Abstract
The microbial community dynamics of a functionally stable, well-mixed, methanogenic reactor fed with glucose were analyzed over a 605-day period. The reactor maintained constant pH and chemical oxygen demand removal during this period. Thirty-six rrn clones from each of seven sampling events were analyzed by amplified ribosomal DNA restriction analysis (ARDRA) for the Bacteria and Archaea domains and by sequence analysis of dominant members of the community. Operational taxonomic units (OTUs), distinguished as unique ARDRA patterns, showed reproducible distribution for three sample replicates. The highest diversity was observed in the Bacteria domain. The 16S ribosomal DNA Bacteria clone library contained 75 OTUs, with the dominant OTU accounting for 13% of the total clones, but just 21 Archaea OTUs were found, and the most prominent OTU represented 50% of the clones from the respective library. Succession in methanogenic populations was observed, and two periods were distinguished: in the first, Methanobacterium formicicum was dominant, and in the second, Methanosarcina mazei and a Methanobacterium bryantii-related organism were dominant. Higher variability in Bacteria populations was detected, and the temporal OTU distribution suggested a chaotic pattern. Although dominant OTUs were constantly replaced from one sampling point to the next, phylogenetic analysis indicated that inferred physiologic changes in the community were not as dramatic as were genetic changes. Seven of eight dominant OTUs during the first period clustered with the spirochete group, although a cyclic pattern of substitution occurred among members within this order. A more flexible community structure characterized the second period, since a sequential replacement of a Eubacterium-related organism by an unrelated deep-branched organism and finally by a Propionibacterium-like species was observed. Metabolic differences among the dominant fermenters detected suggest that changes in carbon and electron flow occurred during the stable performance and indicate that an extremely dynamic community can maintain a stable ecosystem function.
Understanding the factors that determine ecosystem stability has been one of the main challenges for ecologists for many years. Knowledge of the conditions that affect stability is needed to determine the effects of external parameters on habitats and is valuable for environmental management and biotechnological applications. Nevertheless, definitions of ecosystem stability are not precise and in many cases are referenced either to measurable parameters describing the function of the whole system or to the community composition (15). Most of the studies performed on microbial population dynamics have focused on the detection of a shift in bacterial populations when an open ecosystem is exposed to a natural disturbance, such as seasonal changes (12, 33), or to an artificial perturbation (2, 4, 11, 12). However, one of the main questions that still remains unanswered is whether, in undisturbed ecosystems, functional stability implies a persistent community. In the present study, a methanogenic bioreactor was chosen as the ecosystem to address this question. The reactor had experienced the same controlled environmental parameters for 1,505 days, and functional stability after day 400 was confirmed by a constant performance with respect to chemical oxygen demand (COD) reduction, pH, and methane production (39).
Methanogenic ecosystems constitute a bacterial food chain where three trophic levels can be distinguished as fermentable substrates are anaerobically degraded in the absence of inorganic electron acceptors. The concerted activity of fermenting populations, acetogenic hydrogen-producing bacteria, and methanogens is required for the complete degradation of glucose to CH4 and CO2 (40). Disturbances in populations from one trophic level affect the entire community and cause an imbalance that is reflected in the bioreactor performance by accumulation of intermediates, pH changes, or reduced efficiency (28).
Because of the limitations of culture methods, it has not been possible to retrieve most of the bacteria present in complex communities (1) to elucidate microbial community structure. Molecular methods, particularly those involving the 16S rRNA gene, are currently widely used to suggest the identity of uncultured microorganisms. Comparison of PCR-amplified 16S ribosomal DNA (rDNA) sequences complemented with screening strategies such as restriction profile analysis (21, 37) constitutes a rapid method that provides improved but not complete information on microbial community composition.
The aim of this work was to evaluate how stable the microbial community was over a nearly 2-year period (day 900 to day 1505) in a methanogenic reactor operated under constant conditions and exhibiting stable performance. Populations from Bacteria and Archaea domains were analyzed by amplified rDNA restriction analysis (ARDRA). The composition of the former group varied considerably while the latter group was more stable, but a shift in the latter coincided with a major shift in the Bacteria community members.
MATERIALS AND METHODS
Control and operation of the reactor.
A 1.5-liter continuously stirred tank reactor, described previously (39), was operated anaerobically at 35°C for 1,500 days. The substrate (glucose) was supplied in a 2-day cycle: 16 g/liter one day and 0 g/liter the following day. A constant dilution rate (0.1 day−1) was maintained, and steady state was achieved after day 400. No perturbations were applied during this period except for an accidental temperature drop to room temperature during 1 day (day 977) and an increase in glucose feeding for 1 day (day 983). Performance was monitored with effluent COD and volatile fatty acids, methane production, and pH measurement (39).
Extraction and purification of total genomic DNA.
Samples (10 ml) were collected from the continuously stirred tank reactor after manual shaking, immediately concentrated by centrifugation at 6,000 × g for 15 min, washed with a solution of 0.9 g of NaCl per liter, and frozen at −20°C. Cells were resuspended in 3 ml of a high-salt lysis buffer (1 M NaCl, 5 mM MgCl2, and 10 mM Tris [pH 8.0]) to reduce shearing of DNA (3) and vortexed to homogenize the samples. Suspended cells were passed three times through a French press at a pressure of 15,000 lb/in2. This procedure gave a high DNA yield, and it is particularly critical for the difficult-to-lyse methanogenic bacteria (3).
