·5 |
| Mother-to-child | 9 (3·1) | 1·7 |
| Same-sex sexual transmission | 2 (0·7) | 0·4 |
| Unknown | 13 (4·5) | 4·6 |
Corrected by the geographical distribution in the population.
Distribution of HIV-1 subtypes according to age, gender, and ethnicity
Table 3 shows the distribution of HIV subtypes with respect to age, gender, and ethnic group. There was no significant difference in the distribution of HIV-1 subtypes between various age groups (P = 0·189) or between females and males (P = 0·817). The three major HIV-1 subtypes in the Han group were CRF08_BC (43·5%), CRF_07_BC (25·0%), and CRF01_AE (18·5%). Similar distributions were observed in the Hui and Yi groups. The three major HIV-1 subtypes in the Dai group were CRF01_AE (47·1%), B (17·6%), and CRF_07_BC (11·8%). The two major HIV-1 subtypes in the Bai group were CRF08_BC (63·6%) and CRF01_AE (18·2%).
Table 3.
Distribution of HIV-1 subtypes according to age, gender, and race
| HIV-1 subtypes | P value | ||||||
|---|---|---|---|---|---|---|---|
| CRF01_AE | CRF08_BC | CRF07_BC | B | C | URFs | ||
| Age (years) | 0·189 | ||||||
| 1–18 | 3 (27·3%) | 4 (36·4%) | 1 (9·1%) | 0 (0·0%) | 0 (0·0%) | 3 (27·3%) | |
| 19–25 | 11 (16·9%) | 26 (40·0%) | 14 (21·5%) | 2 (3·1%) | 6 (9·2%) | 6 (9·2%) | |
| 26–35 | 21 (17·4%) | 58 (47·9%) | 25 (20·7%) | 3 (2·5%) | 5 (4·1%) | 9 (7·4%) | |
| 36–45 | 14 (23·0%) | 30 (49·2%) | 13 (21·3%) | 3 (4·9%) | 0 (0·0%) | 1 (1·6%) | |
| ⩾ 46 | 8 (25·0%) | 11 (34·4%) | 8 (25·0%) | 0 (0·0%) | 0 (0·0%) | 5 (15·6%) | |
| Gender | 0·817 | ||||||
| Female (n = 112) | 21 (18·8%) | 48 (42·9%) | 24 (21·4%) | 5 (4·5%) | 4 (3·6%) | 10 (8·9%) | |
| Male (n = 178) | 36 (20·2%) | 81 (45·5%) | 37 (20·8%) | 3 (1·7%) | 7 (3·9%) | 14 (7·9%) | |
| Ethnic group | <0·001* | ||||||
| Han (n = 200) | 37 (18·5%) | 87 (43·5%) | 50 (25·0%) | 4 (2·0%) | 6 (3·0%) | 16 (8·0%) | |
| Dai (n = 17) | 8 (47·1%) | 1 (5·9%) | 2 (11·8%) | 3 (17·6%)† | 1 (5·9%) | 2 (11·8%) | |
| Hui (n = 16) | 1 (6·3%) | 12 (75·0%)‡ | 3 (18·8%) | 0 | 0 | 0 | |
| Yi (n = 14) | 1 (7·1%) | 9 (64·3%)‡ | 4 (28·6%) | 0 | 0 | 0 | |
| Bai (n = 11) | 2 (18·2%) | 7 (63·6%)‡ | 1 (9·1%) | 1 (9·1%) | 0 | 0 | |
| Zhuang (n = 8) | 3 (37·5%) | 3 (37·5%) | 0 | 0 | 0 | 2 (25·0%) | |
| Jingpo (n = 7) | 1 (14·3%) | 0 (0·0%) | 1 (14·3%) | 0 | 3 (42·9%)† | 2 (28·6%) | |
| Hani (n = 5) | 1 (20·0%) | 4 (80·0%)‡ | 0 (0·0%) | 0 | 0 | 0 | |
| Other§ (n = 12) | 3 (25·0%) | 6 (50·0%) | 0 | 0 | 1 (8·3%) | 2 (16·7%) | |
Significant association by Fisher's exact test.
Significant difference in the corresponding proportion compared to Han.
