Abstract
Objectives
To develop a robust phenotypic antimicrobial susceptibility testing (AST) method with a correctly set breakpoint for pretomanid (Pa), the most recently approved anti-tuberculosis drug.
Methods
The Becton Dickinson Mycobacterial Growth Indicator Tube™ (MGIT) system was used at six laboratories to determine the MICs of a phylogenetically diverse collection of 356 Mycobacterium tuberculosis complex (MTBC) strains to establish the epidemiological cut-off value for pretomanid. MICs were correlated with WGS data to study the genetic basis of differences in the susceptibility to pretomanid.
Results
We observed ancient differences in the susceptibility to pretomanid among various members of MTBC. Most notably, lineage 1 of M. tuberculosis, which is estimated to account for 28% of tuberculosis cases globally, was less susceptible than lineages 2, 3, 4 and 7 of M. tuberculosis, resulting in a 99th percentile of 2 mg/L for lineage 1 compared with 0.5 mg/L for the remaining M. tuberculosis lineages. Moreover, we observed that higher MICs (≥8 mg/L), which probably confer resistance, had recently evolved independently in six different M. tuberculosis strains. Unlike the aforementioned ancient differences in susceptibility, these recent differences were likely caused by mutations in the known pretomanid resistance genes.
Conclusions
In light of these findings, the provisional critical concentration of 1 mg/L for MGIT set by EMA must be re-evaluated. More broadly, these findings underline the importance of considering the global diversity of MTBC during clinical development of drugs and when defining breakpoints for AST.
Introduction
Shorter, more-effective and less-toxic regimens are urgently required to treat the nearly half a million people who develop rifampicin-resistant TB annually.1 To this end, EMA and FDA have approved pretomanid, in combination with bedaquiline and linezolid for the treatment of pulmonary XDR TB, based on the pre-2021 WHO definition, or treatment-intolerant or non-responsive MDR TB.2,3 This oral 26 week once-daily regimen cured 90% of patients enrolled in the Nix-TB trial in South Africa.4 More recently, similar results were obtained in ZeNix, a follow-up randomized trial examining varying doses and durations of linezolid in combination with bedaquiline and pretomanid in South Africa, Russia, Georgia and Moldova.5 Pretomanid is now also being studied in combination with other anti-TB drugs for the treatment of drug-susceptible (DS) and drug-resistant (DR) TB in other trials (SimpliciTB and TB-PRACTECAL).6
Loss-of-function mutations in the ddn, fbiA, fbiB, fbiC, fbiD, and fgd1 genes, the products of which are responsible for the activation of the nitroimidazoles pretomanid and delamanid, typically result in large MIC increases and, consequently, probably clinical resistance.7,8 Importantly, intrinsic resistance to delamanid (i.e. resistance not due to selection by nitroimidazole treatment) is known to have evolved recently and independently in individual strains due to mutations in at least five of these resistance genes.9,10 The role of ndh, the seventh delamanid resistance gene, in pretomanid susceptibility is not yet established.11 Even though not all mutations in these genes affect pretomanid and delamanid susceptibility equally, this means that some intrinsic resistance to pretomanid likely also exists in treatment-naive patients.12 This highlights the need for widely available, robust AST for treatment optimization/follow-up and surveillance purposes.13
Phenotypic AST of Mycobacterium tuberculosis for pretomanid has been the subject of several previous studies, using various liquid and solid media methodologies.14–17 In most studies, only the reference strain H37Rv or H37Rv-derived mutants were tested. The few analyses that included strains of other members of the M. tuberculosis complex (MTBC) were not aimed at setting an epidemiological cut-off value (ECOFF).16,18 More recently, pretomanid MIC testing from 56 baseline isolates from Nix-TB (the data from which are included in this study) showed MICs between 0.03 and 1 mg/L using the BACTEC™ Mycobacterial Growth Indicator Tube™ (MGIT) system by Becton Dickinson. These results informed the provisional critical concentration (CC; see Supplementary data at JAC Online for an overview of the different terminology used for breakpoints in the TB field) of 1 mg/L included in the Summary of Product Characteristics by EMA.2
In this study, we developed a standardized protocol for routine phenotypic AST of MTBC for pretomanid, using MGIT with the EpiCenter™/TB eXiST™ software.19 We defined a quality control (QC) range for this assay and determined the pretomanid MICs for a phylogenetically diverse collection of 356 MTBC strains from pretomanid-naive and likely delamanid-naive patients to re-evaluate the provisional CC of 1 mg/L.2
Methods
Study design and strains
This study was designed to establish the pretomanid MIC range and the inter-laboratory variability for the M. tuberculosis reference strain H37Rv (ATCC 27294); and to determine the phenotypically wild-type (pWT) MIC distribution, using the MGIT method, to provide the data required to define an ECOFF. As per the EUCAST SOP 10.2, MICs for at least 100 strains from five laboratories, each contributing at least 15 strains, are needed to set an ECOFF.20 Phylogenetically diverse MTBC strains (Table 1) were tested at six laboratories: University College London Centre for Clinical Microbiology, London; Emerging Bacterial Pathogens Unit, IRCCS San Raffaele Scientific Institute, Milan; Research Center, Borstel; Supranational Reference Laboratory for TB, Stockholm; Instituto de Higiene e Medicina Tropical, Universidade Nova de Lisboa, Lisbon; and Stellenbosch University, Cape Town. At the London laboratory, a preliminary pretomanid MIC validation included the testing of 11 known pretomanid-resistant strains (Table S1). For the main study, the London laboratory tested a total of 148 strains including those from baseline cultures of patients participating in TB Alliance trials for pretomanid-containing regimens, for which it was serving as the central microbiology laboratory: Nix-TB (n = 56); ZeNix (n = 11), STAND (n = 35) and SimpliciTB (n = 15).4–6,21 These were supplemented with additional strains provided by B. Kreiswirth, Centre for Discovery and Innovation, Hackensack Meriden Health (n = 24), and strains obtained from BCCM/ITM (n = 7).22 For the first part of the study, the Borstel, Lisbon, Milan and Stockholm laboratories each tested a minimum of 20 strains (15 DS-TB and 5 DR-TB) from their own collections, including at least five H37Rv replicates. Analysis of these data suggested lower pretomanid susceptibility for lineage 1 (L1) strains, and thus a further round of testing was carried out in Borstel (n = 24), Cape Town (n = 22), Milan (n = 18), and Stockholm (n = 26), focusing on L1 strains to ensure the study included MICs for at least 20 L1 strains from five laboratories.
