Abstract
Venoms have evolved >100 times in all major animal groups, and their components, known as toxins, have been fine-tuned over millions of years into highly effective biochemical weapons. There are many outstanding questions on the evolution of toxin arsenals, such as how venom genes originate, how venom contributes to the fitness of venomous species, and which modifications at the genomic, transcriptomic, and protein level drive their evolution. These questions have received particularly little attention outside of snakes, cone snails, spiders, and scorpions. Venom compounds have further become a source of inspiration for translational research using their diverse bioactivities for various applications. We highlight here recent advances and new strategies in modern venomics and discuss how recent technological innovations and multi-omic methods dramatically improve research on venomous animals. The study of genomes and their modifications through CRISPR and knockdown technologies will increase our understanding of how toxins evolve and which functions they have in the different ontogenetic stages during the development of venomous animals. Mass spectrometry imaging combined with spatial transcriptomics, in situ hybridization techniques, and modern computer tomography gives us further insights into the spatial distribution of toxins in the venom system and the function of the venom apparatus. All these evolutionary and biological insights contribute to more efficiently identify venom compounds, which can then be synthesized or produced in adapted expression systems to test their bioactivity. Finally, we critically discuss recent agrochemical, pharmaceutical, therapeutic, and diagnostic (so-called translational) aspects of venoms from which humans benefit.
Keywords: venom, modern venomics, genomics, spatial -omics, evolution, translational research, bioassays, envenomation, antivenom, toxin production
Background—Why Venoms Matter
Venomous animals fascinate and affect humankind from time immemorial and influence—often unnoticed—many cultural, ecological, and economical aspects of our life [1,2]. Venom is such a successful adaptation that is critical for the fitness of many species, that it has evolved independently >100 times, across all major animal lineages, where it is predominately used for defense or predation [3–5]. Venomous species play key roles in ecological networks in almost all natural habitats. We are just starting to understand many of these relationships, through recent advances in the knowledge of the biology of many venomous animals, the ecological implications of such a complex trait as venom, and its dynamic composition [5–7] (see Fig. 1).
Figure 1:
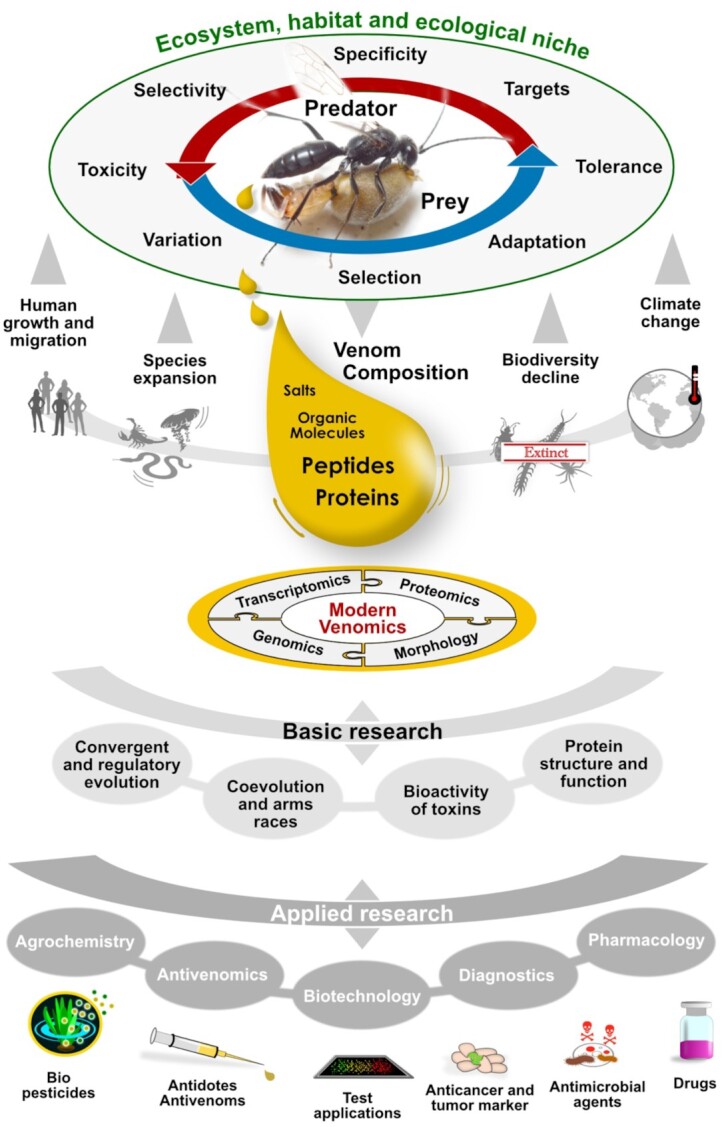
The importance and impact of venom. The biology and ecology of venomous species prompt diversity of venoms, which are constituted of highly specific toxin components that were adaptively produced over time. Predator-prey interactions are major evolutionary forces that often trigger arms races of venom toxicity and resistance. Extrinsic factors that affect venomous species and their interaction with humans include species expansion or decline (linked to the biodiversity crisis and climate change) but also the increasing human growth and migration. Basic venom research investigates why and how venoms and toxin genes evolve based on modern “omics” methods. Translational research exploits these basic studies for developing various applications, ranging from pharmacology (e.g., anti-pain and anti-cancer drugs, diagnostic markers, antivenom development) to agrochemistry (pesticides, antiparasitic compounds for crop and livestock protection) and biotechnology (e.g., nanopore sensing).
Venom is predominantly used in interspecific interaction, including both predation (such as in spiders, scorpions, centipedes, snakes) and defence (typical examples include bees, sea urchins, and fishes) [5]. In each lineage, venom components are refined—often presumed by arms races—making them highly effective disruptors of physiological processes. The co-evolutionary processes that shape venom diversity and specificity are still being studied, and especially larger, comparative studies are lacking. For few snake species first research results are available that focus on the evolution of venom resistance of rattlesnake prey, which support the arms race theory [8–12]. The remarkable target specificity of many venom compounds stimulated early interest in their potential uses for applied and translational research. As a result, molecules from a few selected taxa such as cone snails, snakes, spiders, and scorpions have been characterized in complex studies aiming at exploring their bioactivity over the course of decades [1,2]. Today, toxins are used in a variety of translational sectors including therapeutics, sustainable bioinsecticides in agrochemistry, and clinical markers in diagnostics [1,2,13–15] (see Fig. 1).
The research field in which all aspects of animal venoms such as evolution, ecology, and translational research, including antivenomics, are integratively studied is modern venomics [16]. Clinical effects of envenomations are frequently untreated because effective and cheap antivenoms, even for the most notorious snakes, spiders, scorpions, and bees, are often lacking. This is one of the urgent humanitarian challenges, especially in countries where envenomations are frequent [17–22]. Moreover, many venomous neobiota that invade new ecosystems facilitated by climate change pose threats not only to humans by increasing envenomations but also to native species and livestock [23,24].
Here we summarize current challenges and approaches on the most relevant theoretical, basic, and applied research disciplines of modern venomics (see Fig. 1). In addition, we highlight future directions and most promising innovations in methods, technology, and platforms that can contribute to animal venom research. The structure of this review reflects the typical workflow of venom studies, from the collection of venomous organisms to applied research, with the aim to be easily used as a blueprint for future venomics studies and a roadmap towards new methodological perspectives.
Collection of Venomous Organisms
Taxonomic expertise on venomous animals
Most studies on venomous animals start by sampling, identifying, and collecting specimens of venomous species. Until recently, studies almost exclusively focused on taxa that were harmful to humans, such as snakes, spiders, and scorpions, driven partially by the need to mitigate the effects of envenomations [1,5,6] (see Fig. 2). The increasing collection of so far understudied species, particularly invertebrates, raises particular attention to a general, persisting impediment that affects all branches of modern zoology. For many animal groups taxonomic expertise has been declining for decades, precluding the precise assessment of global biodiversity and its trends [25–27]. Understanding the diversity of venomous animals is also crucial to understanding venom diversity and novel protein functions. New strategies to maintain and nurture taxonomic expertise in a biodiversity-driven biodiscovery approach have relevant impact on the field of venomics, especially because venom composition can vary between even closely related species [28].
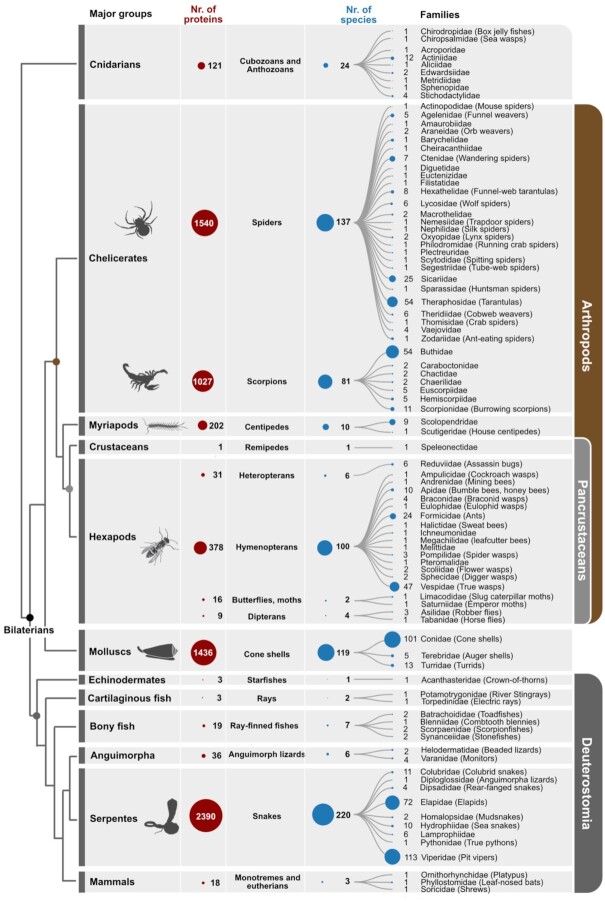
Figure 2:
Studied venomous metazoan species and available venom proteins. The numbers of studied venomous metazoans and their reviewed venom proteins that are provided in UniProt's animal venom database (ToxProt) are illustrated. Accessed on 1 April 2022, we mined 7,230 entries for venomous species. Venom protein numbers are only given for the larger taxonomic groups; the red circles are proportional to each other. Only taxa with a described venom protein are included; other metazoan species are pruned. The blue circles that show the species numbers are in proportion to each other.
Legal collection aspects
A long overdue awareness of equally shared bioresources and responsible collection of species emphasizes old and new legal aspects linked to fieldwork. Naturally, researchers obtain official permissions for fieldwork, depending on collection locality and conservation status of the target species. More demanding, however, are the rather novel rules established by the international agreement on “Access and Benefit Sharing, ABS” of the Convention of Biological Diversity. This agreement aims to standardize a legal framework for the access, transfer, utilization, and benefits of organisms (genetic resources) in a fair and equitable way for the providing country in which samples are collected [29]. The resulting Nagoya protocol is currently enforced in 131 countries worldwide [30], leading on one hand to obvious benefits, such as a legal framework that prevents biopiracy and protects biodiversity, scientists, and traditional medicines in the countries of origin. On the other hand it also implies technocratic hurdles that often hinder collaborative research and in particular translational applications [31–33]. One difficulty is that the rules for knowledge transfer or (financial) benefit from collected organisms (or molecules from these) change from member state to member state if research is published or finally translated into a commercial application. These issues should be more explicitly addressed in the framework of the critical debate around the Nagoya protocol and its implementation.
Various venom systems require different methods to obtain crude venom
The tremendously diverse venom systems in most animals and the complex anatomy of their venom apparatus [5] require different approaches to collect crude venom. A well-known venom collection method is the milking of front-fanged snakes, where the animals are forced to bite through a thin membrane and release their venom into a clean glass vessel. In contrast, rear-fanged snakes are usually injected with pilocarpine to increase salivation and the released venom is collected manually from the fangs [34]. Similar pilocarpine-based methods have been established for venomous lizards, mammals, and amphibians [34–36].
Fish venoms are often extracted from living or frozen specimens by dissecting their venom glands. Many fishes do not have distinct venom glands but clustered, venom-producing, secretory cells that end in a spine groove [37]. For those species, protocols were developed in which crude venoms are extracted through a syringe or by a forced sting into a sponge contained in a tube [38,39]. Chemical extraction from partial- and whole-body samples represents the predominant way of venom collection in many marine invertebrates including echinoderms and several cnidarians [40–43]. For cnidarians, however, alternative protocols that are based on chemically induced discharge have likewise been designed [44]. Cone snail venom can be collected by using live prey as lure or a predator as threat, which stimulates the cones to shoot their venom harpoon into microcentrifuge tubes [45,46]. Venoms of most arthropods such as centipedes, chelicerates, crustaceans, and insects are obtained by electrical, mechanical, or chemical stimulation of venom ejection or dissection of the venom system [47–52]. All these protocols have their pros and cons in terms of convenience and venom yield, but whenever possible, the most “natural” collection method should be preferred. For instance, electrostimulation is known to reveal differing venom profiles compared to manually collected venoms, calling for a cautionary interpretation of putative ecological roles of venoms without the support of further evidence (see, e.g., [53,54]). A comprehensive overview of major venom collection protocols is given in Supplementary Table S1. After collection, obtained samples of crude venom are often pre-filtered from tissue remains, then lyophilized and stored in freezers [34,55] for later analyses (see Fig. 3).
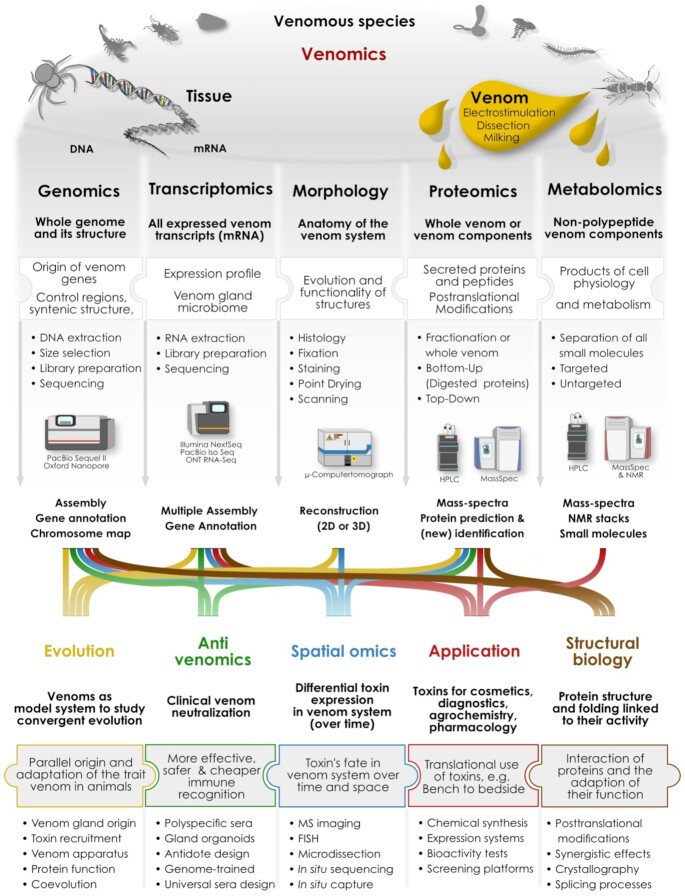
Figure 3:
The major interdisciplinary research areas in venomics. The basic, interlinked, modern research fields in venomics are shown in the first row, and linked through simplified workflows with the final output(s). The main applied and evolutionary questions addressed are shown in the bottom, and integrated in the relevant topics. The flow diagrams that connect most research areas with each other illustrate the highly integrative nature of modern venomics. FISH: fluorescence in situ hybridization.
Venom Metabolomics
Metabolitic molecules are often neglected
Metabolic profiling of venom refers to targeted and untargeted analysis of its composition of small molecular weight compounds (metabolites). These are sugars, sugar alcohols, sugar phosphates, amino acids, lipids, and nucleotides, covering primary and secondary metabolism intermediates. Small molecules in biological systems act as reactants or products in the metabolic reactions or as signalling molecules for the initiation of certain biological processes and regulatory molecules of protein function. Recent studies have indicated a surprising richness of the venom metabolic profiles across species, which need to be further explored with respect to both its biological role and potential biotechnological impact [56]. So far, venom metabolic profiling has been carried out in a targeted way, searching for molecules that have been known toxins or of potential pharmaceutical interest. However, holistic quantitative analyses of the venom metabolic composition in various species, the parameters affecting it, how the metabolite profile is related to the protein content, and commonalities and differences in the venom metabolic profile between species have not been carried out yet. In this sense, metabolic profiling of venoms is still a rather young research field, requiring further investigation [56]. It is expected that at least some of these metabolites can act as regulatory molecules or direct metabolic intermediates of biological processes in the target species of venom-producing organisms [56–58].
In the case of snake venom, the information about small molecule composition remains largely qualitative rather than quantitative. Only in the past 15 years has the presence of tens to hundreds of small molecules been reported in these venoms [59,60]. Recent studies reported ∼200 metabolites [61] or ∼50 lipids [62] in snake venoms. On the basis of these and upcoming studies, citrate has been the most abundant molecule in snake venom. It has been considered that this is the case in all venoms because citrate can protect the animal from its own toxins. Recent studies in scorpions have added an additional aspect to the high abundance of citrate because the low pH contributes to the high pain that the prey feels from the bite [63]. Moreover, all 20 amino acids have been identified in snake venom, which is another aspect to be further explored. It is expected that the concerted action of many molecules actually affects prey or predator, rather than the specific activity of certain molecules. So far, venom metabolic profiles have been mainly analysed with respect to toxin content, as in the case of acylpolyamines. This group of small neurotoxins with a molecular weight <1 kDa that inhibit glutamatergic synapses are structurally characterized in several spider genera using nuclear magnetic resonance (NMR) spectroscopy and liquid chromatography–tandem mass spectrometry (LC-MS/MS) approaches [64,65], and subsequently identified also in snake venom [60]. It has been postulated that polyamines induce hypotension and direct paralysis, facilitating prey hunting. Following the optimized workflow of these initial studies, Schroeder et al. used an untargeted NMR- and LC-MS/MS–based metabolomics approach for widespread identification of thus far undescribed small molecules in venoms of >70 different spider species [66]. They identified small-molecular polyamines, neurotransmitters, nucleosides, amino acid derivatives, and organic acids. Known and novel low molecular mass compounds from spiders are provided in VenMS, a newly available database [67].
Metabolomics studies have also been carried out on insect venoms. The venom of several species of fire ants (genus Solenopsis) contains a characteristic group of piperidine alkaloids [68], in both cis and trans stereoisomers, the trans isomer being dominant as retrieved in gas chromatography–mass spectrometry (GC-MS) [69,70]. More recent studies have been conducted on honeybees [71, 72] and wasps [73] where untargeted and targeted LC–MS(/MS) analyses identified and quantified several organic acids, amines, amino acids, and carbohydrates. Comparison between the venoms among various species and strains of the same species could provide significant insight to the evolution of venom, the actual role of the molecules inside the venom, and the effect of the season, sex, predators, and preys in its composition.
Proteome Analyses of Crude Venoms
To describe animal venoms, which are predominantly proteinaceous, state-of-the-art mass spectrometry (MS) instruments are used, even for small organisms that deliver minute amounts of venom [54,74]. MS-based venom proteomics are used for (i) general characterization of venom proteomes at the protein family level, (ii) partial or full sequencing of (purified) venom peptides and proteins, (iii) accurate mass determination of peptides and proteins either in crude venom (mass fingerprinting) or after purification, (iv) relative or absolute quantitation of venom proteins and peptides, (v) effective antivenom production (antivenomics), and (vi) 3D structure elucidation by hydrogen deuterium exchange-MS and/or cross-linking MS methods [28, 54, 75–79].
