Abstract
Fluorescence in situ hybridization (FISH) with rRNA-targeted oligonucleotide probes was used to investigate the phylogenetic composition of bacterioplankton communities in several freshwater and marine samples. An average of about 50% of the cells were detected by probes for the domains Bacteria and Archaea, and of these, about half could be identified at the subdomain level with a set of group-specific probes. Beta subclass proteobacteria constituted a dominant fraction in freshwater systems, accounting for 16% (range, 3 to 32%) of the cells, although they were essentially absent in the marine samples examined. Members of the Cytophaga-Flavobacterium cluster were the most abundant group detected in the marine systems, accounting for 18% (range, 2 to 72%) of the 4′,6-diamidino-2-phenylindole (DAPI) counts, and they were also important in freshwater systems (7%, range 0 to 18%). Furthermore, members of the alpha and gamma subclasses of Proteobacteria as well as members of the Planctomycetales were detected in both freshwater and marine water in abundances <7%.
In recent years, molecular methods based on the comparative analysis of 16S rRNA sequences have yielded new insights into the diversity of marine and freshwater bacterioplankton communities (see, e.g., references 4, 17, 25, 35, 37, and 49). Numerous new rRNA sequences have been found, indicating that the vast majority of bacterioplankton species are not yet represented in the collections of marine and freshwater strains. It has even been shown that the “bacterioplankton” contains members of the Archaea (11, 13, 15, 39). Whereas microbial diversity can be readily studied by 16S rRNA gene libraries, it is difficult, if not impossible, to deduce the community composition from them (3), especially when they are PCR based (59). Quantitative slot blot hybridization or fluorescence in situ hybridization (FISH) with rRNA-targeted oligonucleotide probes is better suited for this task (3). FISH has the potential to supplement the total cell counts, which are routinely determined in aquatic samples by the 4′,6-diamidino-2-phenylindol (DAPI) membrane filter technique (48), with counts on specific phylogenetic groups. With the recently described improved FISH protocol for aquatic samples (18), FISH seems no longer to be limited to systems with high nutrient concentrations. Consequently, it was the aim of this study to test the general applicability of this improved protocol to bacterioplankton and to gain the first insights into differences in the community compositions of marine and freshwater systems with domain- and group-specific oligonucleotide probes.
Sampling and fixation.
Data on the sampling time and locations are summarized in Table 1. Important characteristics like the trophic state (61), P concentration (in micrograms per liter (38), area (in square kilometers), and maximum depth (in meters) of the lakes are the following: Lake Gossenköllesee, oligotrophic, 1 to 7, 0.065, and 9.9; Lake Lago di Cadagno, mesotrophic, meromictic, 20 to 30, 0.357, 21; Lake Grosser Ostersee, mesotrophic, 20 to 25, 1.78, 29.7; Lake Baikal, oligotrophic, 2 to 11, 3.1 × 104, 1,741. Fixation was done by the method of Glöckner et al. (18). Cells were concentrated from water samples (1 to 100 ml) on white polycarbonate filters (diameter, 47 mm; pore size, 0.2 μm; type GTTP 4700 [Millipore, Eschborn, Germany]) by applying a vacuum of <25 kPa. They were subsequently fixed by covering the filter with 3 ml of a freshly prepared, phosphate-buffered saline (pH 7.2)–4% paraformaldehyde (Sigma, Deisenhofen, Germany) solution for 30 min at room temperature. The fixative was removed by applying vacuum, and the filter was subsequently covered with 3 ml each of phosphate-buffered saline and distilled water. Both were immediately removed by applying a vacuum. Air-dried filters are ready for hybridization and can be stored at −20°C or room temperature for several months without showing apparent changes.
TABLE 1.
Sampling sites used in this study
| Sampling site | Location | Date of sampling (mo/yr) |
|---|---|---|
| Lake Gossenköllesee | Austrian Alps near Innsbruck; 2,417 m a.m.s.l.a | 12/95 |
| Lake Lago di Cadagno | Piora Valley in the south of Switzerland; 1,923 m a.m.s.l. | 8/95 |
| Lake Grosser Ostersee | Southern Bavaria near Munich; 580 m a.m.s.l. | 7/96 |
| Lake Baikal | Southeastern Siberia near Irkutsk; 456 m a.m.s.l. | 8/95 and 8/96 |
| North Sea | “Kabeltonne,” offshore Helgoland | 10/97 |
| Baltic Sea | 57°18.18′N, 20°04.36′E | 8/96 |
| Pacific Ocean | Playa del Rey pier in Venice, Calif. | 10/96 |
| Antarctic Ocean | 68°50.57′S, 06°01.08′E; 58°56.52′S, 03°58.44′E; 49°29.40′S, 11°23.58′E; 50°28.62′S, 08°08.82′E | 1/96 |
a.m.s.l., above mean sea level.
FISH and probe-specific cell counts.
CY3-labeled oligonucleotides were purchased from Interactiva (Ulm, Germany). The probe sequences, hybridization conditions, and references are given in Table 2. Each filter was cut into 12 sections. The filter sections were placed on glass slides and covered with 20 μl of hybridization solution containing 0.9 M NaCl, 20 mM Tris-HCl (pH 7.4), 35% formamide, 0.01% sodium dodecyl sulfate, and 50 ng of CY3-labeled oligonucleotide and incubated at 46°C for 90 min in an equilibrated chamber. Probes BET42a, GAM42a, and PLA886 were used with competitor oligonucleotides as described previously (32, 40). The filters were transferred to a vial containing 50 ml of prewarmed (48°C) washing solution (70 mM NaCl, 20 mM Tris-HCl [pH 7.4], 5 mM EDTA, 0.01% sodium dodecyl sulfate) and incubated freely floating without shaking at 48°C for 15 min. The filter sections were dried on Whatman 3M paper (Whatman Ltd., Maidstone, United Kingdom), placed back on a glass slide, and covered with 50 μl of DAPI solution (1 μg/ml in distilled water filtered through at 0.2-μm filter) for 5 min at room temperature in the dark. Subsequently, they were gently washed in 50 ml of 0.2-μm-filtered distilled water, dried on Whatman 3M paper, and mounted on glass slides with Citifluor AF1 (Citifluor Ltd., Canterbury, United Kingdom). Glöckner et al. (18) have shown previously that due to firm adhesion of the cells to the polycarbonate filters, cumulative cell losses are below 10%.