Table 1.
Overview of MTBC members included in this study
| Group | MTBC membera | Lineageb | Lineage synonymc | Major spoligotype(s)c | No. | Resistance (%) by resistance type | |||
|---|---|---|---|---|---|---|---|---|---|
| DS-TB | DR-TB | MDR-TB | XDR-TB | ||||||
| A | M. africanum | 5 | West African I | AFRI2 | 2 | 100 | 0 | 0 | 0 |
| 6 | West African II | AFRI1 | 3 | 100 | 0 | 0 | 0 | ||
| M. bovis d | 2 | 0 | 100 | 0 | 0 | ||||
| M. bovis BCGe | 1 | 0 | 100 | 0 | 0 | ||||
| M. caprae | 1 | 100 | 0 | 0 | 0 | ||||
| M. microti | 1 | 100 | 0 | 0 | 0 | ||||
| M. pinnipedii | 1 | 100 | 0 | 0 | 0 | ||||
| B | M. tuberculosis | 2 | East Asian | Beijing | 63 | 17 | 2 | 51 | 31 |
| 3 | East African-Indian | CAS | 12 | 92 | 0 | 8 | 0 | ||
| 4 | Euro-American | Haarlem, LAM, T, X | 113 | 42 | 9 | 36 | 13 | ||
| 7 | 3 | 100 | 0 | 0 | 0 | ||||
| C | M. tuberculosis | 1 | Indo-Oceanic | EAI | 127 | 64 | 10 | 24 | 2 |
| D | M. canettii f | 21 | 0 | 100 | 0 | 0 | |||
| Probably resistantg | M. tuberculosis | 1 | Indo-Oceanic | EAI | 4 | 100 | 0 | 0 | 0 |
| 2 | East Asian | Beijing | 2 | 50 | 0 | 50 | 0 | ||
In accordance with current CLSI guidelines, we considered M. canettii to be part of MTBC, although this classification is disputed.54,55
Based on Coll et al.29
Based on Gagneux & Small.56
Largely intrinsically resistant to pyrazinamide.31
Intrinsically resistant to pyrazinamide and cycloserine.52
Intrinsically resistant to pyrazinamide.31
See Table 2 for more details.
Antimicrobial agents
Pretomanid powder was provided by the TB Alliance to all participating laboratories. Stock solutions of 4 mg/mL were prepared in DMSO, and frozen at −20°C in aliquots for up to 6 months in either glass or polypropylene tubes. Frozen stock solutions were thawed once and discarded afterwards; working solutions were not stored. For routine pretomanid MIC testing a range of serial 2-fold drug dilutions were prepared in DMSO from 0.03 to 1 mg/L. If the MIC obtained was outside of this range, the test was repeated at higher (0.5–8 mg/L) or lower (0.004–0.06 mg/L) concentrations to avoid truncating the pWT MIC distribution. In some cases, higher range repeats were performed up to 64 mg/L. For H37Rv, the concentrations tested were serial 2-fold dilutions from 0.03–0.5 mg/L, based on validation data obtained from the London laboratory.
MIC in MGIT
MICs were determined in all laboratories using MGIT 960 connected to an EpiCenter™ equipped with TBeXIST module (Becton Dickenson), according to the manufacturer’s instructions.23 Preparation of seed cultures (from liquid or solid media) and inocula, and the inoculation of MGIT tubes for MIC testing were performed as described previously.24 The drug-free growth control tube was inoculated with a 1:100 dilution of the bacterial suspension used for the drug-containing tubes (no DMSO was added to the drug-free control tube). The pretomanid MIC was defined as the lowest drug concentration for which the growth units (GU) were <100, at the point when the drug-free growth control reached 400 GU and therefore the test was completed. H37Rv was included as a control in each batch of clinical strains. For any tests where the MIC result was ≥1 mg/L, a blood agar culture and acid-fast stain was prepared to confirm the absence of contamination, and the test was repeated for confirmation of the result. In addition, MGIT tubes and supplement batches were subjected to QC as outlined in the MGIT 960 System User’s Manual.25
DNA extraction and WGS analysis
Genomic DNA was extracted using the CTAB or phenol/chloroform methods and prior to sequencing, DNA was quantified using Qubit dsDNA kits (Life Technologies).26,27 WGS was performed using different sequencing platforms (Illumina; MiSeq, HiSeq or NextSeq; Ion Torrent) according to manufacturer’s instructions and local validated protocols in participating laboratories. All sequences have been deposited (Table S2).