Lysate was diluted 1:5 in TE buffer (10 mM Tris [pH 8.0], 1 mM EDTA [pH 8.0]) to a final volume of 15 ml. Deproteinization was achieved by addition of sodium dodecyl sulfate (2% [wt/vol]) and proteinase K (50 μg/ml) (Boehringer Mannheim, Indianapolis, Ind.) and incubation at 65°C for 2 h. After cooling, nucleic acids were extracted by adding 10 ml of phenol-chloroform (50/50 ratio) followed by centrifugation at 3,000 × g for 20 min. After a second extraction with 10 ml of chloroform, nucleic acids were precipitated overnight with 10 ml of ice-cold isopropanol at −20°C. The precipitate was washed with 70% (vol/vol) ice-cold ethanol and dissolved in 400 μl of warm (50°C) deionized water. The solution was incubated for 2 h at 37°C with DNAse-free (27) RNase A (Boehringer Mannheim) at a final concentration of 1.2 μg/μl. After protein extraction with 500 μl of chloroform and centrifugation at 14,000 × g for 10 min, DNA was precipitated by addition of a 1/10 volume of 3 M sodium acetate and ice-cold ethanol to a final concentration of 70% (vol/vol). DNA was dissolved in deionized water, and its purity and concentration were checked by spectrophotometry. DNA was purified with Wizard Plus miniprep columns (Promega, Madison, Wis.).
ARDRA.
Amplification of 16S rDNA from purified genomic DNA from each sample was carried out with primers for the Bacteria and Archaea domains. The Bacteria clone library was prepared from amplification products produced with a forward primer which corresponds to nucleotide positions 19 to 38 (5′-AGAGTTTGATCCTGGCTCAG-3′; primer A) of the Escherichia coli rRNA and a reverse primer which corresponds to the complement of positions 1581 to 1541 (5′-AAGGAGGTGATCCAGCCGCA-3′; primer H) (34). All primers were synthesized with an Applied Biosystems DNA synthesizer at the Macromolecular Structure and Sequencing Facility, Michigan State University. Amplification was done in a 25-μl reaction volume containing 100 ng of DNA, 0.5 μM (each) primer, 0.2 mM (each) deoxynucleoside triphosphate, 1.5 mM MgCl2, 0.2 mg of bovine serum albumin per ml, 2.5 μl of 10× Taq buffer, and 4 U of Taq DNA polymerase (Perkin-Elmer Cetus, Norwalk, Conn.) (21). PCR amplification was performed in an automated thermal cycler (Model 9600; Perkin-Elmer) with an initial denaturation (94°C for 5 min) followed by 30 cycles of denaturation (94°C for 1 min), annealing (55°C for 1 min), and extension (72°C for 3 min) and a single final extension (72°C for 7 min).
The Archaea clone library was prepared from amplification products produced with a forward primer which corresponds to nucleotide positions 19 to 38 (5′-TAAGCCATGCRAGTCGAAYG-3′; primer 69F) of the E. coli rRNA (18) and a reverse primer which corresponds to the complement of positions 1581 to 1541 (5′-[C/T]CCGGCGTTGA[A/C]TCCAATT-3′; primer 958R) (9). Amplification was optimized by checking different conditions with Methanosarcina barkeri (ATCC 43241) and Methanobrevibacter smithii (ATCC 35061) DNA as positive controls and DNA of several isolates from this reactor belonging to the Bacteria domain as negative controls. The following modifications of the previous conditions were used: 1.75 mM MgCl2, 35 cycles, and an annealing temperature of 52°C for 1 min. The products were electrophoresed on a 1% agarose gel to evaluate the quality and concentration of the amplified fragments.
All amplicons were cloned with the TA cloning kit (Invitrogen, Carlsbad, Calif.) according to the manufacturer’s instructions. Direct amplification of plasmid DNA from positive clones was accomplished with the following conditions: initial denaturation (92°C for 2 min) followed by 35 cycles of denaturation (94°C for 30 s), annealing (67°C for 1 min), and extension (72°C for 3 min) and a single final extension (72°C for 7 min). Primers were 5′-GCCGCCAGTGTGCTGGAATT-3′ (TAf) and 5′-TAGATGCATGCTCGAGCGGC-3′ (TAr) (43). The final reaction volume was 20 μl, and modifications of the above Bacteria 16S rDNA amplification conditions were 1.2 mM MgCl2, and 3 U of Taq. The fragment size of these amplicons was checked by electrophoresis in 1% agarose.
PCR products (16 μl) of the right size (1,500 bp for Bacteria and 900 bp for Archaea) were digested simultaneously for 12 h with two restriction enzymes (HaeIII and HhaI; Gibco BRL, Gaithersburg, Md.) according to the manufacturer’s specifications. The restriction fragments from each clone were separated by electrophoresis on a 3.5% (wt/vol) MetaPhor agarose gel (FMC, Indianapolis, Ind.) in fresh 1× Tris-borate-EDTA buffer at 4 V/cm in a cold room and observed after staining with ethidium bromide. Restriction patterns were normalized and compared with GelCompar software (version 3.1; Applied Maths, Kortrijk, Belgium). Pattern clustering was done by the unweighted pair group method with averages with application of the Dice coefficient. A maximum tolerance for band position of 3.0% ± 0.5% was used, and each specific pattern was defined as an operational taxonomic unit (OTU). Thirty-six ARDRA patterns were analyzed for each sample, and frequencies were calculated as the percentages of the clones showing the same OTU. OTU labeling includes the domain A (Archaea) and E (Bacteria, formerly Eubacteria), and the OTU relative abundance in the respective library expressed in roman numerals, e.g., AO-I, dominant OTU in the Archaea library.
Sequencing and sequence analysis.
The 16S rDNA gene fragments were amplified for sequencing directly from whole cells containing the correct plasmid and insert with the TA cloning kit (Invitrogen). The amplicons were then purified with Microcon-100 spin columns (Millipore Corp.). Sequences were obtained via the dye terminator method by the Michigan State University DNA Sequencing Facility with modified versions of previously described sequencing primers targeting conserved regions of the 16S rRNA gene (35, 36, 38, 42). Alignment of sequences, mask construction, and chimera check were performed with software provided by the Ribosomal Database Project (20). Dendrograms were constructed with PAUP (31) and MacClade 3.0 (19) software.
