Abstract
Objectives
The standard of care (SOC) for the treatment of pulmonary Mycobacterium avium complex (MAC) disease (clarithromycin, rifabutin, and ethambutol) achieves sustained sputum conversion rates of only 54%. Thus, new treatments should be prioritized.
Methods
We identified the omadacycline MIC against one laboratory MAC strain and calculated drug half life in solution, which we compared with measured MAC doubling times. Next, we performed an omadacycline hollow fibre system model of intracellular MAC (HFS-MAC) exposure–effect study, as well as the three-drug SOC, using pharmacokinetics achieved in patient lung lesions. Data was analysed using bacterial kill slopes (γ-slopes) and inhibitory sigmoid Emax bacterial burden versus exposure analyses. Monte Carlo experiments (MCE) were used to identify the optimal omadacycline clinical dose.
Results
Omadacycline concentration declined in solution with a half-life of 27.7 h versus a MAC doubling time of 16.3 h, leading to artefactually high MICs. Exposures mediating 80% of maximal effect changed up to 8-fold depending on sampling day with bacterial burden versus exposure analyses, while γ-slope-based analyses gave a single robust estimate. The highest omadacycline monotherapy γ-slope was −0.114 (95% CI: −0.141 to −0.087) (r2 = 0.98) versus −0.114 (95% CI: −0.133 to −0.094) (r2 = 0.99) with the SOC. MCEs demonstrated that 450 mg of omadacycline given orally on the first 2 days followed by 300 mg daily would achieve the AUC0-24 target of 39.67 mg·h/L.
Conclusions
Omadacycline may be a potential treatment option for pulmonary MAC, possibly as a back-bone treatment for a new MAC regimen and warrants future study in treatment of this disease.
Introduction
The currently recommended standard of care (SOC) for pulmonary Mycobacterium avium complex (MAC), which consists of a combination of a macrolide, rifamycin, and ethambutol, achieves sustained sputum conversion rates of only 54% at 6 months.1,2 Moreover, the median therapy duration is 18 months; at durations longer than 12 months the sputum conversion rates fall steadily at 1% per month.1,2 Furthermore, across published cohorts, up to 70% of all treated patients reported a treatment-related adverse event and 30%–70% of patients receiving daily antimicrobial treatment permanently discontinued at least one drug in their initial regimen because of adverse events.3–5 This is reflected by patients’ disgruntlement: patients’ most important concerns with SOC are the high rate of adverse events, the long duration of therapy, and poor infection eradication rates. They often view treatment as being worse than the disease itself.6 Several years ago, one of us (T.G.) received a sad e-mail from a patient from California who had read our papers on repurposed antibiotics.7–11 She stated that given intolerance to the SOC, she would likely be dead by the time new therapies that are more tolerable were found. This illustrated a pressing issue in the minds and lives of patients. New drugs that have a high efficacy [defined here as maximal kill (Emax) based on colony forming units per mL (cfu/mL)] in pulmonary MAC, at doses easily tolerated by patients, are an emergency necessity. Recent searches have revealed that β-lactams, oxazolidinones, and next generation tetracyclines, first tested in the hollow fibre system model of pulmonary intracellular MAC (HFS-MAC), could play such a role.7–11 The pathological features of the disease that affect drug choice include: (i) pulmonary nodules and/or cavities as barriers; (ii) intracellular MAC in monocyte-lineage cells in alveoli and infected multinucleated giant cells in necrotic lesions; and (iii) a median MAC burden (range) of 1.5 × 105 (1.7 × 104–1.6 × 106) cfu/mL in cavitary lesions versus 1.0 × 103 (3.0 × 101–7.1 × 103) cfu/mL for nodular/bronchiectasis lesions.12,13 This means that to be effective, new therapies must have: (i) high intrapulmonary/intralesional penetration ratios; (ii) high intracellular penetration; and (iii) achieve a target profile efficacy ≥105 cfu/mL kill in cavitary lesions and ≥104 cfu/mL kill in nodular/bronchiectasis lesions.
Omadacycline is a tetracycline with an aminomethyl group at the C9 position, which overcomes microbial ribosomal protection proteins and efflux pump resistance mechanisms.14–16 It has been approved for the treatment of community-acquired bacterial pneumonia and acute bacterial skin and skin structure infections, and has both oral and intravenous formulations.17 Omadacycline may have fewer adverse events than other tetracyclines such as tigecycline, based on limited early data.18 Here we wanted to identify the omadacycline potency (defined as concentration or exposure mediating 50% of Emax or EC50).19 There have been a few studies on omadacycline MICs for MAC, with one study showing poor activity.20 On the other hand, omadacycline achieves higher epithelial lining fluid (ELF) concentrations than other tetracyclines: the 0–24 h area under the concentration–time curve (AUC0-24) after 100 mg/day of omadacycline was 17.23 mg·h/L versus 6.32 mg·h/L for standard dose tigecycline.21 The omadacycline penetration into alveolar macrophages versus plasma was 25.8-fold.21 Here, we tested the stability of omadacycline and determined MAC MICs in parallel, followed by exposure–effect studies in the intracellular HFS-MAC, in which we compared monotherapy effect to that of SOC. Moreover, data was also modelled using novel γ-slopes, further described in the Methods section.22–24
Materials and methods
Materials and cell lines
A full description of materials and cell lines and their catalogue numbers is available as Supplementary data at JAC Online.
Omadacycline MICs and omadacycline stability
The Praedicare standard operating procedure and quality control steps for broth microdilution assays, based on the CLSI, were used to identify the omadacycline MIC.25 We also examined the stability of omadacycline. Details of methods used for MIC determination, omadacycline stability, and drug concentrations, are shown in Supplementary data and Tables S1, S2 and S3.
HFS-MAC studies
The HFS-MAC construction has been previously described in detail in the past.2,7–11,26–28 In brief, MAC was grown in log phase to reach a bacterial density of approximately 106 log10 cfu/mL. Next, THP-1 cells were infected overnight at a multiplicity of infection of 1:10, washed to remove extracellular bacteria, and 20 mL inoculated to the peripheral compartment of each HFS-MAC with RPMI 1640 plus 2% fetal bovine serum (FBS) circulating. The targeted bacterial burden was ∼5 log10 cfu/mL on day 0. Drug treatment for HFS-MAC started 24 h later. There were two HFS-MAC replicates for each dose/condition. Omadacycline intrapulmonary pharmacokinetics of Gotfried et al.21 were used. The concentration–time profiles achieved were examined in the HFS-MAC using a half-life of 16 h and oral dosing pharmacokinetics. The exposures used in the dose–effect study are shown in Table 1. The SOC of rifabutin plus clarithromycin plus ethambutol was based on the following intrapulmonary pharmacokinetic assumptions: (i) clarithromycin 1000 mg/day AUC0-24 of 60 mg·h/L and Cmax of 8 mg/L; (ii) rifabutin 300 mg/day Cmax of 0.15 mg/L and an AUC0-24 of 1.5 mg·h/mL based on equivalence of lesion penetration which we measured for rifampicin; and (iii) the ethambutol concentrations we measured inside tuberculosis lung cavities of 39 mg·h/L and Cmax 4.0 mg/L.29–33 Peripheral compartment sampling for bacterial burden was performed on days 3, 5, 7, 10, 14, 21 and 28, and cultures were spread on Middlebrook 7H10 agar with 10% OADC. Omadacycline-resistant burden on the same day was established by the same cultures on agar supplemented with 3 × MIC of omadacycline. The central compartment sampling was performed at nine timepoints over the first 48 h, pre-dose, then 1, 8, 16, 23.5, 25, 32, 40 and 47.5 h after first dose, and a peak and trough every time bacteria were sampled (i.e. 23 timepoints).