Advantages and challenges of bottom-up and top-down approaches
In general, the methodological roadmap for any proteomic analysis in venom research is split into 2 major approaches: bottom-up and top-down proteomics [80–82] (Fig. 3). In a bottom-up experiment, intact polypeptides are cleaved by proteases (generally trypsin) and the resulting peptide fragments are analysed by tandem MS. Top-down approaches in contrast describe the native form of venom proteins without any prior degradation. Thereafter, internal fragmentation processes by built-in collision cells of the MS instrument allow for toxin identification, which are well covered in other reviews and therefore not further elaborated here [77,82].
Bottom-up proteomics, achieved by in-solution digestion and direct MS analysis without prior decomplexation (shotgun proteomics), allows for a fast qualitative overview but is hindered by the critical “protein inference problem” that often hinders the differentiation of the numerous toxin isoforms [83]. Therefore, a decisive factor for an extensive quantitative venom analysis involves usually an upstream decomplexation and/or purification (clean-up) of the crude venoms applying several complementary separation methods, either by liquid chromatography (LC), gel electrophoresis, or a combination of both [80]. The existing decomplexation protocols can be adapted to many different instrumental set-ups and provide a detailed quantitative overview to characterize manifold toxin families. Nevertheless, sample preparation is less suitable for high-throughput analyses because it requires large quantities of venom samples and is more prone to contamination that results in false-positive identification of venom peptides [81]. Furthermore, trypsin digestion often prevents the clear identification of different toxin variants, like isoforms, proteoforms, or complex multimer formations [84,85]. To bypass these limitations, a logical step is to eliminate the digestion step and directly analyse intact toxin proteins by tandem MS, in a top-down proteomic approach [86].
In top-down methods crude venom samples are directly loaded to a front-end LC system coupled to the MS instrument. This set-up enables intact toxin mass profiling (MS1) and resolves toxin proteoforms and native posttranslational modifications (PTMs), which are not detectable by bottom-up approaches [86]. To identify the toxin proteins, information by tandem MS (MS2) in data-dependent acquisition mode is acquired. Therefore, a specific peptide ion is delivered for fragmentation to obtain its MS/MS spectrum. The established workflow reduced the needed venom amount as well as operational time, and it is associated with a much lower contamination risk [87]. However, top-down venom proteomics requires a highly specific set-up of high-resolution MS instruments that are only available in specialized laboratories [88]. In the case of high molecular mass toxin proteins, top-down analysis remains challenging and only provides a few observable fragments in tandem MS owing to inefficient ionization by denaturing electrospray ionization (ESI) [89].
Shortcomings in bottom-up and top-down approaches
Until today, most of the venom proteome studies use one of the well-established bottom-up strategies [54]. A shortcoming of this approach is the bias in protein quantification, arising from many experimental factors, such as instrumental set-up, applied protocols, or databases, which highly affects the protein characterization and prevents quantitative comparison between different studies [90]. This fundamental problem has general validity and also applies to the top-down approach, which is similarly influenced by a number of experimental parameters.
In addition to the various experimental factors, data interpretation and bioinformatic analysis are also important aspects [81,90]. The basic concept for search algorithms fall into 2 broad classes: database-depending and de novo. An increasing number of software and packages are now available for peptide/protein identification [91,92]. However, some tools remain challenging for inexperienced end-users owing to lack of appropriate documentation or poor GUIs and show a limited robustness for the output of the same proteomic dataset [93,94]. Experience in handling such proteomic software tools and in partially manual assessment of the data is therefore usually required to properly evaluate the analytical outputs. For all approaches, well-annotated genome and/or transcriptome data are an essential prerequisite to enhance the annotation performance of venom proteomes especially in understudied venomous organisms [95]. Although databases are still limited in terms of taxonomic coverage and do not include species-specific venom protein sequences, close evolutionary relationships within a particular taxonomic group allow the identification of protein families of even totally unexplored venom organisms, reflected by protein sequence homology [54]. However, identifying homologs of venom proteins from totally unexplored venomous taxa with few covered sistergroup species remains difficult and requires a variety of analysed and closely related species, which is possible, e.g., for snakes, but limited for larger taxons such as cnidarians and arthropods.
Future perspectives for high-throughput venom proteomics
Owing to the limitations summarized above, the current gold standard and good practice for venom proteome analyses consists of application of both complementary proteomic approaches. An overarching future goal for venom proteomics studies is to improve the existing methods to allow faster and even more precise analyses of larger sample sets [54, 86]. A top-down protocol, overcoming some of the aforementioned limitations, was recently developed [96]. This approach enables rapid and detailed profiling of multiple individual venom samples, along with statistical correlation tests for different factors, allowing population-scale analyses for a better understanding of regional and intraspecific venom protein variations.
Nonetheless, for high molecular mass toxin proteins (>30 kDa), current top-down analyses run into technical limits [87,97]. A future application to overcome these limitations in terms of ionization could be native electrospray ionization (nESI). However, native MS requires a specific platform with extended mass range, which is again associated with a loss of speed due to more extensive sample preparation, making this type of analysis still unfavourable for high throughput [98,99].
The application of a hybrid element approach and molecular MS configuration is another powerful concept to decipher venom proteomes in their entirety. The parallel absolute quantification of ultra-high-performance liquid chromatography–separated intact sulfur-containing venom proteins by inductively coupled plasma triple quadrupole MS and 32S/34S isotope dilution analysis, combined with bottom-up and top-down molecular MS, allows for both the exact quantification and the identification of proteins [79]. Another upcoming MS-based method that offers molecular information on the spatial distribution of toxins and new insights into the biology of venoms, as well as their highly functionalized storage and delivery systems, is further discussed in section “Production of Venom Components.”
Transcriptome Analyses of the Venom System
The recent advances in high-throughput proteomics to analyse novel venoms are also fostered by the fast development of next-generation nucleic acid sequencing technologies [54,74, 100, 101] (see Fig. 4). De novo venom protein analyses, as described above, depend on specific sequence databases of proteins to match masses of native or fragmented (novel) venom proteins. Because many venom proteins, especially of unstudied species, are unknown, high-throughput mRNA sequencing (RNA-seq) of venom glands is often coupled to the proteomics analysis to provide a custom sample-specific database. RNA-seq represents consequently an important and growing core-pillar of venomics to describe the expression of venom genes and proteins even in the smallest venom systems because the required RNA quantities for library preparation range from 100 ng down to 1 µg [102]. However, maybe even more importantly, RNA-seq allows the description of differentially expressed genes in venom-producing tissues, aiding in the identification of putative toxins and their possible origin and evolution from ancestral gene variants in body tissues [103–107]. These aspects are covered in sections “Spatial venomics” and “Significance of genomic data.” Diverse workflows of RNA-seq (also for venomics) have been addressed and reviewed previously [54,100, 108–112].
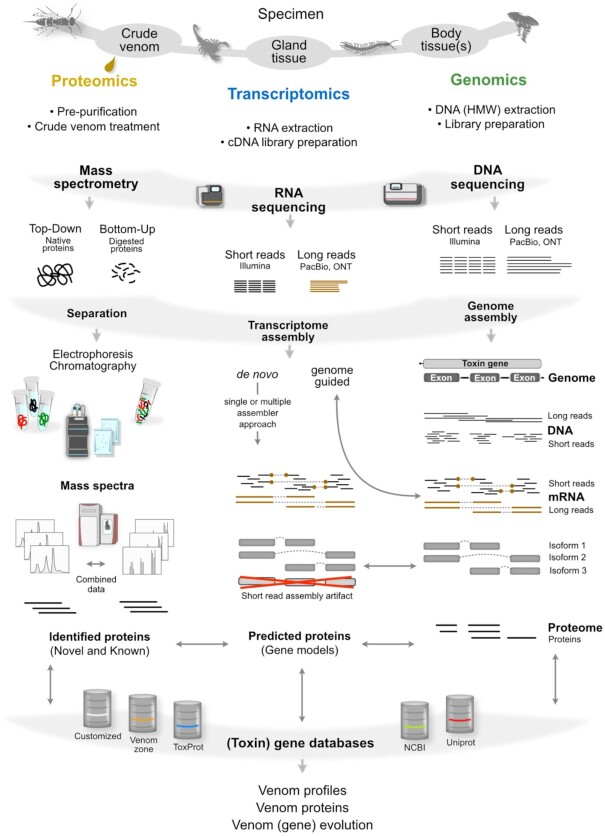
Figure 4:
The integration of proteomics, transcriptomics, and genomics in venom research. The general workflow for proteomics is shown on the left. Transcriptome analysis steps are illustrated in the middle. Please note that for state-of-the-art genomics multiple RNA samples from both sexes and different tissues (not only venom glands) are sequenced to perform differential gene expression analyses and to predict gene models more precisely. The genome sequencing steps are condensed and focused on the RNA read mapping. For more details please refer to the references given in the text. cDNA: complementary DNA; HMW: high molecular weight; ONT: Oxford Nanopore Technologies; PacBio: Pacific Biosciences.
Advantages and challenges of transcriptomics
Several venomous animals harbour such minute venom systems that several specimens must be pooled to obtain sufficient amounts of tissue material for RNA-seq. For some particularly small and difficult-to-rear organisms (e.g., remipedes, pseudoscorpions, smaller spiders) the sensitivity of transcriptomics is indeed the last resort to grasp an idea of their supposed venom compositions because crude venom is difficult to obtain [100]. The downside of the sensitivity of modern RNA-seq is that, even if carefully prepared, venom system tissues can be contaminated by other body tissues; in addition, they also contain transcripts of proteins with normal, non-venom-related functions [101]. The best practice is generally to avoid transcriptome-only studies, which should always be integrated with proteomic analyses—a strategy that is now commonly referred to as proteo-transcriptomics [100,101].
For many species a physiological normalization of the venom system, e.g., by milking specimens at the same time to synchronize the replenishment cycle of their gland tissues, is not applicable in the laboratory because milking, rearing, or keeping them alive in the laboratory is difficult [100]. Examples are small solitary bees, marine remipedes, small spiders, marine molluscs and other species. As a consequence, many studies describe venom transcripts and venom gene populations as a snapshot, without the statistical power of differential gene expression analyses with multiple replicates applied in ecological and clinical studies [113,114]. Increasing the sample size of the specimen pool could level heterogeneity by including a larger mix of different “wild-type” venom gland states.
Novel RNA-seq strategies
The specificity of assembly algorithms implies that diverse assemblers predict venom protein transcripts very differently and that single-assembler approaches might underestimate transcript populations and isoforms [108,111,115,116]. As a consequence, the identification and prediction of proteins via MS might be affected when using these assemblies as specific databases. Recently developed de novo assembly packages for short-read data generated by Illumina sequencing platforms thus apply multiple-assembler strategies that combine different assemblers and output a merged assembly [117–119]. New versions even include the annotation steps in the automatized process. One downside is that these programs currently require advanced bioinformatics expertise and often a familiarity with either virtual or physical, often Linux-based, environments, such as Docker or Conda. One future direction is to transform these approaches to more usable mainstream solutions and to link these to genome data to perform genome-guided transcriptome assembly and to foster more hands-on training of venom researchers in bioinformatics. The commercial software packages such as Geneious or CLC Genomic Workbench can cover some bioinformatics aspects. However, they run with expensive subscriptions and are methodologically often less suitable. Henceforward, direct RNA sequencing with novel sequencing platforms, such as ONT Nanopore or PacBio IsoSeq, with long reads and improved accuracy, will be increasingly applied, minimizing artificial transcripts or gene predictions [110,120].
The sequenced snapshots of expressed mRNA protein precursor molecules from tissue of the venom systems reveal not only transcripts of toxins and other venom proteins but identify as well house-keeping genes that assist venom secretion. As a consequence, RNA-seq of multiple tissues (differential gene expression) in combination with genome data and spatial -omics techniques is an important tool to reconstruct cellular pathways and mechanisms through which venom proteins and toxins are processed and translated [110]. A future direction will be to apply single-cell RNA-seq (scRNA-seq) methods to differentiate expressed toxins in diverse gland cell populations to reveal spatial and temporal venom variations [114,121, 122]. Single-cell transcriptomics has been successfully applied in general to a variety of diverse animals including sponges, ctenophores, placozoans, cnidarians, planarians, nematodes, arthropods, ascidians, and vertebrates (see, e.g., [123]). This breakthrough method simultaneously measures gene expression from thousands of individual cells. Clustering cells that share similar expression profiles allows for the identification and characterization of cell types that can be even more nuanced than traditional morphological characterizations. Cnidarians are a phylum typified by their venom-producing cells called nematocytes, whose biochemical and structural components have been successfully identified using scRNA-seq analysis [123–126]. Not only is this method capable of answering essential biological questions related to venom, but it is also likely capable of being implemented in non-model organisms and should be further tested in other venomous taxa. Unlike the use of transgenics to generate reporter lines and then sorting positive cells to generate a cell type–specific transcriptome, virtually any non-model organism can now be explored at a cellular resolution. Beyond mRNA, other techniques can reveal genomic features at the cellular level. For example, ATAC-seq sequences portions of DNA to assess genome-wide chromatin accessibility and identifies key gene regulation mechanisms such as transcription factor binding sites [127]. Because ATAC-seq is highly sensitive it requires only minute amounts of chromatin and it can be used to sequence even single cells, allowing the integration of transcriptomics and epigenomics at cellular resolution [128]. Such insights into the gene regulation of venom-secreting cells are essential to understand the evolution and development of venom systems (see also section “Critical and future aspects on current bioassays”). A recent study on rattlesnakes used this (in venomics) relatively new approach to reveal that a complex genotype underlies a simple venom phenotype [129].
Future perspectives of proteo-transcriptomics
Future directions for further developments of proteo-transcriptomics consist mainly in the development of more sophisticated and user-friendly data analysis strategies in combined, integrative interfaces. The most comprehensively assembled transcript libraries are generated with multiple-assembler pipelines including long-read RNA-seq data and are ideally guided by available genome data. The predicted gene models and annotated protein genes are then used to identify proteins, using the outputs of MS approaches in which bottom-up and top-down methods are applied to fragmented and native protein samples (see Fig. 4). An even more holistic design is achieved if complementary spatial transcriptomics (ST) and mass spectroscopy imaging (MSI) methods are applied (see section “Spatial venomics: non-targeted, high-throughput methods to visualize toxins”).
Integrating Molecular Venomics with Functional Morphology
Challenges in connecting morphology, function, and molecular data
Beyond classical compositional and structural toxin analyses by proteo-transcriptomic approaches, in recent years there has been a steadily growing interest in the connection of these data to morphology, to elucidate the localization and mechanisms of venom toxin production, storage, and delivery [36,130]. Obtaining information on the morphological aspects of venom systems provides mechanistic insight into how and where venom is produced and expelled, which is often not in a uniform manner [45, 122, 130, 131]. Thus the morphology of venom systems is crucial to understanding venom function [5]. Furthermore, integration of morphological and molecular aspects of a venom system—e.g., through information on the spatial distribution of toxins—can provide an important functional context for understanding both toxin function [45,132] and evolution [45,130], as well as the intricacies of the venom system itself [133].
Venom gland morphology is extremely variable: glands with a pronounced secretory function can have different numbers of cells, shapes and secretory modes [134]. Unicellular glands are mostly located in the epithelium of, e.g., aquatic vertebrates, annelids, and molluscs. Multicellular glands are usually located beneath the epithelium. In terms of shape, glands can be defined as globular (acinous) or tubular. Composite glands can result from the association of several acinous and/or tubular glands [134]. Below the cuticle of arthropods, sunken uni- or multicellular glands are present that possess a specialized canal cell, which develops a conducting canal lined by a cuticle [135]. In terms of secretion mode, 3 types can be distinguished. In apocrine secretion, secretory grana or a liquid secretion is released. However, this secretion also contains organelles and mitochondrial as well as nuclear proteins [136]. In merocrine secretion, parts of the gland cells are released with the secretion. In holocrine secretion, the whole cell is released (e.g., in mammal sebaceous glands). Because the loss of cell material in merocrine and apparently holocrine secretion is large, regenerative cells are present, e.g., in cnidocytes of Cnidaria or in the midgut of insects [134]. Thus, in different animal taxa glandular structures can range from single cells to large composite glands. Visualization as well as anatomical analysis methods have to be chosen according to the level of interest, ranging from ultrathin sectioning to analyse subcellular anatomy to micro-CT analysis to visualize general gland morphology [137,138]. Novel technological innovations towards 4D tomography, which includes dynamic data from samples that undergo change during scanning, might enhance our functional understanding of venom systems [139]. Nevertheless, integration of molecular data in the context of morphological or functional aspects is still challenging, and the classical venomics approaches (proteomics, transcriptomics, genomics), used to examine spatial information of toxin production in various insect-feeding species, only allow limited resolution [5,122, 140]. The glandular origin of the venoms in these studies was investigated by dissecting the secretory portions of the venom apparatus into a series of multiple segments and analysing respective sections by proteo-transcriptomic methods for variable toxin profiles [141–144]. Although macrodissection of venom glandular apparatus gives new insights into the biology of venoms, it has several drawbacks including the laborious preparation, low resolution, loss of morphological structures, and averaging effects across the section samples.
Targeted methods for the inference of spatial toxin distribution
As outlined above, the difficulties derived from the intrinsic nature of some venomous organisms and from the technological limitations of most commonly applied analysis methods have hampered a comprehensive integration of functional, morphological, and molecular data. To obtain molecular information on the subcellular level, techniques such as in situ hybridization and immunocytochemistry have been used to map the spatial distribution of toxins and venom components directly on tissue sections [145,146]. These methods have demonstrated great potential in venom research, for instance, revealing previously unknown parts of the venom apparatus [133] or heterogeneity of toxin expression in venom glands [121,147]. However, these techniques allow only a few previously knowSTn targets to be mapped simultaneously, providing limited molecular information.
Spatial venomics: non-targeted, high-throughput methods to visualize toxins
Advances in imaging technologies, proteomic analyses, and high-throughput sequencing have facilitated the development of non-targeted techniques, such as MSI and ST, termed under the name “spatial venomics.” MSI has become popular in recent years and as a non-targeted approach, which is ideally suited to interrogate the spatial distribution of multiple toxin proteins, peptides, or small molecules without prior knowledge of their identities [54]. The spatial resolution for different MSI instrumentation spans several orders of magnitude from 1 mm to 30 nm [148]. While several modes of ionization exist, matrix-assisted laser desorption/ionization (MALDI) remains the most appropriate for mapping proteinaceous toxins within venom gland systems. To date MSI has been used to explore the distribution of venom components in a variety of venomous organisms including cnidarians, arthropods, and reptiles [130,149–152]. The MSI workflow in all studies acquires individual mass spectra in a regular raster (usually ∼50 µm) across venom gland sections, which allows their distribution to be displayed on the basis of single toxins in a 2D density map. Recently, a new approach, named functional MSI (fMSI), allowed indirect detection of phospholipase A2 (PLA2) proteins by on-tissue enzymatic activity screening, which underlines the great potential of MSI for future in situ approaches [150,153].
ST is a novel technology that allows the visualization and quantitative analysis of whole transcriptomes, creating gene expression maps within individual histological sections [154,155]. Tissue cryosections are placed on glass slides that contain an array of poly-T capture probes uniquely identified by spatial barcodes that allow the determination of the origin of each mRNA molecule within the tissue. Therefore, ST allows the generation of complementary DNA (cDNA) libraries with accurate positional information for RNA-seq, adding a spatial dimension to transcriptome data that enables analyses of gene expression within a morphological context. It is thus an ideal technique to investigate poorly known or challenging venomous organisms because it allows the identification of toxin genes and their spatial expression patterns within the tissue, thus simultaneously characterizing the molecular composition of the venom and the morphological and functional organization of the venom-producing tissue. Additionally, the sensitivity and high spatial resolution of up to 55 µm (equivalent to 5–10 cells) of the ST array allows the study of very small venomous organisms while circumventing common obstacles encountered in bulk RNA-seq differential gene expression analyses. For instance, ST eliminates the need to pool small specimens, losing the statistical power of biological replicates, and avoids contamination from tissues not related to venom production [100,101]. Furthermore, ST can also be combined with single-cell RNA-seq [156], offering the possibility of simultaneously identifying different venom secretory cell types and their specific spatial location in the venom system.