Significant difference in the corresponding proportion compared to Dai.
The 12 patients classified as ‘other’ include two each of Yao, Wa, Naxi, and Miao groups, and one each of Du, Lisu, and Lahu groups, and one whose race was unknown.
Significant differences were observed between various ethnic groups with respect to the distributions of CRF08_BC, B, and C subtypes. The proportion of subtype CRF08_BC in the Dai group was significantly lower than in the Hui, Yi, Bai, or Han groups (5·9% vs. 75·0%, 64·3%, 63·6%, and 80·0%, respectively). The proportion of subtype B was significantly higher in the Dai group than the Han group (17·6% vs. 2·0%). The proportion of subtype C in the Jinpo group was significantly higher than in the Han group (42·9% vs. 3·0%).
In addition, subtype B was only observed in the Han, Dai, and Bai ethnic groups; subtype C was only observed in Han, Dai, Jinpo, and Wa groups. URFs were only observed in Han, Dai, Zhuang, Jinpo, Du, and Lisu groups.
Geographical distribution of HIV-1 subtypes
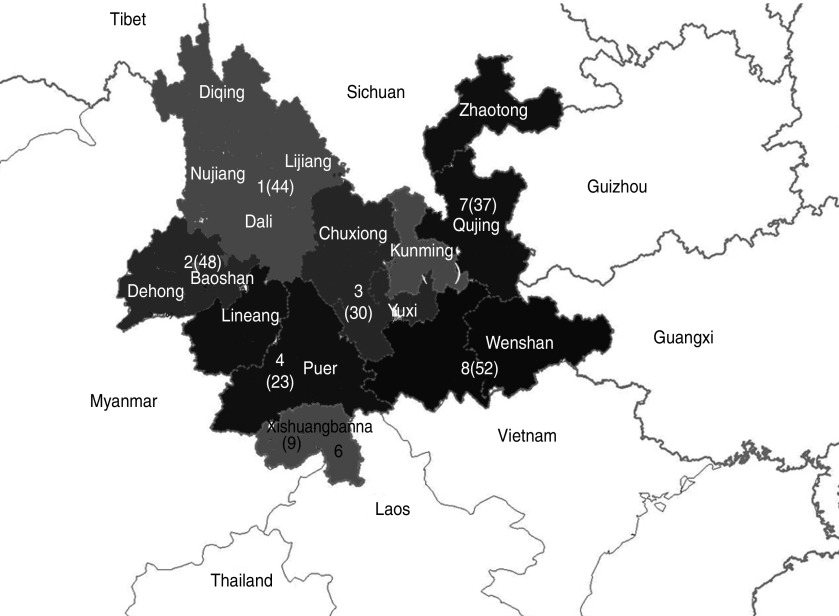
The population of Yunnan province was divided into eight geographical sections according to the location of the 16 prefectures in Yunnan, as indicated in Figure 1 and Table 4. CRF01_AE, CRF08_BC, and CRF07_BC subtypes were present in patients from all sections. The proportion of CRF08_BC, the most prevalent HIV-1 subtype, was over 50% in sections 1, 3, 4, 7, and 8. The proportion of CRF01_AE was over 50% in section 6. The proportion of CRF07_BC was 48·9% in section 5. Subtype B was only present in sections 1, 2, and 3. Subtype C was only present in sections 1, 2, 5, 7, and 8. URFs were present in all sections except for section 3.
Fig. 1.
Geographical distribution of HIV-1 in Yunnan province divided into eight sections. The number of successfully amplified samples in each region is shown in parentheses on the map.
Table 4.