Table 1.
Omadacycline HFS-MAC target versus achieved intrapulmonary concentrations using non-compartmental analyses (NCA)
| Regimen | Dosing frequency | Target Cmax (mg/L) | Target AUC0-24 (mg·h/L) | NCA-based Cmax (mg/L) achieved first 24 h | NCA-based AUC0-24 (mg·h/L) achieved first 24 h | First 24 h AUC/MIC achieved |
|---|---|---|---|---|---|---|
| 1 | Daily | 0.0 | 0.0 | 0 | 0 | 0.00 |
| 2 | Daily | 0.5 | 4.29 | 0.28 | 4.57 | 1.14 |
| 3 | Daily | 1.0 | 8.58 | 1.17 | 15.42 | 3.86 |
| 4 | Daily | 2.0 | 17.15 | 3.15 | 47.08 | 11.77 |
| 5 | Daily | 4.0 | 34.3 | 3.86 | 60.08 | 15.02 |
| 6 | Daily | 8.0 | 68.6 | 8.40 | 125.95 | 31.49 |
| 7 | Daily | 16.0 | 137.2 | 17.96 | 158.20 | 39.55 |
| 8 | Daily | 32.0 | 274.4 | 34.46 | 367.51 | 91.88 |
| 10 +SOC | Daily | 3.0 | 25.76 | 2.20 | 21.54 | 5.39 |
SOC, standard of care.
Pharmacokinetics/pharmacodynamics analyses
Drug concentration–time profiles in the HFS-MAC were analysed using ADAPT 5 software,34 as described in detail in Supplementary data. First, we analysed the total bacterial burden (log10 cfu/mL) in each HFS-MAC on each sampling day using an inhibitory sigmoid Emax model:
| (1) |
where Econ is bacterial burden in non-treated controls, Emax is maximal effect or kill as defined earlier, H is the Hill slope, EC is exposure (either AUC0-24 or AUC0-24/MIC), and EC50 is the exposure mediating 50% of Emax. This is defined here as the ‘traditional’ pharmacokinetics/pharmacodynamics approach (traditional here means long-established), and which we have also used in the past for HFS-MAC.19,35
Next, we introduced a new form pharmacodynamic outcome. We derived and have used a system of ordinary differential equations (ODE) to estimate bacterial decline in the HFS model for tuberculosis (HFS-TB) which was followed by application of the same ODEs in patients sputa, since trajectories of bacterial clearance are neither linear nor monotonic nor simple exponential kill slopes.22,24 For the same reasons, the ODE were then applied to another slow-growing mycobacteria (SGM), Mycobacterium kansasii.22–24 Similarly, in the past at Praedicare we have noticed that MAC trajectories on therapy are neither linear nor monotonic, and will hereby use the same form of the ODE. The MAC grow and are killed by chemotherapy as described by the ODE:
| (2) |
where r and γ are bacterial growth rate and kill rate in untreated controls and treated HFS-MAC, respectively, B is MAC bacterial burden in log10 cfu/mL and the HFS-MAC carrying capacity K = 1−B/Kmax, while time (t) on treatment is in days. This model does not presuppose a pattern of kill (linear or monotonic or otherwise) and is integrative of different sampling time-points. Equation 2 was custom fitted to the omadacycline-treated experimental HFS-MAC and untreated control data, with initial conditions derived from the model fit and carrying capacity. In Monte Carlo Experiments (MCE), stochasticity and random fluctuations in population sizes that occur with low bacterial populations were accounted for when we estimated time-to-extinction (TTE) for each therapy regimen. TTE was defined exactly as derived for HFS-TB, as time to achieve a MAC burden of 10−2 cfu/mL.22
MCE for dose selection
Steps in MCE for dose selection are described in detail in Supplementary Methods. Published population pharmacokinetic parameters and ELF concentrations identified by others were used.21,36,37
Results
MIC results and drug stability in solution
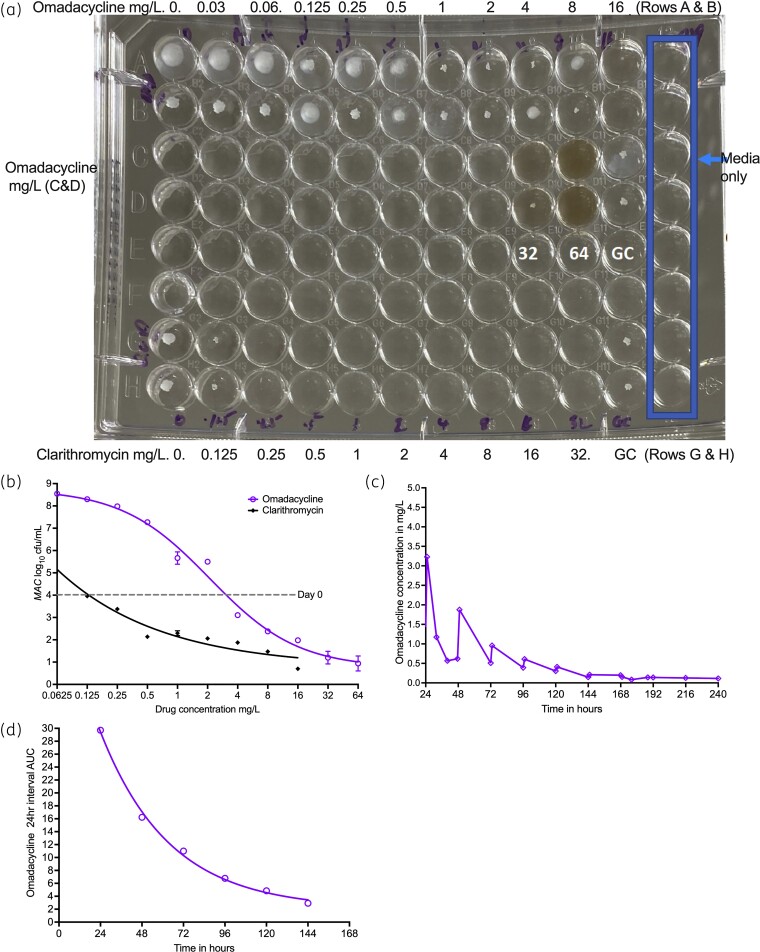
Figure 1(a) shows the broth microdilution MIC readout for omadacycline and clarithromycin QC, in Middlebrook 7H9 broth. There was a ‘trailing effect’ for the omadacycline MIC, with an MIC of 16 mg/L. Figure 1(b) shows that when the cultures were grown on agar for cfu/mL counts, and MICs were read as the lowest concentration associated with a bacterial burden below that on day 0 with and without an inhibitory sigmoid Emax-based regression line, the clarithromycin MIC remained at 0.25 mg/L, similar to the naked eye readout. Using these same criteria, the omadacycline MIC in Figure 1(b) was read as 4.0 mg/L. One possible explanation for the trailing effect could be rapid omadacycline degradation in the face of slow-growing mycobacteria, with a doubling time of similar magnitude to drug degradation. Thus, we calculated the MAC doubling time during the MIC study and identified a doubling time of 16.3 h (95% CI: 10.9–105.1), based on the exponential growth model (r2 = 0.98) of untreated controls from day 0 to day 7. Next, we repeated the MIC study using cation-adjusted Muller Hinton broth, and identified an MIC of >64 mg/L.
