Abstract
As the size of genome-wide association studies increase, the number of associated trait loci identified inevitably increase. One welcomes this if it allows the better delineation of the pathways to disease and increases the accuracy of genetic prediction of disease risk through polygenic risk score analysis. However, there are several problems in the continuing increase in the genome-wide analysis of ‘Alzheimer’s disease’. In this review, we have systematically assessed the history of Alzheimer’s disease genome-wide association studies, including their sample sizes, age and selection/assessment criteria of cases and controls and heritability explained by these disease genome-wide association studies. We observe that nearly all earlier disease genome-wide association studies are now part of all current disease genome-wide association studies. In addition, the latest disease genome-wide association studies include (i) only a small fraction (∼10%) of clinically screened controls, substituting for them population-based samples which are systematically younger than cases, and (ii) around 50% of Alzheimer’s disease cases are in fact ‘proxy dementia cases’. As a consequence, the more genes the field finds, the less the heritability they explain. We highlight potential caveats this situation creates and discuss some of the consequences occurring when translating the newest Alzheimer’s disease genome-wide association study results into basic research and/or clinical practice.
Keywords: Alzheimer’s disease, genome-wide association study, heritability
Escott-Price and John Hardy report that large genome-wide association studies are reported as identifying loci for Alzheimer’s disease loci. They suggest that many are incorrectly designated because of misdiagnoses inherent in data collection. After genome-wide association studies of dementia have been performed, significant loci should be tested in neuropathologically confirmed data sets before they are designated as Alzheimer’s disease loci.
Graphical Abstract
Graphical abstract.
Introduction
As the size of genome-wide association studies (GWASs) increase, the number of associated trait loci identified inevitably increase.1 One welcomes this if it allows the better delineation of the pathways to disease and increases the accuracy of genetic prediction of disease risk through polygenic risk score (PRS) analysis. However, there are several problems in the continuing increase in the genome-wide analysis of ‘Alzheimer’s disease’. The first is that the diagnostic accuracy for Alzheimer’s disease is poor: of the order of 80% in clinic-based series based both on neuropathological criteria2 and on genetic analysis3 and this is certain to be worse in the case of the proxy cases used in the larger and more recent GWAS. The second is that, while for many rare diseases, age matching of controls makes little difference to the results obtained, because Alzheimer’s disease is a common cause of mortality, the risk gene APOE also has the greatest effect on longevity.4,5 This makes age-matching essential for accurate risk assessment. In addition, a simple inclusion of age as a covariate in the GWAS creates a robust but biologically spurious association between Alzheimer’s disease and age-associated variants, similar to the association identified between sex- and height-associated variants.6 Thus, in case of Alzheimer’s disease, the appropriate use of age-matched controls is important.7 A final major problem in the published GWAS is that for most of them, only summary statistics are made available.
These problems are systemic in nearly all the ‘Alzheimer’ GWASs, including ones in which we have been co-authors, except those using neuropathologically defined disease samples8, 9 and as data from different studies are meta-analysed together, these systematic errors get baked into the updated analyses. An indicator of diluting the true Alzheimer’s disease associations is the reported heritability estimates. If in a small clinically assessed GWAS (N = 11 789 with 3 genome-wide significant loci identified), the heritability was estimated as h2 = 17% (SE = 3%)10, 11: the latest GWAS with a sample size of more than 1.1M people with 38 independent genome-wide significant loci, accounts only for 3% (SE = 0.6%) of heritability.1 These errors then get incorporated into PRS analyses and also, perhaps, incorrectly contribute to the suggestion that neurodegenerative diseases share disease mechanisms. In this regard, for example, the designation of TMEM106B and GRN loci as Alzheimer’s disease loci (both are known frontotemporal dementia loci12) is of particular concern, even though they appear in both clinic-based and proxy GWAS data sets. A related problem is likely to be the reported evidence of APOE association with clinical frontotemporal dementia (FTD).13 What is needed is larger GWAS of Alzheimer’s cases based on either neuropathological or on good biomarker data as, at present, such studies are underpowered. Neuropathological GWAS should give definitive risk loci for disease, whereas GWAS based on biomarker data perhaps give information on disease progression.14,15 The danger is that as larger and larger studies of cases with unsatisfactory diagnoses are analysed, the statistical weight behind unwarranted conclusions will become stronger.