Nucleotide sequence accession numbers.
The sequences determined in this study have been deposited in the GenBank database under accession no. AF149878 to AF149893.
RESULTS
Reactor performance.
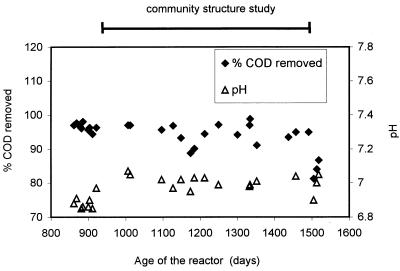
The methanogenic reactor exhibited a constant performance as measured by effluent pH and COD removal from day 900 through day 1505, the period in which the community composition was studied (Fig. 1). The pH was in the range of 6.85 to 7.07, and the COD removed was between 89 and 99% until the reactor began to fail on day 1491.
FIG. 1.
Functional performance of the methanogenic reactor during the 605-day period of the community structure study. The reactor was operated for 1,505 days, steady state was achieved at day 400, and the community analysis study was started at day 900.
ARDRA reproducibility.
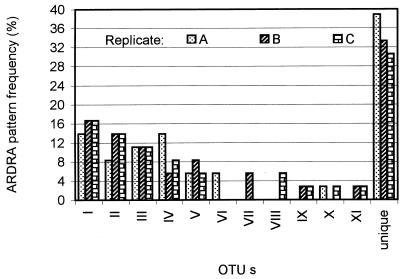
Three samples (A, B, and C) from a similar reactor were taken simultaneously and analyzed separately. DNA extraction, Bacteria 16S rDNA amplification, cloning, and restriction analysis were applied to DNA from each replicate, and OTU profiles from the replicates were compared. The three replicates showed a similar richness. Twenty-one OTUs were detected in samples B and C, and 22 OTUs were detected in sample A (Fig. 2).
FIG. 2.
Bacteria ARDRA pattern distribution in three samples processed separately. OTUs were defined by restriction with two enzymes. Unique OTUs (14, 12, and 11 in replicates A, B, and C, respectively) appeared once in this clone library.
Clone distribution in the different OTUs was also very similar among the replicates. Patterns were organized in three groups according to their presence and abundance in each replicate. The first group included OTUs common to all the samples (I, II, III, IV, and V). These represented 52.7% of the total clones analyzed in sample A and 55.5% of the total clones analyzed in samples B and C. The pattern distribution within this group was consistent in all the samples: the dominant OTU (I) had a frequency of 13.9% in sample A and of 16.7% in samples B and C. The distribution of the rarest pattern (V) was 5.5% in replicates A and C and 8.3% in replicate B (Fig. 2).
The second group comprised less than 14% of the clones analyzed in any sample. It contained OTUs which were observed twice (frequency, 5.5%) in the same sample but were undetected in other samples (VI, VII, and VIII) and OTUs which were observed once in two of three samples (IX, X, and XI) (Fig. 2). Finally, the third group comprised the unique OTUs: those that were observed just once in a sample and were not detected in any other sample. The rare OTUs, which constituted a significant fraction of the community, represented 30.5% (sample C), 33.3% (sample B), and 38.9% (sample A) of the total clones. These results imply that in this community, the proportion of rare OTUs detected was consistent among the replicates (Fig. 2). Based on these results, ARDRA reproducibility was assumed for (i) pattern distribution and identity of the most abundant OTUs (frequency equal to or higher than 8.3%) and (ii) proportion of rare patterns.
Archaea and Bacteria clone distribution.
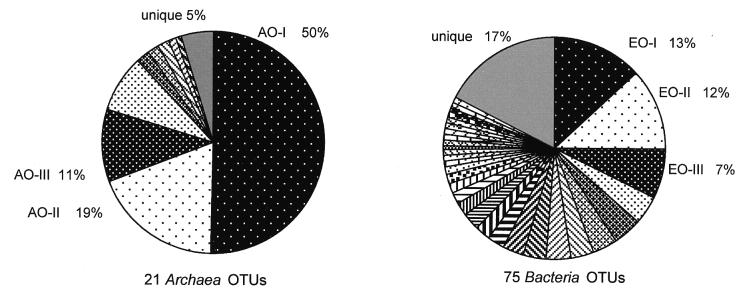
The reactor was sampled seven times during 605 days of steady-state operation for ARDRA. Archaea and Bacteria from 36 clones for each sampling point resulted in 252 clones analyzed from each domain. The diversity of Archaea and Bacteria populations is described on the basis of the total number of OTUs detected (richness) and the number and distribution of clones showing the same OTU (evenness). Higher richness in the Bacteria populations was detected; 75 OTUs were found in this domain, whereas only 21 Archaea OTUs appeared during the same period (Fig. 3). Clone distribution data also shows that diversity was higher in Bacteria populations. The three most frequent OTUs accounted for 80% of the clones in the Archaea library but for only 32% of the clones in the Bacteria library. In addition, the number of unique OTUs in both domains was significantly different: 5 and 17% for Archaea and Bacteria, respectively (Fig. 3).
FIG. 3.
OTU distribution in Archaea and Bacteria domains detected in the methanogenic reactor during 605 days of stable performance.
Archaea population dynamics.