Analysis of the WGS data was performed as described previously.28 Only strains showing coverage of at least 20× (mean read depth) were included in the analysis. All variant calls relative to the H37Rv reference genome were determined for the six (ddn, fbiA, fbiB, fbiC, fbiD and fgd1) known pretomanid resistance genes. Site statistics were generated using SAMtools mpileup and gene annotation generated using snpEff software. In addition, lineages based on Coll et al.29 and key resistance-determining genes for first- and second-line drugs were determined using MTBseq (v1.0.3).30 To standardize drug resistance data across laboratories, strains were classified as DS-TB, MDR-TB, XDR-TB (using the pre-2021 WHO definition), or DR-TB (i.e. resistant to at least one agent without meeting the definition for MDR-TB or XDR-TB) according to the genotypic resistance data generated by MTBseq, taking the intrinsic resistance of Mycobacterium canettii to pyrazinamide into consideration (Table S2).3,31 All sequences were also used to reconstruct a study-wide, maximum likelihood phylogenetic tree using IQ-TREE (v2.0.3) with a General Time Reversible model of nucleotide substitution (model selection restricted to those supported by RAxML); branch support values were determined using 1000 bootstrap replicates (Figure 2c).28
Figure 2.
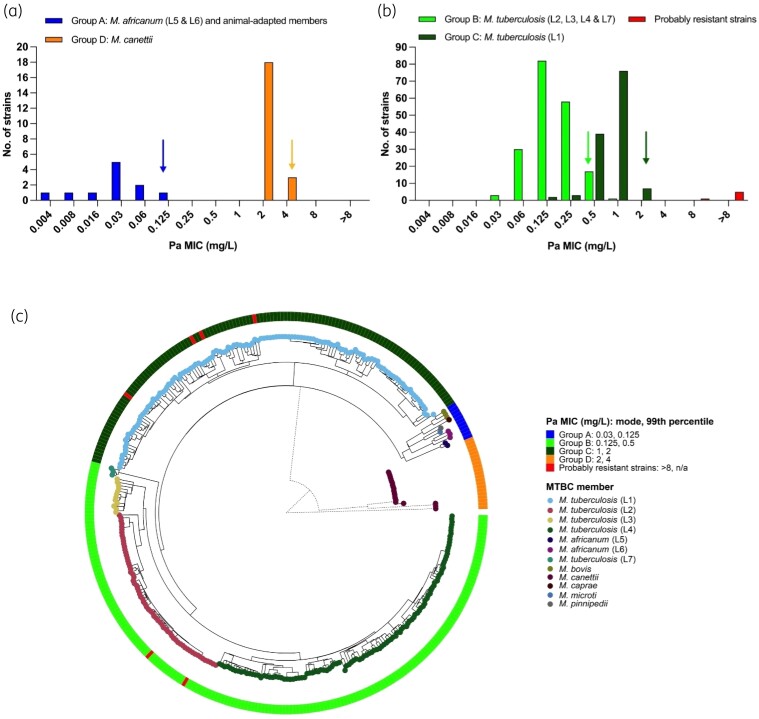
Pretomanid (Pa) MIC distributions for (a) Groups A and D, and (b) Groups B and C (excluding H37Rv). The 99th percentiles for Groups B and C based on ECOFFinder and visual estimation for Groups A and D are indicated with arrows. (c) The relationship between these MICs, represented by the heat map on the outer circle, and the underlying MTBC phylogeny (BCG is shown as part of M. bovis). The phylogenetic tree was rooted at the midpoint. The M. canettii branch lengths (dotted lines) were scaled to be 5× shorter for the purpose of visualization. An unscaled tree is shown in Figure S1.
Results and discussion
H37Rv QC range and in vitro control mutants
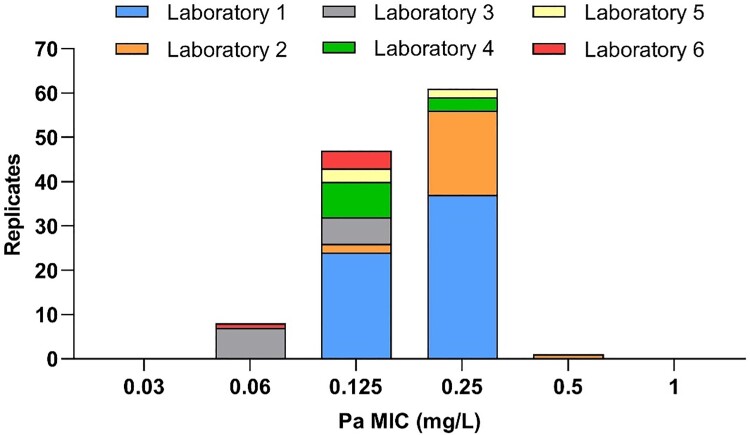
H37Rv was included in every batch of clinical strains tested in the MGIT system throughout this study. Based on 117 replicates of H37Rv from six laboratories, testing was found to be reproducible with 99% (95% CI 95%–100%) of replicates falling in a three-dilution range (0.06–0.25 mg/L; Figure 1). In addition, 11 in vitro resistant control strains all had MICs of at least 16 mg/L (Table S1), consistent with previous results obtained when testing these mutants with delamanid or pretomanid.8,16,32,33
Figure 1.
Pretomanid (Pa) MIC distribution for the M. tuberculosis H37Rv laboratory reference strain (n = 117).