Figure 1.
MICs and omadacycline stability. (a) Visual inspection of plates demonstrates omadacycline trailing effect and an MIC of 16 mg/L. (b) The wells were cultured for colony forming units and results modelled using inhibitory sigmoid Emax model. The lowest concentration associated with a bacterial burden below that on day 0, which corresponded with clarithromycin MIC, was used to read the omadacycline MIC. Since the x-axis is a log-scale, and log 0 does not exist (or is undefined), the lowest concentration shown is at 0.0625 mg/L and not 0 mg/L. (c) Decline in omadacycline concentration with time when infused from the same solution in a syringe. (d) The drug AUC decline followed an exponential decline model. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
The omadacycline concentration–time profiles in the HFS-MAC, based on a solution prepared on day 1 (injected daily), are shown in Figure 1(c). The omadacycline concentrations successively dropped over time. AUC is a measure of the total amount of drug over a dosing interval (24 h in this case) and is shown in Figure 1(d). The omadacycline AUCs fell in an exponential fashion with time, with a half-life of 27.7 h (95% CI: 20.0–40.5; r2 > 0.99), in the same range as doubling time of MAC. Based on this, for subsequent HFS-MAC studies, we made the drug fresh for each day of infusion.
Drug concentrations and pharmacokinetics measured in the HFS-MAC
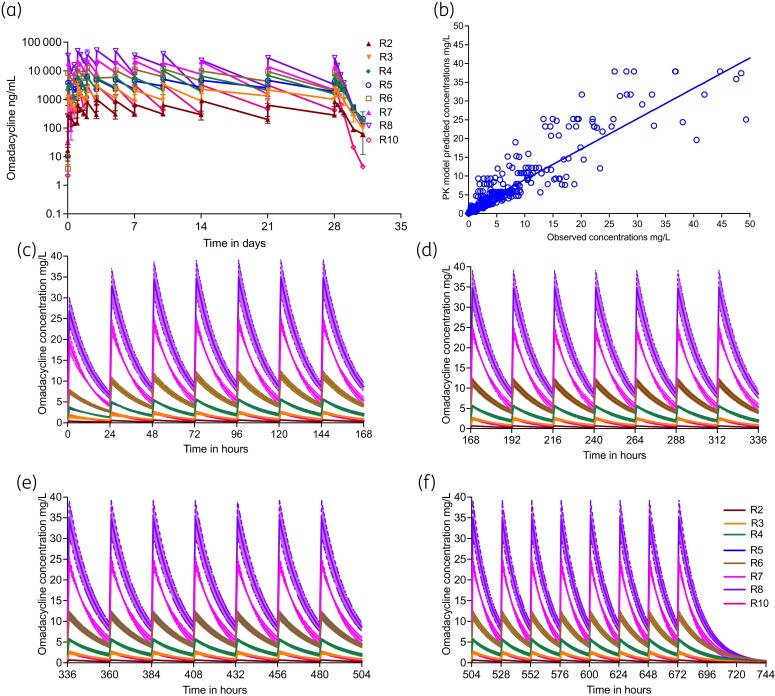
Since bacteria respond to actual drug concentrations achieved (and not to the intended drug concentrations), we measured omadacycline concentrations over 31 days. Drug concentrations were measured within 24 h of the HFS-MAC experiments with results shown in Figure 2(a) and in Table 1. Results shown in Table 1 calculate to an overestimation bias of the observed versus intended AUC0-24 of 33% (95% CI: −16% to 83%) and Cmax of 5% (95% CI: −27% to 17%); since 95% CI crosses zero the bias was statistically insignificant. This means that the strategy of making fresh doses each day worked. Compartmental pharmacokinetic analyses using a population pharmacokinetic approach demonstrated that a one-compartment model best described the HFS-MAC data. The mean pharmacokinetic parameter estimates ±SD were a clearance of 7.501 ± 8.07 × 10−3 L/h, volume 0.135 ± 0.112 L, and thus a half-life of 14.39 ± 3.39 h. The model predicted versus observed concentrations are shown in Figure 2(b). The pharmacokinetic model concentration–time profiles are broken into four 7 day intervals, which shows build up to steady-state (Figure 2c–f).
Figure 2.
Drug pharmacokinetics achieved in the HFS-MAC, equivalent to site of infection. (a) Non-compartmental pharmacokinetic analysis of omadacycline concentrations measured in each HFS-MAC unit. Because of the dynamic range, the concentrations are shown as ng/mL. The last dose was given on day 28, and then allowed to decline. (b) Compartmental pharmacokinetic model predicted versus observed concentrations [r2 = 0.84]. (c to f) The lines show the mean concentration and shaded areas 95% CIs for a compartmental pharmacokinetic model where each HFS-MAC unit was taken as a patient in a population. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
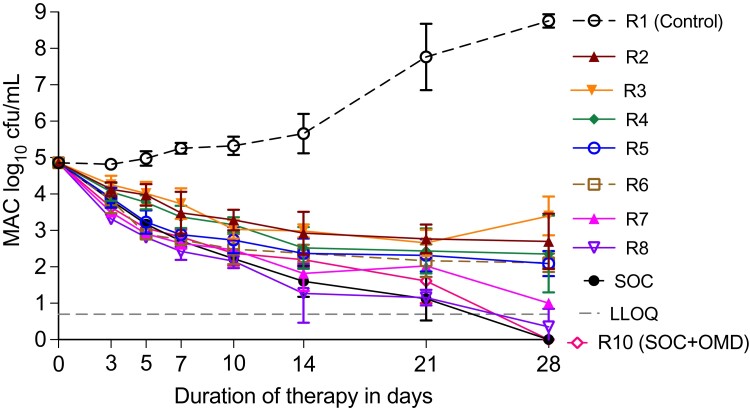
Time–kill curves for different doses and regimens
Changes in THP-1 counts are shown in Figure S1, and could be accounted for by several processes.38 Figure 3(a) shows time–kill curve for all regimens, including the three-drug SOC and SOC plus omadacycline at an AUC0-24 of 21.5 mg·h/L. Figure 3(a) shows that the highest omadacycline exposure killed ∼5.0 log10 cfu/mL as monotherapy and matched the kill of the three-drug combination SOC. Thus, despite the signal of a high MIC, omadacycline achieved high efficacy. Second, regimen 2 (R2) which has an AUC/MIC ratio of ∼1.0, killed about 2.0 log10 cfu/mL below day 0; this observed killing would not be possible unless the MIC is artefactually high. Resistance data is shown in online Supplementary data and Figure S2.
Figure 3.
Change of MAC burden with treatment in the HFS-MAC. Symbols show the mean cfu per mL and error bars are standard deviation. Particularly interesting is using the three-drug standard of care (SOC) as the context, and that high dose monotherapy has efficacy that equals that of SOC. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
Modelling the pharmacodynamic effects of each exposure using γ-slopes
Next, we modelled individual bacterial burden trajectories in the HFS-MAC using equation 2, with results shown in Table 2 and Figure S3. In addition, this approach integrates both the drug-susceptible bacterial population as well as drug-resistant subpopulation. In Table 2 for non-treated controls, r, the growth rate, is shown in place of γ-slope. All the omadacycline exposures achieved in the HFS-MAC killed MAC below day 0 and exhibited exposure-dependent γ-slope and TTE in the HFS-MAC. The highest omadacycline exposure had both γ-slope and TTE values identical to the three-drug SOC. On the other hand, adding omadacycline to SOC did not significantly improve the γ-kill slope [mean difference 0.02 (95% CI: −0.01 to 0.05), P = 0.132] in this one isolate.