TABLE 2.
Oligonucleotide probes used in this study
| Probe | Specificity | Sequence (5′-3′) of probe | Targeta site (rRNA positions) | % FAb in situ | Reference |
|---|---|---|---|---|---|
| EUB338 | Bacteria | GCTGCCTCCCGTAGGAGT | 16S (338–355) | 0–35 | 2 |
| NON338 | ACTCCTACGGGAGGCAGC | 16S (338–355) | 0–35 | 64 | |
| ALF968 | Alpha subclass of Proteobacteria | GGTAAGGTTCTGCGCGTT | 16S (968–986) | 35 | 40 |
| BET42a | Beta subclass of Proteobacteria | GCCTTCCCACTTCGTTT | 23S (1027–1043) | 35 | 32 |
| GAM42a | Gamma subclass of Proteobacteria | GCCTTCCCACATCGTTT | 23S (1027–1043) | 35 | 32 |
| CF319a | Cytophaga-Flavobacterium cluster of CFB phylum | TGGTCCGTGTCTCAGTAC | 16S (319–336) | 35 | 31 |
| PLA886 | Planctomycetales | GCCTTGCGACCATACTCCC | 16S (886–904) | 35 | 41 |
| ARCH915 | Archaea | GTGCTCCCCCGCCAATTCCT | 16S (915–935) | 20–35 | 58 |
Escherichia coli numbering (9).
Percentage of formamide (FA) in in situ hybridization buffer.
The filter sections were inspected with an Axioplan epifluorescence microscope (Zeiss, Jena, Germany) equipped with a 50-W high-pressure mercury bulb and specific filter sets (DAPI [Zeiss 01]; CY3 [Chroma HQ 41007; Chroma Tech. Corp., Brattleboro, Vt.]). Each microscopic field was first viewed with the CY3 filter set before switching to the DAPI filter set, to avoid bleaching of CY3 during the DAPI examination. For each sample and probe, more than 500 cells were enumerated; for the DAPI examination, more than 1,500 cells were counted per sample. All probe-specific cell counts are presented as the percentage of cells visualized by DAPI. The mean abundances and standard deviations were calculated from the counts of 10 to 20 randomly chosen fields on each filter section. All counts were corrected by subtracting the counts obtained with the negative control NON338 (Table 3). Concerning statistical analysis, we used nonparametric methods. If necessary, median values and confidence intervals (CI) are given instead of means and standard deviations. For testing statistical significance of differences, the Mann-Whitney U test was used.
TABLE 3.
DAPI and FISH counts
| Location | Sampling site | Depth (m) | Total cell counts (106 ml−1) (mean ± SD) | Fraction (%) of total cells (mean ± SD) detected with probea:
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| EUB338 | ALF968 | BET42a | GAM42a | CF319a | PLA886 | NON338 | ||||
| Lake Gossenköllesee | Above deepest point | 3 | 0.3 ± 0.04 | 51 ± 4 | 10 ± 3 | 18 ± 2 | 1 ± 1 | 16 ± 2 | 0 | 0 ± 0 |
| Lake Lago di Cadagno | Above deepest point | 6 | 3.2 ± 0.07 | 46 ± 9 | 0 ± 3 | 21 ± 4 | 0 ± 3 | 3 ± 5 | 0 | 7 ± 3 |
| 13 | 4.2 ± 0.7 | 61 ± 7 | 2 ± 2 | 17 ± 3 | 5 ± 3 | 8 ± 3 | 1 | 5 ± 1 | ||
| 18 | 4.7 ± 0.1 | 46 ± 5 | 0 ± 1 | 20 ± 4 | 2 ± 3 | 12 ± 3 | 1 | 6 ± 1 | ||
| Lake Grosser Ostersee | Above deepest point | 0 | 3.0 ± 0.3 | 46 ± 10 | 3 ± 5 | 14 ± 4 | 1 ± 4 | 2 ± 4 | 3 ± 3 | 3 ± 2 |
| 3.5 | 2.8 ± 0.3 | 46 ± 4 | 1 ± 1 | 5 ± 4 | 0 ± 1 | 1 ± 2 | 1 | 2 ± 1 | ||
| 7 | 3.5 ± 0.4 | 49 ± 4 | 2 ± 2 | 3 ± 2 | 0 ± 2 | 2 ± 2 | 1 ± 2 | 5 ± 3 | ||
| 18 | 2.2 ± 0.4 | 59 ± 7 | 1 ± 1 | 16 ± 3 | 1 ± 0 | 6 ± 3 | 2 ± 1 | 0 ± 0 | ||
| 27 | 3.3 ± 0.5 | 61 ± 12 | 3 ± 1 | 11 ± 2 | 0 ± 1 | 7 ± 5 | 1 ± 1 | 1 ± 1 | ||
| Lake Baikal | Southern basin, above deepest point | 0 | 0.6 ± 0.05 | 44 ± 5 | 1 ± 4 | 4 ± 4 | 0 ± 2 | 4 ± 1 | 1 | 12 ± 3 |
| 25 | 0.7 ± 0.04 | 55 ± 3 | 1 | 17 ± 2 | 1 | 10 ± 2 | 2 | 4 ± 1 | ||
| 1,000 | 0.9 ± 0.03 | 43 ± 3 | ND | 15 ± 1 | 4 ± 2 | ND | ND | 12 ± 2 | ||
| 1,200 | 0.3 ± 0.07 | ND | 1 ± 2 | ND | ND | 5 ± 2 | ND | 12 ± 2 | ||
| Middle basin, above deepest point | 0 | 0.8 ± 0.2 | 49 ± 10 | 1 | 18 ± 2 | 1 | 12 ± 3 | 1 | 7 ± 1 | |
| 25 | 0.9 ± 0.03 | 53 ± 3 | 1 | 11 ± 2 | 1 | 18 ± 2 | 2 ± 2 | 6 ± 2 | ||
| 1,255 | 0.3 ± 0.01 | ND | 1 ± 1 | ND | ND | 0 ± 1 | ND | 2 ± 1 | ||
| 1,455 | 0.08 ± 0.002 | 59 ± 4 | ND | 32 ± 4 | 0 ± 1 | ND | ND | 1 ± 1 | ||