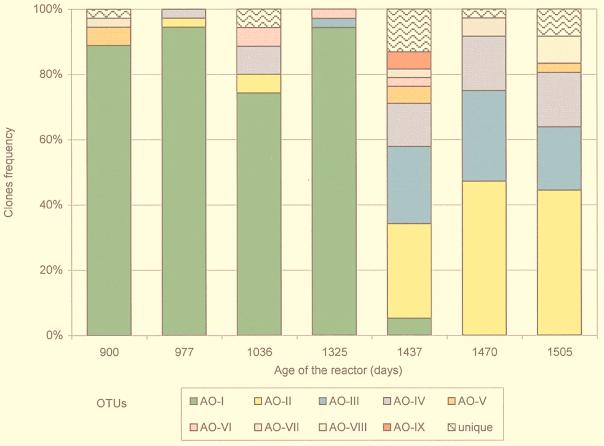
Changes in the Archaea populations were observed by comparing ARDRA pattern frequencies during the course of reactor operation. Seven samples were taken at time intervals ranging from 33 days to 289 days. Archaea population succession was observed, and two remarkably different periods were distinguished (Fig. 4). The first period, from day 900 to day 1325, was characterized by low diversity due to the prevalence of the OTU AO-I (representing between 72 and 94% of the clones) and by the small proportion of different OTUs (less than seven OTUs at any sampling point). A notable shift was observed between days 1325 and 1427. Three codominant OTUs (AO-II, AO-III, and AO-IV) now accounted for 69% of the clones, and their frequencies were quite constant between days 1437 and 1505 (Fig. 4). The highest diversity during this interval was observed immediately after the shift, at day 1437, when 12 different patterns were detected.
FIG. 4.
Dynamics of distribution of Archaea OTUs in the methanogenic reactor. OTUs were defined by ARDRA with two restriction enzymes, and frequency distribution was calculated over 36 clones analyzed from each sample. Unique OTUs appeared once in the entire clone library.
Population succession could be recognized in samples taken closer to the population shift that occurred after day 1325. Dominant patterns from one period were present in a low frequency in the other period. Nevertheless, differences in persistence were observed: two prevalent OTUs (AO-II and AO-IV) from the second period were detected for up to 460 days before they became dominant, but the dominant OTU in the first period (AO-I) was quickly displaced; it was undetectable by day 1470 after the community shift (Fig. 4). These results suggest that dominant OTUs from the second period had a higher fitness for the operational conditions.
Bacteria population dynamics.
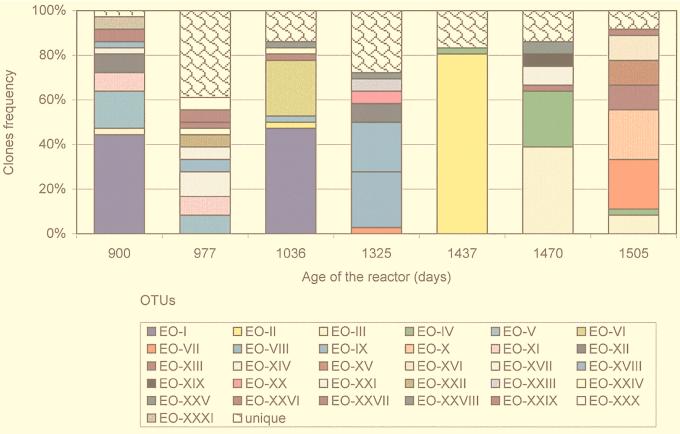
The Bacteria community was more variable than the Archaea community, as would be expected because of the higher diversity detected in this domain (Fig. 3). Rapid changes occurred, and no long periods of particular OTU persistence could be distinguished (Fig. 5). Further, almost no trends in population succession could be detected, even in a period as short as 33 days. Dominance was not maintained by any OTU except for OTU EO-I, which predominated on days 900 and 1036 but was undetectable between these dates and after day 1036. Some dominant OTUs (EO-II, EO-III, EO-IV, EO-V, and EO-VII) were detected sporadically at low frequency (Fig. 5).
FIG. 5.
Dynamics of distribution of Bacteria OTUs in the methanogenic reactor. OTUs were defined by ARDRA with two restriction enzymes, and frequency distribution was calculated over 36 clones analyzed from each sample. Unique OTUs appeared once in the entire clone library.
The diversity, measured as the ARDRA profile richness, was also chaotic. The total number of OTUs at each sampling point ranged between 8 and 24 with a mean of 13. The highest diversity was detected at day 977, when 24 OTUs were observed and the three most abundant profiles accounted for only 27.8% of the clones. However, dominance was observed at the closest sampling dates: 77 days before and 59 days after, EO-I OTU represented 44.4 and 47.2% of the total clones, respectively. Subsequently, similar or more gradual evenness was maintained, and one OTU (day 1437) or two OTUs (days 1325, 1470, and 1505) represented more than 40% of the clones. The lowest diversity was observed concomitant with the shift in Archaea populations, at day 1437, when the OTU EO-II accounted for 80.5% of the clones and only eight OTUs (EO-II, EO-IV, and six unique OTUs) were distinguished (Fig. 5).
Phylogenetic analysis of dominant OTUs.
rRNA sequence and phylogenetic analysis was performed on the two most abundant OTUs from each sampling point that exhibited a frequency equal to or higher than 8.3%, a value determined from the replication study. Several clones with identical ARDRA profiles from the two domains were sequenced, and the sequences were compared (data not shown). A higher probability of sequence divergence between clones in the same OTU was expected for the Archaea domain because a shorter fragment (900 bp) was amplified. However, 100% sequence similarity was found in the seven ARDRA patterns analyzed from the dominant Archaea OTU (AO-I). The remaining patterns analyzed (three of EO-I, two of EO-V, and two of AO-III) showed minor sequence divergence (0.5% or lower). These results demonstrate that, although only two restriction enzymes were used, rRNA diversity was not underestimated by ARDRA.
(i) Archaea populations.
Two of four dominant OTUs from this domain were characterized phylogenetically by partial sequencing of 16S rDNA. The sequence of AO-I, dominant during the first period of reactor operation, was 100% similar to the 16S rDNA sequence of Methanobacterium formicicum, and the sequence of AO-II, the most frequent OTU in the second period, exhibited a 99.7% similarity to the Methanosarcina mazei sequence. This shift in dominance revealed an important metabolic change, since a hydrogen- and formate-utilizing methanogen was replaced by a methylotrophic methanogen. The two codominant profiles during the second period (AO-III and AO-IV) were closely related to each other by sequence (99.7% similarity), indicating that subtle sequence differences were detected by ARDRA. The sequence of this OTU was 8.0% dissimilar to the closest known methanogen, Methanobacterium bryantii.