Materials and methods
We have reviewed the GWAS for Alzheimer’s disease derived from analysis of populations of historical European ancestry and assessed their samples sizes, diagnosis and age distributions of cases and controls where possible, as well as the number of genome-wide associated loci they report. The numbers of clinically assessed cases and controls were calculated from the numbers of cases and controls reported in the publication, excluding cases with family-history-based diagnosis (proxy) and controls from the population cohorts in all previous studies contributed to the publication via meta-analysis.
We have extracted the single-nucleotide polymorphisms (SNP)-based heritability estimates for the GWAS from the publications where available and calculated the heritability ourselves if the authors did not provide them in the paper. For the latter, we have downloaded the corresponding summary statistics and used the Linkage Disequilibrium Score (LDSC) regression approach.16 We estimated heritability ourselves for fix studies8,17–21 using the default settings of the LDSC regression software and pre-calculated LDSCs from the 1000 Genomes European reference population, supplied with the LDSC software. Although Jansen et al.19 provided heritability estimate for Phase 1 in their Supplemental Note, we have also downloaded the study’s summary statistics, which included the UK Biobank (UKBB; combining Phases 1 and 2). Wightman et al.1 provided their own heritability estimate, with the same approach, reference population and software options. For the pathology confirmed sample of 1011 cases and 583 controls, we used the summary statistics as reported in Escott-Price et al.9 Due to the relatively small sample size, the LDSC heritability estimates were negative for these summary statistics when default LDSC parameters were used. Since in the pathology confirmed sample, there were no confounders (such as age mismatching or misdiagnosis), we estimated the heritability for this sample by constraining the intercept using theno-intercept flag.22 All heritability estimates were (re)calculated on a liability scale assuming a population prevalence of 5%.
Data availability
Data sharing is not applicable to this article as no new data were created or analysed in this study.
Results
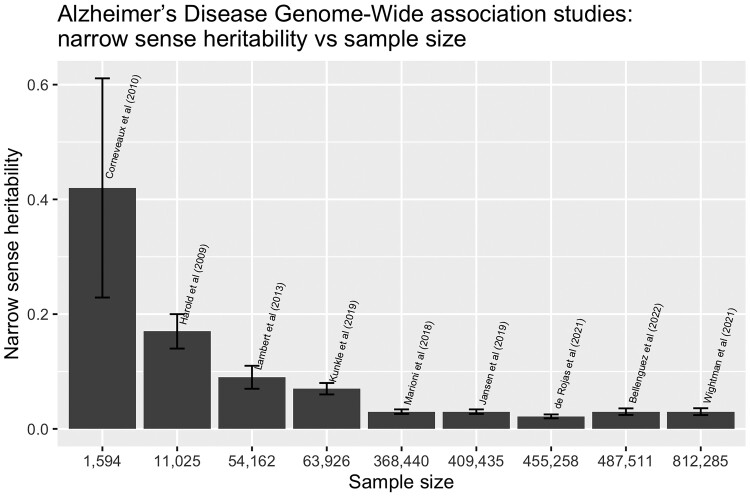
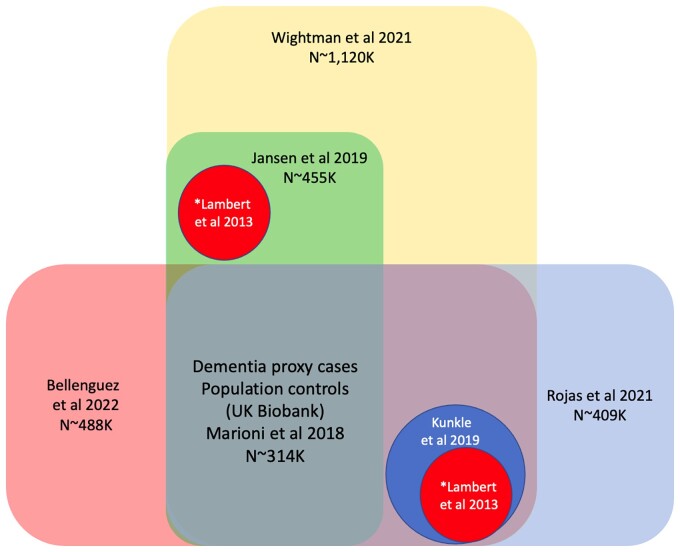