| Baltic Sea | 57°18.18′N, 20°04.36′E | 18 | 3.6 ± 0.4 | 72 ± 8 | 14 ± 2 | 15 ± 1 | 3 ± 2 | 14 ± 3 | 0 ± 1 | 1 ± 1 |
| 78 | 1.4 ± 0.2 | 64 ± 5 | 8 ± 2 | 29 ± 6 | 0 ± 1 | 2 ± 1 | 0 | 4 ± 1 | ||
| 121 | 2.3 ± 0.2 | 62 ± 4 | 5 ± 2 | 15 ± 2 | 1 | 10 ± 2 | 0 | 5 ± 2 | ||
| 229 | 3.0 ± 0.3 | 66 ± 7 | 4 ± 1 | 6 ± 3 | 1 ± 0 | 12 ± 1 | 0 | 1 ± 1 | ||
| North Sea | “Kabeltonne” off Helgoland | 1 | 7.0 ± 0.2 | 39 ± 3 | 1 | 0 | 6 ± 2 | 18 ± 2 | 1 ± 2 | 1 ± 2 |
| Pacific Ocean | Playa del Rey, near shore | 3.0 ± 0.03 | 42 ± 3 | 5 ± 2 | 4 ± 3 | 4 ± 2 | 12 ± 2 | 0 ± 1 | 3 ± 1 | |
| Antarctic Ocean | 68°50.57′S, 06°01.08′E | 0 | 1.3 ± 0.09 | 96 ± 3 | 8 ± 2 | 0 ± 1 | 7 ± 2 | 72 ± 5 | 0 | 1 ± 1 |
| 58°56.52′S, 03°58.44′E | 0 | 0.7 ± 0.07 | 85 ± 4 | 6 ± 1 | 0 ± 1 | 9 ± 2 | 40 | 0 | 1 ± 1 | |
| 49°29.40′S, 11°23.58′E | 20 | 0.9 ± 0.05 | 57 ± 10 | 4 ± 2 | 0 ± 1 | 0 ± 1 | 19 ± 3 | 0 | 1 ± 1 | |
| 40 | 1.6 ± 0.1 | 61 ± 8 | 11 ± 1 | 0 ± 0 | 1 ± 1 | 24 ± 2 | 0 | 1 ± 0 | ||
| 50°28.62′S, 08°08.82′E | 60 | 0.8 ± 0.08 | 72 ± 8 | 11 ± 2 | 0 ± 0 | 0 ± 0 | 19 ± 4 | 0 | 0 ± 0 | |
Percent detection compared to DAPI. Numbers have been corrected by subtracting NON338 counts. Mean and standard deviation were calculated from the counts of 10 to 20 randomly chosen fields on each filter section.
ND, not done.
Total cell counts.
The total cell counts found in our samples (105 to 107 cells ml−1) were in the normal range reported for oligotrophic and mesotrophic aquatic systems (28). The more oligotrophic systems usually contained between 3 × 105 and 9 × 105 cells ml−1, and the mesotrophic systems contained between 1.4 × 106 and 7.0 × 106 cells ml−1 (Table 3). Exceptions were the middle basin of Lake Baikal at 1,455 m, where only 8 × 104 cells ml−1 were present and samples from the oligotrophic Antarctic Ocean at 0 and 40 m, with total cell counts between 1.3 × 106 and 1.6 × 106 cells ml−1.
Domain-specific probing.
Detection yields relative to DAPI with the EUB338 probe, complementary to a signature site present in most members of the Bacteria, ranged from 39% in the North Sea to 96% in the Antarctic Ocean at 0 m, with a median of 56% (CI, 46 to 61%; n = 26) (Table 3). All samples examined showed bright hybridization signals and a clear distinction between probe-conferred signals and the background. The fraction of autofluorescent and nonspecifically stained cells as determined with the negative control probe NON338 was moderate in all samples, with a median of 5% (CI, 2 to 7%; n = 17) in the freshwater and 1% (CI, 1 to 4%; n = 11) in the marine systems. Results obtained with controls without the addition of CY3-labeled probes showed that the background signals were derived mainly from chlorophyll-containing cells (e.g., algae and cyanobacteria) and inorganic particles and to a much lesser extent from nonspecifically stained cells.
Consequently, with the recently described protocol (18), FISH seems no longer limited to hypereutrophic and eutrophic aquatic ecosystems (1, 3, 24). Around 50% of the cells could be detected. A closer microscopic examination of the exceptional Antarctic Ocean samples revealed a Phaeocystis algal bloom, which coincided with a quite uniform and clearly detectable bloom of members of the Cytophaga-Flavobacterium group. Members of the Bacteria were more abundant in all samples than were members of the Archaea. Although applied to all samples, probe ARCH915, specific for most members of the Archaea, detected cells only in the samples from Lake Gossenköllesee (6%), the North Sea (3%), and the Pacific Ocean (2%). This supports earlier findings on the widespread occurrence of Archaea in the water column (11, 43), even though they did not reach the high abundances described previously (13, 39). Methodological limitations or temporal and spatial variations could have been the reason for not detecting Archaea in the Antarctic Ocean, where they have been found previously (13, 39).