Geographical distribution of HIV-1 subtypes
| HIV-1 subtypes | P value | ||||||
|---|---|---|---|---|---|---|---|
| CRF01_AE | CRF08_BC | CRF07_BC | B | C | URFs | ||
| Section 1: Dali, Nujiang, Lijiang, Diqing (n = 44) | 4 (9·1%) | 27 (61·4%) | 4 (9·1%) | 1 (2·3%) | 1 (2·3%) | 7 (15·9%) | <0·001* |
| Section 2: Baoshan, Dehong (n = 48) | 18 (37·5%)† | 4 (8·3%)† | 7 (14·6%) | 6 (12·5%) | 6 (12·5%) | 7 (14·6%) | |
| Section 3: Chuxiong, Yuxi (n = 30) | 1 (3·3%)‡ | 20 (66·7%)‡ | 8 (26·7%) | 1 (3·3%) | 0 | 0 | |
| Section 4: Lincang, Pu'er (n = 23) | 5 (21·7%) | 14 (60·9%)‡ | 2 (8·7%) | 0 | 0 | 2 (8·7%) | |
| Section 5: Kunming (n = 47) | 10 (21·3%) | 10 (21·3%)†§¶ | 23 (48·9%)†ce | 0 | 2 (4·3%) | 2 (4·3%) | |
| Section 6: Xishuangbanna (n = 9) | 6 (66·7%)†§ | 1 (11·1%) | 1 (11·1%) | 0 | 0 | 1 (11·1%) | |
| Section 7: Qujing, Zhaotong (n = 37) | 5 (13·5%)# | 23 (62·2%)‡∥ | 7 (18·9%) | 0 | 1 (2·7%) | 1 (2·7%) | |
| Section 8: Honghe, Wenshan (n = 52) | 8 (15·4%)# | 30 (57·7%)‡∥ | 9 (17·3%)f | 0 | 1 (1·9%) | 4 (7·7%) | |
Significant association by Fisher's exact test.
Significant difference in the corresponding proportion compared to section 1.
Significant difference in the corresponding proportion compared to section 2.
Significant difference in the corresponding proportion compared to section 3.
Significant difference in the corresponding proportion compared to section 4.
Significant difference in the corresponding proportion compared to section 5.
Significant difference in the corresponding proportion compared to section 6.
There were significant differences in the distribution of HIV-1 subtypes between the various sections of Yunnan (P < 0·001). The proportion of CRF_01AE in sections 2 (37·5%) and 6 (66·7%) was significantly higher than in sections 1 (9·1%) and 3 (3·3%). The proportion of CRF_01AE in sections 7 (13·5%) and 8 (15·4%) was significantly lower than in section 6 (66·7%). The proportion of CRF08_BC in sections 1 (61·4%), 3 (66·7%), and 4 (60·9%) was significantly higher than in sections 2 (8·3%) and 5 (21·3%). The proportion of CRF08_BC in sections 7 (62·2%) and 8 (57·7%) was significantly higher than in sections 2 (8·3%) and 5 (11·1%).
The proportion of CRF_07_BC in section 5 (48·9%) was significantly higher than in sections 1 (9·1%), 2 (14·6%), 4 (8·7%), and 8 (17·3%). No significant geographical differences were observed in the proportion of subtypes B, C, or URFs.
Association of HIV-1 subtypes with transmission routes
After adjustment for the geographical distribution in the population, sexual transmission was the most common route of infection (54·8%), followed by IDU (38·5%), unknown transmission route (4·6%), mother-to-child transmission (1·7%), and same-sex sexual transmission (0·4%) (Table 2).
Our data indicate a correlation between HIV-1 subtype and transmission route (P = 0·007) (Table 5). The prevalence of CRF01_AE in those infected through IDU was significantly lower than in those infected through sexual transmission (8·0% vs. 25·9%). The major subtypes of those infected through sexual transmission were CRF08_BC (38·0%) and CRF01_AE (25·9%). The major subtypes of those infected through IDU were CRF08_BC (52·0%) and CRF07_BC (27·0%). The major subtypes of those infected through mother-to-child transmission were CRF08_BC (33·3%) and CRF01_AE (33·3%); and the two patients infected through same-sex sexual transmission were both CRF01_AE.
Table 5.
Association of HIV-1 subtypes with transmission route
| Transmission route | HIV-1 subtype | P value | |||||
|---|---|---|---|---|---|---|---|
| CRF01_AE | CRF08_BC | CRF07_BC | B | C | URFs | ||
| Heterosexual (n = 166) | 43 (25·9%) | 63 (38·0%) | 32 (19·3%) | 5 (3·0%) | 7 (4·2%) | 16 (9·6%) | 0·007* |
| Injecting drug use (n = 100) | 8 (8·0%)† | 52 (52·0%) | 27 (27·0%) | 3 (3·0%) | 4 (4·0%) | 6 (6·0%) | |
| Mother-to-child (n = 9) | 3 (33·3%) | 3 (33·3%) | 1 (11·1%) | 0 | 0 | 2 (22·2%) | |
| Homosexual (n = 2) | 2 (100·0%) | 0 | 0 | 0 | 0 | 0 | |
| Unknown (n = 13) | 1 (7·7%) | 11 (84·6%) | 1 (7·7%) | 0 | 0 | 0 | |
Significant association by Fisher's exact test.