Apart from four early GWASs (2009–2011), none of the current GWASs are independent (see Fig. 1 and Table 1). The latest GWASs (2019 onwards) include a large proportion of ‘cases’ are based upon the reported impression of offspring that their parent had dementia (usually referred to as ‘proxy Alzheimer’s disease cases’). The accuracy of these impressions is suspect, but, even assuming that 80% of parents have dementia, only 60% of them are likely to have had Alzheimer’s disease. This will introduce significant noise into the data set resulting in about 50% of parental cases having a different form of dementia or no dementia at all. This and any other diagnostic imprecision may specifically limit the detection of variants of small effect, which are the basis of the polygenic architecture of Alzheimer’s disease.
Figure 1.
Overlap of the AD GWAS. *Lambert et al. (2013) and Kunkle et al. (2019) are included to Wightman et al. (2021) only once.
Table 1.
History of AD GWAS and their SNP-based heritability assuming 5% disease prevalence estimated with LDSC regression16
| Year | Author | Sample size (Stage 1) | Mean age at assessmenta | Clinical/pathology assessment (%) | SNP-based heritability on liability scale (5% prevalence) | Number of GWAS significant locib | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | Cases | Controls | Cases | Controls | Cases | Controls | Total | Novel | |||
| 2010 | Corneveaux et al.8 | 1594 | 1011 | 583 | 81.9 | 80.8 | 100c | 100c | 0.42 (0.19)d | 1 | 0 |
| 2009 | Harold et al.10 | 11 025 | 3177 | 7848 | 78.6 | 51 | 100 | 26.5 | 0.17 (0.03)e | 3 | 2 |
| 2009 | Lambert et al.23 | 8260 | 2243 | 6017 | 68.5 | 74 | 100 | 100 | NA | 3 | 2 |
| 2010 | Seshadri et al.24 | 14 283 | 1315 | 12 968 | 82.7 | 72.8 | 100 | 100 | NA | 5 | 2 |
| 2011 | Naj et al.25 | 21 165 | 10 273 | 10 892 | 74.7 | 76.3 | 100 | 100 | 0.25 (0.02)e,f | 9 | 4 |
| 2013 | Lambert et al.26 | 54 162 | 17 008 | 37 154 | 76.6 | 70.5 | 100 | 84.5 | 0.09 (0.02)e | 20 | 11 |
| 2019 | Kunkle et al.17 | 63 926 | 21 982 | 41 944 | 72.9 | 72.4 | 100 | 86.2 | 0.07 (0.01) | 25 | 5 |
| 2018 | Marioni et al.18 | 368 440 | 70 306 | 298 134 | Not known | 67.3 | 48.0 | 18.6 | 0.03 (0.004) | 26 | 7 |
| 2019 | Jansen et al.19 | 455 258 | 71 880 | 383 378 | Not known | 67.3 | 33.5 | 14.4 | 0.06 (0.01)/0.02 (0.003)g | 29 | 13 |
| 2021 | Rojas et al.20 | 409 435 | 81 611 | 308 979 | Not known | 67.3 | 34.4 | 13.4h | 0.03 (0.004) | 35 | 6 |
| 2022 | Bellenguez et al.21 | 487 511 | 85 934 | 401 577 | 67.2 | 57.9 | 45.5 | 14.0 | 0.03 (0.003)i | 75 | 42 |
| 2021 | Wightman et al.1 | 1 126 563 | 90 338 | 1 036 225 | NA | NA | 51.6 | 9.8 | 0.03 (0.006)j | 38 | 7 |
Mean age at assessment (if not reported) was estimated as weighted (by the sample sizes) average of the ages at assessments reported in the contributing studies.