Clinical strains
We conducted a multicentre study based on the EUCAST standard operating procedures for setting ECOFFs (SOP 10.2).20 Each of the six laboratories tested between 20 and 148 strains. A total of 356 strains were tested that featured all major lineages of MTBC and key animal-adapted members, as well as Mycobacterium canettii (Table 1). Only lineages 8 and 9, which are exceptionally rare and had not been described when this study started, and some uncommon animal-adapted variants were not represented.34 46% of strains were DS, 13% DR, 30% MDR, and 10% XDR. We found that MTBC can be divided in four groups with respect to pretomanid susceptibility, from more to less susceptible: Group A = Mycobacterium africanum lineages L5 and L6, and the animal-adapted members (Mycobacterium bovis, M. bovis BCG, Mycobacterium caprae, Mycobacterium microti, and Mycobacterium pinnipedii); Group B = M. tuberculosis lineages L2, L3, L4 and L7; Group C = M. tuberculosis lineage L1; Group D = M. canettii (Figure 2 and Table S2); with modal MICs of 0.03, 0.125, 1 and 2 mg/L, respectively. The mechanism(s) for these ancient MIC differences could not be explained by polymorphisms in the six canonical resistance genes (Table S3) and did not follow an obvious evolutionary trend (Figure 2c). Interestingly, this pattern, at least with respect to L1, is not shared by delamanid.35 By contrast, the higher pretomanid MICs (≥8 mg/L), which we assume to probably confer clinical resistance, arose more recently on six independent occasions in four L1 and two L2 strains (Figure 2c), likely due to different mutations in three of the known resistance genes, of which one mutation has not been described in the literature before (Table 2).
Table 2.
Likely mechanisms of pretomanid resistance in six clinical M. tuberculosis strains [shown as probably resistant strains in red in Figure 2(b) and in the outer ring of Figure 2(c)]
| ID | Year and country of isolation | Resistance type | Pa MIC (mg/L) | Lineage | Gene | Previous reports describing mutation, if available | ||
|---|---|---|---|---|---|---|---|---|
| ddn | fbiC | fgd1 | ||||||
| 295/0857 | 2008, Portugalb | MDR-TB | 64 | 2.2.1 | Trp88Stop | Selected in vitro with Pa58; observed in three DLM-resistant clinical strains35; DLM-resistant control strain.33 | ||
| PRE_007a | 2015, Ukraine | DS-TB | >16 | 2.2.1 | Gln58Stop | Observed in three DLM-resistant L2.2.1 MDR-TB strains from Ukraine.10 | ||
| TBA_067 | 2019, Tanzania | DS-TB | 16 | 1.1.2 | Lys19Glu | Novel mutation. | ||
| PRE_004a | 2015, USAc | DS-TB | >8 | 1.1.3 | Trp27Stop | Observed in DLM-resistant L1.1.3 MDR-TB strain from Bangladesh.10 | ||
| 10213-1259 | 2012, Germany | DS-TB | 8 | 1.1.1.1 | Ala349Val | Mutation observed in seven L1.1.1 strains and predicted to be functionally significant.60 | ||
| 8844-1059 | 2010, Germany | DS-TB | >32 | 1.1.1.1 | Glu230Lys | Selected in vitro with Pa.17 | ||
DLM, delamanid; Pa, pretomanid.
Lyophilized strains available from BCCM/ITM [PRE_004 (TN34503) is ITM 501092; PRE_007 is ITM 501095].22
Patient from Moldova.
Patient from Myanmar.
Based on ECOFFinder analyses of re-weighted distributions, the 99th percentile of the MIC distribution for Group B was 0.5 mg/L compared with 2 mg/L for Group C (Figure 2b). Our study was not powered to define 99th percentile for Groups A and D, both in terms of the number of strains tested and contributing laboratories. Based on these limited data, the upper end by visual inspection of Group A was 0.125 mg/L and for Group D was 4 mg/L (Figure 2a). However, given that Group A appeared to be most susceptible and Group D (M. canettii) is exceptionally rare globally, neither group were our focus.36 Feuerriegel et al.16 had previously reported that M. canettii is less susceptible to pretomanid. Here, we tested a total of 21 M. canettii strains across two laboratories and consistently obtained MICs of 2–4 mg/L. This included the two strains tested in Feuerriegel et al.16, which were found to have MICs of 8 mg/L using MGIT in the 2011 study using a less-rigorous testing procedure than the one employed in the present study (e.g. H37Rv was not included in every batch and testing could not be repeated at the time because of the limited amounts of pretomanid available). Retesting of these strains in two laboratories as part of this study consistently yielded MICs of 2 mg/L, suggesting that the results from 2011 were incorrect (i.e. M. canettii still has intrinsically elevated MICs but to a lesser degree than previously reported).16
Implications for setting a pretomanid breakpoint
In 2020, EUCAST announced that, in line with its approach to AST for fungi and other bacteria, it would transition to setting clinical breakpoints (CBs) for MTBC using its non-commercial broth microdilution reference method against which other methods would have to be calibrated.37 This will ensure that MIC, pharmacokinetics/pharmacodynamics (PK/PD), and clinical outcome data from different studies can be compared directly or using a conversion factor when systematic MIC differences exist between media, as is the case with bedaquiline and delamanid.9 However, it is accepted that this reference method will not be implemented routinely in most settings because of the advantages offered by other methods (e.g. biosafety and labour requirements).37 For this study, which started before the endorsement of the EUCAST method, we conducted a multicentre study modelled on EUCAST SOP 10.2 and adopted the MGIT methodology given that it is semi-automated and widely used for routine AST of MTBC globally.20 A QC range spanning four dilutions was obtained, of which 92% of replicates spanned just two concentrations (0.125–0.25 mg/L; Figure 1). Moreover, approximately 95% of samples reached the cut-off for reading the MIC within the standard 13 days (i.e. few samples required an extended incubation using the EpiCenter/TB eXIST software; London laboratory, data not shown). Taken together, these results underline the need for developing a quality-assured, lyophilized pretomanid product given that preparing antibiotic solutions, as required by our procedure, is a known source of error during routine AST. In the meantime, interested laboratories can order pretomanid powder from TB Alliance and a lyophilized panel of control strains with known MGIT pretomanid MICs is available from the BCCM/ITM collection (Table S2).22