These spatial non-targeted methods allow us to conduct data-driven exploratory analyses without preselecting known targets of interest and are excellent tools to investigate cell types and tissues whose organization and functions are not well understood [157], such as many animal venom systems. These 2 technologies add a spatial dimension to venomics, revealing genes and proteins associated with morphological features, providing essential functional information about venom systems, from the genetic to the phenotypic level, from the molecular composition of the venom to the morphological features of the delivery system.
The Significance of Genomic Data
Despite the increasing availability of technologies for generating high-quality genomes, venomous animals are still under-represented in most databases and studies. In particular, comparative genomics studies on the origin and evolution of venoms are very sparse [103]. A major barrier that hinders comparative genomics is the reduced quantity of material obtainable from very small venomous organisms. However, developments in (ultra) low-input protocols may aid in overcoming this hurdle, using amplification techniques. Novel methodologies also allow sequencing of difficult genomes of predominantly small marine invertebrates (e.g., nematodes, molluscs, and others) that are characterized by extensive production of mucus (Mucopolysaccharides) and/or other inhibitory molecules (polyphenolic proteins) [158, 159]. These new methodologies are being exploited by a number of genome consortia that are connected under the umbrella of the Earth Biogenome Project [160], whose ultimate goal is to sequence, within the next decades, the genomes of all animal and plant species to better understand their evolution, ecology, adaptations, and interconnections, and to safeguard—as last resort digitally—the threatened biodiversity and bioresources on earth [160,161, 161]. Linked to these efforts, the numbers of published high-quality, chromosome-level genomes have already substantially increased, allowing for more comprehensive investigation of the origin and evolution of venom genes, predominantly from iconic groups such as snakes, spiders, and cone snails [129,162–171].
Venom gene origin
Genome data are an important reference material to assess the accuracy of transcripts and gene models obtained from RNA-seq data by genome-guided transcriptome assembly approaches. However, high-quality genomic data with good gene annotations are only obtained if multiple tissue samples are mapped on the genome and transcript-based gene predictions, improved by corresponding proteome data, are implemented [103,104] (see Fig. 4). Many available genomes lack a reliable gene annotation because they were automatically annotated [161,172]. Annotation with automated pipelines is prone to both false-positive and false-negative matches because venom genes belong in most cases to multi-gene families, often with high similarity of new toxin copies to their ancestral non-venom-related paralogs [173]. Genomic data are likewise of utmost importance for identifying ortholog genes (especially when short) and for comparing venom genes to their non-toxic homologs in individual genomes [174–176]. One future challenge is to improve the reliability and the speed of the process to predict genes in genomes. Without knowledge of the physical genomic location of a toxin-encoding gene, it is very difficult to identify its orthologs in other species. Genomes also provide information on exons and introns that is crucial to gene structure evolution (Fig. 5A). For instance, gene duplication often results in incomplete sets of exons that can be used to trace back duplication events [165,173] that are impossible to detect otherwise [177]. It has been shown recently that some genomic studies have made erroneous assumptions by overlooking orphan exons [174,177]. At the same time, intronic sequences can provide a more reliable phylogenetic signal when genes evolve under extremely strong positive selection [178]. For example, sometimes a toxin-encoding gene can evolve from a non-toxic gene by deletion or gaining of exons [179].
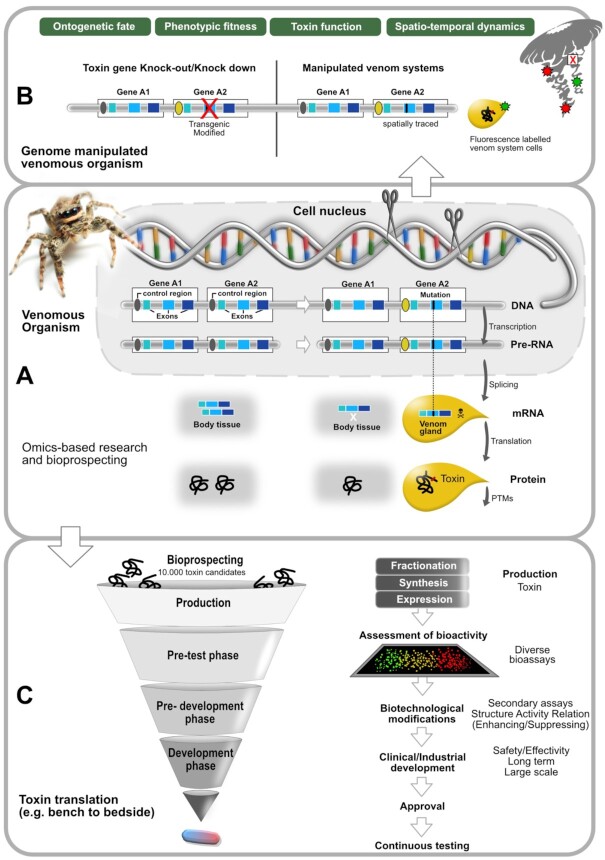
Figure 5:
The integration of -omics-based research to improve translational research but also our basic understanding of venom and toxin gene evolution. (A) Shows the biological process from gene to protein; (B) illustrates genome editing aspects to investigate toxin evolution, function, adaptive value, spatio-temporal variability, and ontogenetic fate; (C) summarizes the major steps in translational research, from bioprospecting to application. PTMs: posttranslational modifications.
The evolutionary history of toxin genes is more realistically reconstructed if their exact genomic location is identified using unrelated, syntenically conserved flanking genes, followed by location of that same genomic region in the outgroup species’ genomes. After that, an exon screening (via BLAST or other sequence similarity tools) should take place to locate all related genes and pseudogenes in that region. A phylogenetic analysis of complete gene sequences subsequently helps to identify gene subclades. Previous knowledge of gene evolution can help to infer the most likely evolutionary history of a given gene [177,180–183]. Several helpful online and standalone software tools have been recently developed (e.g., SimpleSynteny, SynMap, AliTV [184–186]), however, they often rely on previously published de novo genomic annotations, which as explained above are particularly error-prone [172]. One direction for improvement of this step is to train the gene prediction with specific proteo-transcriptomic data from venom proteins (e.g., [104]. With the aforementioned genome sequencing initiatives we will soon be able to apply comparative genomics methods to detect the occurrence of convergent venom gene evolution in larger clades: the inclusion of many non-typical venom taxa is in fact crucial to infer general and lineage-specific patterns of gene evolution.
Venom gene manipulation by knockdown and CRISPR
Advancements in available tools and techniques for genetic manipulations are currently increasing among non-model species, including venomous animals. For instance, parental and embryonic RNA interference (RNAi) are regularly used to investigate the developmental biology of the common house spider, Parasteatoda tepidariorum [187,188]. More advanced CRISPR-mediated mutagenesis has also been developed for some model venomous species, e.g., the jewel wasp, Nasonia vitripennis [189]; the honeybee, Apis mellifera [190]; and the red imported fire ant, Solenopsis invicta [191]. However, it is only in the cnidarian Nematostella vectensis that genetic manipulations, including knockdowns using morpholinos and short hairpin RNA, as well as CRISPR-mediated techniques [192, 193], have been used to address venom-related questions, such as elucidating the factors associated with the biogenesis of venom-secreting cells [122,131] (see Fig. 5B).
Transgenic approaches in N. vectensis have allowed the tracing of spatiotemporal dynamics as well as the localization of 2 distinct venom-secreting cells (nematocysts and gland cells [194,195]). Furthermore, specific toxins were found to be secreted in subpopulations of both cell types [194], adding a new level of complexity lacking in previous analyses. Furthermore, by incorporating fluorescent markers into the structural components of venom-secreting cells using CRISPR/Cas9 techniques followed by FACS sorting of different types of nematocytes, different types of stinging cells were isolated [196]. RNA sequencing of the isolated cells revealed numerous differentially expressed genes, including some transcription factors resulting from lineage-specific duplication and essential for proper cnidocyte differentiation [196].
While these techniques have been instrumental in elucidating the structural components of the venom system, the characteristics of toxin components remain largely unresolved. Particularly of interest is the ability to genetically manipulate toxin-encoding genes in animals and test the impact on the fitness of mutants. Examples of such studies may include the deletion of a functional toxin before exposing the mutant to native predators and prey to test whether defense and predation abilities are affected.
A further expansion of this approach would be deleting multiple different toxins followed by subsequent mutants’ crossings to produce individuals completely lacking venom. Additional assays could include knock into the animal's genome (“gene knockin”) additional toxin domains, to cause overexpression of a toxin, or introduce precise modifications of single nucleotides to recapitulate an ancestral venom profile. The recent development of organoids from snake venom glands represents a new opportunity to test in vitro genetic manipulations [121]. Although this technology will need further developments to be easily applied to other systems, it may provide opportunities to simultaneously knock down toxin genes or edit regulatory regions to perform functional studies.
Production of Venom Components
Challenges of isolation-based venom biodiscovery
Functional characterization of toxins isolated from venom can be conducted directly using the purified peptide or protein. However, the small size of many venomous species, in particular invertebrates, hinders the mechanical manipulation of the venom system and/or the collection of a sufficient amount of venom from a single specimen [6]. In these cases, an extraordinarily large number of specimens must be sampled to accumulate sufficient venom for the isolation of single compounds, raising ethical concerns [6, 100,197]. In these and other cases where toxin sequences can be identified only in silico (transcriptome or genome-based, see Fig. 4 and Section: Challenges in connecting morphology, function, and molecular data), methods of chemical synthesis and recombinant expression, which can produce milligram amounts of single venom components, are becoming increasingly important (Fig. 5).
Chemical synthesis
Chemical synthesis is ideally suited for relatively short peptides (<50 residues) and requires prior knowledge about the peptide sequence, which must be obtained by MS or other methods (Edman degradation, novel NMR-based methods) on the isolated natural toxin or from genome or transcriptome sequencing. In many cases, additional knowledge about the disulfide pattern (as well as other PTMs) is required to ensure the correct native folding of the produced toxin.
While not suited for larger venom proteins, this approach has been instrumental in the functional and structural characterization of toxin peptides. The most common method applied is solid-phase peptide synthesis, which can produce quite large amounts of peptide (generally milligrams or grams, but kilogram yields are possible in industrial settings). Advantages of chemical synthesis include the ability to incorporate, e.g., unnatural amino acids, D-amino acids, reporter groups, and unusual PTMs (such as brominated tryptophan) that are impossible to produce by recombinant production, as well as the regio-selective formation of disulfide bonds and cyclization [199].
Recombinant production
With an increasing number of both prokaryotic and eukaryotic expression systems that support the production of post-translationally modified proteins, recombinant production of toxins is also becoming more accessible. Given that most toxins are endogenously produced in the endoplasmic reticulum of the host and that PTMs can play crucial roles for toxin activity [200–202], eukaryotic host cells for recombinant production generally provide the best chance of producing functional toxins. Most common eukaryotic host systems used for toxin production include the yeast Pichia pastoris, insect cells, and a variety of mammalian cell lines such as HEK293 and CHO. Although prokaryotic, the bacterial host Escherichia coli has been used to express thousands of toxins [203,204] and can yield milligram amounts of toxin in a standard laboratory setting. However, it is prone to a major drawback, the inability to add common PTMs such as glycosylation, C-terminal amidation, and hydroxylation. The availability of a variety of systems, including specialized strains, can allow for the production of disulfide-bound toxins in E. coli (see below). Regardless of the specific expression host, recombinant expression offers the advantage of incorporation of affinity purification tags and the ability to easily produce a large number of variants for functional testing and to obtain proteins that are substantially larger than those commonly made by chemical synthesis.
Future perspectives of toxin production
In vitro refolding of chemically synthesized peptides is often inefficient, especially for peptides containing ≥3 disulfide bridges. On the basis of sequence homology with characterized toxins, the disulfide pattern may well be deduced and thus allow for directed folding strategies. However, for novel, previously uncharacterized sequences, recombinant production may be the best suited or unique option, and as such has been subject to remarkable developments in recent years. To allow disulfide bond formation in E. coli, a variety of methods and strains already exist and many have been used for toxin production [205–210]. Recently, new systems have been introduced [211] and the ability to produce thousands of disulfide-bonded animal toxins in E. coli has been demonstrated in important studies from the Vincentelli lab [204,212,213]. Hundreds of cystine-dense peptides containing ≤5 disulfide bonds have recently been produced in HEK293 for, e.g., structural characterization [214]. Moreover, the same expression system has been used for surface display, which allowed screening of thousands of toxin peptide sequences to identify strong peptide interactors for specific targets [215]. This work demonstrated the potential of cystine-dense peptides to function as binders for transmembrane targets that are otherwise difficult to inhibit. Finally, cell-free synthesis approaches that have been used sporadically to produce venom toxins [216,217] have a great potential for further developments. It is imperative to scale up such techniques to fulfil the potential of the many new sequences now available. The infrastructure needed to produce thousands of peptides is often not available in a typical (academic) laboratory setting. Such undertakings will therefore require larger publicly funded consortia and/or closer collaborations between academia and industry than currently exist.
Applied and Translational Venom Research
After sufficient quantities of crude venom or venom fractions are obtained and/or suitable amounts of a single compound are produced (see sections “Collection of Venomous Organisms” ) their applicative potential can be assessed through a variety of bioassays. This translational perspective of animal venoms has always been a major driver of venom research. However, a large gap remains between the plethora of described animal venom compounds and the much fewer approved drugs, bioinsecticides, and pharmacological and cosmeceutical products based on animal toxins [2,15]. Most candidates are dropped during the trial phase, while the few remaining ones have to be adapted biotechnologically to enhance or improve their properties. Both the testing and the optimization steps are expensive and time-consuming, being a major hurdle for applicative developments. The insights from the aforementioned -omics methods now allow a far more efficient and targeted bioprospecting that also increases the chances of realistically identifying promising candidates and reducing the number of unsuccessful candidates. An equally important aspect of bioassays is that they allow interactions between predator and prey to be tested, e.g., venom resistance and ecological factors such as differing prey that might influence venom complexity [140]. However, in this section we focus now predominantly on applied aspects of bioassays.
Bioassays in pharmacology
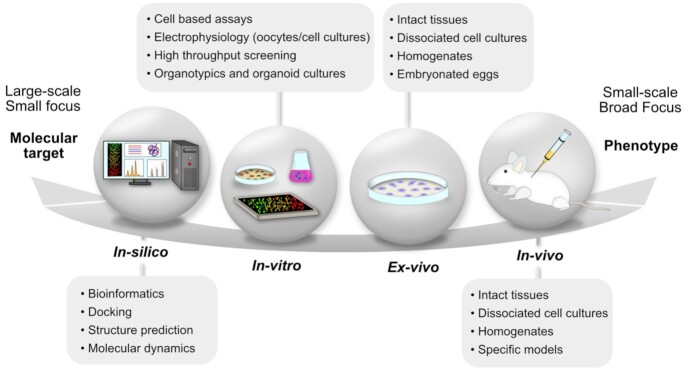
Bioactivity assay systems have been developed over many years to characterize the mechanism of action and pharmacological properties of venoms and have been constantly improved to reveal novel targets [218–220]. In particular, the complexity of venoms is mirrored by an ever-increasing number of bioassays developed to characterize their structural, functional, and pharmacological properties. These bioassays span from in vivo phenotypic screens to ex vivo and in vitro models, as well as in silico analyses (Fig. 6). These approaches are mostly pursued from 2 complementary perspectives: (i) Basic biological characterization of a given venom leads to description of behavioural phenotypes and the identification of underlying cellular and molecular targets. (ii) Target-specific assays may be used to identify venoms and toxins interacting with the molecular target(s) of interest [221,222].
Figure 6.
Approaches to study the activity of venom components span from in vivo, ex vivo, and in vitro to in silico methods. This allows the characterization of a broad spectrum of physiological effects, from whole-organism phenotype to molecular target.
Injection into an organism in vivo can mimic many aspects of naturally occurring bites or stings, thereby reflecting the complexity of physiological and behavioural phenotypes [220]. To understand ecological interaction in vivo assays are thus an important tool, despite the drawback that bioactivity of single components is masked by the use of whole venom [140]. Whole-organism phenotypes and in vivo assays offer a particularly powerful approach when combined with the use of transgenic animal and transient knockdown/expression systems to test putative mechanisms of actions of a venom [194,223].
In vivo assays
Despite in vivo assays on vertebrates posing ethical limitations, being labour intensive, and often not being scalable to high-throughput approaches [222], murine models are widely used in basic and applied venom research for the characterization of effective and/or lethal doses (ED50, LD50). In vivo assays remain especially important to better understand arms races between predator toxicity and natural prey resistance [224], which has especially been described in more detail for rattlesnakes and their prey, such as squirrels [8,9,225,226]. The effects of venom from jellyfish (Chrysaora sp.) were, e.g., analysed in zebrafish (Danio regio) as one of the toxinological model organisms, which revealed toxicological mechanisms, such as hemorrhagin in eyes and hyperpleasia or hypertrophy on other organs [227]. In vivo assays also remain a prerequisite for the clearance of novel therapeutic agents by most regulatory agencies [228]. High-throughput chemical screens are performed using zebrafish embryos and chemical libraries, taking advantage of several available transgenic lines and disease models. Other potential organisms such as Drosophila have also been used in venom-based research with promising outcomes [229].
Ex vivo assays
The complexity of in vivo approaches can be reduced by experimenting on representative ex vivo tissues. Tissues extracted from mice, frogs, electric eels, chicken, and other organisms were instrumental for laying some of the most fundamental cornerstones of basic physiology studies of venom research [222]. Ex vivo methods reduce and refine animal use because multiple samples can be established from a single killed animal. In addition, they offer a more precise control of experimental conditions, allow convenient access for microscopy and biophysical probes, and facilitate the study of venom-induced effects on specific cell types even on subcellular structures and organelles. Similar to in vivo, transgenic or transient transfection/transduction may help to elucidate molecular mechanisms of action. However, freshly isolated tissues from animals may not be well suited for high-throughput analysis or for studying the toxin function at a single-protein level [222,223].
In vitro assays
Broad functional studies can be performed in vitro in cell lines, which allows precise system compositions, efficient pharmacological access, and genetic manipulation for knockout/down, knockin, and mutagenesis, as well as the use of reporter systems [230, 231]. Immortalized cell lines provide valuable insights into the therapeutic potentials of animal venoms and their components as drug candidates. In addition, primary cells obtained from oocytes of the South African clawed frog Xenopus laevis allow exogenous expression of functional ion channels for electrophysiological analysis to evaluate their interactions with venoms and toxins [232,233]. Miniaturization and sensitivity are ever increased to characterize minimal amounts of venom components. Multiwell-plate assays have facilitated high-throughput screening of venom components against cells, enzymes, receptors, and ion channels, many of which are approved drug targets [222,234]. In vitro biophysical methods offer the ability to manipulate both toxins and receptors at a molecular level and record the resulting effects with high spatial and temporal precision [235]. In vitro studies of venom cannot replace in vivo experimentation owing to the inability to reflect the full physiological complexity; however, they are important to reduce and refine animal use by providing mechanistic insights to allow focused and informative in vivo experiments [236].
In silico assays
Modelling of toxin interaction with cell-membrane receptors in silico has emerged as a powerful novel approach for drug discovery [237] that requires detailed structural information of both the toxin and its receptor protein. Structures of numerous toxins derived from animal venom were determined using X-ray crystallography or NMR spectroscopy in the 1970s and 1980s [238,239]. In contrast, nearly 80% of all membrane proteins with known structures were determined only in the past decade, owing to the “resolution revolution” in electron-microscopy technology and the development of advanced crystallographic techniques [240], which has provided numerous structures of venom peptides in complex with their cell-membrane receptors [241–244]. Atomistic simulations of toxin-receptor interactions currently rely on 2 complementary methods, namely, docking and molecular dynamics [245], which may provide realistic representations of the system under study when combined [246]. Molecular dynamics trajectories can capture intricate details such as ion permeation events, binding/unbinding of the toxin, conformational changes of the receptor, and various protein-lipid and protein-solvent interactions at the atomic level [247–249]. The ever-increasing computing power available for research facilities establishes in silico approaches as central parts of venomic analysis pipelines. AI-driven structure predictions provide increasingly high-quality structural models and will gain importance for elucidation of toxin receptor interactions and accelerate the discovery of new promising venom-based therapeutic lead structures [250].