Table 2.
Omadacycline and combination regimen based γ-slopes and time-to-extinction in the HFS-MAC
| Regimens | Number of observations from HFS replicates | B, initial bacterial load (95% CI) | γ-Kill slope (95% CI) | Time-to-extinction, months (95% credible intervals) |
Adjusted R2 |
|---|---|---|---|---|---|
| R1 | 16 | 4.224 (3.704–4.744) | 0.065 (0.046–0.083) | None | 0.991 |
| R2 | 16 | 4.366 (3.954–4.778) | −0.034 (−0.048 to −0.020) | 9.20 (6.13–13.44) | 0.984 |
| R3 | 16 | 4.303 (3.821–4.785) | −0.027 (−0.043 to −0.012) | 12.72 (7.31–21.00) | 0.978 |
| R4 | 16 | 4.349 (3.882–4.817) | −0.043 (−0.059 to −0.027) | 7.01 (4.37–10.36) | 0.978 |
| R5 | 16 | 4.105 (3.635–4.576) | −0.046 (−0.063 to −0.028) | 6.45 (4.19–9.98) | 0.975 |
| R6 | 16 | 3.973 (3.438–4.508) | −0.046 (−0.067 to −0.026) | 6.22 (4.10–9.27) | 0.966 |
| R7 | 16 | 4.212 (3.756–4.668) | −0.078 (−0.101 to −0.055) | 2.83 (2.19–4.10) | 0.978 |
| R8 | 16 | 4.401 (3.939–4.862) | −0.114 (−0.141 to −0.087) | 2.57 (2.10–3.05) | 0.976 |
| R9 (SOC) | 16 | 4.689 (4.345–5.032) | −0.114 (−0.133 to −0.094) | 2.60 (2.30–2.96) | 0.988 |
| R10 (SOC + omadacycline) | 16 | 4.472 (4.006–4.939) | −0.091 (−0.115 to −0.068) | 3.28 (2.64–4.37) | 0.977 |
r, the growth rate, was used instead of γ-kill slopes for the untreated control arm.
Pharmacokinetics/pharmacodynamics modelling using traditional approaches versus γ-slopes and TTE
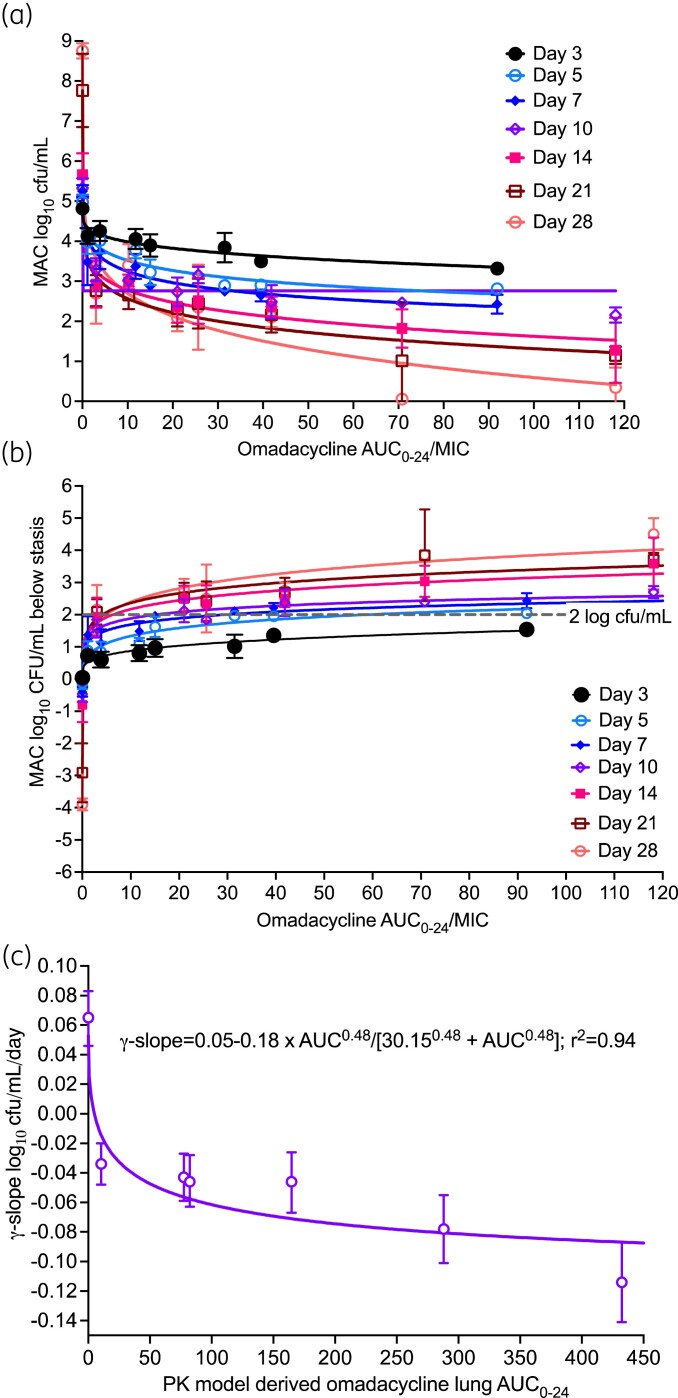
Inhibitory sigmoid Emax model parameter estimates for AUC0-24/MIC versus bacterial burden (equation 1) on each sampling day are shown in Figure 4(a) and Table 3; for week 1 we used AUC0-24 from day 1, while the rest of the weeks used steady-state AUCs in Figure 3(d–f) to calculate the AUC/MIC ratio. Table 3 shows that, as expected, the Econ and Emax will change with sampling time-point; however, so did the EC50 and EC80. Another important pharmacokinetics/pharmacodynamics parameter that is often examined, especially to allow comparisons of the effect of different compounds is microbial kill below day 0 or stasis, with microbial kill of >2.0 log10 cfu/mL indicating ‘cidal’ activity. Inhibitory sigmoid Emax model parameter estimates for AUC0-24/MIC versus microbial kill below stasis are shown in Figure 4(b) and Table 3. Besides the poor model convergence, hence a lot of imprecise pharmacokinetics/pharmacodynamics parameter estimates, it can be seen from Figure 4(b) that all exposures except one achieved microbial kill of >2.0 log10 cfu/mL. However, each one achieved that target on different sampling days. Moreover, the EC50 derived differed dramatically from those in Figure 4(a). Several questions arise from Figures 4(a,b) and Table 3, and the fact that MAC is a slow-growing mycobacteria (SGM) treated over long durations of therapy. (i) Which sampling day should be used for pharmacokinetics/pharmacodynamics parameter values such as EC50 and EC80, potency parameter values used for optimal dose selection, since these vary with time during repetitive sampling by over 800%? (ii) Which microbial kill parameter (bacterial burden or kill below stasis) should be used to define potency? (iii) When, during the 28 day repetitive sampling, should the exposure associated with >2.0 log10 cfu/mL be calculated, since it changes by sampling day? Our solution is use of γ-slopes.
Figure 4.
Traditional versus γ-slope-based pharmacokinetics/pharmacodynamics approaches for omadacycline. (a and b) Symbols are mean log10 cfu/mL and error bars are standard deviation; on days 3 and 7 the datapoints for the higher AUC MIC exposures were automatically eliminated by the program as outliers. Three important findings are the change of Emax, EC50, and Econ with each sampling day. Particularly, in (b), the 2.0 log10 cfu/mL kill is achieved at different exposures depending on sampling day and the exposure value varies almost 50-fold depending on which day is used for the regression. (c) The error bars show the 95% CI. In the γ-slope-base pharmacokinetics/pharmacodynamics modelling, a single equation and single set of parameter estimates summarized all data points. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
Table 3.