Group-specific probing.
When the community composition was further analyzed with a set of oligonucleotide probes targeting larger phylogenetic groups within the domain Bacteria, only about 56% (CI, 44 to 62%; n = 26) of the cells detected with probe EUB338 could be assigned (Table 3). This is certainly due to the incompleteness of the probe set used in this study. This set was developed for biotechnological, more eutrophic systems like activated sludge and obviously needs to be supplemented for use in mesotrophic and oligotrophic aquatic systems. New probes must be developed to target sequences found in the rDNA libraries from freshwater and marine systems (see, e.g., references 4, 25, and 37). In particular, new group-specific probes for the epsilon and delta subclasses of the Proteobacteria and the Chlorobiaceae need to be designed (21, 67).
Beta-subclass proteobacteria.
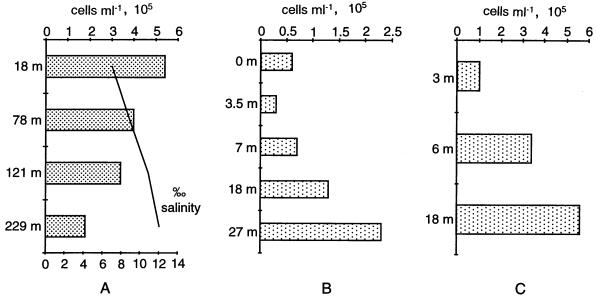
The incomplete set of probes nevertheless allowed the detection of differences between the bacterioplankton composition of marine and freshwater systems. These differences were most pronounced for the beta-subclass proteobacteria. These accounted for 16% (CI, 11 to 18%; n = 15) of DAPI-stained cells in the freshwater samples, equivalent to 1.4 × 105 cells ml−1 (CI, 9.9 × 104 to 4.2 × 105 cells ml−1). In the deep layers of the middle basin of Lake Baikal, they made up 32% of the cells stained with DAPI. The median abundance of beta-subclass proteobacteria in the marine samples investigated was 0% (CI, 0 to 15%; n = 11). No beta-subclass proteobacteria could be found in the Antarctic Ocean and the North Sea. In the sample from the coastal Pacific Ocean, 4% of the total counts were detected with the BET42a probe. The upper layers of the Baltic Sea, which are strongly influenced by freshwater (salinity, 7.2 to 8.0%), were exceptions. Here, the counts ranged between 15 and 29% (Fig. 1A). In most of the samples examined, beta-subclass proteobacteria were straight to curved rods 1.5 to 4.5 μm long and approximately 1 μm wide. Only in Lake Gossenköllesee were short filaments (8 to 13 μm long) visible. Our findings corroborate diversity data obtained by cloning of 16S rRNA gene fragments from freshwater and marine environments. In 16S rDNA libraries from pelagic freshwater and lake snow aggregates, the majority of clones was affiliated with the beta subclass of Proteobacteria (4, 25, 35, 66), whereas beta-proteobacterial sequences were not found in most marine libraries (see, e.g., references 6, 8, 16, 17, 20, 37, and 54) or in marine aggregates (50). The exceptions are a report from Suzuki et al. (60) that beta-subclass proteobacteria were found at the coast of Oregon and reports on PCR detection of ammonium-oxidizing beta-subclass proteobacteria in marine samples (27, 62). Interesting in this respect are the results from the Baltic Sea, a heterogeneous system in which low-salt water (7‰ salinity) overlays a water body (12‰ salinity) influenced by the North Sea (51). At the time of sampling, a halocline between 78 m (8‰ salinity) and 121 m (11‰ salinity) clearly separated an upper water body with high relative abundances of beta-subclass proteobacteria (15 to 29%; 4.1 × 105 to 5.4 × 105 cells ml−1) from a water body with lower abundances (6%; 1.8 × 105 cells ml−1) (Fig. 1A). The higher salinity in the deeper layers might prevent the establishment of this group in large numbers. This idea is supported by recent findings by Painchaud et al. (44), who reported increased mortality of freshwater bacteria from the St. Lawrence River in brackish water with more than 10‰ salinity.
FIG. 1.
(A) Depth profile from the Baltic Sea. The bars show the total abundances of members of the beta subclass of the proteobacteria as revealed with probe BET42a. The line indicates the salinity. (B and C) Depth profiles from Lake Grosser Ostersee (B) and Lake Lago di Cadagno (C). The bars show the total abundances of the Cytophaga-Flavobacterium group as revealed with probe CF319a.
Cytophaga-Flavobacterium group.
Members of the Cytophaga-Flavobacterium group could be found in all marine and freshwater samples investigated and usually formed the largest bacterial group in marine water, with a median of 18% (CI, 10 to 40%; n = 11). The relative abundance ranged from 2% in the Baltic Sea (depth, 78 m) to 72% in the Antarctic Ocean (68°50.57′S 06°01.08′E; depth, 0 m). Members of this group were also present in all freshwater systems studied, with a lower median abundance of 6% (CI, 2 to 12%; n = 15). The average total cell counts were 3.6 × 105 cells ml−1 (CI, 1.5 × 105 to 9.4 × 105 cells ml−1) for marine samples and 7.0 × 104 cells ml−1 (CI, 2.8 × 104 to 1.6 × 105 cells ml−1) for freshwater samples. This is in good agreement with cultivation studies that report frequent isolation of Cytophaga and Flavobacterium spp. from freshwater and marine systems (4, 22, 36, 47, 56, 69), whereas in some 16S rRNA gene libraries, sequences of the Cytophaga-Flavobacterium group seem to be underrepresented (4, 17, 21, 37, 57, 60, 67). Comparative sequence analysis showed that the frequently used reverse “universal” primers OX2, 1518R, and 1522R at around Escherichia coli position 1520 and the forward primer 68F at E. coli positions 48 to 68 (8, 17, 21, 37, 54, 57, 60, 67) discriminate against the amplification of 16S rDNA of members of the Cytophaga-Flavobacterium group (e.g., E. coli position 67, where most members of the Cytophaga-Flavobacterium group have a G instead of a C). The potential of PCR primers to influence the sequence diversity within 16S rRNA gene libraries has been discussed previously (33). Clearly, there is no guarantee that modern molecular methods of biodiversity research are representative, but by using a combination of various methods, biases should be detected.