Critical and future aspects of current bioassays
The door to powerful high-throughput assays available in other biological sciences has been opened by the refinement and miniaturization of test models in combination with recombinant and/or synthetic toxin production, as well as organoid venom-glands [121]. Increasingly, this allows broad screening of large numbers of toxins for action on a focussed biological target. Alternatively, a limited number of toxins may be screened on a large number of cell types or organisms, providing fast access to highly biological active toxins with the potential for novel and surprising applications. Indeed, in vivo large-scale toxin testing for phenotypic changes even on a whole-organism level such as flies, fishes, or nematodes can be performed in a high-throughput screening manner [221]. In vitro high-throughput electrophysiology in mammalian cells and Xenopus oocytes is also gaining importance [222]. Ex vivo, automatized “high-content screening” microscopy offers a versatile approach for testing many toxins, many cell types, or even both together. The automatization allows for simultaneous high-throughput acquisition of a large array of readouts such as various morphological features in combination with multiple immunocytochemical stainings on a single-cell basis [234,251]. Interestingly, classical pharmaceutical in vitro screening platforms with highly sensitive target-optimized cell assays such as fluorometric imaging plate reader (FLIPR), amplified luminescent proximity homogeneous assay (ALPHAscreen), and homogeneous time-resolved fluorescence (HTRF) screens, which are the workhorses for large-scale screening in the pharmacological industry, have so far only scarcely been applied to venom and its components [252,253]. It is anticipated that these methodologies, combined with microfluidic approaches, will propel the biological characterization of venoms and their toxins.
Pharmaceutical applications
Venom compounds have a wide spectrum of pharmacological applications, including analgesic, anti-inflammatory, antimicrobial, and anti-cancer activities that have been used as prototypes for drug design and therapeutic agents and are used in a variety of therapeutical settings [2,218, 254–257]. Currently 11 toxin-based molecules have been approved by the US Food and Drug Administration (FDA) or the European Medicines Agency (EMA) and are on the market [2,15]. These venom-derived drugs are used for the treatment of hypertension, acute coronary syndromes, coagulation during surgery, chronic pain, type 2 diabetes, and perioperative bleeding, while many others are currently in clinical trials or in preclinical development. The original molecules were discovered predominantly in snakes (captopril, enalapril, tirofiban, eptifibatide, batroxobin, and cobratide), lizards (exenatide and lixisenatide), and several marine and terrestrial invertebrates from cone snails to leeches (e.g., ziconotide, bivalirudin, and desirudin) [15, 218,258–260]. However, critically it has to be noted that the whole process from bioprospecting to the final development of a compound for pharmaceutical applications remains challenging (Fig. 5C). In the following sections, we discuss challenges and highlight biological and ecological traits of venomous species that could greatly improve the effectiveness of this process.
Targeting pain
Severe pain is often one of the main symptoms of envenomation, especially in defensive venom, where toxins are instrumental in triggering aversive responses. This ability made venom toxins fundamental tools to investigate the physiology of nociception, which involves a number of receptors located in the peripheral nervous system, including the voltage-gated Nav, Kv, and Cav channels and the ligand-gated transient receptor potential (TRP) channel, acid-sensing ion channel (ASIC), and P2X in the primary afferent neurons. AMPA (α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor), NMDA (glutamate-gated cation channels), NET (norepinephrine transporter), and GPCRs (G-protein-coupled receptors), together with Nav and Cav, affect modulation of pain at the spinal level [261]. Generally, agonists of these channels in nature elicit pain and trigger aversive responses [262], while antagonist toxins are extremely promising as analgesic drugs and indeed their efficacy as antinociceptives has been demonstrated by multiple studies in murine models. This is the case of toxins from the sea anemone Heteractis crispa that act as selective TRPV1 modulators and show analgesic effects in acute and chronic pain models in mice without causing hyperthermia, a common adverse effect of other TRPV1 antagonists [263]. On the contrary, crotalphine from the South American rattlesnake induces a potent and long-lasting analgesic effect in mice by activating and thus desensitizing the ankyrin-type TRPA1, which plays a critical role in the pathogenesis of pain and inflammation [264]. Several spider, snake, and sea anemone–derived toxins, including the well-characterized mambalgins, inhibit the activation of ASICs and are involved in different pain conditions [265–267]. A wide range of venom toxins target the voltage-gated Nav channels, which are crucial in electrical signalling and neuromuscular function. Activators induce rigid paralysis and pain, while inhibitors are able to elicit spastic paralysis and analgesia, in both cases with a remarkable predatory and defensive effectiveness. Among them, the inhibitory cysteine knot (ICK) peptides, produced by spiders, scorpions, and cone snails, have been particularly studied [268,269]. Some ICK peptides also act as blocker of Cav channels including the cone snail o-conotoxin MVIIA (Prialt [ziconotide]), an FDA-approved analgesic for spinal administration in severe chronic pain [270]. Relatively few classes of toxins target GPCRs, including conotoxins, which are active against visceral and post-surgery pain through different mechanisms involving GABAB and k-opioid receptors, NMDA, and NET [271,272]. Others are snake and spider toxins that modulate P2X and AMPA receptors to reduce inflammatory pain [273]. Overall, a variety of venom toxins have a great potential to be developed into novel analgesics that are able to block pain at its source [274].
Anticancer applications
Anticancer properties of animal toxins, which manipulate signalling cascades controlling cell death and tumour growth, are promising therapeutics [256,275–279]. In particular, peptides from spiders and octopus and the crude venom of various snake species (cobras and vipers) have recently been reported to specifically target human melanoma, often with minimal effects on healthy fibroblast cells [278–282]. Other anticancer activities of animal venoms highlight their potentials by inhibiting the proliferation and invasion of cancer cells, through cell cycle arrest and/or induction of apoptosis, as well as by revealing the affected signalling pathways [256, 275,283,284]. However, potent venoms with anticancer activities many times raise concerns regarding their toxicity in healthy, non-targeted cells and tissues [285]. These could be overcome by directly targeting tumour cells (e.g., nanoparticle-based delivery systems). In addition, combination approaches use venom or the active compound coupled with existing chemotherapeutic agents at a low dose [276,285,286]. However, toxic effects that emerge by the combination still need to be evaluated along with the observed anticancer or other therapeutic potential.
Immunomodulation
The potential immunomodulating abilities of venoms and toxins have also started to receive attention [281,287,288]. Immunosuppressive activity has been demonstrated in snake crude venoms. In particular, the activity of the red-bellied black snake Pseudechis porphyriacus venom might translate to therapeutic applications for T-cell–associated conditions including rheumatoid arthritis and inflammatory bowel disease [289]. Venom components from the rattlesnake Crotalus durissus terrificus specifically diminish T-cell proliferation and IL-2 production [290]. They induce a shift in the colonic microenvironment from proinflammatory to anti-inflammatory in mouse models of induced colitis [291]. These effects have been linked to the action of several specific toxins, belonging to different classes, including PLA2, cysteine-rich secretory proteins (CRISPs), metalloproteases, serine proteases, and L-amino acid oxidases (L-AAOs). In addition, many invertebrate venoms have been used by traditional medicine in different cultures to treat, among others, autoimmune diseases, from bees to scorpion [292,293]. Studies have confirmed that the 2 major bee venom components, melittin and apamin, regulate, respectively, TH2-cell–mediated responses [294] and the production of monocytes and macrophages [293]. On the other hand, both margatoxin from Centruroides margaritatus scorpions [295] and the Stichodactyla toxin (ShK) from the anemone Stichodactyla helianthus are able to selectively block the Kv1.3 channel, a key component in autoimmune disease progression, highly expressed in effector memory T cells [296].
Antimicrobial activity
In the context of the current antibiotic crisis and the scarcity of therapeutic alternatives for the treatment of bacterial infections caused by multi-resistant bacteria or to treat viral infections, the search for new therapeutic alternatives is 1 persisting challenge. Antimicrobial peptides (AMPs) derived from animal venoms are biologically active cationic, anionic, or amphipathic peptides of <100 amino acid residues with a wide structural but stable range (α-helices, β-sheets, extended structures, or disordered loops) [297,298]. In this sense, AMPs and other metabolites obtained from animal venoms are a future solution to develop a new generation of synthetic antimicrobial molecules with improved antibacterial and antiviral activity, safety, and a broader spectrum of activity [297,299]. There are multiple examples of approved or promising AMPs described from various taxa such as serrulin or androctonin from scorpions, melittin and derivatives from bees, or L-AAO from snakes, to mention a few [300–303] (see Supplementary Tables S2–S4).
Pore-forming toxins in sensing applications
One of the most interesting recent applications of venom proteins is their use in nanopore biosensing, which allows detection of various small molecules, peptides, proteins, DNA and RNA, sequencing, and analysis of enzymatic reactions at the single-molecule level (Fig. 7A). Although prokaryotic channels or pore-forming proteins are most commonly used for nanopore biosensing [304], cytolytic venom proteins are also very attractive candidates owing to some advantageous properties [305]. These include channel stability, ease of insertion into artificial hydrophobic supports [306], and the ability to alter channel size through mutations that affect oligomerization [307]. Although the current nanopore sequencing set-up of Oxford Nanopore Technologies involves a prokaryotic transport channel [308], it is foreseeable that venom protein channels will be developed for use in MinION® devices, whether for long-read sequencing or other biosensing applications (Fig. 7B).
Figure 7.
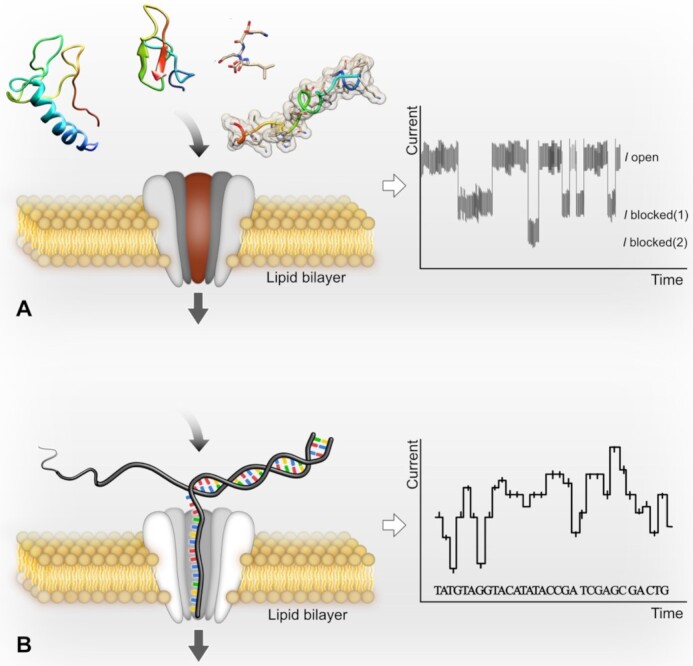
Schematic representation of nanopore biosensing. (A) Nanopore biosensing uses minute changes in electric current caused by the translocation of an analyte through the pore; each analyte is characterized by the percentage of current blockage and its duration. (B) The most widely used application of nanopore biosensing is DNA/RNA sequencing. The present trace is adapted from [312].
Using the classical patch-clamp method, channels formed by a toxin from the sea anemone Actinia fragacea have been used so far to detect DNA, peptides, proteins, and small molecules [309–311]. However, nanopore biosensing may require more or less extensive mutagenesis of protein residues to enable analyte capture or translocation. Prior structural and biochemical knowledge is therefore paramount for the adaptation of venom proteins for biosensing experiments. It is important to note that nanopore-based identification and sequencing is performed at the single-molecule level, which means that it can enable the discovery of new or rare (macro)molecules and create opportunities for the development of highly sensitive diagnostic devices.
Agrochemical applications
The main approach to control pest species in agricultural and public health contexts relies on chemical pesticides. However, the continuous use of specific classes of insecticides has inevitably led to resistance in various pest species. In addition, current pesticides have a devastating impact on biodiversity [14,313–317] and have often raised concerns regarding human safety; owing to improved health legislation, many previously successful insecticides have been de-registered [318].
Altogether, these circumstances led to a renewed interest in the development of novel, eco-friendly bioinsecticides. Animal venoms, especially from predators that feed on insects, may be extremely promising for identifying novel natural insecticides, with strict species-specific action. The venom-derived insecticidal compounds tested to date have revealed a rich repertoire of bioactive compounds that specifically target ion channels of prey insects [14,319–321].
Novel spider peptides derived from the African and Australian Theraphosidae spiders, as well as the African Augacepahuls ezendami, have shown insecticidal capabilities for further development [322–324]. In line with this research approach, in 2017 the US-based company Vestaron launched the first peptide-based pesticide based on a knottin from a funnel web spider, thus validating the immense potential of animal venom-derivatives as bioinsecticides. The innovative aspect of this knottin is that it is sprayed on plants and then orally taken in by pest species. This makes conventional genetic modifications of plants by incorporating toxin genes in their genomes obsolete, and the creation of recently more critically seen gene modified organisms is avoided [14].
Diagnostics
As a result of extensive toxin investigations mostly from the late 1980s, several in vitro diagnostic tests were developed, commercialized, and adopted as routine applications in hematology laboratories to be used for assessing hemostatic disorders. Many hemostatic parameters such as fibrinogen breakdown products, activation/inhibition of various clotting factors, protein C activation, von Willebrand factor–related disorders, and lupus anticoagulants can be assayed by using snake venom proteins, mostly proteinases [327, 328]. These tests have some advantages over other common assays with their unique mechanisms of action. For example, snake venom thrombin-like enzymes are generally not inhibited by thrombin inhibitors such as heparin, allowing the test to be performed with samples containing these inhibitors [325]. Detailed information about this topic can be obtained from cited references.
A more recent venom-based diagnostic tool was developed from a species of scorpion, Leiurus quinquestriatus (death stalker). One of its major venom components, chlorotoxin, which blocks chlorine channels, can also bind to matrix metalloprotease-2 (MMP-2), which is specifically upregulated on the membrane of cancer cells but not in normal cells. This unique feature has led to a diagnostic reagent, so-called “tumor paint,” which can be used for monitoring tumors. Chlorotoxin peptide is labelled with a fluorescent cyanine dye, which when subsequently bound to cancer cells selectively helps to visualize the borders of tumor tissue precisely. This is particularly useful in treating brain tumors because it is critical to be as precise as possible when excising tumors during surgery to prevent irreparable brain damage. Chlorotoxin is additionally being evaluated in clinical trials as an in vivo diagnostic imaging agent for various cancers, including glioma [218,329], and recently was granted a fast-track designation from the US FDA for pediatric brain tumors. We predict that with more detailed knowledge on toxins and their distribution and developmental fate within the venomous organisms further promising candidates with specific activities suitable for diagnostic applications will be identified.
Envenomation therapy: antivenoms (in a nutshell)
Animal envenomation by several key taxa such as spiders, scorpions, and snakes is a major public health concern worldwide; however, most dramatic are effects from snake bites. Millions of individuals are at risk owing to their geographic location, which is inhabited by various lethal snakes, especially in Africa, the Middle East, India, Mexico, and South America [330]. Approximately 1.8 million people are annually bitten by snakes, of which 138,000 people die as a result of envenoming while up to 500,000 snake bite survivors experience permanent physical or psychological disabilities worldwide [19,28]. As a consequence, the World Health Organization included snakebite envenoming in the list of category A Neglected Tropical Diseases [331] and developed a strategy to reduce mortality and disability by 50% before 2030 [332].
Antivenom is the only specific and effective therapy for victims of envenomation. The active compounds reported are whole immunoglobulins G, their F(ab’)2 or Fab fragments extracted and purified from the hyperimmune plasma of large animals (mostly horse) and prepared by their immunization with a single venom or a mixture of several of them [333,334]. Unfortunately, we have a current, serious crisis in antivenom availability in such most endangered regions, like sub-Saharan Africa and tropical and subtropical Asia [335]. It is determined by cost, frequent scarcity, and poor distribution because only a few countries are the antivenom manufacturers (and only 3 in Europe). In addition, it may require a cold-chain for transport and storage, which is problematic for rural areas of low-to-middle income countries (LMIC). Additionally, some major antivenom manufacturers (Syntex, Behringwerke, and Sanofi Pasteur) have stopped antivenom production over the past two decades for commercial reasons, creating a noticeable deficit of antivenom in the countries that they previously supplied, especially in Africa [19]. Even Europe faces current antivenom shortages, due to the low financial sustainability of their production and lack of compliance to good manufacturing practice (GMP) regulations. Further, recent analyses revealed the lack of comparative information on available antivenoms against European vipers (see Supplementary Table S5 and cited references for more details).
Several promising new technologies have been presented in recent years for the manufacturing of therapeutic antibodies on an industrial scale as antivenoms. Some of these molecules include monoclonal antibodies, scfv (single-chain fraction variable fragments), and nanobodies among others that could form the basis for future treatments [334, 336]. However, to develop these new antivenomics platforms the most detailed knowledge of venom composition is crucial. The herein-discussed methods and future perspectives facilitate an unprecedented understanding of the ecology and biology of venomous animals and their venoms, which allows in consequence the production of more effective antivenoms.
Conclusions
Fast advancements in genomics, transcriptomics, and proteomics technologies increase our knowledge of convergently evolved venoms across the tree of life.
A more detailed knowledge on toxins and their distribution and developmental fate within the venomous organisms will reveal new insights on their evolutionary origins while also identifying compounds with novel bioactivity and targets.
Venom toxins possess a great translational potential, with applications in the therapeutic, diagnostic, agrochemical, and biosensing fields. More detailed biological insights on venomous species facilitate a more targeted identification of new promising candidates with specific activities suitable for known and novel applications.
In particular, in the context of the current antibiotic crisis and the scarcity of therapeutic alternatives for the treatment of multidrug-resistant bacterial and viral infections, the search for new therapeutics is 1 persisting challenge that could be addressed by venom research.
Owing to their devastating impact on biodiversity and concerns for human safety, there is great interest in replacing conventional pesticides with eco-friendly bioinsecticides. Animal venoms, especially from predators that feed on insects, may be extremely promising for identifying novel natural insecticides, with strict species-specific action.
The whole process from bioprospecting to the final development of a compound for translational applications remains challenging. The approaches here outlined combining multiple aspects of animal venoms, including the biological and ecological traits of venomous species, would greatly improve the effectiveness of this process.
Data Availability
Not applicable.
Abbreviations
AMP: antimicrobial peptide; AMPA: α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor; ASIC: acid-sensing ion channel; BLAST: Basic Local Alignment Search Tool; cDNA: complementary DNA; CRISPR: clustered regularly interspaced short palindromic repeats; ESI: electrospray ionization; FDA: Food and Drug Administration; GPCR: G-protein-coupled receptors; GUI: graphical user interface; ICK: inhibitory cysteine knot; L-AAO: L-amino acid oxidase; LC-MS/MS: liquid chromatography–tandem mass spectrometry; mRNA: messenger RNA; MS: mass spectrometry; MSI: mass spectrometry imaging; NET: norepinephrine transporter; NMDA: glutamate-gated cation channels; NMR: nuclear magnetic resonance; PLA2: phospholipase A2; PTM: posttranslational modification; scRNA-seq: single-cell RNA-sequencing; TRP: transient receptor potential.
Competing Interests
The authors declare that they have no competing interests.