Inhibitory sigmoid Emax parameter estimates using two different MAC burden endpoints for each sampling day
| E con (log10 cfu/mL) | E max (log10 cfu/mL) | H | EC50 (AUC0-24/MIC) | EC80 (AUC0-24/MIC) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Bacterial burden on each sampling day | Mean | 95% CI | Mean | 95% CI | Mean | 95% CI | Mean | 95% CI | Mean | 95% CI | r 2 |
| Day 3 | 4.67 | 4.30–5.02 | 1.5 | 1.36–1.64 | 0.61 | 0.26–1.49 | 11.23 | 0.77–55.69 | 108.98 | 7.47–540.46 | 0.83 |
| Day 5 | 4.78 | 4.39–5.14 | 2.17 | 1.54–2.80 | 0.73 | 0.42–1.24 | 5.54 | 1.17–19.8 | 37.00 | 7.81–132.25 | 0.89 |
| Day 7 | 5.11 | 4.63–5.55 | 2.83 | 2.09–3.17 | 0.46 | 0.22–0.83 | 1.47 | 0.07–9.29 | 29.93 | 1.43–189.17 | 0.95 |
| Day 10 | 5.24 | 4.80–5.64 | 3.17 | 2.31–4.03 | 0.48 | 0.22–0.92 | 1.56 | 0.06–6.88 | 28.02 | 1.08–123.56 | 0.81 |
| Day 14 | 5.52 | 4.77–6.21 | 4.39 | 3.84–4.94 | 0.45 | 0.17–0.83 | 2.49 | 0.03–16.36 | 54.21 | 0.65–356.20 | 0.89 |
| Day 21 | 7.58 | 6.58–8.45 | 6.62 | 4.42–8.82 | 0.42 | 0.10–1.17 | 0.50 | <0.01–3.77 | 13.57 | <0.02–102.29 | 0.92 |
| Day 28 | 8.41 | 7.10–9.49 | 8.41 | 7.06–9.76 | 0.47 | 0.17–0.85 | 1.37 | 0.01–8.56 | 31.99 | 0.23–199.89 | 0.92 |
| Burden decline below stasis | |||||||||||
| Day 3 | 0.07 | 0–0.42 | −458.80 | Imprecise | 0.26 | 0.12–0.88 | Imprecise | Imprecise | Imprecise | Imprecise | 0.83 |
| Day 5 | −0.10 | −0.53 to 0.34 | −4.40 | Imprecise | 0.36 | 0.13–1.04 | 74.69 | Imprecise | 3512 | 283–3.1 × 106 | 0.89 |
| Day 7 | −0.39 | −0.86 to 0.08 | −6.76 | Imprecise | 0.19 | 0.07–0.66 | 693.00 | Imprecise | Imprecise | Imprecise | 0.90 |
| Day 10 | −0.47 | −0.85 to 0.08 | −5.28 | Imprecise | 0.21 | 0–0.87 | 27.83 | Imprecise | Imprecise | Imprecise | 0.95 |
| Day 14 | −0.79 | 0–0.01 | −264.50 | Imprecise | 0.13 | 0–0.73 | Imprecise | Imprecise | Imprecise | Imprecise | 0.89 |
| Day 21 | −2.91 | Imprecise, −1.88 | −109.10 | Imprecise | 0.08 | 0.03–0.99 | Imprecise | Imprecise | Imprecise | Imprecise | 0.92 |
| Day 28 | −3.89 | Imprecise, −2.95 | −634.50 | Imprecise | 0.09 | 0.05–0.42 | Imprecise | Imprecise | Imprecise | Imprecise | 0.94 |
Since our γ-slope is integrative of bacterial burden changes with time upon repetitive sampling and incorporates both omadacycline-susceptible and omadacycline-resistant bacterial burden trajectories, we analysed exposure versus γ-slope using a pharmacokinetic-model-derived AUC0-24, with results shown in Figure 4(c). Thus, all results were described by one inhibitory sigmoid Emax model for all 320 samples/observations, and a single EC50 AUC0-24 of 30.15 ± 37.6 mg·h/L which translates to an AUC0-24/MIC of 7.53. This exposure mediates a γ-slope able to kill >2.0 log10 cfu/mL in about 34 days as monotherapy.
Monte Carlo experiments
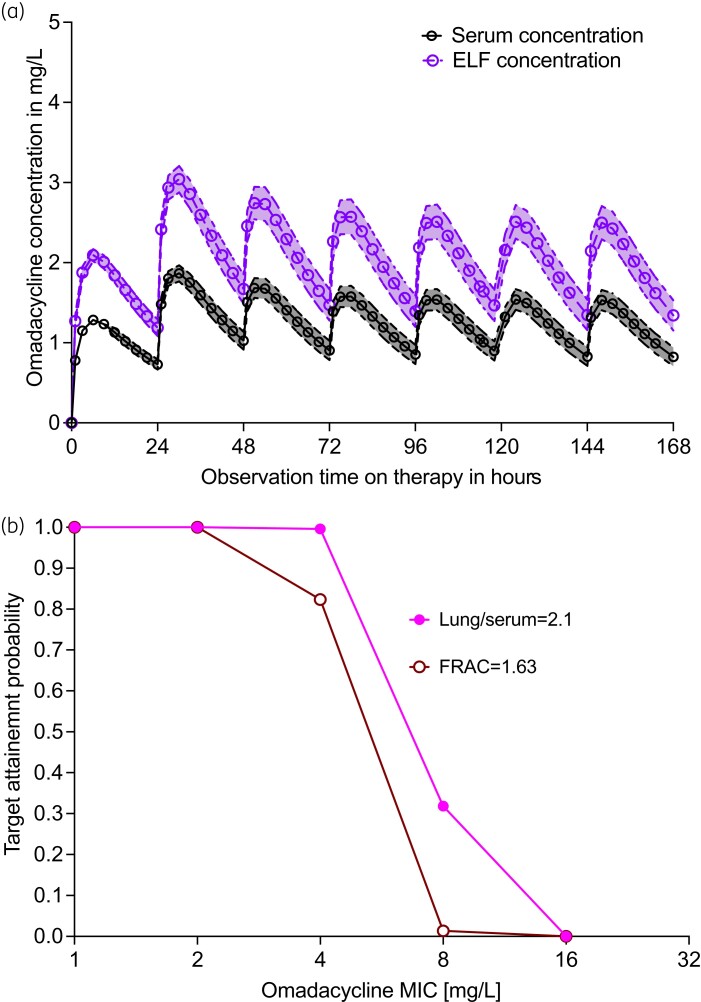
MCE were performed for the dose of 450 mg a day for 2 days, followed by 300 mg daily, administered to 10 000 patients to achieve the AUC0-24/MIC ratio of 9.92 ± 10.28 or AUC0-24 of 39.67 mg·h/L. The population pharmacokinetic parameter estimates, and variance outputs are compared with those used in the domain of input in Table 4, as part of the internal model validation step. The concentration–time profiles over 1 week (until steady-state) for these doses were as shown in Figure 5(a). When the doses were tested based on the AUC0-24 = 39.67 mg·h/L target, the probability of target attainment (PTA) was 99.61%. When the AUC/MIC target was used, the PTA at each MIC was as shown in Figure 5(b), which shows good target attainment until an MIC >8 mg/L even when two assumptions of drug penetration into the lung were used. However, because of the imprecision of current MIC assays, and since the omadacycline MIC distribution in MAC is unknown, the AUC/MIC-based PTAs should be interpreted with caution.