Significant difference in the corresponding proportion compared to those infected by sexual transmission.
Phylogenetic tree analysis of epidemic HIV-1 strains
We conducted phylogenetic analysis of sequences longer than 800 bp in the gag region and sequences longer than 380 bp in the env region; a total of 214 samples were included in the analysis. Of these samples, 168 were analysed in both the env and gag regions, 26 in the env region only, and 20 in the gag region only. Thus, a total of 188 gag sequences and 194 env sequences were analysed. The resulting phylogenetic trees are shown in Supplementary Figure S1 (online).
In further analysis, the association between sampling region and transmission route was not significant (P = 0·175, data not shown). It also indicated that the transmission routes among these eight regions did not differ from each other significantly (data not shown).
Supplementary Figure S1A shows the phylogenetic tree based on gag region gene fragments. This tree has four large branches; from the top to the bottom, the branches are CRF07_BC, CRF08_BC, B, and CRF01_AE. Strains dh3, h9, n3, h17, and the Indian subtype C reference strain gathered in one branch located within the cluster of the CRF07_BC subtype. The strains from the same area are scattered on different branches of the phylogenetic tree. For example, the CRF08_BC subtype from Red River (yellow dots), Dali (blue triangles), and Qujing (red dots) are scattered in various branches of the phylogenetic tree, and are not clustered together. The CRF07_BC and CRF01_AE subtypes in other areas showed the same results, indicating that the three predominant recombinant subtypes had province-wide distribution.
Our data indicate that strains Banna bn1, Nujiang n1, and Dali d17 and d16 are independent from the CRF07_BC, CRF08_BC, and C branches; instead, they are located below CRF08_BC in the phylogenetic tree. Dali d10 was located between CRF07_BC and CRF08_BC (Supplementary Fig. S1A). This relationship could be due to recombination of the once prevalent CRF08_BC, CRF07_BC, B, and C in local areas, or might indicate the inflow of URFs. Below the BC branch, strain m2 (derived from Burmese living in Dehong) was located below the BC branch alone, which could represent the inflow of remote CRF07_BC or B/C recombinant subtypes. Further away from the CRF01_AE branch, strains such as g3 and l2 (Supplementary Fig. S1A) might also be recombinants, involving CRF01_AE and other subtypes.
The phylogenetic tree based on gene fragments in the env region is shown in Supplementary Figure S1B. The epidemic subtypes in Yunnan are mainly CRF07_BC and CRF08_BC. The genes in the env region of these two recombinant subtypes are homologous to the genes of subtype C, preventing differentiation between CRF07_BC, CRF08_BC, and subtype C. These subtypes are thus located within the same cluster in the phylogenetic tree (Fig. 1).
This phylogenetic tree indicates that various subtype strains were not completely isolated by geographical area. The samples in this study are divided into three major clusters in the phylogenetic tree: C/CRF07/08_BC, B, and CRF01_AE, but some strains were not in the same cluster as the reference strains. For example, strains dh14, h1, and q12, located above the CRF01_AE branch, were located within the same branch. The d10, dh3 and q27 strains were located separately, below C/CRF07/08_BC, having a more distant phylogenetic relationship with the C/CRF07/08_BC cluster reference sequence. The gag region of the h1 strain from Red River indicated that it was subtype CRF07_BC, and the d10 strain was located between CFR07_BC and CRF08_BC in the gag gene phylogenetic tree.