Funding
This work is funded by the European Cooperation in Science and Technology (COST, www.cost.eu) and based upon work from the COST Action CA19144 – European Venom Network (EUVEN, see https://euven-network.eu/). This review is an outcome of EUVEN Working Group 2 (“Best practices and innovative tools in venomics”) led by B.M.v.R. As coordinator of the group Animal Venomics until end 2021 at the Institute for Insectbiotechnology, JLU Giessen, B.M.v.R. acknowledges the Centre for Translational Biodiversity Genomics (LOEWE-TBG) in the programme “LOEWE – Landes-Offensive zur Entwicklung Wissenschaftlich-ökonomischer Exzellenz” of Hesse's Ministry of Higher Education, Research, and the Arts. B.M.v.R. and I.K. further acknowledge funding on venom research by the German Science Foundation to B.M.v.R. (DFG RE3454/6-1). A.C., A.V., and G.Z. were supported by the European Union's Horizon 2020 Research and Innovation program through Marie Sklodowska-Curie Individual Fellowships (grant agreements No. A.C.: 896849, A.V.: 841576, and G.Z.: 845674). M.P.I. is supported by the TALENTO Program by the Regional Madrid Government (2018-T1/BIO-11262). T.H.'s venom research is funded by the DFG projects 271522021 and 413120531. L.E. was supported by grant No. 7017-00288 from the Danish Council for Independent Research (Technology and Production Sciences). N.I. acknowledges funding on venom research by the Research Fund of Nevsehir Haci Bektas Veli University (project Nos. ABAP20F28, BAP18F26). M.I.K. and A.P. acknowledge support from GSRT National Research Infrastructure structural funding project INSPIRED (MIS 5002550). G.A. acknowledges support from the Slovenian Research Agency grants P1-0391, J4-8225, and J4-2547. G.G. acknowledges support from the Institute for Medical Research and Occupational Health, Zagreb, Croatia. E.A.B.U. is supported by a Norwegian Research Council FRIPRO-YRT Fellowship No. 287462.
Authors' Contributions
Lead, major conceptualization, first draft, graphics, and illustration by B.M.v.R.; all authors wrote the main text and edited the final manuscript. Except for the first author, authors are listed alphabetically with respect to the last name. All authors have read and agreed to the published version of the manuscript.
Supplementary Material
Mark J. Margres -- 2/17/2022 Reviewed
Matthew Holding -- 2/21/2022 Reviewed
Jason Macrander, Ph. D. -- 3/8/2022 Reviewed
ACKNOWLEDGEMENTS
We thank Ronald Jenner and Stuart Ainsworth for commenting and editing the final manuscript version. M.D. is grateful to Prof. R. D. Süßmuth for the supervision and support during the Ph.D. time, in which this manuscript was achieved. B.M.v.R. and I.K. thank Andreas Vilcinskas for support and work space at the Institute of Insectbiotechnology within the group Animal Venomics. B.M.v.R. finally thanks Ingo Ebersberger for his help and support.
Contributor Information
Bjoern M von Reumont, Goethe University Frankfurt, Institute for Cell Biology and Neuroscience, Department for Applied Bioinformatics, 60438 Frankfurt am Main, Germany; LOEWE Centre for Translational Biodiversity Genomics, Senckenberg Frankfurt, Senckenberganlage 25, 60235 Frankfurt, Germany; Justus Liebig University Giessen, Institute for Insectbiotechnology, Heinrich Buff Ring 26-32, 35396 Giessen, Germany.
Gregor Anderluh, Department of Molecular Biology and Nanobiotechnology, National Institute of Chemistry, 1000 Ljubljana, Slovenia.
Agostinho Antunes, CIIMAR/CIMAR, Interdisciplinary Centre of Marine and Environmental Research, University of Porto, Terminal de Cruzeiros do Porto de Leixões, Av. General Norton de Matos, s/n, 4450–208 Porto, Portugal; Department of Biology, Faculty of Sciences, University of Porto, Rua do Campo Alegre, 4169-007 Porto, Portugal.
Naira Ayvazyan, Orbeli Institute of Physiology of NAS RA, Orbeli ave. 22, 0028 Yerevan, Armenia.
Dimitris Beis, Developmental Biology, Centre for Clinical, Experimental Surgery and Translational Research, Biomedical Research Foundation Academy of Athens, Athens 11527, Greece.
Figen Caliskan, Department of Biology, Faculty of Science and Letters, Eskisehir Osmangazi University, TR-26040 Eskisehir, Turkey.
Ana Crnković, Department of Molecular Biology and Nanobiotechnology, National Institute of Chemistry, 1000 Ljubljana, Slovenia.
Maik Damm, Technische Universität Berlin, Department of Chemistry, Straße des 17. Juni 135, 10623 Berlin, Germany.
Sebastien Dutertre, IBMM, Univ Montpellier, CNRS, ENSCM, 34095 Montpellier, France.
Lars Ellgaard, Department of Biology, University of Copenhagen, DK-2200 Copenhagen, Denmark.
Goran Gajski, Institute for Medical Research and Occupational Health, Mutagenesis Unit, Ksaverska cesta 2, 10000 Zagreb, Croatia.
Hannah German, Amsterdam Institute of Molecular and Life Sciences, Division of BioAnalytical Chemistry, Faculty of Science, Vrije Universiteit Amsterdam, De Boelelaan 1085, 1081HV Amsterdam, The Netherlands.
Beata Halassy, University of Zagreb, Centre for Research and Knowledge Transfer in Biotechnology, Trg Republike Hrvatske 14, 10000 Zagreb, Croatia.
Benjamin-Florian Hempel, BIH Center for Regenerative Therapies BCRT, Charité - Universitätsmedizin Berlin, Augustenburger Platz 1, 13353 Berlin, Germany.
Tim Hucho, Translational Pain Research, Department of Anesthesiology and Intensive Care Medicine, Faculty of Medicine and University Hospital Cologne, University of Cologne, 50931 Cologne, Germany.
Nasit Igci, Nevsehir Haci Bektas Veli University, Faculty of Arts and Sciences, Department of Molecular Biology and Genetics, 50300 Nevsehir, Turkey.
Maria P Ikonomopoulou, Madrid Institute for Advanced Studies in Food, Madrid,E28049, Spain; The University of Queensland, St Lucia, QLD 4072, Australia.
Izhar Karbat, Department of Biomolecular Sciences, Weizmann Institute of Science, Rehovot 76100, Israel.
Maria I Klapa, Metabolic Engineering and Systems Biology Laboratory, Institute of Chemical Engineering Sciences, Foundation for Research & Technology Hellas (FORTH/ICE-HT), Patras GR-26504, Greece.
Ivan Koludarov, Justus Liebig University Giessen, Institute for Insectbiotechnology, Heinrich Buff Ring 26-32, 35396 Giessen, Germany.
Jeroen Kool, Amsterdam Institute of Molecular and Life Sciences, Division of BioAnalytical Chemistry, Faculty of Science, Vrije Universiteit Amsterdam, De Boelelaan 1085, 1081HV Amsterdam, The Netherlands.
Tim Lüddecke, LOEWE Centre for Translational Biodiversity Genomics, Senckenberg Frankfurt, Senckenberganlage 25, 60235 Frankfurt, Germany; Department of Bioresources, Fraunhofer Institute for Molecular Biology and Applied Ecology, 35392 Gießen, Germany.
Riadh Ben Mansour, Department of Life Sciences, Faculty of Sciences, Gafsa University, Campus Universitaire Siidi Ahmed Zarrouk, 2112 Gafsa, Tunisia.
Maria Vittoria Modica, Dept. of Biology and Evolution of Marine Organisms (BEOM), Stazione Zoologica Anton Dohrn, Via Po 25c, I-00198 Roma, Italy.
Yehu Moran, Department of Ecology, Evolution and Behavior, Alexander Silberman Institute of Life Sciences, Faculty of Science, The Hebrew University of Jerusalem, Jerusalem 9190401, Israel.
Ayse Nalbantsoy, Department of Bioengineering, Faculty of Engineering, Ege University, 35100 Bornova, Izmir, Turkey.
María Eugenia Pachón Ibáñez, Unit of Infectious Diseases, Microbiology, and Preventive Medicine, Virgen del Rocío University Hospital, Institute of Biomedicine of Seville, 41013 Sevilla, Spain; CIBER de Enfermedades Infecciosas, Instituto de Salud Carlos III, Madrid, Spain.
Alexios Panagiotopoulos, Metabolic Engineering and Systems Biology Laboratory, Institute of Chemical Engineering Sciences, Foundation for Research & Technology Hellas (FORTH/ICE-HT), Patras GR-26504, Greece; Animal Biology Division, Department of Biology, University of Patras, Patras, GR-26500, Greece.
Eitan Reuveny, Department of Biomolecular Sciences, Weizmann Institute of Science, Rehovot 76100, Israel.
Javier Sánchez Céspedes, Unit of Infectious Diseases, Microbiology, and Preventive Medicine, Virgen del Rocío University Hospital, Institute of Biomedicine of Seville, 41013 Sevilla, Spain; CIBER de Enfermedades Infecciosas, Instituto de Salud Carlos III, Madrid, Spain.
Andy Sombke, Department of Evolutionary Biology, University of Vienna, Djerassiplatz 1, 1030 Vienna, Austria.
Joachim M Surm, Department of Ecology, Evolution and Behavior, Alexander Silberman Institute of Life Sciences, Faculty of Science, The Hebrew University of Jerusalem, Jerusalem 9190401, Israel.
Eivind A B Undheim, University of Oslo, Centre for Ecological and Evolutionary Synthesis, Postboks 1066 Blindern 0316 Oslo, Norway.
Aida Verdes, Department of Biodiversity and Evolutionary Biology, Museo Nacional de Ciencias Naturales, José Gutiérrez Abascal 2, 28006 Madrid, Spain.
Giulia Zancolli, Department of Ecology and Evolution, University of Lausanne, 1015 Lausanne, Switzerland; Swiss Institute of Bioinformatics, 1015 Lausanne, Switzerland.
References
- 1. Holford M, Daly M, King GF, et al. Venoms to the rescue. Science. 2018;361(6405):842–4. [DOI] [PubMed] [Google Scholar]
- 2. McDermott A. News Feature: Venom back in vogue as a wellspring for drug candidates. Proc Natl Acad Sci U S A. 2020;117(19):10100–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fry BG, Roelants K, Champagne DE, et al. The toxicogenomic multiverse: convergent recruitment of proteins into animal venoms. Annu Rev Genomics Hum Genet. 2009;10(1):483–511. [DOI] [PubMed] [Google Scholar]
- 4. Casewell NR, Wüster W, Vonk FJ, et al. Complex cocktails: the evolutionary novelty of venoms. Trends Ecol Evol. 2013;28(4):219–29. [DOI] [PubMed] [Google Scholar]
- 5. Schendel V, Rash LD, Jenner RA, et al. The diversity of venom: the importance of behavior and venom system morphology in understanding its ecology and evolution. Toxins. 2019;11(11):666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. von Reumont BM, Campbell LI, Jenner RA. Quo vadis venomics? A roadmap to neglected venomous invertebrates. Toxins. 2014;6(12):3488–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sunagar K, Morgenstern D, Reitzel AM, et al. Ecological venomics: how genomics, transcriptomics and proteomics can shed new light on the ecology and evolution of venom. J Proteomics. 2016;135:62–72. [DOI] [PubMed] [Google Scholar]
- 8. Robinson KE, Holding ML, Whitford MD, et al. Phenotypic and functional variation in venom and venom resistance of two sympatric rattlesnakes and their prey. J Evol Biol. 2021;34(9):1447–65. [DOI] [PubMed] [Google Scholar]
- 9. Holding ML, Biardi JE, Gibbs HL. Coevolution of venom function and venom resistance in a rattlesnake predator and its squirrel prey. Proc Biol Sci. 2016;283(1829):20152841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Holding ML, Drabeck DH, Jansa SA, et al. Venom resistance as a model for understanding the molecular basis of complex coevolutionary adaptations. Integr Comp Biol. 2016;56(5):1032–43. [DOI] [PubMed] [Google Scholar]
- 11. Smiley-Walters SA, Farrell TM, Gibbs HL. The importance of species: pygmy rattlesnake venom toxicity differs between native prey and related non-native species. Toxicon. 2018;144:42–7. [DOI] [PubMed] [Google Scholar]
- 12. Smiley-Walters SA, Farrell TM, Gibbs HL. High levels of functional divergence in toxicity towards prey among the venoms of individual pigmy rattlesnakes. Biol Lett. 2019;15(2):20180876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. King G. Venoms to Drugs: Venom as a source for the development of human therapeutics. Cambridge: Royal Society of Chemistry; 2015. [Google Scholar]
- 14. King GF. Tying pest insects in knots: the deployment of spider-venom-derived knottins as bioinsecticides. Pest Manag Sci. 2019;75(9):2437–45. [DOI] [PubMed] [Google Scholar]
- 15. Bordon KCF, Cologna CT, Fornari-Baldo EC, et al. From animal poisons and venoms to medicines: achievements, challenges and perspectives in drug discovery. Front Pharmacol. 2020;11:1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Modica MV, Ahmad R, Ainsworth S, et al. The new COST Action European Venom Network (EUVEN)—synergy and future perspectives of modern venomics. Gigascience. 2021;10(3):giab019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rodrigo C, Gnanathasan A. Management of scorpion envenoming: a systematic review and meta-analysis of controlled clinical trials. Syst Rev. 2017;6(1):74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pla D, Rodríguez Y, Calvete JJ. Third generation antivenomics: pushing the limits of the in vitro preclinical assessment of antivenoms. Toxins. 2017;9(5):158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gutiérrez JM, Calvete JJ, Habib AG, et al. Snakebite envenoming. Nat Rev Dis Primers. 2017;3:17063. [DOI] [PubMed] [Google Scholar]
- 20. Needleman RK, Neylan IP, Erickson T. Potential environmental and ecological effects of global climate change on venomous terrestrial species in the wilderness. Wilderness Environ Med. 2018;29(2):226–38. [DOI] [PubMed] [Google Scholar]
- 21. Dias-Lopes C, Paiva AL, Guerra-Duarte C, et al. Venomous arachnid diagnostic assays, lessons from past attempts. Toxins. 2018;10(9):365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pucca MB, Cerni FA, Oliveira IS, et al. Bee updated: current knowledge on bee venom and bee envenoming therapy. Front Immunol. 2019;10:2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Linardich C, Brookson CB, Green SJ. Trait-based vulnerability reveals hotspots of potential impact for a global marine invader. Global Change Biol. 2021;27(18):4322–38. [DOI] [PubMed] [Google Scholar]
- 24. Giallongo G, Douek J, Harbuzov Z, et al. Long-term changes in population genetic features of a rapidly expanding marine invader: implication for invasion success. Biol Invasions. 2021;23(8):2541–52. [Google Scholar]
- 25. Wägele H, Klussmann-Kolb A, Kuhlmann M, et al. The taxonomist—an endangered race. A practical proposal for its survival. Front Zool. 2011;8(1):25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Britz R, Hundsdörfer A, Fritz U. Funding, training, permits—the three big challenges of taxonomy. Megataxa. 2020;1(1):doi: 10.11646/MEGATAXA.1.1.10. [DOI] [Google Scholar]
- 27. Coleman CO, Radulovici AE. Challenges for the future of taxonomy: talents, databases and knowledge growth. Megataxa. 2020;1(1):doi: 10.11646/MEGATAXA.1.1.5. [DOI] [Google Scholar]
- 28. Casewell NR, Jackson TNW, Laustsen AH, et al. Causes and consequences of snake venom variation. Trends Pharmacol Sci. 2020;41(8):570–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ambler J, Diallo AA, Dearden PK, et al. Including digital sequence data in the Nagoya Protocol can promote data sharing. Trends Biotechnol. 2021;39(2):116–25. [DOI] [PubMed] [Google Scholar]
- 30. UNEP-CBD Secretariat : Convention on Biological Diversity - The Access and Benefit-Sharing Clearing-House. https://www.cbd.int/. 2021. Accessed 1 November 2021. [Google Scholar]
- 31. Prathapan KD, Pethiyagoda R, Bawa KS, et al. 172 co-signatories from 35 countries. When the cure kills—CBD limits biodiversity research. Science. 2018;360(6396):1405–6. [DOI] [PubMed] [Google Scholar]
- 32. Heinrich M, Scotti F, Andrade-Cetto A, et al. Access and benefit sharing under the Nagoya protocol—Quo Vadis? Six Latin American case studies assessing opportunities and risk. Front Pharmacol. 2020;11:765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Karger EJ, Scholz AH. DSI, the Nagoya Protocol, and stakeholders’ concerns. Trends Biotechnol. 2021;39(2):110–2. [DOI] [PubMed] [Google Scholar]
- 34. Fry BG, Undheim EAB, Jackson TNW, et al. Research methods. In: Venomous Reptiles and Their Toxins Evolution, Pathophysiology and Biodiscovery. Oxford University Press; 2015:153–214. [Google Scholar]
- 35. Low DHW, Sunagar K, Undheim EAB, et al. Dracula's children: molecular evolution of vampire bat venom. J Proteomics. 2013;89:95–111. [DOI] [PubMed] [Google Scholar]
- 36. Mailho-Fontana PL, Antoniazzi MM, Alexandre C, et al. Morphological evidence for an oral venom system in caecilian amphibians. iScience. 2020;23(7):101234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Harris RJ, Jenner RA. Evolutionary ecology of fish venom: adaptations and consequences of evolving a venom system. Toxins. 2019;11(2):60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Almada F, Duarte AG, Besson S, et al. A simple and practical technique for fish venom extraction - protein content analysis for future biotechnological applications. Front Mar Sci. 2016;3:doi: 10.3389/conf.FMARS.2016.04.00124. [DOI] [Google Scholar]
- 39. Saggiomo SL, Zelenka C, Seymour J. Relationship between food and venom production in the estuarine stonefish Synanceia horrida. Toxicon. 2017;125:19–23. [DOI] [PubMed] [Google Scholar]
- 40. Mac̆ek P, Senc̆ic̆ L, Lebez D. Isolation and partial characterisation of three lethal and hemolytic toxins from the sea anemone Actinia cari. Toxicon. 1982;20(1):181–5. [DOI] [PubMed] [Google Scholar]
- 41. Kimura A, Nakagawa H, Hayashi H, et al. Seasonal changes in contractile activity of a toxic substance from the pedicellaria of the sea urchin Toxopneustes pileolus.Toxicon. 1984;22(3):353–8. [DOI] [PubMed] [Google Scholar]
- 42. Kem WR, Parten B, Pennington MW, et al. Isolation, characterization, and amino acid sequence of a polypeptide neurotoxin occurring in the sea anemone Stichodactyla helianthus. Biochemistry. 1989;28(8):3483–9. [DOI] [PubMed] [Google Scholar]
- 43. Purushottama G, Venkateshvaran K, Nalini P. Bioactivities of extracts from the marine sponge Halichondria panicea. J Venom Anim Toxins Incl Trop Dis. 2009;15(3):444–59. [Google Scholar]
- 44. Jouiaei M, Casewell NR, Yanagihara AA, et al. Firing the sting: chemically induced discharge of cnidae reveals novel proteins and peptides from box jellyfish (Chironex fleckeri) venom. Toxins. 2015;7(3):936–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dutertre S, Jin A, Vetter I, et al. Evolution of separate predation- and defence-evoked venoms in carnivorous cone snails. Nat Commun. 2014;5:3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hopkins C, Grilley M, Miller C, et al. A new family of Conus peptides targeted to the nicotinic acetylcholine receptor. J Biol Chem. 1995;270(38):22361–7. [DOI] [PubMed] [Google Scholar]
- 47. Gonçalves Paterson Fox E, Russ Solis D, Delazari dos Santos L, et al. A simple, rapid method for the extraction of whole fire ant venom (Insecta: Formicidae: Solenopsis). Toxicon. 2013;65:5–8. [DOI] [PubMed] [Google Scholar]
- 48. Garb JE. Extraction of venom and venom gland microdissections from spiders for proteomic and transcriptomic analyses. J Vis Exp. 2014;(93):e51618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Undheim EAB, Fry BG, King GF. Centipede venom: recent discoveries and current state of knowledge. Toxins. 2015;7(3):679–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. von Reumont BM, Blanke A, Richter S, et al. The first venomous crustacean revealed by transcriptomics and functional morphology: remipede venom glands express a unique toxin cocktail dominated by enzymes and a neurotoxin. Mol Biol Evol. 2014;31(1):48–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Walker AA, Rosenthal M, Undheim EEA, et al. Harvesting venom toxins from assassin bugs and other heteropteran insects. J Vis Exp. 2018;(134):57729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Piek T. Methods for the Collection of Venoms. Venoms of the Hymenoptera. Oxford: Academic; 1986:45–54. [Google Scholar]
- 53. Aili SR, Touchard A, Petitclerc F, et al. Combined peptidomic and proteomic analysis of electrically stimulated and manually dissected venom from the South American bullet ant Paraponera clavata. J Proteome Res. 2017;16(3):1339–51. [DOI] [PubMed] [Google Scholar]
- 54. Walker AA, Robinson SD, Hamilton BF, et al. Deadly proteomes: a practical guide to proteotranscriptomics of animal venoms. Proteomics. 2020;20(17-18):1900324. [DOI] [PubMed] [Google Scholar]