(ii) Bacteria populations.
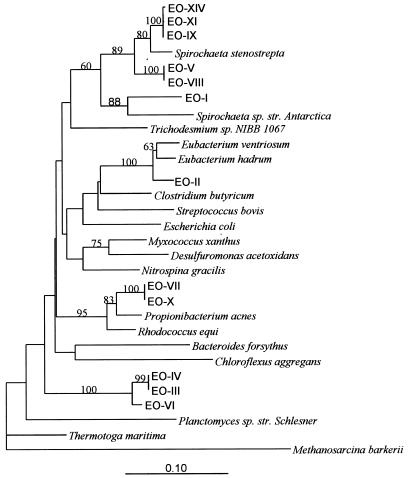
Most of the 12 sequences of Bacteria OTUs were not closely related to sequences of other known organisms (Fig. 6). The high diversity detected by ARDRA in this domain was quite apparent since several OTUs exhibited a high degree of sequence similarity.
FIG. 6.
Phylogram of representative Bacteria 16S rDNA cloned genes from the methanogenic reactor. A distance matrix was calculated by the maximum-likelihood method with the region of the 16S rRNA gene corresponding to E. coli positions 111 through 465. The phylogram shown was constructed from this distance matrix by the neighbor-joining method. Parsimony bootstrap values from 100 bootstrap replicates are shown on each node. Values below 50 are not shown. The scale bar represents an estimated 10% difference in nucleotide sequence.
The organization of OTUs according to their sequence similarity showed four main clusters revealing an interesting temporal pattern. Six of seven dominant and codominant OTUs in the first period clustered within the spirochete phylum. Each of the remaining three clusters corresponded mostly to OTUs occurring at one of the three sampling dates during the second period. One cluster with OTU EO-II includes bacteria belonging to the Eubacterium genus. The coexistent OTUs EO-VII and EO-X showed 0.5% sequence divergence and were related to the Propionibacterium genus. The rrn sequence of the third cluster was 20% dissimilar to that of any known division. It included the simultaneously occurring OTUs EO-III and EO-IV with 100% (partial) sequence similarity and OTU EO-VI (97.9% similar to the former), the dominant non-spirochete-related OTU present in the first period.
The spirochete cluster showed three main similarity groups (Fig. 6). OTU EO-I was closely related to the Spirochaeta sp. strain Antarctic, which belongs to the cluster of spirochetes of marine origin (group 3). The remaining two groups branched within the Treponema-spirochete cluster and showed a 15% sequence dissimilarity to group 3 spirochetes. Group 1 spirochetes, whose closest relative is a thermophilic spirochete, Spirochaeta caldaria, included OTUs EO-XIV, EO-XI, and EO-IX. The group 2 spirochetes, which had sequences 4% divergent from those of group 1, included OTUs EO-VIII and EO-V.
DISCUSSION
Ecosystem stability encompasses a broad spectrum of definitions, which resolve into two components: the measurable functional properties of the ecosystem and the population composition. Currently, stability is more commonly measured by the dynamics of a property of the whole ecosystem while information about population stability is often dismissed. However, population persistence is the most important property for ecosystem stability since it is closely related to fitness of individuals, and it is the fitness parameters that provide the information needed to manage natural communities (15). The relation between ecosystem stability and population stability has recently been studied most intensively in grassland plots. Population variability, caused by interspecific competition, produced compensatory effects over the total community, increasing ecosystem stability as measured by primary production (32). The results from our work indicate that an extremely dynamic community sustains a functionally stable ecosystem. Our data showed that prevalent members and diversity, within both Archaea and Bacteria domains, changed dramatically even over short periods, 3.3 retention times. These results indicated that functional parameters like pH and COD are inadequate to reveal community structure variations.
Direct correlation between OTU distribution and species distribution is uncertain because of multiple-copy rRNA genes (10, 25) and PCR and cloning biases (8, 24, 26, 30). Despite these limitations, molecular methods can reveal the presence of microorganisms that are undetectable by classical cultivation techniques. In our case, this approach was particularly valuable since most of the sequences of dominant OTUs correspond to difficult-to-cultivate microorganisms, like spirochetes. Furthermore, the effects of these biases are minimized when relative changes are studied in the same ecosystem and when the ARDRA profile distribution replicates, as was the case in this study.
Diversity in the whole Bacteria community was considerably higher than that among Archaea in our reactor. Likewise, extremely high diversity in the Bacteria domain has been observed in a fluidized bed reactor treating vinasse (14). In that study, 16S rDNA sequence analysis of 460 clones revealed the presence of 133 OTUs, with the most frequent OTU representing less than 5% of the clones. On the other hand, the Archaea domain revealed low variability, with six OTUs found after analysis of 98 clones, three of them representing 95% of the total clones (14). Most of the energy available in methanogenesis from glucose is spent during the first step of degradation (29). Consequently, higher yields in bacteria from this trophic level can be expected; hence, replacement among populations and higher diversity would be more feasible. Although syntrophism contributed to the carbon and electron flow in our reactor, especially during the second period when the main fermenters were butyrate or propionate producers, acetogenic hydrogen-producing bacteria were not detected. This is to be predicted, however, since they constitute a small fraction of the community in anaerobic reactors (14, 16, 28) and would not be expected given the size of our Bacteria clone library.