In two of the water columns studied, Lake Grosser Ostersee and Lake Lago di Cadagno, the relative abundance and total counts of Cytophaga-Flavobacterium members increased with depth (Fig. 1B and C). From pure-culture studies, it is known that members of the Cytophaga-Flavobacterium group are able to degrade aerobically a large spectrum of substrates ranging from various proteins, carbohydrates, pesticides, and insecticides to complex macromolecules (7). It is now tempting to speculate that this group is involved in the mineralization of the more slowly degradable macromolecules which accumulate in deeper water. The absolute cell counts determined by FISH will in the future allow us to relate the size of specific populations to nutrient availability and other factors such as mortality due to grazing or virus infections. Most of the cells detected with probe CF319a were filaments 20 to 300 μm long and 0.5 to 1 μm wide. Some samples (Lake Gossenköllesee and Antarctic Ocean) also contained rods 2 to 3 μm long and 0.5 to 1 μm width.
Alpha-subclass proteobacteria.
The medians of relative abundances and absolute cell numbers of alpha-subclass proteobacteria detected with the probe ALF968 were significantly higher in the marine samples (6%; CI, 4 to 11%; 1.1 × 105 cells ml−1; CI, 4.2 × 104 to 1.8 × 105 cells ml−1 [n = 11]) than in the freshwater samples used (1%; CI, 1 to 2%; 9.0 × 103 cells ml−1; CI, 3.0 × 103 to 7.0 × 104 cells ml−1 [n = 15]). In lakes the variation was between 0% in Lake Lago di Cadagno at 3 and 18 m and 10% in Lake Gossenköllesee. In marine systems, the range of relative abundance of alpha-subclass proteobacteria was between 1% in the North Sea and 14% in the Baltic Sea (18 m). The morphology of the alpha subclass of Proteobacteria was diverse and included rods, vibrios, and filaments of various size. Clone sequences affiliated with the alpha and gamma subclasses of Proteobacteria are commonly retrieved from marine aquatic ecosystems (16, 17, 20, 37, 60). Our FISH results support the widespread occurrence of members of the alpha subclass of Proteobacteria with larger numbers in the marine samples than in the freshwater samples. In no sample were the detectable alpha-subclass proteobacteria more abundant than members of the Cytophaga-Flavobacterium group. It remains to be investigated with, e.g., genus-level probes which groups of alpha-subclass proteobacteria are frequent in aquatic systems.
Gamma-subclass proteobacteria.
There were no clear-cut trends for gamma-subclass proteobacteria. In Lake Lago di Cadagno, the distribution of probe GAM42a-positive cells was strongly influenced by a layer of Chromatium sp. and Amoebobacter sp. in the metalimnion (13 m; 5%, 2.1 × 105 cells ml−1). Even though these cells showed a red autofluorescence, they could be unambiguously assigned to the gamma subclass of Proteobacteria by the yellow-orange signal conferred by CY3-labeled probe GAM42a. In the marine samples, only those from the North Sea and the surface layers of the Antarctic Ocean showed increased abundances of gamma-subclass proteobacteria, with 6% (4.2 × 105 cells ml−1) and 9% (6.3 × 104 cells ml−1), respectively. In all other samples, the cells detected with GAM42a were at or below 4% of the total counts. Traditional cultivation methods attributed a high importance to members of the gamma subclass of Proteobacteria (5, 30, 69). Our current FISH data reveal relative abundances of <4%, suggesting that members of this group may, at least numerically, be only a minor part of the bacterioplankton. The positive selection for gamma-subclass proteobacteria on agar plates is a well-known phenomenon which has recently been analyzed by FISH (63). Many members of this group are typical copiotrophs, adapted to high nutrient concentrations, and therefore grow well under laboratory conditions (68). Moreover, it has been reported recently that the 23S rRNA-targeted probe GAM42a did not detect all deep-branching bacteria in the gamma subclass of Proteobacteria (10, 19). In a time of a rapidly increasing rRNA database, the set of rRNA-targeted oligonucleotide probes needs to be continuously refined, and therefore it might be necessary to supplement probe GAM42a with some additional probes.
Planctomycetales.
Planctomycetes are known to be typical and widespread members of freshwater and marine ecosystems (26, 53), and rRNA clones related to Planctomyces spp. have also been found associated with marine macroaggregates (12, 50). We found them in small numbers in all freshwater samples (1 to 3%; 2.8 × 104 to 9.0 × 104 cells ml−1) and also in the North Sea sample (1%; 7.0 × 104 cells ml−1). The cells detected were usually large cocci with diameters of >1 μm; there was frequently an uneven distribution of the fluorescent signal in the cell, due to the lack of probe hybridization to the nucleoid (14). An explanation for their absence in the marine samples investigated, except for the North Sea, could be the low relative abundances (ca. 1%), which is close to the present detection limits of FISH.
Limitations.