To interpret the MICs obtained from the current MGIT method, a CB has to be set to distinguish susceptible strains with a high likelihood of treatment success using the standard dosing regimen from resistant strains that are likely to fail [the susceptible, increased exposure (I) category does not apply to bedaquiline/pretomanid/linezolid (BPaL) treatment, given that dosing is fixed].38 In this context, the ECOFF plays a key role. When the MICs of a novel agent that does not share any resistance mechanism with an existing antibiotic are measured, a unimodal distribution spanning 3–5 two-fold dilutions is typically obtained, provided that the technical variability of testing is minimized.20 This MIC distribution is referred to as pWT because it is devoid of phenotypically detectable resistance to the agent in question. This does not mean that there is no biological variability in this pWT distribution but, rather, that this is masked by the intra- and inter-laboratory variability inherent in MIC testing under routine conditions (at least if the MIC is measured only once as is usually the case).39,40 As a result, any CB that divides the pWT distribution (i.e. is set below the ECOFF that corresponds to the upper end of this distribution) will result in a poor reproducibility of phenotypic AST for pWT strains. Nevertheless, this does not mean that an ECOFF automatically becomes the CB. Instead, PK/PD and clinical evidence is required to demonstrate that the proposed dosing regimen is sufficient to treat pWT strains successfully, in which case the ECOFF can be used as the CB and pWT strains become clinically susceptible.18 In this scenario, the ECOFF represents a conservative CB until sufficient evidence becomes available that phenotypically non-wild-type strains with MICs > ECOFF are treatable (i.e. during the first trials of novel agents such strains are usually too rare to investigate this question).
Unlike the classical paradigm of a unimodal pWT MIC distribution, we observed marked and ancient differences in the susceptibility of MTBC to pretomanid, which fell into four groups from more to less susceptible: Group A = M. africanum and the animal-adapted members; Group B = M. tuberculosis L2, L3, L4 and L7; Group C = M. tuberculosis L1; Group D = M. canettii (Figure 2). Most notably, we report an 8-fold increase in the modal MIC and a 4-fold increase in the ECOFF of Group C compared with Group B. Therefore, taking into consideration these new data, regulators need to reassess for which groups sufficient PK/PD and/or clinical evidence exists to classify them as susceptible and to set a single interim MGIT CB accordingly. Considering the 99th percentiles of the groups, an interim CB of 0.5 mg/L would be suitable for Groups A and B, 2 mg/L for Groups A–C, and 4 mg/L for Groups A–D. The provisional CC of 1 mg/L set by EMA is too high for Groups A and B and too low for Groups C and D.2 Fewer than 200 cases of M. canettii infection (Group D) have been described to date, mostly from patients with links to the Horn of Africa.36 Consequently, it is unlikely that robust clinical evidence for pretomanid treatment in this group will ever become available. In contrast, L1 strains (Group C) are estimated to account for 28% of the global TB burden.41 80% of those strains are in India, the Philippines, Indonesia and Bangladesh, which are amongst the top seven countries with the highest absolute number of MDR/rifampicin resistant-TB globally.41,42 As such, it is essential that data from L1 strains is now considered to re-evaluate the provisional CC set by the EMA.
In terms of clinical data, consistent with L1 strains being rare in South Africa, among the 56 Nix-TB patients for which baseline isolates were tested, only one belonged to Group C (the remaining were from Group B); that patient had a favourable outcome. Now, data are available for 38 additional patients known to be infected with L1 strains and receiving pretomanid-containing regimens in other TB Alliance trials: ZeNix (regimen BPaL; 1 patient); SimpliciTB [regimen BPa plus moxifloxacin (M) plus pyrazinamide (Z); 30 patients]; and STAND (regimen PaMZ; 7 patients).5,6,21 Regardless of the regimen, 29 of the 30 patients who completed treatment (excluding cases of early withdrawals, early death due to hepatotoxicity, and late exclusions) culture converted and had a favourable outcome (J. Timm, unpublished data), suggesting infection with L1 strains has not led to worse outcomes. More data on the effectiveness of pretomanid-containing regimens in the treatment of patients with L1 strains will be collected in the various operational studies underway in Asia (LIFT-TB).43
Target attainment analysis for pretomanid is still preliminary, but several studies in animal models and humans provided insights into its PK.15 The most important studies with human data are a population PK model developed using data from 14 studies in the pretomanid development programme and 1054 patients,44 and PK data from the Nix-TB trial (J. Nedelman, personal communication). The first analysis revealed that the median Cavg, Cmax, and C24h values in plasma for a reference subject administered 200 mg once daily of pretomanid alone in a fed condition to steady-state were 2.4, 3.2, and 1.6 mg/L, respectively. For a subset of 27 Nix-TB patients with full PK profiles at week 16 (i.e. at steady-state), who received 200 mg daily in a fed condition, the geometric means of trough Cmin and Cmax were 1.4 and 2.9 mg/L, respectively. Given that the primary site of MTBC infection is lung tissue, the blood exposure is less important than the exposure achieved at the sites of infection, particularly at the centre of granulomas.45 Studies in mice showed that efficacy of pretomanid in vivo correlated better with lung PK than with plasma PK, thanks to a 3-fold accumulation of pretomanid in the lung.46 Similar pretomanid penetration levels (4-fold increases) have been reported in lungs or lung lesions of rabbits.47 However, the precise PK/PD target of efficacy of pretomanid is not known to date, which means that regulators will have to primarily rely on the above trial data to set an interim CB. When making this decision, regulators will have to take into consideration that pretomanid has been approved as part of a three-drug regimen, which complicates the assessment of the impact of an individual component compared with agents that are used in monotherapy (although for core drugs in regimens, this is possible).48–50
Differences in the intrinsic susceptibility linked to particular genotypes has been reported previously, with intrinsic resistance of M. bovis and M. canettii to pyrazinamide, M. bovis BCG to cycloserine, and a subgroup of M. tuberculosis lineage 4 to capreomycin.51,52 More recently, it has been reported that L1 has intrinsically higher pyrazinamide MICs compared with other M. tuberculosis lineages, which may require the current MGIT CC of 100 mg/L to be raised.53 Conversely, it is also possible for some genotypes to be more susceptible than other MTBC members (e.g. the more recently derived variants of M. bovis BCG are susceptible to macrolides).52 Taken together, these results and our data on pretomanid support the requirement by EUCAST to test phylogenetically diverse MTBC strains to set ECOFFs, and highlight the importance of testing globally representative strain panels during pre-clinical development.37,52 This should not be regarded as an additional expense but an insurance policy, as it may prompt an agent to be abandoned or inform additional preclinical testing and the trial design to ensure that intrinsically less-susceptible genotypes are sampled adequately.37
Supplementary Material
Acknowledgements
We thank B. Kreiswirth, E. Nuermberger and L. Rigouts for contributing strains; the Centre for Disease Control and Prevention (J. Posey) and King Abdullah University of Science and Technology (A. Pain) for sequencing the South African strains; and C. Mendel, J. Nedelman, D. Summers and A. Upton for helpful discussions.