Calculation of the Kimura 2-parameter was performed to determine the genetic distances between different subtypes and different routes of transmission. For the gag gene, the average genetic distance of subtype CRF01_AE was 0·047 ± 0·003, and the average genetic distances of subtypes CRF07_BC and CRF08_BC were 0·027 ± 0·002 and 0·020 ± 0·002, respectively. The average genetic distance of the subtype B virus was 0·072 ± 0·007, and that of the new BC subtype was 0·021 ± 0·003 (data not shown), reflecting the epidemiological history that subtype B was the first subtype to have spread into Yunnan, followed by CRF01_AE.
DISCUSSION
Our phylogenetic analysis revealed six HIV-1 subtypes in Yunnan, including eight new URFs. The four most prevalent subtypes in this province, CRF07_BC, CRF08_BC, CRF01_AE and URFs, were all recombinants. We found significant differences in the distribution of these HIV-1 subtypes geographically, between various ethnic groups, and with respect to transmission routes.
Representative sampling of circulating HIV-1 strains in Yunnan province
Previous molecular epidemiological studies of HIV-1 in Yunnan province have had some limitations. For example, most molecular data are only from IDUs, and many studies have used convenience sampling methods, which can lead to biased results. Our samples were selected by systematic random sampling and covered various routes of transmission, occupation, age, ethnicity, and geographical area. In addition, we corrected the data to account for geographical differences in subtype occurrence. We believe this study is the first complete and systematic province-wide molecular epidemiological investigation of HIV-1 strains in Yunnan. The results are representative of the molecular characteristics of circulating HIV-1 strains in Yunnan.
Epidemic trends in HIV-1 subtypes in Yunnan province
An epidemiological study in 1996 showed that subtype C accounted for 34·3% of the HIV-1 infections in Yunnan province [15]. During 2001–2002, subtype C accounted for only 1·03% of all cases [8]. It was unclear whether this decrease was due to sampling differences or an increase in the proportion of recombinant subtypes appearing after 2000. Our data show that the percentages of subtypes B and C were 2·8% and 3·8%, respectively, in 2006. Recombination analysis based on Thailand subtype B and India subtype C showed that subtypes B and C were not pure, indicating that the prevalence of pure subtype B and C strains had decreased, presumably due to an increase in recombinant strains.
We found that CRF07_BC and CRF08_BC accounted for 65·5% of the total HIV cases. This result confirmed previous reports that B’ and C subtypes had recombined in IDUs in the early 1990s, resulting in the CRF07_BC and CRF08_BC subtypes [4, 16–20]. With stronger transmission advantages and adaptability, these subtypes have become the principal circulating viruses.
In 1993–1994, a Yunnan woman who had been a commercial sex worker in Thailand was confirmed to be infected with subtype CRF01_AE [10]. Between 1997 and 2004, the prevalence of CRF01_AE cases rose from 5% to 40·5% of all HIV cases in Yunnan [10, 21]. Using data from 1989 to 2004, Zhang et al. showed that CRF01_AE was mainly concentrated in the western regions of Yunnan [9], with a smaller fraction residing in the eastern regions. CRF01_AE was shown to account for only 4% of HIV in sex workers in Kunming [10]. However, our study shows that the percentage of CRF01_AE in Kunming has increased significantly to 14·4%. This finding is similar to that of Zhou et al. regarding blood donors in Kunming (20·4% CRF01_AE) [21]. These results indicate that the CRF01_AE subtype has spread to the eastern regions of Yunnan and that its prevalence could continue to increase with the increase in sexually transmitted infections observed in recent years.
Prevalence of URFs in Yunnan
URFs accounted for 9·2% of the total HIV subtypes. This finding reflects the multiple risk factors in the Yunnan population, which has led to the co-circulation of multiple HIV subtypes capable of recombination.
Our study found evidence of unique recombinants in some strains at the distal end of the BC branch that did not belong to CBF07_BC or CRF08_BC (Supplementary Fig. S1A). Other strains were located between clusters of CRF07_BC and CRF08_BC, and the recombination analysis results showed that they were subtype BC recombinant strains (data not shown). Because CRF07_BC and CRF08_BC are highly prevalent in Yunnan, subtype BC recombinants accounted for the highest percentage (3·1%) of all the new recombinant strains. Genetic distance analysis showed a new BC subtype located between CRF07_BC and CRF08_BC (0·021 ± 0·003), suggesting that the BC subtype could be a recombinant generated during the same time period as CRF07_BC and CRF08_BC. Alternatively, it could be the result of another recombination involving CRF08_BC and CRF07_BC.