Table 4.
Monte Carlo simulation population pharmacokinetic parameters in 10 000 subjects
| In subroutine PRIOR | Observed in 10 000 subjects | |||
|---|---|---|---|---|
| Parameter | Median parameter estimate | %CV | Median parameter estimate | %CV |
| Clearance (L/h) | 10.3 | 22.3 | 10.31 | 22.40 |
| Central volume (L) | 21.1 | 94.1 | 21.04 | 95.28 |
| Distributional clearance P1 (L/h) | 101.0 | 65.0 | 100.70 | 65.54 |
| Peripheral Volume 1 (L) | 79.9 | 27.9 | 79.74 | 26.96 |
| Distributional clearance P2 (L/h) | 21.3 | 40 [fixed] | 21.35 | 40.06 |
| Peripheral Volume 2 (L) | 129.0 | 27.5 | 129.10 | 24.40 |
Figure 5.
Monte Carlo experiments in 10 000 subjects treated with standard dose omadacycline. (a) Symbols are mean concentrations and shaded area is the 95% CI. Concentration–time profiles in the first week are shown for serum and in the lung. (b) Target attainment probability for 10 000 patients showing proportion of patients who achieve target AUC/MIC exposures under two different drug penetration criteria. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
Discussion
Omadacycline MICs for rapidly growing mycobacteria (RGMs) have an MIC50 value of 0.004–0.12 mg/L, after 3 days of incubation, which suggests good potency.20,39 On the other hand, for SGM, including MAC strains, the MIC50 values were >16 mg/L after 7–8 days incubation.20 Our first major finding is that these high MICs could be a reflection of the mismatch between omadacycline degradation rate in solution versus the slow doubling time of SGM, whereas RGMs, with much shorter doubling times, are exposed to higher concentrations in the first 48 h. This suggests that in the case of SGM, omadacycline MICs could be unreliable tests of potency.40 This means that newer methods of susceptibility assays for omadacycline effect against SGM need to be developed. Until then, correct AUC/MIC exposure ratios cannot be reliably calculated.
Second, when omadacycline solution was prepared fresh each day and administered into the HFS-MAC, it demonstrated considerable potency and efficacy. The MCE predicted that the standard clinical doses of oral omadacycline of 450 mg a day for 2 days, followed by 300 mg daily, would be bactericidal in the treatment of pulmonary MAC. In Table 5, we compared the microbial kill below stasis of all drugs tested in the HFS-MAC that have been published. Table 5 shows that omadacycline is the most efficacious drug published to date in this model. We propose the next steps as identification of a second drug that can be either additive or synergistic with omadacycline, and can also kill at least 2 log10 cfu/mL, creating a backbone that can kill at least 4.0 log10 cfu/mL at doses tolerable to patients. Rifamycins (rifabutin, rifapentine), associated with >2 log10 cfu/mL kill, as depicted in Table 5, and shown to have concentration-dependent additivity with other tetracyclines in treatment of different infections, even in the setting of biofilm,41–43 are potential companion drugs for omadacycline, especially when rifamycin doses are optimized for the combination. A third agent, preferably dose-optimized for additivity or synergy with both omadacycline and the rifamycin, at doses easily tolerated by patients, can then be added. This approach possibly makes omadacycline the backbone of a new regimen to treat MAC, whose γ-slopes and TTE can then be compared with the SOC in in vivo models and clinical trials.
Table 5.
Comparing the efficacy of omadacycline with those of other drugs in the HFS-MAC
| Drug | E max (log10 cfu/mL) | Kill below stasis (log10 cfu/mL) at maximal effect | Comment |
|---|---|---|---|
| Azithromycin27 | 2.11 | 0.60 | High dose required to achieve Emax.53 |
| Ethambutol54 | 0.79 | – | Bacterial burden does not decrease below day 0 value. |
| Moxifloxacin55 | 3.03 | 3.00 | MICs for clinical isolates too high to be useful.55 |
| Linezolid10 | 2.10 | 1.06 | Poorly tolerated by patients. |
| Tedizolid9 | 3.78 | 2.07 | Tolerability in long therapy duration unknown. |
| Ceftazidime/avibactam7 | 2.71 | 2.44 | Short half-life; requires frequent dosing per day. |
| Rifabutin49 | 4.29 | 3.59 | Potential combination; good intracellular penetration. |
| Rifampicin49 | 2.69 | 2.68 | Potential combination; poor intralesional penetration. |
| Rifapentine49 | 3.14 | 3.03 | Potential combination. |
| Minocycline11 | 7.29 | 3.60 | Microbial ribosomal protection proteins and efflux pumps. |
| Omadacycline [current] | 8.41 | 4.86 | Referent. |
Third, traditional pharmacokinetics/pharmacodynamics approaches to identifying the pharmacokinetics/pharmacodynamics drivers and optimal exposures were developed as an extension of work with rapidly growing Gram-positive cocci and Gram-negative bacilli, which have doubling times of about 20 min.35,44,45 Such experiments last only a day to a few days and provide unequivocal pharmacokinetics/pharmacodynamics exposures, in a reproducible fashion. In SGM, the bacterial doubling time is often in the same range as the dosing interval of 24 h, and treatment duration lasts months. In the HFS-MAC, as in patients, repetitive sampling from the same units is the norm, which results in multiple opportunities to explore pharmacokinetics/pharmacodynamics relationships. This presents a problem with SGM since the pharmacokinetics/pharmacodynamics driver and potency could shift with therapy duration, as do decisions for synergy or antagonism in combination therapy.46–49 In addition, the pharmacokinetics/pharmacodynamics drivers and exposures often differ between resistance emergence and microbial kill. Therefore, we have developed integrative models that take into account all these factors, and are summarized using the γ-slope and TTE in patients and the HFS.23,24,32,50,51 These newly proposed pharmacodynamic outcomes are tractable, and have the virtue of being applicable to both monotherapy and combination therapies, making them ideal tools for building combination therapies. Moreover, we have mapped these between patients and the HFS, allowing us to translate findings in the HFS to the expected patient rates of change of bacterial burden and time to relapse-free cure.23,24,32,50,51
There are several limitations to our study. First, we used the laboratory ATCC strain of MAC; different clinical isolates show different responses to monotherapy and combination therapy compared with the ATCC strain, while some show the same responses as that strain. Thus, our omadacycline exposures require further generalization across different clinical isolates. Second, even though we combined a single dose of omadacycline with the SOC, a more factorial design that explores several different doses of omadacycline would be required since additivity, or synergy, or antagonism, are concentration-dependent. Third, a recent HFS-MAC study by Ruth et al.52 also examined the efficacy of a three-drug combination (azithromycin/rifampicin/ethambutol) SOC against MAC pulmonary disease in HFS-MAC and concluded that the current regimen against MAC was ineffective; indeed we also demonstrated similar poor effectiveness with azithromycin-containing SOC a few years earlier.8 In multiple HFS-MAC studies with up to 10 pulmonary MAC clinical isolates that used intra-cavitary AUCs (rifabutin, ethambutol) plus clarithromycin ELF concentrations demonstrated that this regimen worked in half the isolates, consistent with our findings here, and consistent with response in our patients on the clarithromycin/ethambutol/rifabutin SOC, but not in a one-to-one fashion (manuscripts in preparation). Fourth, it goes without saying that no pre-clinical model (mouse, HFS) translates one-to-one with patients’ responses; indeed, that is why we employ morphism mapping for translation.22–24 In fact, all preclinical models (HFS, mouse, macaque, etc) are ‘liars’, but some contain enough truth that can be quantitatively translated to the clinic. That is why we caution clinicians to not directly extrapolate current omadacycline data till clinical trials demonstrate the same.