Using meta-analysis of Stages 1 and 2 (replication) data.
Pathology confirmed.
Heritability is estimated using summary statistics of imputed GWAS.9
Transformation to liability scale with 5% prevalence is reported by Zhang et al.11
Estimated with GCTA software.27
Without/with UK Biobank data.
Reported in Moreno-Grau et al.28
With UK Biobank data.
Without UK Biobank data.
The number of clinically assessed controls drops down to ∼10% as the majority of them are population based, and consequently not age matched. If in the pathology assessed GWAS8 and (mostly) clinically assessed GWAS,26 the average age difference was about 1 year, in the latest GWAS, it is about 10 years or simply impossible to trace (Table 1).
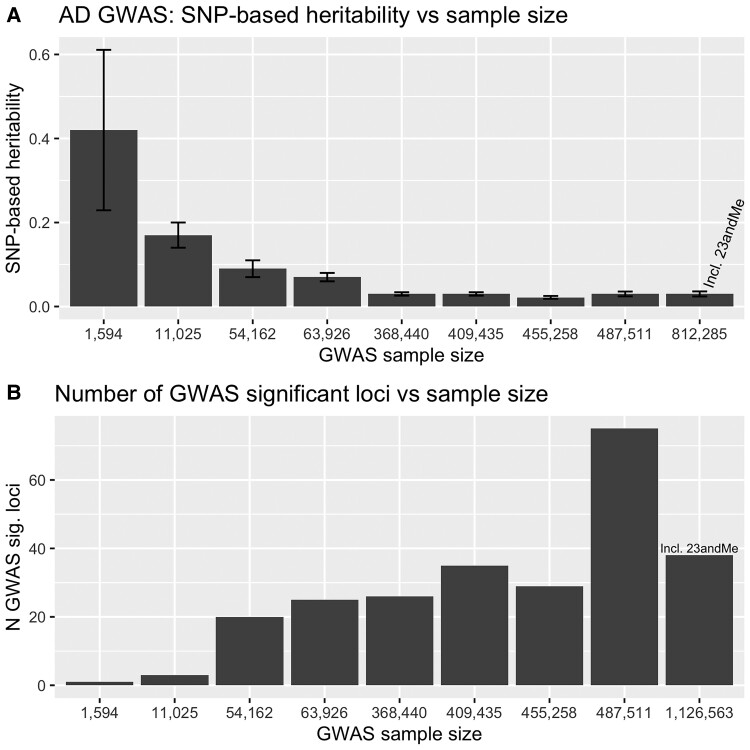
Counterintuitively, the exponential increase in sample size provides only marginal increases in the identification of novel GWAS significant loci: 2 in the samples of ∼10 000 people,10,23 and 7 in the sample of ∼1 126 563 people.1 Remarkably, the heritability estimates drop from ∼408,9 to 2–3%1,18–21 (see Table 1, Fig. 2) as the sample size increases, despite the fact that all earlier GWAS are included to the latest ones (see Fig. 1).
Figure 2.
Relationship between the GWAS sample size and the genetic findings. (A) Heritability. (B) The number of novel loci. For Wightman et al. (2021) and Bellenguez et al. (2022) studies, the heritability was estimated using summary statistics, excluding UK Biobank data.
Discussion
Why the heritability estimates are not accurate?