This study also found multiple other types of recombinant strains, resulting from recombination of CRF01_AE with subtype C, CRF08_BC and CRF07_BC with CRF01_AE, subtype B with CRF01_AE, and a BC recombinant strain (involving CRF07_BC and CRF08_BC) with CRF01_AE. The population of recombinants is thus diverse and complex.
We analysed the sources of these recombinant strains and found that they were mostly distributed in the western regions of Yunnan with the initial epidemic of HIV including Dehong, Baoshan, Dali, and Xishuangbanna. URFs were also prevalent in Honghe, Kunming, and Qujing, indicating that URFs are spreading to the interior of the province.
Changing HIV-1 subtypes in populations and geographical areas
The transmission pathway of HIV in Yunnan involves horizontal transmission from IDUs (introducers) to sex workers (bridge population) to brothel clients, their sexual partners, and vertical transmission to infants – gradually spreading to the general population. The characteristics of circulating strains trace this transmission pathway.
Our results indicate that the percentage of CRF07_BC and CRF08_BC subtypes in the IDU population has decreased since the 2004 molecular epidemiological study of Yunnan province [17], but has not reached statistical significance (P > 0·05). The CRF01_AE subtype from the South East Asian countries spread to the western part of Yunnan province via cross-border prostitution and then inland via sexual transmission. This increase in sexual transmission of HIV has led to a province-wide epidemic of the CRF01_AE subtype. Our study found two cases of infection via male-to-male sexual transmission, both caused by subtype CRF01_AE. We speculate that the CRF01_AE subtype might have spread into the male homosexual population via female-to-male sexual transmission, leading to spread within that population. URFs accounted for a larger percentage of infections acquired through mother-to-child transmission and heterosexual contact than those acquired through IDU. This result is surprising, as it is well known that IDUs are far more promiscuous than other transmission groups and are thus more likely to become dually infected, subsequently giving rise to recombinant strains. The low proportion of URFs in the IDU and men who have sex with men populations might be caused by selection bias due to small sample size. Another possible explanation is that transmission route statistics are approximate, as many IDUs and sex workers will not admit to the transmission route because drug abuse and prostitution are illegal in China.
This study was limited by the inconsistent preservation of patient samples. The quality of the samples thus varied between different prefectures and might have inadvertently influenced the quantification of HIV subtypes. Future studies could be made more inclusive and accurate by devising consistent means of sample storage. A further limitation of our study was that our use of the bulk PCR/sequencing approach made it impossible to detect dual infections in our samples. The number of possible recombinants detected was limited in this study because we sequenced just an 800 bp fragment of gag and a 380 bp fragment of env; the rest of the genome (∼90%) was not assessed. In addition, some samples had only one or the other fragment sequenced, thereby further reducing the ability to detect recombinant strains.
The unique geographical and demographic characteristics of Yunnan have made it the origin of the major HIV-1 subtypes now circulating in China. For this reason, national policy on HIV control should be made with an eye on Yunnan Province. The finding that the four most prevalent subtypes in Yunnan, CRF07_BC, CRF08_BC, CRF01_AE and URFs, are recombinants indicates that future treatment and vaccine options in China will be challenged by a diverse population of HIV subtypes. Comprehensive measures should include strengthening interventions within high-risk populations and enhancing HIV testing and antiretroviral treatment to prevent further recombination and the development of more complex strains.
Supplementary Material
Supplementary information supplied by authors.
Supplementary information supplied by authors.
ACKNOWLEDGEMENTS
This work was supported by the Major Project of China's ‘Eleventh Five-Years Plan’ for Science and Technologies Development (2009ZX10004-902 and 2008ZX10001-004).
Supplementary material
For supplementary material accompanying this paper visit https://doi.org/10.1017/S0950268812002713.
click here to view supplementary material
DECLARATION OF INTEREST
None.
REFERENCES
- 1.Xiao Y, et al. Expansion of HIV/AIDS in China: lessons from Yunnan Province. Social Science & Medicine 2007; 64: 665–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.China Daily. HIV infections exceed 90,000 in SW China's Yunnan, 30 November 2011.