- 55. Jesupret C, Baumann K, Jackson TNW, et al. Vintage venoms: proteomic and pharmacological stability of snake venoms stored for up to eight decades. J Proteomics. 2014;105:285–94. [DOI] [PubMed] [Google Scholar]
- 56. Klupczynska A, Pawlak M, Kokot Z, et al. Application of metabolomic tools for studying low molecular-weight fraction of animal venoms and poisons. Toxins. 2018;10(8):306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gorrochategui E, Jaumot J, Lacorte S, et al. Data analysis strategies for targeted and untargeted LC-MS metabolomic studies: overview and workflow. TrAC Trends Anal Chem. 2016;82:425–42. [Google Scholar]
- 58. Lee DY, Bowen BP, Northen TR. Mass spectrometry—based metabolomics, analysis of metabolite-protein interactions, and imaging. Biotechniques. 2010;49(2):557–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hutchinson DA, Savitzky AH, Burghardt GM, et al. Chemical defense of an Asian snake reflects local availability of toxic prey and hatchling diet. J Zool. 2013;289(4):270–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Aird S, Villar Briones A, Roy M, et al. Polyamines as snake toxins and their probable pharmacological functions in envenomation. Toxins. 2016;8(10):279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Villar-Briones A, Aird S. Organic and peptidyl constituents of snake venoms: the picture is vastly more complex than we imagined. Toxins. 2018;10(10):392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Acunha T, Nardini V, Faccioli LH. A lipidomics approach reveals new insights into Crotalus durissus terrificus and Bothrops moojeni snake venoms. Arch Toxicol. 2021;95(1):345–53. [DOI] [PubMed] [Google Scholar]
- 63. Evans ERJ, McIntyre L, Northfield TD, et al. Small molecules in the venom of the scorpion Hormurus waigiensis. Biomedicines. 2020;8(8):259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Palma MS, Itagaki Y, Fujita T, et al. Structural characterization of a new acylpolyaminetoxin from the venom of Brazilian garden spider Nephilengys cruentata. Toxicon. 1998;36(3):485–93. [DOI] [PubMed] [Google Scholar]
- 65. Hisada M, Fujita T, Naoki H, et al. Structures of spider toxins: hydroxyindole-3-acetylpolyamines and a new generalized structure of type-E compounds obtained from the venom of the Joro spider, Nephila clavata. Toxicon. 1998;36(8):1115–25. [DOI] [PubMed] [Google Scholar]
- 66. Schroeder FC, Taggi AE, Gronquist M, et al. NMR-spectroscopic screening of spider venom reveals sulfated nucleosides as major components for the brown recluse and related species. Proc Natl Acad Sci U S A. 2008;105(38):14283–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Forster YM, Reusser S, Forster F, et al. VenoMS—a website for the low molecular mass compounds in spider venoms. Metabolites. 2020;10(8):327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lai L-C, Huang R-N, Wu W-J. Venom alkaloids of monogyne and polygyne forms of the red imported fire ant, Solenopsis invicta, in Taiwan. Insect Soc. 2008;55(4):443–9. [Google Scholar]
- 69. Chen L, Fadamiro HY. Re-investigation of venom chemistry of Solenopsis fire ants. II. Identification of novel alkaloids in S. invicta. Toxicon. 2009;53(5):479–86. [DOI] [PubMed] [Google Scholar]
- 70. Chen L, Fadamiro HY. Re-investigation of venom chemistry of Solenopsis fire ants. I. Identification of novel alkaloids in S. richteri. Toxicon. 2009;53(5):469–78. [DOI] [PubMed] [Google Scholar]
- 71. Pawlak M, Klupczynska A, Kokot ZJ, et al. Extending metabolomic studies of Apis mellifera venom: LC-MS-based targeted analysis of organic acids. Toxins. 2020;12(1):14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Klupczynska A, Plewa S, Dereziński P, et al. Identification and quantification of honeybee venom constituents by multiplatform metabolomics. Sci Rep. 2020;10(1):21645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Torres VDO, Piva RC, Antonialli WF Jr, et al. Free amino acids analysis in the venom of the social wasp Polistes lanio under different forms of preservation. Orbital. 2018;10(1):doi: 10.17807/orbital.v10i1.1005. [DOI] [Google Scholar]
- 74. Yates JR, Ruse CI, Nakorchevsky A. Proteomics by mass spectrometry: approaches, advances, and applications. Annu Rev Biomed Eng. 2009;11(1):49–79. [DOI] [PubMed] [Google Scholar]
- 75. Damm M, Hempel B-F, Nalbantsoy A, et al. Comprehensive snake venomics of the Okinawa Habu pit viper, Protobothrops flavoviridis, by complementary mass spectrometry-guided approaches. Molecules. 2018;23(8):1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Bastos VA, Gomes-Neto F, Rocha SLG, et al. The interaction between the natural metalloendopeptidase inhibitor BJ46a and its target toxin jararhagin analyzed by structural mass spectrometry and molecular modeling. J Proteomics. 2020;221:103761. [DOI] [PubMed] [Google Scholar]
- 77. Mouchbahani-Constance S, Sharif-Naeini R. Proteomic and transcriptomic techniques to decipher the molecular evolution of venoms. Toxins. 2021;13(2):154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Calvete JJ, Lomonte B, Saviola AJ, et al. Mutual enlightenment: a toolbox of concepts and methods for integrating evolutionary and clinical toxinology via snake venomics and the contextual stance. Toxicon: X. 2021;9-10:100070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Calvete JJ, Pla D, Els J, et al. Combined molecular and elemental mass spectrometry approaches for absolute quantification of proteomes: application to the venomics characterization of the two species of desert black cobras, Walterinnesia aegyptia and Walterinnesia morgani. J Proteome Res. 2021;20(11):5064–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Lomonte B, Calvete JJ. Strategies in “snake venomics” aiming at an integrative view of compositional, functional, and immunological characteristics of venoms. J Venom Anim Toxins Incl Trop Dis. 2017;23:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Dupree EJ, Jayathirtha M, Yorkey H, et al. A critical review of bottom-up proteomics: the good, the bad, and the future of this field. Proteomes. 2020;8(3):14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Slagboom J, Kaal C, Arrahman A, et al. Analytical strategies in venomics. Microchem J. 2022;175:107187. [Google Scholar]
- 83. Melani RD, Goto-Silva L, Nogueira FCS, et al. Shotgun approaches for venom analysis. In: Gopalakrishnakone P, Calvete JJ, eds. Venom Genomics and Proteomics. Dordrecht: Springer; 2016. doi: 10.1007/978-94-007-6649-5_26-1. [DOI] [Google Scholar]
- 84. Huang T, Wang J, Yu W, et al. Protein inference: a review. Brief Bioinform. 2012;13(5):586–614. [DOI] [PubMed] [Google Scholar]
- 85. Toby TK, Fornelli L, Kelleher NL. Progress in top-down proteomics and the analysis of proteoforms. Annu Rev Anal Chem. 2016;9(1):499–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Melani RD, Nogueira FCS, Domont GB. It is time for top-down venomics. J Venom Anim Toxins Incl Trop Dis. 2017;23:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Hempel B-F, Damm M, Mrinalini, et al. Extended snake venomics by top-down in-source decay: investigating the newly discovered Anatolian meadow viper subspecies, Vipera anatolica senliki. J Proteome Res. 2020;19(4):1731–49. [DOI] [PubMed] [Google Scholar]
- 88. Donnelly DP, Rawlins CM, DeHart CJ, et al. Best practices and benchmarks for intact protein analysis for top-down mass spectrometry. Nat Methods. 2019;16(7):587–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Ghezellou P, Garikapati V, Kazemi SM, et al. A perspective view of top-down proteomics in snake venom research. Rapid Commun Mass Spectrom. 2019;33(S1):20–7. [DOI] [PubMed] [Google Scholar]
- 90. Damm M, Hempel B-F, Süssmuth RD. Old World Vipers—a review about snake venom proteomics of Viperinae and their variations. Toxins. 2021;13(6):427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Vaudel M, Burkhart JM, Zahedi RP, et al. PeptideShaker enables reanalysis of MS-derived proteomics data sets. Nat Biotechnol. 2015;33(1):22–4. [DOI] [PubMed] [Google Scholar]
- 92. Chen C, Hou J, Tanner JJ, et al. Bioinformatics methods for mass spectrometry-based proteomics data analysis. Int J Mol Sci. 2020;21(8):2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Hus KK, Marczak Ł, Petrilla V, et al. Different research approaches in unraveling the venom proteome of Naja ashei. Biomolecules. 2020;10(9):1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Yang H, Chi H, Zeng W-F, et al. pNovo 3: precise de novo peptide sequencing using a learning-to-rank framework. Bioinformatics. 2019;35(14):i183–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Brahma RK, McCleary RJR, Kini RM, et al. Venom gland transcriptomics for identifying, cataloging, and characterizing venom proteins in snakes. Toxicon. 2015;93:1–10. [DOI] [PubMed] [Google Scholar]
- 96. Petras D, Hempel B-F, Göçmen B, et al. Intact protein mass spectrometry reveals intraspecies variations in venom composition of a local population of Vipera kaznakovi in Northeastern Turkey. J Proteomics. 2019;199:31–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Catherman AD, Skinner OS, Kelleher NL. Top-down proteomics: facts and perspectives. Biochem Biophys Res Commun. 2014;445(4):683–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Wang CR, Bubner ER, Jovcevski B, et al. Interrogating the higher order structures of snake venom proteins using an integrated mass spectrometric approach. J Proteomics. 2020;216:103680. [DOI] [PubMed] [Google Scholar]
- 99. Melani RD, Skinner OS, Fornelli L, et al. Mapping proteoforms and protein complexes from king cobra venom using both denaturing and native top-down proteomics. Mol Cell Proteomics. 2016;15(7):2423–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. von Reumont BM. Studying smaller and neglected organisms in modern evolutionary venomics implementing RNASeq (transcriptomics)—a critical guide. Toxins. 2018;10(7):292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Smith JJ, Undheim EAB. True lies: using proteomics to assess the accuracy of transcriptome-based venomics in centipedes uncovers false positives and reveals startling intraspecific variation in Scolopendra subspinipes. Toxins. 2018;10(3):96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Schuierer S, Carbone W, Knehr J, et al. A comprehensive assessment of RNA-seq protocols for degraded and low-quantity samples. BMC Genomics. 2017;18(1):442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Drukewitz SH, von Reumont BM. The significance of comparative genomics in modern evolutionary venomics. Front Ecol Evol. 2019;7:doi: 10.3389/fevo.2019.00163. [DOI] [Google Scholar]
- 104. Drukewitz SH, Bokelmann L, Undheim EAB, et al. Toxins from scratch? Diverse, multimodal gene origins in the predatory robber fly Dasypogon diadema indicate a dynamic venom evolution in dipteran insects. Gigascience. 2019;8(7):doi: 10.1093/gigascience/giz081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Modica MV, Lombardo F, Franchini P, et al. The venomous cocktail of the vampire snail Colubraria reticulata (Mollusca, Gastropoda). BMC Genomics. 2015;16(1):441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Fassio G, Modica MV, Mary L, et al. Venom diversity and evolution in the most divergent cone snail genus Profundiconus. Toxins. 2019;11(11):623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Macrander J, Broe M, Daly M. Tissue-specific venom composition and differential gene expression in sea anemones. Genome Biol Evol. 2016;8(8):2358–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Earl D, Bradnam K, St John J, et al. Assemblathon 1: a competitive assessment of de novo short read assembly methods. Genome Res. 2011;21(12):2224–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Hara Y, Tatsumi K, Yoshida M, et al. Optimizing and benchmarking de novo transcriptome sequencing: from library preparation to assembly evaluation. BMC Genomics. 2015;16:977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Conesa A, Madrigal P, Tarazona S, et al. A survey of best practices for RNA-seq data analysis. Genome Biol. 2016;17:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Holding ML, Margres MJ, Mason AJ, et al. Evaluating the performance of de novo assembly methods for venom-gland transcriptomics. Toxins. 2018;10(6):249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Hölzer M, Marz M. De novo transcriptome assembly: a comprehensive cross-species comparison of short-read RNA-Seq assemblers. Gigascience. 2019;8(5):doi: 10.1093/gigascience/giz039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Schurch NJ, Schofield P, Gierliński M, et al. How many biological replicates are needed in an RNA-seq experiment and which differential expression tool should you use?. RNA. 2016;22(6):839–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Van den Berge K, Hembach KM, Soneson C, et al. RNA sequencing data: hitchhiker's guide to expression analysis. Annu Rev Biomed Data Sci. 2019;2(1):139–73. [Google Scholar]
- 115. Steijger T, Abril JF, Engström PG, et al. Assessment of transcript reconstruction methods for RNA-seq. Nat Methods. 2013;10(12):1177–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Venturini L, Caim S, Kaithakottil GG, et al. Leveraging multiple transcriptome assembly methods for improved gene structure annotation. Gigascience. 2018;7(8):doi: 10.1093/gigascience/giy093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Cerveau N, Jackson DJ. Combining independent de novo assemblies optimizes the coding transcriptome for nonconventional model eukaryotic organisms. BMC Bioinformatics. 2016;17(1):525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. MacManes MD. The Oyster River Protocol: a multi-assembler and kmer approach for de novo transcriptome assembly. PeerJ. 2018;6:e5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Rivera-Vicéns RE, Escudero CG, Conci N, et al. TransPi – a comprehensive TRanscriptome ANalysiS PIpeline for de novo transcriptome assembly. Mol Ecol Resour. 2022:doi: 10.1111/1755-0998.13593. [DOI] [PubMed] [Google Scholar]
- 120. Ramberg S, Høyheim B, Østbye T-KK, et al. A de novo full-length mRNA transcriptome generated from hybrid-corrected PacBio long-reads improves the transcript annotation and identifies thousands of novel splice variants in Atlantic salmon. Front Genet. 2021;12:656334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Post Y, Puschhof J, Beumer J, et al. Snake venom gland organoids. Cell. 2020;180(2):233–47.e21. [DOI] [PubMed] [Google Scholar]
- 122. Surm JM, Moran Y. Insights into how development and life-history dynamics shape the evolution of venom. Evodevo. 2021;12(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. García-Castro H, Kenny NJ, Iglesias M, et al. ACME dissociation: a versatile cell fixation-dissociation method for single-cell transcriptomics. Genome Biol. 2021;22(1):89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Levy S, Elek A, Grau-Bové X, et al. A stony coral cell atlas illuminates the molecular and cellular basis of coral symbiosis, calcification, and immunity. Cell. 2021;184(11):2973–87.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Sebé-Pedrós A, Saudemont B, Chomsky E, et al. Cnidarian cell type diversity and regulation revealed by whole-organism single-cell RNA-Seq. Cell. 2018;173(6):1520–34.e20. [DOI] [PubMed] [Google Scholar]
- 126. Siebert S, Farrell JA, Cazet JF, et al. Stem cell differentiation trajectories in Hydra resolved at single-cell resolution. Science. 2019;365(6451):eaav9314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Yan F, Powell DR, Curtis DJ, et al. From reads to insight: a hitchhiker's guide to ATAC-seq data analysis. Genome Biol. 2020;21(1):22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Buenrostro JD, Giresi PG, Zaba LC, et al. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10(12):1213–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Margres MJ, Rautsaw RM, Strickland JL, et al. The Tiger Rattlesnake genome reveals a complex genotype underlying a simple venom phenotype. Proc Natl Acad Sci U S A. 2021;118(4):e2014634118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Undheim EAB, Hamilton BR, Kurniawan ND, et al. Production and packaging of a biological arsenal: evolution of centipede venoms under morphological constraint. Proc Natl Acad Sci U S A. 2015;112(13):4026–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Zancolli G, Casewell NR. Venom systems as models for studying the origin and regulation of evolutionary novelties. Mol Biol Evol. 2020;37(10):2777–90. [DOI] [PubMed] [Google Scholar]
- 132. Ashwood LM, Undheim EAB, Madio B, et al. Venoms for all occasions: the functional toxin profiles of different anatomical regions in sea anemones are related to their ecological function. Mol Ecol. 2022;31(3):866–83. [DOI] [PubMed] [Google Scholar]
- 133. Richter S, Helm C, Meunier FA, et al. Comparative analyses of glycerotoxin expression unveil a novel structural organization of the bloodworm venom system. BMC Evol Biol. 2017;17(1):64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Wurmbach H. Lehrbuch der Zoologie - Zoologie und Ökologie. Stuttgart: Gustav Fischer;1970:72–136. [Google Scholar]
- 135. Müller CHG, Rosenberg J, Hilken G. Ultrastructure, functional morphology and evolution of recto-canal epidermal glands in Myriapoda. Arthropod Struct Dev. 2014;433(1):43–61. [DOI] [PubMed] [Google Scholar]
- 136. Farkaš R. Apocrine secretion: new insights into an old phenomenon. Biochim Biophys Acta. 2015;1850(9):1740–50. [DOI] [PubMed] [Google Scholar]
- 137. Ritman EL. Current status of developments and applications of micro-CT. Annu Rev Biomed Eng. 2011;13(1):531–52. [DOI] [PubMed] [Google Scholar]
- 138. Gutiérrez Y, Ott D, Töpperwien M, et al. X-ray computed tomography and its potential in ecological research: a review of studies and optimization of specimen preparation. Ecol Evol. 2018;8(15):7717–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Hunter L, Dewanckele J. Evolution of micro-CT: Moving from 3D to 4D. Micros Today. 2021;29(3):28–34. [Google Scholar]
- 140. Arbuckle K. From molecules to macroevolution: venom as a model system for evolutionary biology across levels of life. Toxicon: X. 2020;6:100034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Robinson SD, Mueller A, Clayton D, et al. A comprehensive portrait of the venom of the giant red bull ant, Myrmecia gulosa, reveals a hyperdiverse hymenopteran toxin gene family. Sci Adv. 2018;4(9):eaau4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Walker Robinson, Undheim Jin, Han Fry, et al. Missiles of mass disruption: composition and glandular origin of venom used as a projectile defensive weapon by the assassin bug Platymeris rhadamanthus. Mol Cell Proteomics. 2012;11(3):M111.013987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Walker AA, Mayhew ML, Jin J, et al. The assassin bug Pristhesancus plagipennis produces two distinct venoms in separate gland lumens. Nat Commun. 2018;9(1):755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Arvidson R, Kaiser M, Lee SS, et al. Parasitoid jewel wasp mounts multipronged neurochemical attack to hijack a host brain. Mol Cell Proteomics. 2019;18(1):99–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Escalante T, Shannon J, Moura-da-Silva AM, et al. Novel insights into capillary vessel basement membrane damage by snake venom hemorrhagic metalloproteinases: a biochemical and immunohistochemical study. Arch Biochem Biophys. 2006;455(2):144–53. [DOI] [PubMed] [Google Scholar]
- 146. Baldo C, Ferreira MJ, Lopes DS, et al. Action of neuwiedase, a metalloproteinase isolated from Bothrops neuwiedi venom, on skeletal muscle: an ultrastructural and immunocytochemistry study. J Venom Anim Toxins Incl Trop Dis. 2010;16(3):462–9. [Google Scholar]
- 147. Lachumanan R, Armugam A, Durairaj P, et al. In situ hybridization and immunohistochemical analysis of the expression of cardiotoxin and neurotoxin genes in Naja naja sputatrix. J Histochem Cytochem. 1999;47(4):551–60. [DOI] [PubMed] [Google Scholar]