In the context of Alzheimer’s disease, heritability itself is a complex concept since it is possible that everyone would develop Alzheimer’s disease if they lived long enough (but see Morris29); and genetic risk appears to determine when this occurs, not if it will occur30,31: thus heritability estimates are exquisitely age dependent. Twin studies report heritability of Alzheimer’s disease 59–78%32 usually referred as broad-sense heritability. The SNP-based (narrow-sense) heritability estimates are varied across different data sets between 31 and 31%.33–35
Different approaches are used for heritability estimates [genome-wide complex trait analysis (GCTA)27 and LDSC16] with the latter gaining more popularity as it requires only summary statistics. However, the two approaches disagree in their estimates even for the same Alzheimer’s disease data sets, while for neurodevelopmental disorders, the heritability estimates are consistent.33 For example, in the same data set,10 the estimate is 31% with GCTA and 17% with LDSC.11 As LDSC uses only summary statistics, it will not pick up the relatedness between the study participants, specific to neurodegenerative disorders. In particular, there could be a different genetic architecture of APOE-ɛ4 carriers when compared with non-carriers.36 Indeed, it is known that the APOE-ɛ4 allele frequency decreases with age,5,37 while Alzheimer’s disease prevalence increases with age. In neurodevelopmental disorders (where the methodologies agree), the diagnosis is likely to be more precise since the disorder’s age at onset is early in life.38
Other traits such as Parkinson’s disease and major depressive disorder (MDD) have incorporated data sets from both UKBB and 23andMe and have not observed a corresponding decrease in heritability or the discovery of few GWS loci than expected.39,40 The reason for Parkinson’s disease is likely due to the clinical diagnosis being more precise, than for Alzheimer’s disease. In addition, Parkinson’s disease has lower prevalence in the population, so the addition of unscreened controls does not add much noise. While in MDD the prevalence it is similar to Alzheimer’s disease, it is an earlier onset disorder. Finally, for both disorders, there is no known genetic factor that modifies the age at onset and the rate of mortality (the latter changes the allele frequencies in an age-dependent way).
Longevity
Potential bias in estimates of the GWAS effect sizes and significance of a locus (and consequently of the heritability) can be introduced, as SNPs are associated with both Alzheimer’s disease and age. The APOE is the prime suspect as it is associated with a shorter lifespan41 and with other ‘killers’ in the population such as heart disease and stroke.42–44 It has been reported that APOE-ɛ4ɛ4 carriers have an age at onset of Alzheimer’s disease of about 16 years earlier than APOE-ɛ4 non-carriers, and that the frequency of APOE-ɛ4 reduces with age from ∼0.18 in the general population to 0.1 in the age group 85+.37 Despite this reduced APOE-ɛ4 frequency in the very old (85+), Alzheimer’s disease prevalence is higher in this latter age group.
Lack of study independence
We argue that Russian-doll-like GWAS, where larger studies include all smaller ones, carrying the imperfections and amplifying them, does not bring clarity in understanding the Alzheimer’s disease genetic architecture. This GWAS set up with only summary statistics available for the researchers (i) does not allow the exploration of further hypothesis in the substudies, e.g. Alzheimer’s disease predictability by the hypothesis-driven-specific (gene-network) PRS, and (ii) masks the understanding of the Alzheimer’s disease heritability estimates.
Consequences
Nearly all the ‘Alzheimer’ GWAS suffer from all the criticisms we make, in particular, lack of age matching, poor diagnostic accuracy and lack of data transparency.
This is leading to potentially serious issues (for example drug trials targeted at FTD genes in Alzheimer’s disease cases45). This problem relates not only to the primary ‘new’ studies, but also the ones in which they are meta-analysed. If earlier GWAS studies have shown that genetics of Alzheimer’s disease and Parkinson’s disease is distinct,46 now papers appear discussing genetic overlap between ‘Alzheimer’s disease’ and Parkinson’s disease. However, ‘Alzheimer’s disease’ cohorts certainly include dementia with Lewy body (DLB) cases and overlap between Parkinson’s disease and DLB is well established.47 Thus, in many ways, this genetic sloppiness is having consequences both in terms of the loci associated with disease and therefore passed on to cell biologists and for the utility of PRS analyses for clinical prediction of disease. For example, one of the consequences of the reported low SNP-based heritability is the conclusion that late onset Alzheimer’s disease is oligogenic (∼100 genes),11 where the authors assumed 9% heritability in their simulation study, whereas earlier publications suggest that the disease is polygenic (thousands of genes).9, 48
What is needed?