Supplementary Material
Funding
This work was supported by an Investigator Initiated Research grant to T.G. from Paratek Pharmaceuticals (number 2020-0008-Gumbo).
Transparency declarations
All authors are employees of Praedicare Inc.
Author contributions
M.C.: design and execution of experiments. J.G.P.: γ-slope modelling and MCE. S.A.: hollow fibre system and MIC experiments. C.B. III: bioanalytics. D.H.: PK modelling and simulations. T.G.: Study design, pharmacokinetics/pharmacodynamics modelling, and dose finding MCE. All authors contributed to the manuscript, edited it and approved the final version of the manuscript.
Supplementary data
Additional Methods, resistance emergence results, PK/PD analyses, MCE for dose selection, plus Figures S1 to S3 and Tables S1 and S2 are available as Supplementary data at JAC Online.
References
- 1. Daley CL, Iaccarino JM, Lange Cet al. Treatment of nontuberculous mycobacterial pulmonary disease: An official ATS/ERS/ESCMID/IDSA clinical practice guideline: Executive summary. Clin Infect Dis 2020; 71: e1–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pasipanodya JG, Ogbonna D, Deshpande Det al. Meta-analyses and the evidence base for microbial outcomes in the treatment of pulmonary Mycobacterium avium-intracellulare complex disease. J Antimicrob Chemother 2017; 72: i3–19. [DOI] [PubMed] [Google Scholar]
- 3. Wallace RJ Jr, Brown-Elliott BA, McNulty Set al. Macrolide/Azalide therapy for nodular/bronchiectatic mycobacterium avium complex lung disease. Chest 2014; 146: 276–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jeong B-H, Jeon K, Park HYet al. Intermittent antibiotic therapy for nodular bronchiectatic Mycobacterium avium complex lung disease. Am J Respir Crit Care Med 2015; 191: 96–103. [DOI] [PubMed] [Google Scholar]
- 5. Zweijpfenning S, Kops S, Magis-Escurra Cet al. Treatment and outcome of non-tuberculous mycobacterial pulmonary disease in a predominantly fibro-cavitary disease cohort. Respir Med 2017; 131: 220–4. [DOI] [PubMed] [Google Scholar]
- 6. Shteinberg M, Boyd J, Aliberti Set al. What is important for people with NTM? An EMBARC-ELF patient survey. ERJ Open Research 2020; 7: 00807-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Deshpande D, Srivastava S, Chapagain MLet al. The discovery of ceftazidime/avibactam as an anti-Mycobacterium avium agent. J Antimicrob Chemoth 2017; 72: i36–42. [DOI] [PubMed] [Google Scholar]
- 8. Deshpande D, Srivastava S, Pasipanodya JGet al. A novel ceftazidime/avibactam, rifabutin, tedizolid and moxifloxacin (CARTM) regimen for pulmonary Mycobacterium avium disease. J Antimicrob Chemother 2017; 72: i48–53. [DOI] [PubMed] [Google Scholar]
- 9. Deshpande D, Srivastava S, Pasipanodya JGet al. Tedizolid is highly bactericidal in the treatment of pulmonary Mycobacterium avium complex disease. J Antimicrob Chemoth 2017; 72: i30–35. [DOI] [PubMed] [Google Scholar]
- 10. Deshpande D, Srivastava S, Pasipanodya JGet al. Linezolid as treatment for pulmonary Mycobacterium avium disease. J Antimicrob Chemother 2017; 72: i24–29. [DOI] [PubMed] [Google Scholar]
- 11. Ruth MM, Magombedze G, Gumbo Tet al. Minocycline treatment for pulmonary Mycobacterium avium complex disease based on pharmacokinetics/pharmacodynamics and Bayesian framework mathematical models. J Antimicrob Chemother 2019; 74: 1952–61. [DOI] [PubMed] [Google Scholar]
- 12. Ushiki A, Yamazaki Y, Koyama Set al. Bronchoscopic microsampling for bacterial colony counting in relevant lesions in patients with pulmonary Mycobacterium avium complex infection. Intern Med 2011; 50: 1287–92. [DOI] [PubMed] [Google Scholar]
- 13. Hibiya K, Shigeto E, Iida Ket al. Distribution of mycobacterial antigen based on differences of histological characteristics in pulmonary Mycobacterium avium infectious diseases–consideration of the extent of surgical resection from the pathological standpoint. Pathol Res Pract 2012; 208: 53–8. [DOI] [PubMed] [Google Scholar]
- 14. Tanaka SK, Steenbergen J, Villano S. Discovery, pharmacology, and clinical profile of omadacycline, a novel aminomethylcycline antibiotic. Bioorg Med Chem 2016; 24: 6409–19. [DOI] [PubMed] [Google Scholar]
- 15. Heidrich CG, Mitova S, Schedlbauer Aet al. The novel aminomethylcycline omadacycline has high specificity for the primary tetracycline-binding site on the bacterial ribosome. Antibiotics (Basel) 2016; 5: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Honeyman L, Ismail M, Nelson MLet al. Structure-activity relationship of the aminomethylcyclines and the discovery of omadacycline. Antimicrob Agents Chemother 2015; 59: 7044–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stets R, Popescu M, Gonong JRet al. Omadacycline for community-acquired bacterial pneumonia. N Engl J Med 2019; 380: 517–27. [DOI] [PubMed] [Google Scholar]
- 18. Gallagher JC. Omadacycline: A modernized tetracycline. Clin Infect Dis 2019; 69: S1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Deshpande D, Srivastava S, Gumbo T. A programme to create short-course chemotherapy for pulmonary Mycobacterium avium disease based on pharmacokinetics/pharmacodynamics and mathematical forecasting. J Antimicrob Chemother 2017; 72: i54–60. [DOI] [PubMed] [Google Scholar]
- 20. Brown-Elliott BA, Wallace RJ Jr. In Vitro Susceptibility Testing of Omadacycline against Nontuberculous Mycobacteria. Antimicrob Agents Chemother 2021; 65: e01947-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gotfried MH, Horn K, Garrity-Ryan Let al. Comparison of omadacycline and tigecycline pharmacokinetics in the plasma, epithelial lining fluid, and alveolar cells of healthy adult subjects. Antimicrob Agents Chemother 2017; 61: e01135-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Magombedze G, Pasipanodya JG, Srivastava Set al. Transformation morphisms and time-to-extinction analysis that map therapy duration from preclinical models to patients with tuberculosis: Translating from apples to oranges. Clin Infect Dis 2018; 67: S349–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Srivastava S, Wang J-Y, Magombedze Get al. Nouveau short-course therapy and morphism mapping for clinical pulmonary Mycobacterium kansasii. Antimicrob Agents Chemother 2021; 65: e01553-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Magombedze G, Pasipanodya JG, Gumbo T. Bacterial load slopes represent biomarkers of tuberculosis therapy success, failure, and relapse. Commun Biol 2021; 4: 664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. CLSI . Susceptibility Testing of Mycobacteria, Nocardiae, and Other Aerobic Actinomycetes–Third Edition: M24. 2018. [PubMed] [Google Scholar]
- 26. Deshpande D, Srivastava S, Musuka Set al. Thioridazine as chemotherapy for Mycobacterium avium complex diseases. Antimicrob Agents Chemother 2016; 60: 4652–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schmalstieg AM, Srivastava S, Belkaya Set al. The antibiotic resistance arrow of time: efflux pump induction is a general first step in the evolution of mycobacterial drug resistance. Antimicrob Agents Chemother 2012; 56: 4806–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Srivastava S, Deshpande D, Gumbo T. Failure of the azithromycin and ethambutol combination regimen in the hollow-fibre system model of pulmonary Mycobacterium avium infection is due to acquired resistance. J Antimicrob Chemother 2017; 72: i20–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kikuchi E, Yamazaki K, Kikuchi Jet al. Pharmacokinetics of clarithromycin in bronchial epithelial lining fluid. Respirology 2008; 13: 221–6. [DOI] [PubMed] [Google Scholar]
- 30. Traunmuller F, Zeitlinger M, Zeleny Pet al. Pharmacokinetics of single- and multiple-dose oral clarithromycin in soft tissues determined by microdialysis. Antimicrob Agents Chemother 2007; 51: 3185–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. van Ingen J, Egelund EF, Levin Aet al. The pharmacokinetics and pharmacodynamics of pulmonary Mycobacterium avium complex disease treatment. Am J Respir Crit Care Med 2012; 186: 559–65. [DOI] [PubMed] [Google Scholar]
- 32. Ordonez AA, Wang H, Magombedze Get al. Dynamic imaging in patients with tuberculosis reveals heterogeneous drug exposures in pulmonary lesions. Nat Med 2020; 26: 529–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dheda K, Lenders L, Magombedze Get al. Drug-Penetration Gradients Associated with Acquired Drug Resistance in Patients with Tuberculosis. Am J Respir Crit Care Med 2018; 198: 1208–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. D’Argenio DZ, Schumitzky A, Wang X. ADAPT 5 user's guide: Pharmacokinetic/pharmacodynamic systems analysis software. 2009. https://bmsr.usc.edu/files/2013/02/ADAPT5-User-Guide.pdf.