For most regular water samples, we could not use FISH to assign 40 to 50% of the particles stained with DAPI to one of the three domains Archaea, Bacteria, or Eucarya. A comparison of DAPI and EUB338 pictures showed that the particles stained with DAPI but not with EUB338 probe were usually very small, at about the resolution of the light microscope (0.2 μm). Considering this, an affiliation of the undetected DAPI-stained particles with the domain Eucarya can almost certainly be excluded. This leaves us with two other likely explanations. (i) Bacteria or Archaea remained undetected due to the low ribosome content often found in starved cells (29, 52), to limited probe penetration caused by, e.g., thick cell walls, or to absence of the bacterial and archaeal signatures targeted by the probes. (ii) The DAPI-positive particles which cannot be stained with rRNA-targeted probes are viruses. Viruses are abundant in freshwater and marine aquatic ecosystems (23); in particular, the large ones with sizes up to 150 nm (34, 65) might be detected by sensitive epifluorescence microscopy. Attempts to increase the detection yields by, e.g., the application of highly sensitive enzyme-based signal amplification systems (55) or changes in the cell fixation to increase cell permeability (3) failed (data not shown). Other authors suggested recently that short-term incubations with chloramphenicol increased detection rates (42).
A further limitation of the present study originates from the fact that our probes target quite large phylogenetic groups. The alpha subclass of Proteobacteria, for example, encompasses photosynthetic anaerobes like Rhodobacter spp. as well as aerobic heterotrophs like Caulobacter and Hyphomicrobium spp. Furthermore, as a result of the rapid growth of the 16S rRNA sequence database, the coverage of the target groups by the current probe set has been shown to be rather incomplete. This is true for both the domain- and group-specific probes.
Conclusions.
We have demonstrated the wide applicability of FISH to studies of bacterioplankton composition. FISH can, of course, also be used to observe the development of specific genera or species within the bacterioplankton (45). Additional information on changes in cell size and morphology can be readily obtained. This information is of great ecological importance and allows, e.g., the calculation of biomass distribution to defined groups or the study of the development of grazing-resistant populations (45, 46). FISH studies with high spatial, temporal, and phylogenetic resolution may considerably increase our knowledge of bacterioplankton ecology in the future.
Acknowledgments
This work has been supported by grant Am73/2-4 of the Deutsche Forschungsgemeinschaft.
We thank Albin Alfreider, Natalia Belkova, Meinhard Simon, Pierre Rossi, and Tamara Zemskaya for making samples available; Alexander Neef for providing access to probe ALF968 before publication; and Günter Jost and Jakob Pernthaler for carefully reading the manuscript and helping with the statistics.
REFERENCES
- 1.Amann R, Glöckner F O, Neef A. Modern methods in subsurface microbiology—in situ identification of microorganisms with nucleic acid probes. FEMS Microbiol Rev. 1997;20:191–200. [Google Scholar]
- 2.Amann R I, Binder B J, Olson R J, Chisholm S W, Devereux R, Stahl D A. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann R I, Ludwig W, Schleifer K H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bahr M, Hobbie J E, Sogin M L. Bacterial diversity in an arctic lake—a freshwater SAR11 cluster. Aquat Microb Ecol. 1996;11:271–277. [Google Scholar]
- 5.Baumann L, Baumann P, Mandel M, Allen R D. Taxonomy of aerobic marine eubacteria. J Bacteriol. 1972;110:402–429. doi: 10.1128/jb.110.1.402-429.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benlloch S, Rodriguez Valera F, Martinez Murcia A J. Bacterial diversity in two coastal lagoons deduced from 16S rDNA PCR amplification and partial sequencing. FEMS Microbiol Ecol. 1995;18:267–279. [Google Scholar]
- 7.Bernardet J F, Segers P, Vancanneyt M, Berthe F, Kersters K, Vandamme P. Cutting a gordian knot: emended classification and description of the genus Flavobacterium, emended description of the family Flavobacteriaceae, and proposal of Flavobacterium hydatis nom. nov. (basonym, Cytophaga quatilis Stohl and Tait 1978. Int J Syst Bacteriol. 1996;46:128–148. [Google Scholar]
- 8.Britschgi T B, Giovannoni S J. Phylogenetic analysis of a natural marine bacterioplankton population by rRNA gene cloning and sequencing. Appl Environ Microbiol. 1991;57:1707–1713. doi: 10.1128/aem.57.6.1707-1713.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brosius J, Dull T J, Sleeter D D, Noller H F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981;148:107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- 10.Buchholz-Cleven B, Rattunde B, Straub K. Screening for genetic diversity of isolates of anaerobic Fe(II)-oxidizing bacteria using DGGE and whole-cell hybridization. Syst Appl Microbiol. 1997;20:301–309. [Google Scholar]
- 11.DeLong E F. Archaea in coastal marine environments. Proc Natl Acad Sci USA. 1992;89:5685–5689. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeLong E F, Franks D G, Alldredge A L. Phylogenetic diversity of aggregate-attached vs. free-living marine bacterial assemblages. Limnol Oceanogr. 1993;38:924–934. [Google Scholar]
- 13.DeLong E F, Wu K Y, Prezelin B B, Jovine R V M. High abundance of Archaea in Antarctic marine picoplankton. Nature. 1994;371:695–697. doi: 10.1038/371695a0. [DOI] [PubMed] [Google Scholar]
- 14.Fuerst J A. The planctomycetes: emerging models for microbial ecology, evolution and cell biology. Microbiology. 1995;141:1493–1506. doi: 10.1099/13500872-141-7-1493. [DOI] [PubMed] [Google Scholar]