Funding
This work was supported by TB Alliance with funding from Australia’s Department of Foreign Affairs and Trade, the Bill & Melinda Gates Foundation, Germany’s Federal Ministry of Education and Research through KfW, Irish Aid, Netherlands Ministry of Foreign Affairs, United Kingdom Department of Health, United Kingdom Foreign, Commonwealth and Development Office, and the United States Agency for International Development. R.M.W. was supported by South African Medical Research Council baseline funding. R.M.W. and A.D. acknowledge the support from the Tuberculosis Omics Research Consortium headed by A. Van Rie [Research Foundation Flanders (FWO), under grant No. G0F8316N (FWO Odysseus)]. C.U.K. is a visiting scientist at the Department of Genetics, University of Cambridge, and a research associate at Wolfson College, University of Cambridge. C.U.K. received an observership from the European Society of Clinical Microbiology and Infectious Diseases.
Transparency declarations
C.U.K. is a consultant for the TB Alliance and the Foundation for Innovative New Diagnostics. C.U.K.’s consulting work for Becton Dickinson involves a collaboration with Janssen and Thermo Fisher Scientific. C.U.K. is collaborating with PZA Innovation. C.U.K. worked as a consultant for QuantuMDx, the Stop TB Partnership, the WHO Global TB Programme, and the WHO Regional Office for Europe. C.U.K. gave a paid educational talk for Oxford Immunotec. Hain Lifescience covered C.U.K.’s travel and accommodation to present at a meeting. C.U.K. is an unpaid advisor to BioVersys and GenoScreen. All other authors: none to declare.
Supplementary data
Additional methods detail plus Figure S1 and Tables S1 to S3 are available as Supplementary data at JAC Online.
References
- 1. World Health Organization . Global tuberculosis report 2020. https://apps.who.int/iris/handle/10665/336069.
- 2. European Medicines Agency . Dovprela: EPAR – product information. https://www.ema.europa.eu/en/documents/product-information/dovprela-epar-product-information_en.pdf.
- 3. World Health Organization . Meeting report of the WHO expert consultation on the definition of extensively drug-resistant tuberculosis. 27–29 October 2020. https://apps.who.int/iris/handle/10665/338776.
- 4. Conradie F, Diacon AH, Ngubane Net al. Treatment of highly drug-resistant pulmonary tuberculosis. N Engl J Med 2020; 382: 893–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Conradie F, Everitt D, Olugbosi Met al. High rate of successful outcomes treating highly resistant TB in the ZeNix study of pretomanid, bedaquiline and alternative doses and durations of linezolid. 11th IAS Conference on HIV Science, 2021. Late breaker OALB01LB02.
- 6. Guglielmetti L, Varaine F. The coming-of-age of bedaquiline: a tale with an open ending. Eur Respir J 2021; 57: 2100066. [DOI] [PubMed] [Google Scholar]
- 7. Kadura S, King N, Nakhoul Met al. Systematic review of mutations associated with resistance to the new and repurposed Mycobacterium tuberculosis drugs bedaquiline, clofazimine, linezolid, delamanid and pretomanid. J Antimicrob Chemother 2020; 75: 2031–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rifat D, Li S-Y, Ioerger Tet al. Mutations in fbiD (Rv2983) as a novel determinant of resistance to pretomanid and delamanid in Mycobacterium tuberculosis. Antimicrob Agents Chemother 2020; 65: e01948-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. World Health Organization . Technical report on critical concentrations for drug susceptibility testing of medicines used in the treatment of drug-resistant tuberculosis. https://apps.who.int/iris/handle/10665/260470.