- 148. Han J, Permentier H, Bischoff R, et al. Imaging of protein distribution in tissues using mass spectrometry: an interdisciplinary challenge. TrAC Trends Anal Chem. 2019;112:13–28. [Google Scholar]
- 149. Madio B, Peigneur S, Chin YKY, et al. PHAB toxins: a unique family of predatory sea anemone toxins evolving via intra-gene concerted evolution defines a new peptide fold. Cell Mol Life Sci. 2018;75(24):4511–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Hamilton BR, Marshall DL, Casewell NR, et al. Mapping enzyme activity on tissue by functional mass spectrometry imaging. Angew Chem Int Ed. 2020;59(10):3855–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Ghezellou P, Heiles S, Kadesch P, et al. Venom gland mass spectrometry imaging of saw-scaled viper, Echis carinatus sochureki, at high lateral resolution. J Am Soc Mass Spectrom. 2021;32(4):1105–15. [DOI] [PubMed] [Google Scholar]
- 152. Hempel B-F, Damm M, Petras D, et al. Spatial venomics - Cobra venom system reveals spatial differentiation of snake toxins by mass spectrometry imaging. bioRxiv 2022;doi: 10.1101/2022.01.31.478453. [DOI] [PubMed] [Google Scholar]
- 153. Spraker JE, Luu GT, Sanchez LM. Imaging mass spectrometry for natural products discovery: a review of ionization methods. Nat Prod Rep. 2020;37(2):150–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Ståhl PL, Salmén F, Vickovic S, et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science. 2016;353(6294):78–82. [DOI] [PubMed] [Google Scholar]
- 155. Giacomello S, Salmén F, Terebieniec BK, et al. Spatially resolved transcriptome profiling in model plant species. Nat Plants. 2017;3:17061. [DOI] [PubMed] [Google Scholar]
- 156. Mantri M, Scuderi GJ, Abedini-Nassab R, et al. Spatiotemporal single-cell RNA sequencing of developing chicken hearts identifies interplay between cellular differentiation and morphogenesis. Nat Commun. 2021;12(1):1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Giacomello S. A new era for plant science: spatial single-cell transcriptomics. Curr Opin Plant Biol. 2021;60:102041. [DOI] [PubMed] [Google Scholar]
- 158. Vargas S, Caglar C, Büttner G, et al. (2021): Slime away: a simple CTAB-based high molecular weight DNA and RNA extraction protocol for “difficult” invertebrates. protocols.io. 10.17504/protocols.io.bwcwpaxe. [DOI] [Google Scholar]
- 159. Adema CM. Sticky problems: extraction of nucleic acids from molluscs. Philos Trans R Soc Lond B Biol Sci. 2021;376(1825):20200162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Lewin HA, Robinson GE, Kress WJ, et al. Earth BioGenome Project: sequencing life for the future of life. Proc Natl Acad Sci U S A. 2018;115(17):4325–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Rhie A, McCarthy SA, Fedrigo O, et al. Towards complete and error-free genome assemblies of all vertebrate species. Nature. 2021;592(7856):737–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Yin W, Wang Z-J, Li Q-Y, et al. Evolutionary trajectories of snake genes and genomes revealed by comparative analyses of five-pacer viper. Nat Commun. 2016;7:13107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Gendreau KL, Haney RA, Schwager EE, et al. House spider genome uncovers evolutionary shifts in the diversity and expression of black widow venom proteins associated with extreme toxicity. BMC Genomics. 2017;18(1):178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Shibata H, Chijiwa T, Oda-Ueda N, et al. The habu genome reveals accelerated evolution of venom protein genes. Sci Rep. 2018;8(1):11300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Casewell NR, Petras D, Card DC, et al. Solenodon genome reveals convergent evolution of venom in eulipotyphlan mammals. Proc Natl Acad Sci U S A. 2019;116(51):25745–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Suryamohan K, Krishnankutty SP, Guillory J, et al. The Indian cobra reference genome and transcriptome enables comprehensive identification of venom toxins. Nat Genet. 2020;52(1):106–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Almeida DD, Viala VL, Nachtigall PG, et al. Tracking the recruitment and evolution of snake toxins using the evolutionary context provided by the Bothrops jararaca genome. Proc Natl Acad Sci U S A. 2021;118(20):e2015159118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Sheffer MM, Hoppe A, Krehenwinkel H, et al. Chromosome-level reference genome of the European wasp spider Argiope bruennichi: a resource for studies on range expansion and evolutionary adaptation. Gigascience. 2021;10(1):doi: 10.1093/gigascience/giaa148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Pardos-Blas JR, Irisarri I, Abalde S, et al. The genome of the venomous snail Lautoconus ventricosus sheds light on the origin of conotoxin diversity. Gigascience. 2021;10(5):doi: 10.1093/gigascience/giab037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Schield DR, Card DC, Hales NR, et al. The origins and evolution of chromosomes, dosage compensation, and mechanisms underlying venom regulation in snakes. Genome Res. 2019;29(4):590–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Peng C, Huang Y, Bian C, et al. The first Conus genome assembly reveals a primary genetic central dogma of conopeptides in C. betulinus. Cell Discovery. 2021;7(1):11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Salzberg SL. Next-generation genome annotation: we still struggle to get it right. Genome Biol. 2019;20(1):92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Koludarov I, Jackson TN, Suranse V, et al. Reconstructing the evolutionary history of a functionally diverse gene family reveals complexity at the genetic origins of novelty. Mol Biol. 2019; 10.1101/583344. [DOI] [Google Scholar]
- 174. Dowell NL, Giorgianni MW, Kassner VA, et al. The deep origin and recent loss of venom toxin genes in rattlesnakes. Curr Biol. 2016;26(18):2434–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Dowell NL, Giorgianni MW, Griffin S, et al. Extremely divergent haplotypes in two toxin gene complexes encode alternative venom types within rattlesnake species. Curr Biol. 2018;28(7):1016–26.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Barua A, Koludarov I, Mikheyev AS. Co-option of the same ancestral gene family gave rise to mammalian and reptilian toxins. BMC Biol. 2021;19(1):268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Jackson TNW, Koludarov I. How the toxin got its toxicity. Front Pharmacol. 2020;11:574925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Malhotra A, Creer S, Harris JB, et al. The importance of being genomic: non-coding and coding sequences suggest different models of toxin multi-gene family evolution. Toxicon. 2015;107:344–58. [DOI] [PubMed] [Google Scholar]
- 179. Kini RM. Accelerated evolution of toxin genes: exonization and intronization in snake venom disintegrin/metalloprotease genes. Toxicon. 2018;148:16–25. [DOI] [PubMed] [Google Scholar]
- 180. Bergthorsson U, Andersson DI, Roth JR. Ohno's dilemma: evolution of new genes under continuous selection. Proc Natl Acad Sci U S A. 2007;104(43):17004–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Patthy L. Protein Evolution. Wiley; 2009. [Google Scholar]
- 182. Innan H, Kondrashov F. The evolution of gene duplications: classifying and distinguishing between models. Nat Rev Genet. 2010;11(2):97–108. [DOI] [PubMed] [Google Scholar]
- 183. Espinosa-Cantú A, Ascencio D, Barona-Gómez F, et al. Gene duplication and the evolution of moonlighting proteins. Front Genet. 2015;6:227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Veltri D, Wight MM, Crouch JA. SimpleSynteny: a web-based tool for visualization of microsynteny across multiple species. Nucleic Acids Res. 2016;44(W1):W41–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Haug-Baltzell A, Stephens SA, Davey S, et al. SynMap2 and SynMap3D: web-based whole-genome synteny browsers. Bioinformatics. 2017;33(14):2197–8. [DOI] [PubMed] [Google Scholar]
- 186. Ankenbrand MJ, Hohlfeld S, Hackl T, et al. AliTV—interactive visualization of whole genome comparisons. PeerJ Comput Sci. 2017;3:e116. [Google Scholar]
- 187. Hilbrant M, Damen WGM, McGregor AP. Evolutionary crossroads in developmental biology: the spider Parasteatoda tepidariorum. Development. 2012;139(15):2655–62. [DOI] [PubMed] [Google Scholar]
- 188. Oda H, Akiyama-Oda Y. Evodevo. 2020;11:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Li M, Au LYC, Douglah D, et al. Generation of heritable germline mutations in the jewel wasp Nasonia vitripennis using CRISPR/Cas9. Sci Rep. 2017;7(1):901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Hu XF, Zhang B, Liao CH, et al. High-efficiency CRISPR/Cas9-mediated gene editing in honeybee (Apis mellifera) embryos. G3 (Bethesda). 2019;9(5):1759–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Chiu Y-K, Hsu J-C, Chang T, et al. Mutagenesis mediated by CRISPR/Cas9 in the red imported fire ant. Insect Soc. 2020;67(2):317–26. [Google Scholar]
- 192. Karabulut A, He S, Chen C-Y, et al. Electroporation of short hairpin RNAs for rapid and efficient gene knockdown in the starlet sea anemone, Nematostella vectensis. Dev Biol. 2019;448(1):7–15. [DOI] [PubMed] [Google Scholar]
- 193. Layden MJ, Rentzsch F, Röttinger E. The rise of the starlet sea anemone Nematostella vectensis as a model system to investigate development and regeneration. Wiley Interdiscip Rev Dev Biol. 2016;5(4):408–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Columbus-Shenkar YY, Sachkova MY, Macrander J, et al. Dynamics of venom composition across a complex life cycle. Elife. 2018;7:e35014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Moran Y, Genikhovich G, Gordon D, et al. Neurotoxin localization to ectodermal gland cells uncovers an alternative mechanism of venom delivery in sea anemones. Proc Biol Sci. 2012;279(1732):1351–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Sunagar K, Columbus-Shenkar YY, Fridrich A, et al. Cell type-specific expression profiling unravels the development and evolution of stinging cells in sea anemone. BMC Biol. 2018;16(1):108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Herzig V, King GF, Undheim EAB. Can we resolve the taxonomic bias in spider venom research?. Toxicon X. 2019;1:100005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Lüddecke T, Vilcinskas A, Lemke S. Phylogeny-guided selection of priority groups for venom bioprospecting: harvesting toxin sequences in tarantulas as a case study. Toxins. 2019;11(9):488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Jin A-H, Muttenthaler M, Dutertre S, et al. Conotoxins: chemistry and biology. Chem Rev. 2019;119(21):11510–49. [DOI] [PubMed] [Google Scholar]
- 200. Wang Y-M, Tsai I-H, Chen J-M, et al. Correlation between the glycan variations and defibrinogenating activities of acutobin and its recombinant glycoforms. PLoS One. 2014;9(6):e100354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Luna-Ramirez K, Csoti A, McArthur JR, et al. Structural basis of the potency and selectivity of Urotoxin, a potent Kv1 blocker from scorpion venom. Biochem Pharmacol. 2020;174:113782. [DOI] [PubMed] [Google Scholar]
- 202. Lee H-K, Zhang L, Smith MD, et al. A marine analgesic peptide, Contulakin-G, and neurotensin are distinct agonists for neurotensin receptors: uncovering structural determinants of desensitization properties. Front Pharmacol. 2015;6:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Saikia C, Ben-Nissan G, Reuveny E, et al. Production of recombinant venom peptides as tools for ion channel research. Methods Enzymol. 2021;654:169–201. [DOI] [PubMed] [Google Scholar]
- 204. Turchetto J, Sequeira AF, Ramond L, et al. High-throughput expression of animal venom toxins in Escherichia coli to generate a large library of oxidized disulphide-reticulated peptides for drug discovery. Microb Cell Fact. 2017;16(1):6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Derman AI, Prinz WA, Belin D, et al. Mutations that allow disulfide bond formation in the cytoplasm of Escherichia coli. Science. 1993;262(5140):1744–7. [DOI] [PubMed] [Google Scholar]
- 206. Bessette PH, Aslund F, Beckwith J, et al. Efficient folding of proteins with multiple disulfide bonds in the Escherichia coli cytoplasm. Proc Natl Acad Sci U S A. 1999;96(24):13703–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. de Marco A. Strategies for successful recombinant expression of disulfide bond-dependent proteins in Escherichia coli. Microb Cell Fact. 2009;8(1):26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Hatahet F, Nguyen VD, Salo KE, et al. Disruption of reducing pathways is not essential for efficient disulfide bond formation in the cytoplasm of E. coli. Microb Cell Fact. 2010;9:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Klint JK, Senff S, Saez NJ, et al. Production of recombinant disulfide-rich venom peptides for structural and functional analysis via expression in the periplasm of E. coli. PLoS One. 2013;8(5):e63865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Bertelsen AB, Hackney CM, Bayer CN, et al. DisCoTune: versatile auxiliary plasmids for the production of disulphide-containing proteins and peptides in the E. coli T7 system. Microb Biotechnol. 2021;14(6):2566–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Nielsen LD, Foged MM, Albert A, et al. The three-dimensional structure of an H-superfamily conotoxin reveals a granulin fold arising from a common ICK cysteine framework. J Biol Chem. 2019;294(22):8745–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Nozach H, Fruchart-Gaillard C, Fenaille F, et al. High throughput screening identifies disulfide isomerase DsbC as a very efficient partner for recombinant expression of small disulfide-rich proteins in E. coli. Microb Cell Fact. 2013;12(1):37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. Sequeira AF, Turchetto J, Saez NJ, et al. Gene design, fusion technology and TEV cleavage conditions influence the purification of oxidized disulphide-rich venom peptides in Escherichia coli. Microb Cell Fact. 2017;16(1):4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Correnti CE, Gewe MM, Mehlin C, et al. Screening, large-scale production and structure-based classification of cystine-dense peptides. Nat Struct Mol Biol. 2018;25(3):270–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Crook ZR, Sevilla GP, Friend D, et al. Mammalian display screening of diverse cystine-dense peptides for difficult to drug targets. Nat Commun. 2017;8(1):2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Wang Y, Xu W, Kou X, et al. Establishment and optimization of a wheat germ cell-free protein synthesis system and its application in venom kallikrein. Protein Expr Purif. 2012;84(2):173–80. [DOI] [PubMed] [Google Scholar]
- 217. Vlasak R, Kreil G. Nucleotide sequence of cloned cDNAs coding for preprosecapin, a major product of queen-bee venom glands. Eur J Biochem. 1984;145(2):279–82. [DOI] [PubMed] [Google Scholar]
- 218. Pennington MW, Czerwinski A, Norton RS. Peptide therapeutics from venom: current status and potential. Bioorg Med Chem. 2018;26(10):2738–58. [DOI] [PubMed] [Google Scholar]
- 219. Robinson SD, Undheim EAB, Ueberheide B, et al. Venom peptides as therapeutics: advances, challenges and the future of venom-peptide discovery. Exp Rev Proteomics. 2017;14(10):931–9. [DOI] [PubMed] [Google Scholar]
- 220. Theakston RDG, Reid HA. Development of simple standard assay procedures for the characterization of snake venoms. Bull World Health Organ. 1983;61(6):949–56. [PMC free article] [PubMed] [Google Scholar]
- 221. Giacomotto J, Ségalat L. High-throughput screening and small animal models, where are we?. Br J Pharmacol. 2010;160(2):204–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222. Vetter I, Hodgson WC, Adams DJ, et al. Chap. 4: Venoms-based drug discovery: bioassays, electrophysiology, high-throughput screens and target identification. In: King GF, ed. Venoms to Drugs: Venom as a Source for the Development of Human Therapeutics. Cambridge: Royal Society of Chemistry; 2015:97–128. [Google Scholar]
- 223. Herzig V, Cristofori-Armstrong B, Israel MR, et al. Animal toxins — Nature's evolutionary-refined toolkit for basic research and drug discovery. Biochem Pharmacol. 2020;181:114096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Arbuckle K, Rodríguez de la Vega RC, Casewell NR. Coevolution takes the sting out of it: evolutionary biology and mechanisms of toxin resistance in animals. Toxicon. 2017;140:118–31. [DOI] [PubMed] [Google Scholar]
- 225. Holding ML, Drabeck DH, Jansa SA, et al. Venom resistance as a model for understanding the molecular basis of complex coevolutionary adaptations. Integr Comp Biol. 2016;56(5):1032–43. [DOI] [PubMed] [Google Scholar]
- 226. Biardi JE, Coss RG. Rock squirrel (Spermophilus variegatus) blood sera affects proteolytic and hemolytic activities of rattlesnake venoms. Toxicon. 2011;57(2):323–31. [DOI] [PubMed] [Google Scholar]
- 227. Becerra-Amezcua MP, Guerrero-Legarreta I, González-Márquez H, et al. In vivo analysis of effects of venom from the jellyfish Chrysaora sp. in zebrafish (Danio rerio). Toxicon. 2016;113:49–54. [DOI] [PubMed] [Google Scholar]
- 228. Gutiérrez JM, Vargas M, Segura Á, et al. In vitro tests for assessing the neutralizing ability of snake antivenoms: toward the 3Rs principles. Front Immunol. 2021;11:617429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229. Mejia M, Heghinian MD, Busch A, et al. A novel approach for in vivo screening of toxins using the Drosophila giant fiber circuit. Toxicon. 2010;56(8):1398–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230. Freshney IR. Culture of Animal Cells: A manual of basic technique and specialized applications. Wiley; 2010. [Google Scholar]
- 231. Mathie A, Veale EL, Holden RG. Heterologous expression of ion channels in mammalian cell lines. In: Dallas M, Bell D, eds. Patch Clamp Electrophysiology: Methods and Protocols. New York, NY: Springer; 1995. [DOI] [PubMed] [Google Scholar]
- 232.. In: Penner R., Sakmann B, Neher E A practical guide to patch clamping, eds. Single-Channel Recording. Boston, MA: Springer; 1995. [Google Scholar]
- 233. Schreibmayer W, Lester HA, Dascal N. Voltage clamping of Xenopus laevis oocytes utilizing agarose-cushion electrodes. Pflugers Arch. 1994;426(5):453–8. [DOI] [PubMed] [Google Scholar]
- 234. van Cann M, Kuzmenkov A, Isensee J, et al. Scorpion toxin MeuNaTxα-1 sensitizes primary nociceptors by selective modulation of voltage-gated sodium channels. FEBSJ. 2021;288(7):2418–35. [DOI] [PubMed] [Google Scholar]
- 235. Broichhagen J, Frank JA, Trauner D. A roadmap to success in photopharmacology. Acc Chem Res. 2015;48(7):1947–60. [DOI] [PubMed] [Google Scholar]
- 236. Russell WMS, Burch RL. The Principles of Humane Experimental Technique. Wheathampstead: Universities Federation for Animal Welfare; 1992. [Google Scholar]
- 237. Gordon D, Chen R, Chung S-H. Computational methods of studying the binding of toxins from venomous animals to biological ion channels: theory and applications. Physiol Rev. 2013;93(2):767–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238. Mouhat S, Jouirou B, Mosbah A, et al. Diversity of folds in animal toxins acting on ion channels. Biochem J. 2004;378(3):717–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239. Lavergne V, Alewood PF, Mobli M, et al. Chap. 2: The structural universe of disulfide-rich venom peptidesIn: King GF, ed. Venoms to Drugs: Venom as a Source for the Development of Human Therapeutics. Cambridge: Royal Society of Chemistry; 2015:37–79. [Google Scholar]
- 240. Callaway E. Revolutionary cryo-EM is taking over structural biology. Nature. 2020;578(7794):201. [DOI] [PubMed] [Google Scholar]
- 241. Baconguis I, Bohlen CJ, Goehring A, et al. X-ray structure of acid-sensing ion channel 1–Snake toxin complex reveals open state of a Na+-selective channel. Cell. 2014;156(4):717–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242. Shen H, Li Z, Jiang Y, et al. Structural basis for the modulation of voltage-gated sodium channels by animal toxins. Science. 2018;362(6412):eaau2596. [DOI] [PubMed] [Google Scholar]