The GWASs have clearly made an enormous contribution to our understanding of Alzheimer’s disease, chiefly through the identification of microglial and brain lipid metabolism49 as important risk components, and have focussed attention on the way the brain responds to amyloid deposition.50 Larger and larger GWASs now display the law of diminishing returns. A clear distinction needs to be introduced between Alzheimer’s disease GWAS and GWAS for dementia to avoid sending the misleading messages to molecular biologists: the latest big GWAS needs to be labelled as dementia GWAS, not Alzheimer’s disease GWAS. In these dementia GWAS, the Russian doll needs to be unpacked so that the summary statistics for each of them can be made available without an application process.
The consensus on the heritability of Alzheimer’s disease captured by the SNPs needs to be reached. If there is extensive missing heritability, as is widely believed, then epistatic interactions are likely candidates for this missing heritability where risks at unlinked loci act multiplicatively rather than additively. The possibility to detect epistatic loci is widely debated (28). However, this possibility is impaired if the case/control definition is inaccurate, and is forever lost if all that is available are summary statistics.
We need to understand more subtle phenotypic variability within the disease and the genetic factors which influence the rate of decline in disease. In this context, more genotyping of deeply phenotyped sample series and of cases with pathological confirmation are needed. In both cases, consents and protocols are required which permit academic access to individual level data to allow post hoc informed cleaning of these data. This would be preferable to ever larger GWAS of poorly characterized individuals. In parallel, we certainly need to understand the architecture of disease in non-European populations, and, within the genes we have already found, the identification of variability which would help disease modelling.
One way forward would be to develop a framework where the ever larger dementia GWAS hits were systematically evaluated in GWAS derived solely from Alzheimer’s disease pathologically confirmed samples, independent from the dementia GWAS. The current research trajectory will lead to ever more confusion, especially amongst those who are not aware of the problems we outline.
Abbreviations
- FTD =
frontotemporal dementia
- GWAS =
genome-wide association study
- LDSC =
linkage disequilibrium score
- MDD =
major depressive disorder
- PRS =
polygenic risk score.
Funding
The authors thank the Dementia Research Institute [UKDRI supported by the Medical Research Council (UKDRI-3003), Alzheimer’s Research UK and Alzheimer’s Society].
Competing interests
The authors report no competing interests.
References
- 1. Wightman DP, Jansen IE, Savage JE, et al. . A genome-wide association study with 1,126,563 individuals identifies new risk loci for Alzheimer’s disease. Nat Genet. 2021;53:1276–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Beach TG, Monsell SE, Phillips LE, Kukull W. Accuracy of the clinical diagnosis of Alzheimer disease at national institute on aging Alzheimer disease centers. 2005-2010. J Neuropath Exp Neur. 2012;71:266–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Escott-Price V, Baker E, Shoai M, et al. . Genetic analysis suggests high misassignment rates in clinical Alzheimer’s cases and controls. Neurobiol Aging. 2019;77:178–182. [DOI] [PubMed] [Google Scholar]
- 4. Deelen J, Evans DS, Arking DE, et al. . A meta-analysis of genome-wide association studies identifies multiple longevity genes. Nat Commun. 2019;10:3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schachter F, Fauredelanef L, Guenot F, et al. . Genetic associations with human longevity at the APOE and ACE loci. Nat Genet. 1994;6:29–32. [DOI] [PubMed] [Google Scholar]