- 10. Battaglia S, Spitaleri A, Cabibbe AMet al. Characterization of genomic variants associated with resistance to bedaquiline and delamanid in naive Mycobacterium tuberculosis clinical strains. J Clin Microbiol 2020; 58: e01304-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hayashi M, Nishiyama A, Kitamoto Ret al. Adduct formation of delamanid with NAD in mycobacteria. Antimicrob Agents Chemother 2020; 64: e01755-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lee BM, Harold LK, Almeida DVet al. Predicting nitroimidazole antibiotic resistance mutations in Mycobacterium tuberculosis with protein engineering. PLoS Pathog 2020; 16: e1008287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Köser CU, Maurer FP, Kranzer K. ‘Those who cannot remember the past are condemned to repeat it’: Drug-susceptibility testing for bedaquiline and delamanid. Int J Infect Dis 2019; 80S: S32–5. [DOI] [PubMed] [Google Scholar]
- 14. Stover CK, Warrener P, VanDevanter DRet al. A small-molecule nitroimidazopyran drug candidate for the treatment of tuberculosis. Nature 2000; 405: 962–6. [DOI] [PubMed] [Google Scholar]
- 15. Ahmad Z, Peloquin CA, Singh RPet al. PA-824 exhibits time-dependent activity in a murine model of tuberculosis. Antimicrob Agents Chemother 2011; 55: 239–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Feuerriegel S, Köser CU, Baù Det al. Impact of fgd1 and ddn diversity in Mycobacterium tuberculosis complex on in vitro susceptibility to PA-824. Antimicrob Agents Chemother 2011; 55: 5718–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Haver HL, Chua A, Ghode Pet al. Mutations in genes for the F420 biosynthetic pathway and a nitroreductase enzyme are the primary resistance determinants in spontaneous in vitro-selected PA-824-resistant mutants of Mycobacterium tuberculosis. Antimicrob Agents Chemother 2015; 59: 5316–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kahlmeter G. The 2014 Garrod Lecture: EUCAST - are we heading towards international agreement? J Antimicrob Chemother 2015; 70: 2427–39. [DOI] [PubMed] [Google Scholar]
- 19. Keller PM, Homke R, Ritter Cet al. Determination of MIC distribution and epidemiological cutoff values for bedaquiline and delamanid in Mycobacterium tuberculosis using the MGIT 960 System equipped with TB eXiST. Antimicrob Agents Chemother 2015; 59: 4352–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. European Committee for Antimicrobial Susceptibility Testing . MIC distributions and the setting of epidemiological cut-off (ECOFF) values. EUCAST SOP 10.2. https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/EUCAST_SOPs/2021/EUCAST_SOP_10.2_MIC_distributions_and_epidemiological_cut-off_value__ECOFF__setting_20211202.pdf.
- 21. Tweed CD, Wills GH, Crook AMet al. A partially randomised trial of pretomanid, moxifloxacin and pyrazinamide for pulmonary TB. Int J Tuberc Lung Dis 2021; 25: 305–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Belgian Coordinated Collections of Microorganisms . BCCM/ITM Mycobacteria Collection. https://bccm.belspo.be/about-us/bccm-itm.
- 23. Beckton Dickinson . BD BACTEC™ MGIT™ eXtended individual susceptibility testing for BD EpiCenter™ user manual. 2014.
- 24. World Health Organization . Technical manual for drug susceptibility testing of medicines used in the treatment of tuberculosis. https://apps.who.int/iris/handle/10665/275469.
- 25. Beckton Dickinson . Instrument user’s manual BD BACTEC MGIT 960 and BD BACTEC MGIT 320. 2017.
- 26. van Soolingen D, de Haas PE, Hermans PWet al. DNA fingerprinting of Mycobacterium tuberculosis. Methods Enzymol 1994; 235: 196–205. [DOI] [PubMed] [Google Scholar]
- 27. Warren R, de Kock M, Engelke Eet al. Safe Mycobacterium tuberculosis DNA extraction method that does not compromise integrity. J Clin Microbiol 2006; 44: 254–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Witney AA, Bateson ALE, Jindani Aet al. Use of whole-genome sequencing to distinguish relapse from reinfection in a completed tuberculosis clinical trial. BMC Med 2017; 15: 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Coll F, McNerney R, Guerra-Assuncao JAet al. A robust SNP barcode for typing Mycobacterium tuberculosis complex strains. Nat Commun 2014; 5: 4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kohl TA, Utpatel C, Schleusener Vet al. MTBseq: a comprehensive pipeline for whole genome sequence analysis of Mycobacterium tuberculosis complex isolates. PeerJ 2018; 6: e5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Loiseau C, Brites D, Moser Iet al. Revised interpretation of the Hain Lifescience GenoType MTBC to differentiate Mycobacterium canettii and members of the Mycobacterium tuberculosis complex. Antimicrob Agents Chemother 2019; 63: e00159-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Manjunatha UH, Boshoff H, Dowd CSet al. Identification of a nitroimidazo-oxazine-specific protein involved in PA-824 resistance in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 2006; 103: 431–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rancoita PMV, Cugnata F, Gibertoni Cruz ALet al. Validating a 14-drug microtitre plate containing bedaquiline and delamanid for large-scale research susceptibility testing of Mycobacterium tuberculosis. Antimicrob Agents Chemother 2018; 62: e00344-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Coscolla M, Gagneux S, Menardo Fet al. Phylogenomics of Mycobacterium africanum reveals a new lineage and a complex evolutionary history. Microb Genom 2021; 7: 000477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schena E, Nedialkova L, Borroni Eet al. Delamanid susceptibility testing of Mycobacterium tuberculosis using the resazurin microtitre assay and the BACTEC MGIT 960 system. J Antimicrob Chemother 2016; 71: 1532–9. [DOI] [PubMed] [Google Scholar]
- 36. Briquet A, Vong R, Roseau J-Bet al. Clinical features of Mycobacterium canettii infection: a retrospective study of 20 cases among French soldiers and relatives. Clin Infect Dis 2019; 69: 2003–10. [DOI] [PubMed] [Google Scholar]
- 37. Schön T, Köser CU, Werngren Jet al. What is the role of the EUCAST reference method for MIC testing of the Mycobacterium tuberculosis complex? Clin Microbiol Infect 2020; 26: 1453–5. [DOI] [PubMed] [Google Scholar]
- 38. Kahlmeter G, Giske CG, Kirn TJet al. Point-counterpoint: Differences between the European Committee on Antimicrobial Susceptibility Testing and Clinical and Laboratory Standards Institute recommendations for reporting antimicrobial susceptibility results. J Clin Microbiol 2019; 57: e01129-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mouton JW, Muller AE, Canton Ret al. MIC-based dose adjustment: facts and fables. J Antimicrob Chemother 2018; 73: 564–8. [DOI] [PubMed] [Google Scholar]
- 40. Mouton JW, Meletiadis J, Voss Aet al. Variation of MIC measurements: the contribution of strain and laboratory variability to measurement precision. J Antimicrob Chemother 2018; 73: 2374–9. [DOI] [PubMed] [Google Scholar]
- 41. Netikul T, Palittapongarnpim P, Thawornwattana Yet al. Estimation of the global burden of Mycobacterium tuberculosis lineage 1. Infect Genet Evol 2021; 91: 104802. [DOI] [PubMed] [Google Scholar]
- 42. World Health Organization . WHO global lists of high burden countries for tuberculosis (TB), TB/HIV and multidrug/rifampicin-resistant TB (MDR/RR-TB), 2021–2025. https://apps.who.int/iris/handle/10665/341980.