Our finding is in contrast to temporal population constancy that has been observed in other microbial environments even when they are disturbed. Populations from several fluidized bed reactors treating aromatic hydrocarbons converged to the same climax community irrespective of seeding, time of start-up, or substrate, and they remained stable for a 4-month period (22). It is likely that the strong selection pressure provided by related substrates and the biofilm formation contributed considerably to stabilizing the community in these reactors. Additionally, seasonal permanence of dominant cyanobacterial populations from a hot spring microbial mat was measured by denaturing gradient gel electrophoresis, but the authors suggested that minor population changes might have been undetected (12). Both of these habitats had a stable physical matrix, while ours did not.
The fluctuations of dominant populations observed in our reactor indicate that a climax community was not achieved in the 900 days previous to the community analysis study. Due to the constancy of the environmental parameters and functional performance throughout the analysis period, it may be reasoned that population succession was driven by the interactions among the members of the community. Three main trophic levels are recognized in methanogenic reactors fed with a fermentable substrate (40). Microorganisms in these groups are strongly interrelated, and an imbalance at any level could cause a failure that would be reflected in the reactor function (28). Stable performance implies steady-state production and consumption of metabolites along the trophic chain; consequently, a population shift at one trophic level would likely require a concerted change in the remaining populations to maintain the steady state. Subsequent changes in populations could be caused by variations in particular metabolites as well as their rates of production or consumption.
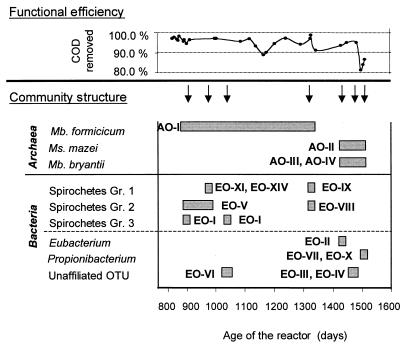
Phylogenetic analysis revealed that the genetic changes detected by ARDRA suggested metabolic variations in the community. However, if similar physiology is assumed for phylogenetically related taxa, the metabolic variation was less dramatic than were the genetic changes observed. Figure 7 summarizes the phylogenetic affiliation of dominant members of the community and the functional performance of our reactor. Two periods were distinguished based on the community composition in both domains during the 605 days of reactor operation. The first period, days 900 to 1325, was characterized by the persistence and prevalence of Methanobacterium formicicum and alternating ribotypes belonging to the spirochete cluster. During the second period, days 1437 to 1505, the main members of the Archaea community were Methanosarcina mazei and a Methanobacterium bryantii-related OTU. Monthly substitution of metabolically different fermentative bacteria characterized this period. This suggests a strong dependence between the Bacteria and the Archaea members of the community. However, the methanogenic groups present in the second period were metabolically more versatile, and therefore, the dependence among members from the two trophic levels was likely more moderate.
FIG. 7.
Summary of functional performance and community dynamics in the methanogenic reactor. Coexistence of dominant OTUs of the Archaea and Bacteria domains is shown. Phylogenetic affiliations of the OTUs are indicated in the left column. Sampling dates for community structure analysis are indicated by arrows.
Although it is not possible to infer whether the driving force behind the community shift resides in one particular trophic level, it seems that, to achieve stability, a given arrangement among populations is more important than any one specific population. On the other hand, coexistence of the highest diversity in the Archaea domain with the lowest diversity in the Bacteria domain argues against the notion that increased diversity at one trophic level necessarily favors increased diversity for a functionally linked trophic level.
Figure 7 shows that two levels of population change can be discerned among the Bacteria 16S rDNA-defined populations: a first level where taxa from different genera (Spirochaeta, Eubacterium, and Propionibacterium) alternated in dominance and a second level where a succession within the Spirochaeta-related cluster occurred. The latter is a cyclic pattern of population substitution among group 3 spirochetes and group 1 and group 2 spirochetes. The dominant spirochetes (group 3) at days 900 and 1036 were undetected at day 977, and the prevalent OTUs in days 977 (group 1) and 1325 (group 2) were rare at day 1036.
The order Spirochaetales constitutes an expanded phylogenetic group exhibiting an rrn range of relatedness of 81.3 to 96.4% among its members (41). However, succession and coexistence of very closely related members of this order (group 1 and group 2 spirochetes) were observed in our reactor. This situation has been reported for other bacteria in natural environments, and it has been suggested that highly related populations could coexist by adaptation to environmental parameters (12).
Prevalence of spirochetes constitutes an unusual community structure for methanogenic bioreactors. Although spirochetes have been detected at a low frequency in an anaerobic reactor (14), dominance has been observed in only a few environments, notably the termite hindgut (5). All isolated spirochetes ferment glucose mainly to acetate, H2, and CO2, along with lesser and variable amounts of ethanol or lactate depending on the species (7, 23, 41). In the presence of powerful hydrogen scavengers, like methanogens, it would be expected that most of the electron flow would be routed to H2, and thus acetate and CO2 production would be favored at the expense of lactate or ethanol. Since methanogenic substrates would be produced directly from glucose, syntrophic bacteria would have an insignificant role, and the food chain would be completed with just two trophic levels. Although this particular electron flow has been proposed for natural environments where the energy input is low (28), to our knowledge it has not been described for methanogenic reactors.
Some spirochetes exhibit interesting strategies of survival during starvation that could explain their success in our reactor. The presence of purine interconversion enzymes (6) and the ability to ferment amino acids to obtain energy for functions other than growth (17) enhance their capability to survive low levels of available nutrients such as intermittent substrate feeding.
It is remarkable that a single simple substrate can maintain such a high population diversity. Presumably, the source of the emergent microorganisms is from within the reactor, since most of the prevalent microorganisms found are unlikely environmental contaminants. In addition, it is improbable that invaders could overgrow the dominant microorganisms present at such high density.