- 6. Day FR, Loh PR, Scott RA, Ong KK, Perry JR. A robust example of collider bias in a genetic association study. Am J Hum Genet. 2016;98:392–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Leonenko G, Baker E, Stevenson-Hoare J, et al. . Identifying individuals with high risk of Alzheimer’s disease using polygenic risk scores. Nat Commun. 2021;12:4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Corneveaux JJ, Myers AJ, Allen AN, et al. . Association of CR1, CLU and PICALM with Alzheimer's disease in a cohort of clinically characterized and neuropathologically verified individuals. Hum Mol Genet. 2010;19:3295–3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Escott-Price V, Myers AJ, Huentelman M, Hardy J. Polygenic risk score analysis of pathologically confirmed Alzheimer disease. Ann Neurol. 2017;82:311–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Harold D, Abraham R, Hollingworth P, et al. . Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat Genet. 2009;41:1088–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang Q, Sidorenko J, Couvy-Duchesne B, et al. . Risk prediction of late-onset Alzheimer’s disease implies an oligogenic architecture. Nat Commun. 2020;11:4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Van Deerlin VM, Sleiman PMA, Martinez-Lage M, et al. . Common variants at 7p21 are associated with frontotemporal lobar degeneration with TDP43 inclusions. Nat Genet. 2010;42:234–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ferrari R, Hernandez DG, Nalls MA, et al. . Frontotemporal dementia and its subtypes: A genome-wide association study. Lancet Neurol. 2014;13:686–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Altmann A, Scelsi MA, Shoai M, et al. . A comprehensive analysis of methods for assessing polygenic burden on Alzheimer's disease pathology and risk beyond APOE. Brain Commun. 2020;2:fcz047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Leonenko G, Shoai M, Bellou E, et al. . Genetic risk for Alzheimer disease is distinct from genetic risk for amyloid deposition. Ann Neurol. 2019;86:427–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bulik-Sullivan BK, Loh PR, Finucane H, et al. . LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47:291–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kunkle BW, Grenier-Boley B, Sims R, et al. . Genetic meta-analysis of diagnosed Alzheimer's disease identifies new risk loci and implicates Abeta, tau, immunity and lipid processing. Nat Genet. 2019;51:414–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Marioni RE, Harris SE, Zhang Q, et al. . GWAS on family history of Alzheimer’s disease. Transl Psychiatry. 2018;8:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jansen IE, Savage JE, Watanabe K, et al. . Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat Genet. 2019;51:404–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. de Rojas I, Moreno-Grau S, Tesi N, et al. . Common variants in Alzheimer’s disease and risk stratification by polygenic risk scores. Nat Commun. 2021;12:3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bellenguez C, Küçükali F, Jansen IE, et al. . New insights into the genetic etiology of Alzheimer’s disease and related dementias. Nat Genet. 2022;54:412–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bulik-Sullivan BK. Genomic Analysis of Polygenic Traits. 2016. PhD-Thesis.
- 23. Lambert JC, Heath S, Even G, et al. . Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nat Genet. 2009;41:1094–1099. [DOI] [PubMed] [Google Scholar]
- 24. Seshadri S, Fitzpatrick AL, Ikram MA, et al. . Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA. 2010;303:1832–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Naj AC, Jun G, Beecham GW, et al. . Common variants at MS4A4/MS4A6E. CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nat Genet. 2011;43:436–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lambert JC, Ibrahim-Verbaas CA, Harold D, et al. . Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45:1452–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: A tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Moreno-Grau S, de Rojas I, Hernandez I, et al. . Genome-wide association analysis of dementia and its clinical endophenotypes reveal novel loci associated with Alzheimer's disease and three causality networks: The GR@ACE project. Alzheimers Dement. 2019;15:1333–1347. [DOI] [PubMed] [Google Scholar]
- 29. Morris JC. Is Alzheimer’s disease inevitable with age?: Lessons from clinicopathologic studies of healthy aging and very mild Alzheimer’s disease. J Clin Invest. 1999;104:1171–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Meyer MR, Tschanz JT, Norton MC, et al. . APOE genotype predicts when - not whether - one is predisposed to develop Alzheimer disease. Nat Genet. 1998;19:321–322. [DOI] [PubMed] [Google Scholar]