- 243. Clairfeuille T, Cloake A, Infield DT, et al. Structural basis of α-scorpion toxin action on Nav channels. Science. 2019;363(6433):eaav8573. [DOI] [PubMed] [Google Scholar]
- 244. Maeda S, Xu JN, Kadji FM, et al. Structure and selectivity engineering of the M1 muscarinic receptor toxin complex. Science. 2020;369(6500):161–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245. Pagadala NS, Syed K, Tuszynski J. Software for molecular docking: a review. Biophys Rev. 2017;9(2):91–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246. Hollingsworth SA, Dror RO. Molecular dynamics simulation for all. Neuron. 2018;99(6):1129–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247. Karbat I, Altman-Gueta H, Fine S, et al. Pore-modulating toxins exploit inherent slow inactivation to block K+ channels. Proc Natl Acad Sci U S A. 2019;116(37):18700–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248. Saikia C, Dym O, Altman-Gueta H, et al. A molecular lid mechanism of K+ channel blocker action revealed by a cone peptide. J Mol Biol. 2021;433(17):166957. [DOI] [PubMed] [Google Scholar]
- 249. Yi M, Tjong H, Zhou H-X. Spontaneous conformational change and toxin binding in α7 acetylcholine receptor: insight into channel activation and inhibition. Proc Natl Acad Sci U S A. 2008;105(24):8280–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250. Jumper J, Evans R, Pritzel A, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596(7873):583–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251. Isensee J, van Cann M, Despang P, et al. Depolarization induces nociceptor sensitization by CaV1.2-mediated PKA-II activation. J Cell Biol. 2021;220(10):e202002083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252. Wilson D, Daly NL. Venomics: a mini-review. High Throughput. 2018;7(3):19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253. Prashanth JR, Hasaballah N, Vetter I. Pharmacological screening technologies for venom peptide discovery. Neuropharmacology. 2017;127:4–19. [DOI] [PubMed] [Google Scholar]
- 254. Clark GC, Casewell NR, Elliott CT, et al. Friends or foes? Emerging impacts of biological toxins. Trends Biochem Sci. 2019;44(4):365–79. [DOI] [PubMed] [Google Scholar]
- 255. Vetter I, Davis JL, Rash LD, et al. Venomics: a new paradigm for natural products-based drug discovery. Amino Acids. 2011;40(1):15–28. [DOI] [PubMed] [Google Scholar]
- 256. Bhaswati C. Animal venoms have potential to treat cancer. Curr Top Med Chem. 2018;18(30):2555–66. [DOI] [PubMed] [Google Scholar]
- 257. Chan YS, Cheung RCF, Xia L, et al. Snake venom toxins: toxicity and medicinal applications. Appl Microbiol Biotechnol. 2016;100(14):6165–81. [DOI] [PubMed] [Google Scholar]
- 258. Harvey AL. Toxins and drug discovery. Toxicon. 2014;92:193–200. [DOI] [PubMed] [Google Scholar]
- 259. Lewis RJ, Garcia ML. Therapeutic potential of venom peptides. Nat Rev Drug Discov. 2003;2(10):790–802. [DOI] [PubMed] [Google Scholar]
- 260. Trim CM, Byrne LJ, Trim SA. Chap. 1 - Utilisation of compounds from venoms in drug discovery. In: Witty DR, Cox B, eds. Progress in Medicinal Chemistry. Elsevier; 2021. doi: 10.1016/bs.pmch.2021.01.001. [DOI] [PubMed] [Google Scholar]
- 261. Cardoso FC, Hasan M, Zhao T, et al. Toxins in pain. Curr Opin Support Palliat Care. 2018;12(2):132–41. [DOI] [PubMed] [Google Scholar]
- 262. Geron M, Hazan A, Priel A. Animal toxins providing insights into TRPV1 activation mechanism. Toxins. 2017;9(10):326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263. Andreev YA, Kozlov SA, Korolkova YV, et al. Polypeptide modulators of TRPV1 produce analgesia without hyperthermia. Mar Drugs. 2013;11(12):5100–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264. Koivisto A, Chapman H, Jalava N, et al. TRPA1: a transducer and amplifier of pain and inflammation. Basic Clin Pharmacol Toxicol. 2014;114(1):50–5. [DOI] [PubMed] [Google Scholar]
- 265. Escoubas P, Weille JRD, Lecoq A, et al. Isolation of a tarantula toxin specific for a class of proton-gated Na+ channels. J Biol Chem. 2000;275(33):25116–21. [DOI] [PubMed] [Google Scholar]
- 266. Diochot S, Baron A, Salinas M, et al. Black mamba venom peptides target acid-sensing ion channels to abolish pain. Nature. 2012;490(7421):552–5. [DOI] [PubMed] [Google Scholar]
- 267. Lee JYP, Saez NJ, Cristofori-Armstrong B, et al. Inhibition of acid-sensing ion channels by diminazene and APETx2 evoke partial and highly variable antihyperalgesia in a rat model of inflammatory pain. Br J Pharmacol. 2018;175(12):2204–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268. Postic G, Gracy J, Perin C, et al. KNOTTIN: the database of inhibitor cystine knot scaffold after 10 years, toward a systematic structure modeling. Nucleic Acids Res. 2018;46(D1):D454–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269. Moore SJ, Leung CL, Cochran JR. Knottins: disulfide-bonded therapeutic and diagnostic peptides. Drug Discov Today Technol. 2012;9(1):e3–11. [DOI] [PubMed] [Google Scholar]
- 270. McDowell GC, Pope JE. Intrathecal ziconotide: Dosing and administration strategies in patients with refractory chronic pain. Neuromodulation. 2016;19(5):522–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271. Brust A, Croker DE, Colless B, et al. Conopeptide-derived κ-opioid agonists (Conorphins): potent, selective, and metabolic stable dynorphin A mimetics with antinociceptive properties. J Med Chem. 2016;59(6):2381–95. [DOI] [PubMed] [Google Scholar]
- 272. Castro J, Harrington AM, Garcia-Caraballo S, et al. α-Conotoxin Vc1.1 inhibits human dorsal root ganglion neuroexcitability and mouse colonic nociception via GABAB receptors. Gut. 2017;66(6):1083–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273. Nasiripourdori A, Taly V, Grutter T, et al. From toxins targeting ligand gated ion channels to therapeutic molecules. Toxins. 2011;3(3):260–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274. Nilius B, Szallasi A. Transient receptor potential channels as drug targets: from the science of basic research to the art of medicine. Pharmacol Rev. 2014;66(3):676–814. [DOI] [PubMed] [Google Scholar]
- 275. Heinen TE, da Veiga ABG. Arthropod venoms and cancer. Toxicon. 2011;57(4):497–511. [DOI] [PubMed] [Google Scholar]
- 276. Gajski G, Garaj-Vrhovac V. Melittin: a lytic peptide with anticancer properties. Environ Toxicol Pharmacol. 2013;36(2):697–705. [DOI] [PubMed] [Google Scholar]
- 277. Fernandez-Rojo MA, Deplazes E, Pineda SS, et al. Gomesin peptides prevent proliferation and lead to the cell death of devil facial tumour disease cells. Cell Death Discov. 2018;4:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278. Ikonomopoulou MP, Fernandez-Rojo MA, Pineda SS, et al. Gomesin inhibits melanoma growth by manipulating key signaling cascades that control cell death and proliferation. Sci Rep. 2018;8(1):11519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279. Moral-Sanz J, Fernandez-Rojo MA, Potriquet J, et al. ERK and mTORC1 inhibitors enhance the anti-cancer capacity of the octpep-1 venom-derived peptide in melanoma BRAF(V600E) mutations. Toxins. 2021;13(2):146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280. Panagides N, Jackson TNW, Ikonomopoulou MP, et al. How the cobra got its flesh-eating venom: cytotoxicity as a defensive innovation and its co-evolution with hooding, aposematic marking, and spitting. Toxins. 2017;9(3):103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281. Goldenberg J, Cipriani V, Jackson TNW, et al. Proteomic and functional variation within black snake venoms (Elapidae: Pseudechis). Comp Biochem Physiol C Toxicol Pharmacol. 2018;205:53–61. [DOI] [PubMed] [Google Scholar]
- 282. op den Brouw B, Coimbra FCP, Bourke LA, et al. Extensive variation in the activities of Pseudocerastes and Eristicophis viper venoms suggests divergent envenoming strategies are used for prey capture. Toxins. 2021;13(2):112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283. Duffy C, Sorolla A, Wang E, et al. Honeybee venom and melittin suppress growth factor receptor activation in HER2-enriched and triple-negative breast cancer. NPJ Precis Oncol. 2020;4:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284. Li L, Huang J, Lin Y. Snake venoms in cancer therapy: past, present and future. Toxins. 2018;10(9):346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285. Gajski G, Domijan A-M, Žegura B, et al. Melittin induced cytogenetic damage, oxidative stress and changes in gene expression in human peripheral blood lymphocytes. Toxicon. 2016;110:56–67. [DOI] [PubMed] [Google Scholar]
- 286. Dabbagh Moghaddam F, Akbarzadeh I, Marzbankia E, et al. Delivery of melittin-loaded niosomes for breast cancer treatment: an in vitro and in vivo evaluation of anti-cancer effect. Cancer Nanotechnol. 2021;12:14. [Google Scholar]
- 287. Jimenez R, Ikonomopoulou MP, Lopez JA, et al. Immune drug discovery from venoms. Toxicon. 2018;141:18–24. [DOI] [PubMed] [Google Scholar]
- 288. Minutti-Zanella C, Gil-Leyva EJ, Vergara I. Immunomodulatory properties of molecules from animal venoms. Toxicon. 2021;191:54–68. [DOI] [PubMed] [Google Scholar]
- 289. Ryan RYM, Seymour J, Loukas A, et al. Immunological responses to envenomation. Front Immunol. 2021;12:661082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290. Freitas AP, Favoretto BC, Clissa PB, et al. Crotoxin isolated from Crotalus durissus terrificus venom modulates the functional activity of dendritic cells via formyl peptide receptors. J Immunol Res. 2018;2018:7873257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291. de Souza Almeida C, Andrade-Oliveira V, Câmara NOS. . PLoS One. 2015;10(4):e0121427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292. Ortiz E, Possani LD. The unfulfilled promises of scorpion insectotoxins. J Venom Anim Toxins Incl Trop Dis. 2015;21:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293. Hwang D-S, Kim SK, Bae H. Therapeutic effects of bee venom on immunological and neurological diseases. Toxins. 2015;7(7):2413–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294. An H, Kim J, Kim W, et al. Therapeutic effects of bee venom and its major component, melittin, on atopic dermatitis in vivo and in vitro. Br J Pharmacol. 2018;175(23):4310–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295. Ortiz E, Gurrola GB, Schwartz EF, et al. Scorpion venom components as potential candidates for drug development. Toxicon. 2015;93:125–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296. Asadi M, Ayat H, Ahadi AM, et al. Genomic structure of two Kv1.3 channel blockers from scorpion Mesobuthus eupeus and sea anemone Stichodactyla haddoni and construction of their chimeric peptide as a novel blocker. Biochem Genet. 2022;60(2):504–26. [DOI] [PubMed] [Google Scholar]
- 297. Huan Y, Kong Q, Mou H, et al. Antimicrobial peptides: classification, design, application and research progress in multiple fields. Front Microbiol. 2020;11:582779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298. Datta S, Roy A. Antimicrobial peptides as potential therapeutic agents: a review. Int J Pept Res Ther. 2021;27(1):555–77. [Google Scholar]
- 299. Gan BH, Gaynord J, Rowe SM, et al. The multifaceted nature of antimicrobial peptides: current synthetic chemistry approaches and future directions. Chem Soc Rev. 2021;50(13):7820–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300. de Barros E, Gonçalves RM, Cardoso MH. Snake venom cathelicidins as natural antimicrobial peptides. Front Pharmacol. 2019;10:1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301. Manniello MD, Moretta A, Salvia R, et al. Insect antimicrobial peptides: potential weapons to counteract the antibiotic resistance. Cell Mol Life Sci. 2021;78(9):4259–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302. Moreau SJ. “It stings a bit but it cleans well”: venoms of Hymenoptera and their antimicrobial potential. J Insect Physiol. 2013;59(2):186–204. [DOI] [PubMed] [Google Scholar]
- 303. Mylonakis E, Podsiadlowski L, Muhammed M, et al. Diversity, evolution and medical applications of insect antimicrobial peptides. Philos Trans R Soc Lond B Biol Sci. 2016;371(1695):20150290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304. Zhou W, Qiu H, Guo Y, et al. Molecular insights into distinct detection properties of α-hemolysin, MspA, CsgG, and aerolysin nanopore sensors. J Phys Chem B. 2020;124(9):1611–8. [DOI] [PubMed] [Google Scholar]
- 305. Crnković A, Srnko M, Anderluh G. Biological nanopores: engineering on demand. Life. 2021;11(1):27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306. Morton D, Mortezaei S, Yemenicioglu S, et al. Tailored polymeric membranes for Mycobacterium smegmatis porin A (MspA) based biosensors. J Mater Chem B. 2015;3(25):5080–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307. Huang G, Voet A, Maglia G. FraC nanopores with adjustable diameter identify the mass of opposite-charge peptides with 44 dalton resolution. Nat Commun. 2019;10(1):835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308. Carter J-M, Hussain S. Robust long-read native DNA sequencing using the ONT CsgG Nanopore system. Wellcome Open Res. 2017;2:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309. Wloka C, Mutter NL, Soskine M, et al. Alpha-Helical Fragaceatoxin C Nanopore engineered for double-stranded and single-stranded nucleic acid analysis. Angew Chem Int Ed. 2016;55(40):12494–8. [DOI] [PubMed] [Google Scholar]
- 310. Zernia S, van der Heide NJ, Galenkamp NS, et al. Current blockades of proteins inside nanopores for real-time metabolome analysis. ACS Nano. 2020;14(2):2296–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311. Lucas FLR, Versloot RCA, Yakovlieva L, et al. Protein identification by nanopore peptide profiling. Nat Commun. 2021;12(1):5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312. Laszlo AH, Derrington IM, Ross BC, et al. Decoding long nanopore sequencing reads of natural DNA. Nat Biotechnol. 2014;32(8):829–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313. Schäfer RB, Liess M, Altenburger R, et al. Future pesticide risk assessment: narrowing the gap between intention and reality. Environ Sci Eur. 2019;31:21. [Google Scholar]
- 314. Sharma A, Kumar V, Shahzad B, et al. Worldwide pesticide usage and its impacts on ecosystem. SN Appl Sci. 2019;1:1446. [Google Scholar]
- 315. Zhu YC, Adamczyk J, Rinderer T, et al. Spray toxicity and risk potential of 42 commonly used formulations of row crop pesticides to adult honey bees (Hymenoptera: Apidae). J Econ Entomol. 2015;108(6):2640–7. [DOI] [PubMed] [Google Scholar]
- 316. Desneux N, Decourtye A, Delpuech J-M. The sublethal effects of pesticides on beneficial arthropods. Annu Rev Entomol. 2007;52(1):81–106. [DOI] [PubMed] [Google Scholar]
- 317. Hallmann CA, Sorg M, Jongejans E, et al. More than 75 percent decline over 27 years in total flying insect biomass in protected areas. PLoS One. 2017;12(10):e0185809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318. King GF, Hardy MC. Spider-venom peptides: structure, pharmacology, and potential for control of insect pests. Annu Rev Entomol. 2013;58(1):475–96. [DOI] [PubMed] [Google Scholar]
- 319. Ikonomopoulou M, King G. Natural born insect killers: spider-venom peptides and their potential for managing arthropod pests. Outlook Pest Manag. 2013;24(1):16–9. [Google Scholar]
- 320. Lüddecke T, Herzig V, von Reumont BM, et al. The biology and evolution of spider venoms. Biol Rev. 2022;97(1):163–78. [DOI] [PubMed] [Google Scholar]
- 321. Yu H, Li R, Wang X, et al. Field experiment effect on citrus spider mite Panonychus citri of venom from jellyfish Nemopilema nomurai: the potential use of jellyfish in agriculture. Toxins. 2021;13(6):411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322. Ikonomopoulou MP, Smith JJ, Herzig V, et al. Isolation of two insecticidal toxins from venom of the Australian theraphosid spider Coremiocnemis tropix. Toxicon. 2016;123:62–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323. Smith JJ, Herzig V, Ikonomopoulou MP, et al. Insect-active toxins with promiscuous pharmacology from the African theraphosid spider Monocentropus balfouri. Toxins. 2017;9(5):155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324. Herzig V, Ikonomopoulou M, Smith JJ, et al. Molecular basis of the remarkable species selectivity of an insecticidal sodium channel toxin from the African spider Augacephalus ezendami. Sci Rep. 2016;6:29538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325. Marsh NA. Diagnostic uses of snake venom. Pathophysiol Haemost Thromb. 2001;31(3-6):211–7. [DOI] [PubMed] [Google Scholar]
- 326. Marsh N, Williams V. Practical applications of snake venom toxins in haemostasis. Toxicon. 2005;45(8):1171–81. [DOI] [PubMed] [Google Scholar]
- 327. Perchuc AM, Wilmer M. et al. Diagnostic use of snake venom components in the coagulation laboratory. In: Kini RM, Clemetson KJ, Markland FS et al.et al., ., eds. Toxins and Hemostasis: From Bench to Bedside. Dordrecht: Springer; 2010. [Google Scholar]
- 328. Jay WF, Solange MTS. Approaching the golden age of natural product pharmaceuticals from venom libraries: an overview of toxins and toxin-derivatives currently involved in therapeutic or diagnostic applications. Curr Pharm Des. 2007;13:2927–34. [DOI] [PubMed] [Google Scholar]
- 329. Dardevet L, Rani D, Aziz T, et al. Chlorotoxin: a helpful natural scorpion peptide to diagnose glioma and fight tumor invasion. Toxins. 2015;7(4):1079–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330. Longbottom J, Shearer FM, Devine M, et al. Vulnerability to snakebite envenoming: a global mapping of hotspots. Lancet. 2018;392(10148):673–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331. Chippaux J-P. Snakebite envenomation turns again into a neglected tropical disease!. J Venom Anim Toxins Incl Trop Dis. 2017;23:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332. Williams DJ, Faiz MA, Abela-Ridder B, et al. Strategy for a globally coordinated response to a priority neglected tropical disease: snakebite envenoming. PLoS Negl Trop Dis. 2019;13(2):e0007059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333. León G, Vargas M, Segura Á, et al. Current technology for the industrial manufacture of snake antivenoms. Toxicon. 2018;151:63–73. [DOI] [PubMed] [Google Scholar]
- 334. Pucca MB, Cerni FA, Janke R, et al. History of envenoming therapy and current perspectives. Front Immunol. 2019;10:1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335. Habib AG, Brown NI. The snakebite problem and antivenom crisis from a health-economic perspective. Toxicon. 2018;150:115–23. [DOI] [PubMed] [Google Scholar]
- 336. Laustsen AH, María Gutiérrez J, Knudsen C, et al. Pros and cons of different therapeutic antibody formats for recombinant antivenom development. Toxicon. 2018;146:151–75. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Mark J. Margres -- 2/17/2022 Reviewed
Matthew Holding -- 2/21/2022 Reviewed
Jason Macrander, Ph. D. -- 3/8/2022 Reviewed
Data Availability Statement
Not applicable.
Abbreviations
AMP: antimicrobial peptide; AMPA: α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor; ASIC: acid-sensing ion channel; BLAST: Basic Local Alignment Search Tool; cDNA: complementary DNA; CRISPR: clustered regularly interspaced short palindromic repeats; ESI: electrospray ionization; FDA: Food and Drug Administration; GPCR: G-protein-coupled receptors; GUI: graphical user interface; ICK: inhibitory cysteine knot; L-AAO: L-amino acid oxidase; LC-MS/MS: liquid chromatography–tandem mass spectrometry; mRNA: messenger RNA; MS: mass spectrometry; MSI: mass spectrometry imaging; NET: norepinephrine transporter; NMDA: glutamate-gated cation channels; NMR: nuclear magnetic resonance; PLA2: phospholipase A2; PTM: posttranslational modification; scRNA-seq: single-cell RNA-sequencing; TRP: transient receptor potential.