Considering that this reactor is a chemostat, it could be assumed that the transiently dominant populations grew at subdominant levels. Population redundancy, particularly among glucose fermenters, indicates that selection by competitive exclusion was precluded and suggests that a great variety of microniches subsisted. While spatial heterogeneity could be neglected, other factors like use of energy sources other than glucose, differences in kinetic parameters for substrate degradation, and differences in species capability to survive in the cyclic feed-starve cycle evidently created sufficient niche diversity to support the microbial diversity observed. Xing et al. (39) previously showed that this feeding regimen caused a shift in the predominant glucose fermentative bacterial ribotypes and in the entire-community morphotypes compared to those of a mother reactor constantly receiving the same average amount of glucose per day.
The results of the present work have shown that functional stability does not imply community stability and that an extremely dynamic community can be developed in a simple ecosystem. The large number and diversity of minority populations likely contribute significantly to these dynamics.
ACKNOWLEDGMENTS
We thank Klaus Nüsslein for assistance with ARDRA and Syed Hashsham for running the reactor during the last period.
This work was supported by NSF grant no. DEB 9120006 to the Center for Microbial Ecology. A.F. was partially supported by an OAS grant and by the Comisión Sectorial de Investigación Científica, Universidad de la República, Uruguay.
REFERENCES
- 1.Amann R I, Ludwig W, Schleifer K H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atlas R M, Horowitz A, Krichevsky M, Bej A K. Response of microbial populations to environmental disturbance. Microbiol Ecol. 1991;22:249–256. doi: 10.1007/BF02540227. [DOI] [PubMed] [Google Scholar]
- 3.Bateson M M, Ward D M. Methods for extracting DNA from microbial mats and cultivated microorganisms: high molecular weight DNA from French press lysis. In: Akkermans A D L, van Elsas J D, de Bruijn F J, editors. Molecular microbial ecology manual. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1996. pp. 1.1.4:1–7. [Google Scholar]
- 4.Borneman J, Triplett E C. Molecular microbial diversity in soils from eastern Amazonia: evidence for unusual microorganisms and microbial population shifts associated with deforestation. Appl Environ Microbiol. 1997;63:2647–2653. doi: 10.1128/aem.63.7.2647-2653.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Breznak J A. Hindgut spirochetes of termites and Cryptocercus punctulatus. In: Krieg N R, Holt J G, editors. Bergey’s manual of systematic bacteriology. Vol. 1. Baltimore, Md: The Williams & Wilkins Co.; 1984. pp. 67–70. [Google Scholar]
- 6.Canale-Parola E, Kidder G W. Enzymatic activities for interconversion of purines in spirochetes. J Bacteriol. 1982;152:1105–1110. doi: 10.1128/jb.152.3.1105-1110.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Canale-Parola E. Spirochaeta. In: Krieg N R, Holt J G, editors. Bergey’s manual of systematic bacteriology. Vol. 1. Baltimore, Md: The Williams & Wilkins Co.; 1984. pp. 39–46. [Google Scholar]
- 8.Chandler D P, Fredrickson J K, Brockman J. Effect of PCR template concentration on the composition and distribution of total community 16S rDNA clone libraries. Mol Ecol. 1997;6:475–482. doi: 10.1046/j.1365-294x.1997.00205.x. [DOI] [PubMed] [Google Scholar]
- 9.DeLong E F. Archaea in coastal marine environments. Proc Natl Acad Sci USA. 1992;89:5685–5689. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farrelly V, Rainey F A, Stackebrandt E. Effect of genome size and rrn gene copy number on PCR amplification of 16S rRNA genes from a mixture of bacterial species. Appl Environ Microbiol. 1995;61:2798–2801. doi: 10.1128/aem.61.7.2798-2801.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferris M J, Nold S C, Revsbech N P, Ward D M. Population structure and physiological changes within a hot spring microbial mat community following disturbance. Appl Environ Microbiol. 1997;63:1367–1374. doi: 10.1128/aem.63.4.1367-1374.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferris M J, Ward D M. Seasonal distributions of dominant 16S rRNA-defined populations in a hot spring microbial mat examined by denaturing gradient gel electrophoresis. Appl Environ Microbiol. 1997;63:1375–1381. doi: 10.1128/aem.63.4.1375-1381.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fries M R, Hopkins G D, McCarty P L, Forney L J, Tiedje J M. Microbial succession during a field evaluation of phenol and toluene as the primary substrates for trichloroethene cometabolism. Appl Environ Microbiol. 1997;63:1515–1522. doi: 10.1128/aem.63.4.1515-1522.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Godon J J, Sumstein E, Dabert P, Habouzit F, Moletta R. Molecular microbial diversity of an anaerobic digestor as determined by small-subunit rDNA sequence analysis. Appl Environ Microbiol. 1997;63:2802–2813. doi: 10.1128/aem.63.7.2802-2813.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grimm V, Schmidt E, Wissel C. On the application of stability concepts in ecology. Ecol Model. 1992;63:143–161. [Google Scholar]
- 16.Harmsen H J M, Akkermans A D L, Stams A J M, de Vos W M. Population dynamics of propionate-oxidizing bacteria under methanogenic and sulfidogenic conditions in anaerobic granular sludge. Appl Environ Microbiol. 1996;62:2163–2168. doi: 10.1128/aem.62.6.2163-2168.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harwood C S, Canale-Parola E. Spirochaeta isovalerica sp. nov., a marine anaerobe that forms branched fatty acids as fermentation products. Int J Syst Bacteriol. 1983;33:573–579. [Google Scholar]