- 3.Beyrer C, et al. Overland heroin trafficking routes and HIV-1 spread in south and south-east Asia. AIDS 2000; 14: 75–83. [DOI] [PubMed] [Google Scholar]
- 4.Cheng H, et al. HIV-1 subtype E in Yunnan, China. Lancet 1994; 344: 953–954. [DOI] [PubMed] [Google Scholar]
- 5.Liu Y, et al. Identification and characterization of two new HIV type 1 unique (B/C) recombinant forms in China. AIDS Research and Human Retroviruses 2011; 27: 445–451. [DOI] [PubMed] [Google Scholar]
- 6.Chen L, et al. Near full-length genomic characterization of a novel HIV type 1 CRF07_ BC/CRF08_ BC recombinant strain from Yunnan, China. AIDS Research and Human Retroviruses 2011; 27: 693–699. [DOI] [PubMed] [Google Scholar]
- 7.Saksena NK, et al. Snapshot of HIV pathogenesis in China. Cell Research 2005; 15: 953–961. [DOI] [PubMed] [Google Scholar]
- 8.Yang R, et al. On-going generation of multiple forms of HIV-1 intersubtype recombinants in the Yunnan Province of China. AIDS 2002; 16: 1401–1407. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Y, et al. Dominance of HIV-1 subtype CRF01_AE in sexually acquired cases leads to a new epidemic in Yunnan province of China. PLoS Medicine 2006; 3: e443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li XJ, et al. Molecular epidemiology of the heterosexual HIV-1 transmission in Kunming, Yunnan Province of China suggests origin from the local IDU epidemic. AIDS Research and Human Retroviruses 2005; 21: 977–980. [DOI] [PubMed] [Google Scholar]
- 11.Piyasirisilp S, et al. A recent outbreak of human immunodeficiency virus type1 infection in southern China was initiated by two highly homogeneous, geographically separated strains, circulating recombinant form AE and a novel BC recombinant. Journal of Virology 2000; 74: 11286–11295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xin RL, et al. Primers of gag gene for HIV-1 subtyping in China and application thereof in practice. National Medical Journal of China 2009; 89: 876–880. [PubMed] [Google Scholar]
- 13.Los Alamos National Laboratory HIV databases. (http://www.hiv.lanl.gov/content.index). Accessed March 2011.
- 14.Lole, et al. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. Journal of Virology 1999; 73: 152–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li DQ, Zheng XW, Zhang GY. Study on the distribution HIV-1 C subtype in Ruili and other counties, Yunnan, China [in Chinese]. Zhonghua liu xing bing xue za zhi = Zhonghua liuxingbingxue zazhi 1996; 17: 337–339. [PubMed] [Google Scholar]
- 16.Weniger BG, et al. The molecular epidemiology of HIV in Asia. AIDS 1994; 8 (Suppl. 2): S13–28. [PubMed] [Google Scholar]
- 17.Laeyendecker O, et al. Molecular epidemiology of HIV-1 subtypes in southern China. Journal of Acquired Immune Deficiency Syndromes 2005; 38: 356–362. [PubMed] [Google Scholar]
- 18.Yu XF, et al. Characterization of HIV type 1 heterosexual transmission in Yunnan, China. AIDS research and human retroviruses. 2003; 19: 1051–1055. [DOI] [PubMed] [Google Scholar]
- 19.Luo CC, et al. HIV-1 subtype C in China. Lancet 1995; 345: 1051–1052. [DOI] [PubMed] [Google Scholar]
- 20.Cassol S, et al. Detection of HIV type 1 env subtypes A, B, C, and E in Asia using dried blood spots: a new surveillance tool for molecular epidemiology. AIDS Research and Human Retroviruses 1996; 12: 1435–1441. [DOI] [PubMed] [Google Scholar]
- 21.Zhou YH, et al. Process of molecular epidemiology of HIV-1 in Yunnan. Journal of Dermatology and Venereology 2010; 32: 4. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information supplied by authors.
Supplementary information supplied by authors.
For supplementary material accompanying this paper visit https://doi.org/10.1017/S0950268812002713.
click here to view supplementary material