- 43. TB Alliance . TB Alliance with the support of the Republic of Korea announce initiative to broaden adoption and scale up of new treatments for drug-resistant tuberculosis (TB). https://www.tballiance.org.za/news/lift-tb-press-release-english.
- 44. Salinger DH, Subramoney V, Everitt Det al. Population pharmacokinetics of the antituberculosis agent pretomanid. Antimicrob Agents Chemother 2019; 63: e00907-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dartois V, Barry CE. Clinical pharmacology and lesion penetrating properties of second- and third-line antituberculous agents used in the management of multidrug-resistant (MDR) and extensively-drug resistant (XDR) tuberculosis. Curr Clin Pharmacol 2010; 5: 96–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lakshminarayana SB, Boshoff HIM, Cherian Jet al. Pharmacokinetics-pharmacodynamics analysis of bicyclic 4-nitroimidazole analogs in a murine model of tuberculosis. PLoS One 2014; 9: e105222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ernest JP, Strydom N, Dartois Vet al. Predicting pretomanid penetration into patient lesions of tuberculosis. TB Science 2020. Poster EP-TBS-81.
- 48. Rigouts L, Coeck N, Gumusboga Met al. Specific gyrA gene mutations predict poor treatment outcome in MDR-TB. J Antimicrob Chemother 2016; 71: 314–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nimmo C, Millard J, Brien Ket al. Bedaquiline resistance in drug-resistant tuberculosis HIV co-infected patients. Eur Respir J 2020; 55: 1902383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Van Deun A, Decroo T, Aung KJMet al. Mycobacterium tuberculosis borderline rpoB mutations: emerging from the unknown. Eur Respir J 2021; 58: 2100783. [DOI] [PubMed] [Google Scholar]
- 51. Walker TM, Merker M, Knoblauch AMet al. A cluster of multidrug-resistant Mycobacterium tuberculosis among patients arriving in Europe from the Horn of Africa: a molecular epidemiological study. Lancet Infect Dis 2018; 18: 431–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Köser CU, Feuerriegel S, Summers DKet al. Importance of the genetic diversity within the Mycobacterium tuberculosis complex for the development of novel antibiotics and diagnostic tests of drug resistance. Antimicrob Agents Chemother 2012; 56: 6080–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Battaglia S, Chiacchiaretta M, Stenback Gabro Vet al. Lineage 1 has elevated pyrazinamide minimum inhibitory concentrations compared with other lineages of Mycobacterium tuberculosis complex. 41st Annual Congress of the European Society of Mycobacteriology, 2021. Poster P14.
- 54. CLSI . Susceptibility Testing of Mycobacteria, Nocardia spp., and Other Aerobic Actinomycetes—Third Edition: M24. 2018. [PubMed] [Google Scholar]
- 55. Gagneux S. Ecology and evolution of Mycobacterium tuberculosis. Nat Rev Microbiol 2018; 16: 202–13. [DOI] [PubMed] [Google Scholar]
- 56. Gagneux S, Small PM. Global phylogeography of Mycobacterium tuberculosis and implications for tuberculosis product development. Lancet Infect Dis 2007; 7: 328–37. [DOI] [PubMed] [Google Scholar]
- 57. Perdigão J, Silva H, Machado Det al. Unraveling Mycobacterium tuberculosis genomic diversity and evolution in Lisbon, Portugal, a highly drug resistant setting. BMC Genomics 2014; 15: 991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hu M, Fu L, Wang Bet al. Genetic and virulence characteristics of linezolid and pretomanid dual drug-resistant strains induced from Mycobacterium tuberculosis in vitro. Infect Drug Resist 2020; 13: 1751–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Diel R, Kohl TA, Maurer FPet al. Accuracy of whole-genome sequencing to determine recent tuberculosis transmission: an 11-year population-based study in Hamburg, Germany. Eur Respir J 2019; 54: 1901154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Gómez-González PJ, Perdigao J, Gomes Pet al. Genetic diversity of candidate loci linked to Mycobacterium tuberculosis resistance to bedaquiline, delamanid and pretomanid. Sci Rep 2021; 11: 19431. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.