- 35. Craig WA. Pharmacokinetic/pharmacodynamic parameters: rationale for antibacterial dosing of mice and men. Clin Infect Dis 1998; 26: 1–10. [DOI] [PubMed] [Google Scholar]
- 36. Lakota EA, Van Wart SA, Trang Met al. Population pharmacokinetic analyses for omadacycline using phase 1 and 3 data. Antimicrob Agents Chemother 2020; 64: e02263-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lin W, Flarakos J, Du Yet al. Pharmacokinetics, distribution, metabolism, and excretion of omadacycline following a single intravenous or oral dose of 14C-omadacycline in rats. Antimicrob Agents Chemother 2017; 61: e01784-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Deshpande D, Pasipanodya JG, Srivastava Set al. Minocycline immunomodulates via sonic hedgehog signaling and apoptosis and has direct potency against drug-resistant tuberculosis. J Infect Dis 2019; 219: 975–85. [DOI] [PubMed] [Google Scholar]
- 39. Gumbo T, Cirrincione K, Srivastava S. Repurposing drugs for treatment of Mycobacterium abscessus: a view to a kill. J Antimicrob Chemother 2020; 75: 1212–7. [DOI] [PubMed] [Google Scholar]
- 40. Srivastava S, van Rijn SP, Wessels AMAet al. Susceptibility testing of antibiotics that degrade faster than the doubling time of slow-growing mycobacteria: ertapenem sterilizing effect versus Mycobacterium tuberculosis. Antimicrob Agents Chemother 2016; 60: 3193–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Szczuka E, Grabska K, Kaznowski A. In vitro activity of rifampicin combined with daptomycin or tigecycline on Staphylococcus haemolyticus biofilms. Curr Microbiol 2015; 71: 184–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Szczuka E, Kaznowski A. Antimicrobial activity of tigecycline alone or in combination with rifampin against Staphylococcus epidermidis in biofilm. Folia Microbiol (Praha) 2014; 59: 283–8. [DOI] [PubMed] [Google Scholar]
- 43. Yamazaki Y, Danelishvili L, Wu Met al. The ability to form biofilm influences Mycobacterium avium invasion and translocation of bronchial epithelial cells. Cell Microbiol 2006; 8: 806–14. [DOI] [PubMed] [Google Scholar]
- 44. Ambrose PG, Bhavnani SM, Rubino CMet al. Pharmacokinetics-pharmacodynamics of antimicrobial therapy: it’s not just for mice anymore. Clin Infect Dis 2007; 44: 79–86. [DOI] [PubMed] [Google Scholar]
- 45. Gumbo T. General principles of chemotherapy of infectious diseases. In: Brunton LLChabner B and Knollmann B, eds. Goodman & Gilman's The Pharmacological Basis of Therapeutics. McGraw Hill Medical, 2018. [Google Scholar]
- 46. Musuka S, Srivastava S, Siyambalapitiyage Dona CWet al. Thioridazine pharmacokinetic-pharmacodynamic parameters ‘Wobble’ during treatment of tuberculosis: a theoretical basis for shorter-duration curative monotherapy with congeners. Antimicrob Agents Chemother 2013; 57: 5870–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pasipanodya JG, McIlleron H, Burger Aet al. Serum drug concentrations predictive of pulmonary tuberculosis outcomes. J Infect Dis 2013; 208: 1464–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rockwood N, Pasipanodya JG, Denti Pet al. Concentration-dependent antagonism and culture conversion in pulmonary tuberculosis. Clin Infect Dis 2017; 64: 1350–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Boorgula GD, Jakkula LUMR, Gumbo Tet al. Comparison of rifamycins for efficacy against Mycobacterium avium complex and resistance emergence in the hollow fiber model system. Front Pharmacol 2021; 12: 645264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Deshpande D, Magombedze G, Srivastava Set al. Once-a-week tigecycline for the treatment of drug-resistant TB. J Antimicrob Chemother 2019; 74: 1607–17. [DOI] [PubMed] [Google Scholar]
- 51. Srivastava S, Deshpande D, Magombedze Get al. Duration of pretomanid/moxifloxacin/pyrazinamide therapy compared with standard therapy based on time-to-extinction mathematics. J Antimicrob Chemother 2019; 75: 392–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ruth MM, Raaijmakers J, van den Hombergh Eet al. Standard therapy of Mycobacterium avium complex pulmonary disease shows limited efficacy in an open source hollow fibre system that simulates human plasma and epithelial lining fluid pharmacokinetics. Clin Microbiol Infect 2021: S1198-743X(21)00407-9. [DOI] [PubMed] [Google Scholar]
- 53. Deshpande D, Pasipanodya JG, Gumbo T. Azithromycin dose to maximize efficacy and suppress acquired drug resistance in pulmonary Mycobacterium avium disease. Antimicrob Agents Chemother 2016; 60: 2157–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Deshpande D, Srivastava S, Meek Cet al. Ethambutol optimal clinical dose and susceptibility breakpoint identification by use of a novel pharmacokinetic-pharmacodynamic model of disseminated intracellular Mycobacterium avium. Antimicrob Agents Chemother 2010; 54: 1728–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Deshpande D, Srivastava S, Meek Cet al. Moxifloxacin pharmacokinetics/pharmacodynamics and optimal dose and susceptibility breakpoint identification for treatment of disseminated Mycobacterium avium infection. Antimicrob Agents Chemother 2010; 54: 2534–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.