- 18.Leadbetter J R, Breznak J A. Physiological ecology of Methanobrevibacter cuticularis sp. nov. and Methanobrevibacter curvatus sp. nov., isolated from the hindgut of the termite Reticulitermes flavipes. Appl Environ Microbiol. 1996;62:3620–3631. doi: 10.1128/aem.62.10.3620-3631.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maddison W P, Maddison D R. MacClade 3.0. Cambridge, Mass: W. P. Maddison and D. R. Maddison; 1992. [Google Scholar]
- 20.Maidak B L, Olsen G L, Larsen N, Overbeek R, McCaughey M J, Woese C R. The RDP (Ribosomal Database Project) Nucleic Acids Res. 1997;25:109–111. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Massol-Deyá A, Odelson D A, Hickey R F, Tiedje J M. Bacterial community fingerprinting of amplified 16S and 16-23S ribosomal DNA gene sequences and restriction endonuclease analysis (ARDRA) In: Akkermans A D L, van Elsas J D, de Bruijn F J, editors. Molecular microbial ecology manual. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 3.3.2:1–8. [Google Scholar]
- 22.Massol-Deyá A, Weller R, Rios-Hernandez L, Zhou J Z, Hickey R F, Tiedje J M. Succession and convergence of biofilm communities in fixed-film reactors treating aromatic hydrocarbons in groundwater. Appl Environ Microbiol. 1997;63:270–276. doi: 10.1128/aem.63.1.270-276.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pohlschroeder M, Leschine S B, Canale-Parola E. Spirochaeta caldaria sp. nov., a thermophilic bacterium that enhances cellulose degradation by Clostridium thermocellum. Arch Microbiol. 1994;161:17–24. [Google Scholar]
- 24.Rainey F A, Ward N, Sly L I, Stackebrandt E. Dependence on the taxon composition of clone libraries for PCR amplified, naturally occurring 16S rDNA, on the primer pair and the cloning system used. Experientia. 1994;50:796–797. [Google Scholar]
- 25.Rainey F A, Ward-Rainey N L, Stackebrandt E. Clostridium paradoxum DSM 7308T contains multiple 16S rRNA genes with heterogeneous intervening sequences. Microbiology. 1996;142:2087–2091. doi: 10.1099/13500872-142-8-2087. [DOI] [PubMed] [Google Scholar]
- 26.Reysenbach A L, Giver L J, Wickham G S, Pace N R. Differential amplification of rRNA genes by polymerase chain reaction. Appl Environ Microbiol. 1992;58:3417–3418. doi: 10.1128/aem.58.10.3417-3418.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 28.Schink B. Principles and limits of anaerobic degradation: environmental and technological aspects. In: Zehnder A J B, editor. Biology of anaerobic microorganisms. New York, N.Y: John Wiley & Sons, Inc.; 1988. pp. 771–846. [Google Scholar]
- 29.Schink B. Energetics of syntrophic cooperation in methanogenic degradation. Microbiol Mol Biol Rev. 1997;61:262–280. doi: 10.1128/mmbr.61.2.262-280.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suzuki M T, Giovannoni S J. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol. 1996;62:625–630. doi: 10.1128/aem.62.2.625-630.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Swofford D L. Phylogenetic analysis using parsimony (PAUP), version 3.0s. Champaign, Ill: Illinois Natural History Survey; 1991. [Google Scholar]
- 32.Tilman D. Biodiversity: populations versus ecosystem stability. Ecology. 1996;77:350–363. [Google Scholar]
- 33.Tuomi P, Torsvik T, Heldal M, Bratbak G. Bacterial population dynamics in a meromitic lake. Appl Environ Microbiol. 1997;63:2181–2188. doi: 10.1128/aem.63.6.2181-2188.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ulrike E, Rogall T, Blocker H, Emde M, Bottger E C. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 1989;17:7843–7853. doi: 10.1093/nar/17.19.7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weisburg W G, Barns S M, Pelletier D A, Lane D J. 16S ribosomal DNA amplification for phylogenetic analysis. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weisburg W G, Oyaizu Y, Oyaizu H, Woese C R. Natural relationship between Bacteroides and Flavobacteria. J Bacteriol. 1985;164:230–236. doi: 10.1128/jb.164.1.230-236.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wen-Tso L, Marsh T L, Cheng H, Forney L. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl Environ Microbiol. 1997;63:4516–4522. doi: 10.1128/aem.63.11.4516-4522.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Woese C R, Gutell R, Gupta R, Noller H F. Detailed analysis of the higher order structure of the 16S-like ribosomal ribonucleic acids. Microbiol Rev. 1983;47:621–669. doi: 10.1128/mr.47.4.621-669.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xing J, Criddle C, Hickey R. Long-term adaptive shifts in anaerobic community structure in response to a sustained cyclic substrate perturbation. Microb Ecol. 1997;33:50–58. doi: 10.1007/s002489900007. [DOI] [PubMed] [Google Scholar]
- 40.Zehnder A J B. Ecology of methane formation. In: Mitchell R, editor. Water pollution microbiology. Vol. 2. London, United Kingdom: John Wiley & Sons, Ltd.; 1978. pp. 349–376. [Google Scholar]
- 41.Zhilina T N, Zavarzin G A, Rainey F, Kevbrin V V, Kostrikina N A, Lysenko A M. Spirochaeta alkalica sp. nov., Spirochaeta africana sp. nov., and Spirochaeta asiatica sp. nov., alkaliphilic anaerobes from the continental Soda Lakes in Central Asia and the East African Rift. Int J Syst Bacteriol. 1996;46:305–312. doi: 10.1099/00207713-46-1-305. [DOI] [PubMed] [Google Scholar]
- 42.Zhou J, Fries M R, Chee-Sanford J C, Tiedje J M. Phylogenetic analysis of a new group of denitrifiers capable of anaerobic growth on toluene and description of Azoarcus tolulyticus sp. nov. Int J Syst Bacteriol. 1995;45:500–506. doi: 10.1099/00207713-45-3-500. [DOI] [PubMed] [Google Scholar]
- 43.Zhou J, Davey M E, Figueras J B, Rivkina E, Gilichinsky D, Tiedje J M. Phylogenetic diversity of a bacterial community determined from Siberian tundra soil DNA. Microbiology. 1997;143:3913–3919. doi: 10.1099/00221287-143-12-3913. [DOI] [PubMed] [Google Scholar]