- 15.Fuhrman J A, McCallum K, Davis A A. Novel major archaebacterial group from marine plankton. Nature. 1992;356:148–149. doi: 10.1038/356148a0. [DOI] [PubMed] [Google Scholar]
- 16.Fuhrman J A, McCallum K, Davis A A. Phylogenetic diversity of subsurface marine microbial communities from the Atlantic and Pacific Oceans. Appl Environ Microbiol. 1993;59:1294–1302. doi: 10.1128/aem.59.5.1294-1302.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giovannoni S J, Britschgi T B, Moyer C L, Field K G. Genetic diversity in Sargasso Sea bacterioplankton. Nature. 1990;345:60–63. doi: 10.1038/345060a0. [DOI] [PubMed] [Google Scholar]
- 18.Glöckner F O, Amann R, Alfreider A, Pernthaler J, Psenner R, Trebesius K, Schleifer K-H. An in situ hybridization protocol for detection and identification of planktonic bacteria. Syst Appl Microbiol. 1996;19:403–406. [Google Scholar]
- 19.Glöckner F O, Babenzien H-D, Amann R. Phylogeny and identification in situ of Nevskia ramosa. Appl Environ Microbiol. 1998;64:1895–1901. doi: 10.1128/aem.64.5.1895-1901.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gonzalez J M, Moran M A. Numerical dominance of a group of marine bacteria in the alpha-subclass of the class Proteobacteria in coastal seawater. Appl Environ Microbiol. 1997;63:4237–4242. doi: 10.1128/aem.63.11.4237-4242.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gordon D A, Giovannoni S J. Detection of stratified microbial populations related to Chlorobium and Fibrobacter species in the Atlantic and Pacific Oceans. Appl Environ Microbiol. 1996;62:1171–1177. doi: 10.1128/aem.62.4.1171-1177.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gosink J J, Staley J T. Biodiversity of gas vacuolate bacteria from Antarctic Sea ice and water. Appl Environ Microbiol. 1995;61:3486–3489. doi: 10.1128/aem.61.9.3486-3489.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heldal M, Bratbak G. Production and decay of viruses in aquatic environments. Mar Ecol Prog Ser. 1991;72:205–212. [Google Scholar]
- 24.Hicks R E, Amann R I, Stahl D A. Dual staining of natural bacterioplankton with 4′,6-diamino-2-phenylindole and fluorescent oligonucleotide probes targeting kingdom-level 16S rRNA sequences. Appl Environ Microbiol. 1992;58:2158–2163. doi: 10.1128/aem.58.7.2158-2163.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hiorns W D, Methe B A, Nierzwicki-Bauer S A, Zehr J P. Bacterial diversity in Adirondack mountain lakes as revealed by 16S rRNA gene sequences. Appl Environ Microbiol. 1997;63:2957–2960. doi: 10.1128/aem.63.7.2957-2960.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirsch P, Rheinheimer G. Biology of budding bacteria. V. Budding bacteria in aquatic habitats: occurrence, enrichment and isolation. Arch Mikrobiol. 1968;62:289–306. [PubMed] [Google Scholar]
- 27.Hovanec T A, Delong E F. Comparative analysis of nitrifying bacteria associated with freshwater and marine aquaria. Appl Environ Microbiol. 1996;62:2888–2896. doi: 10.1128/aem.62.8.2888-2896.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kepner R L, Pratt J R. Use of fluorochromes for direct enumaration of total bacteria in environmental samples—past and present. Microbiol Rev. 1994;58:603–615. doi: 10.1128/mr.58.4.603-615.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kolter R, Siegele D A, Tormo A. The stationary phase of the bacterial life cycle. Annu Rev Microbiol. 1993;47:855–874. doi: 10.1146/annurev.mi.47.100193.004231. [DOI] [PubMed] [Google Scholar]
- 30.MacLeod R A. The question of the existence of specific marine bacteria. Bacteriol Rev. 1965;29:9–23. [PMC free article] [PubMed] [Google Scholar]
- 31.Manz W, Amann R, Ludwig W, Vancanneyt M, Schleifer K-H. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum Cytophaga-Flavobacter-Bacteroides in the natural environment. Microbiology. 1996;142:1097–1106. doi: 10.1099/13500872-142-5-1097. [DOI] [PubMed] [Google Scholar]
- 32.Manz W, Amann R, Ludwig W, Wagner M, Schleifer K-H. Phylogenetic oligodeoxynucleotide probes for the major subclasses of Proteobacteria: problems and solutions. Syst Appl Microbiol. 1992;15:593–600. [Google Scholar]
- 33.Marchesi J R, Sato T, Weightman A J, Martin T A, Fry J C, Hiom S J, Wade W G. Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl Environ Microbiol. 1998;64:795–799. doi: 10.1128/aem.64.2.795-799.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mathias C B, Kirschner A K T, Velimirov B. Seasonal variations of virus abundance and viral control of the bacterial production in a backwater system of the Danube River. Appl Environ Microbiol. 1995;61:3734–3740. doi: 10.1128/aem.61.10.3734-3740.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Methé B A, Hiorns W D, Zehr J P. Contrasts between marine and freshwater bacterial community composition—analyses of communities in Lake George and six other Adirondack lakes. Limnol Oceanogr. 1998;43:368–374. [Google Scholar]
- 36.Morita R Y. Low-nutrient environments. In: Lederberg J, editor. Encyclopedia of Microbiology. Vol. 1. San Diego, Calif: Academic Press, Inc.; 1992. pp. 617–624. [Google Scholar]
- 37.Mullins T D, Britschgi T B, Krest R L, Giovannoni S J. Genetic comparisons reveal the same unknown bacterial lineages in Atlantic and Pacific bacterioplankton communities. Limnol Oceanogr. 1995;40:148–158. [Google Scholar]
- 38.Murphy J, Riley J P. A modified solution method for the determination of phosphate in natural waters. Anal Chim Acta. 1962;27:31–36. [Google Scholar]
- 39.Murray A E, Preston C M, Massana R, Taylor L T, Blakis A, Wu K, Delong E F. Seasonal and spatial variability of bacterial and archaeal assemblages in the coastal waters near Anvers Island, Antarctica. Appl Environ Microbiol. 1998;64:2585–2595. doi: 10.1128/aem.64.7.2585-2595.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Neef A. Anwendung der in situ-Einzelzell-Identifizierung von Bakterien zur Populationsanalyse in komplexen mikrobiellen Biozönosen. Ph.D. Thesis. Munich, Germany: Technische Universität München; 1997. [Google Scholar]