- 31. Reiman EM, Arboleda-Velasquez JF, Quiroz YT, et al. . Exceptionally low likelihood of Alzheimer’s dementia in APOE2 homozygotes from a 5,000-person neuropathological study. Nat Commun. 2020;11:667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gatz M, Reynolds CA, Fratiglioni L, et al. . Role of genes and environments for explaining Alzheimer disease. Arch Gen Psychiatry. 2006;63:168–174. [DOI] [PubMed] [Google Scholar]
- 33. Brainstorm C, Anttila V, Bulik-Sullivan B, et al. . Analysis of shared heritability in common disorders of the brain. Science 2018;360:eaap8757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lee SH, Harold D, Nyholt DR, et al. . Estimation and partitioning of polygenic variation captured by common SNPs for Alzheimer's disease, multiple sclerosis and endometriosis. Hum Mol Genet. 2013;22:832–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ridge PG, Mukherjee S, Crane PK, Kauwe JS, Alzheimer’s Disease Genetics Consortium . Alzheimer’s disease: Analyzing the missing heritability. PLoS One. 2013;8:e79771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Frisoni GB, Altomare D, Thal DR, et al. . The probabilistic model of Alzheimer disease: The amyloid hypothesis revised. Nat Rev Neurosci. 2022;23:53–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. McKay GJ, Silvestri G, Chakravarthy U, et al. . Variations in apolipoprotein e frequency with age in a pooled analysis of a large group of older people. Am J Epidemiol. 2011;173:1357–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kirkbride JB, Errazuriz A, Croudace TJ, et al. . Incidence of schizophrenia and other psychoses in England 1950-2009: A systematic review and meta-analyses. PLoS One. 2012:7:e31660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Howard DM, Adams MJ, Shirali M, et al. . Genome-wide association study of depression phenotypes in UK Biobank identifies variants in excitatory synaptic pathways. Nat Commun. 2018;9:1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nalls MA, Blauwendraat C, Vallerga CL, et al. . Identification of novel risk loci, causal insights, and heritable risk for Parkinson's disease: A meta-analysis of genome-wide association studies. Lancet Neurol. 2019;18:1091–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Raghavachari N. The impact of apolipoprotein E genetic variability in health and life span. J Gerontol A Biol Sci Med Sci. 2020;75:1855–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. El-Lebedy D, Raslan HM, Mohammed AM. Apolipoprotein E gene polymorphism and risk of type 2 diabetes and cardiovascular disease. Cardiovasc Diabetol. 2016;15:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lumsden AL, Mulugeta A, Zhou A, Hypponen E. Apolipoprotein E (APOE) genotype-associated disease risks: A phenome-wide, registry-based, case-control study utilising the UK Biobank. EBioMedicine. 2020;59:102954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Xu M, Zhao J, Zhang Y, et al. . Apolipoprotein E gene variants and risk of coronary heart disease: A meta-analysis. Biomed Res Int. 2016;2016:3912175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Minami SS, Min SW, Krabbe G, et al. . Progranulin protects against amyloid beta deposition and toxicity in Alzheimer’s disease mouse models. Nat Med. 2014;20:1157–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Moskvina V, Harold D, Russo G, et al. . Analysis of genome-wide association studies of Alzheimer disease and of Parkinson disease to determine if these 2 diseases share a common genetic risk. JAMA Neurol. 2013;70:1268–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bras J, Guerreiro R, Darwent L, et al. . Genetic analysis implicates APOE, SNCA and suggests lysosomal dysfunction in the etiology of dementia with Lewy bodies. Hum Mol Genet. 2014;23:6139–6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Escott-Price V, Sims R, Bannister C, et al. . Common polygenic variation enhances risk prediction for Alzheimer’s disease. Brain. 2015;138:3673–3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jones L, Holmans PA, Hamshere ML, et al. . Genetic evidence implicates the immune system and cholesterol metabolism in the aetiology of Alzheimer’s disease. PLoS One 2010;5:e13950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Salih DA, Bayram S, Guelfi S, et al. . Genetic variability in response to amyloid beta deposition influences Alzheimer's disease risk. Brain Commun. 2019;1:fcz022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analysed in this study.