- 41.Neef A, Amann R, Schlesner H, Schleifer K-H. Monitoring a widespread bacterial group: in situ detection of planctomycetes with 16S rRNA-targeted probes. Microbiology. 1998;144:3257–3266. doi: 10.1099/00221287-144-12-3257. [DOI] [PubMed] [Google Scholar]
- 42.Ouverney C C, Fuhrman J A. Increase in fluorescence intensity of 16S rRNA in situ hybridization in natural samples treated with chloramphenicol. Appl Environ Microbiol. 1997;63:2735–2740. doi: 10.1128/aem.63.7.2735-2740.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ovreas L, Forney L, Daae F L, Torsvik V. Distribution of bacterioplankton in meromictic Lake Saelenvannet, as determined by denaturing gradient gel electrophoresis of PCR-amplified gene fragments coding for 16S rRNA. Appl Environ Microbiol. 1997;63:3367–3373. doi: 10.1128/aem.63.9.3367-3373.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Painchaud J, Therriault J C, Legendre L. Assessment of salinity-related mortality of freshwater bacteria in the Saint Lawrence estuary. Appl Environ Microbiol. 1995;61:205–208. doi: 10.1128/aem.61.1.205-208.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pernthaler J, Glöckner F O, Unterholzner S, Alfreider A, Psenner R, Amann R. Seasonal community and population dynamics of pelagic Bacteria and Archaea in a high mountain lake. Appl Environ Microbiol. 1998;64:4299–4306. doi: 10.1128/aem.64.11.4299-4306.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pernthaler J, Posch T, Simek K, Vrba J, Amann R, Psenner R. Contrasting bacterial strategies to coexist with a flagellate predator in an experimental microbial assemblage. Appl Environ Microbiol. 1997;63:596–601. doi: 10.1128/aem.63.2.596-601.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pinhassi J, Zweifel U L, Hagstrom A. Dominant marine bacterioplankton species found among colony-forming bacteria. Appl Environ Microbiol. 1997;63:3359–3366. doi: 10.1128/aem.63.9.3359-3366.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Porter K G, Feig Y S. The use of DAPI for identifying and counting aquatic microflora. Limnol Oceanogr. 1980;25:943–948. [Google Scholar]
- 49.Rappé M S, Suzuki M T, Vergin K L, Giovannoni S J. Phylogenetic diversity of ultraplankton plastid small-subunit rRNA genes recovered in environmental nucleic acid samples from the Pacific and Atlantic coasts of the United States. Appl Environ Microbiol. 1998;64:294–303. doi: 10.1128/aem.64.1.294-303.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rath J, Wu K Y, Herndl G J, Delong E F. High phylogenetic diversity in a marine-snow-associated bacterial assemblage. Aquat Microb Ecol. 1998;14:261–269. [Google Scholar]
- 51.Rheinheimer G. Meereskunde der Ostsee. 2nd ed. Berlin, Germany: Springer-Verlag; 1996. [Google Scholar]
- 52.Roszak D B, Colwell R R. Survival strategies of bacteria in the natural environment. Microbiol Rev. 1987;51:365–379. doi: 10.1128/mr.51.3.365-379.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schlesner H. The development of media suitable for the microorganisms morphologically resembling Planctomyces spp., Pirellula spp., and other Planctomycetales from various aquatic habitats using dilute media. Syst Appl Microbiol. 1994;17:135–145. [Google Scholar]
- 54.Schmidt T M, DeLong E F, Pace N R. Analysis of a marine picoplankton community by 16S rRNA gene cloning and sequencing. J Bacteriol. 1991;173:4371–4378. doi: 10.1128/jb.173.14.4371-4378.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schönhuber W, Fuchs B, Juretschko S, Amann R. Improved sensitivity of whole-cell hybridization by the combination of horseradish peroxidase-labeled oligonucleotides and tyramide signal amplification. Appl Environ Microbiol. 1997;63:3268–3273. doi: 10.1128/aem.63.8.3268-3273.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shewan J M, McMeekin T A. Taxonomy (and ecology) of Flavobacterium and related genera. Annu Rev Microbiol. 1983;37:233–252. doi: 10.1146/annurev.mi.37.100183.001313. [DOI] [PubMed] [Google Scholar]
- 57.Snaidr J, Amann R, Huber I, Ludwig W, Schleifer K-H. Phylogenetic analysis and in situ identification of bacteria in activated sludge. Appl Environ Microbiol. 1997;63:2884–2896. doi: 10.1128/aem.63.7.2884-2896.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stahl D A, Amann R. Development and application of nucleic acid probes. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. Chichester, United Kingdom: John Wiley & Sons Ltd.; 1991. pp. 205–248. [Google Scholar]
- 59.Suzuki M T, Giovannoni S J. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol. 1996;62:625–630. doi: 10.1128/aem.62.2.625-630.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Suzuki M T, Rappe M S, Haimberger Z W, Winfield H, Adair N, Strobel J, Giovannoni S J. Bacterial diversity among small-subunit rRNA gene clones and cellular isolates from the same seawater sample. Appl Environ Microbiol. 1997;63:983–989. doi: 10.1128/aem.63.3.983-989.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vollenweider R A. Scientific fundamentals of the eutrophication of lakes and flowing waters, with particular reference to nitrogen and phosphorus as factors in eutrophication. Paris, France: Organisation for Economic Cooperation and Development; 1968. [Google Scholar]
- 62.Voytek M A, Ward B B. Detection of ammonium-oxidizing bacteria of the beta-subclass of the class Proteobacteria in aquatic samples with the PCR. Appl Environ Microbiol. 1995;61:1444–1450. doi: 10.1128/aem.61.4.1444-1450.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wagner M, Amann R, Lemmer H, Schleifer K-H. Probing activated sludge with oligonucleotides specific for proteobacteria: inadequacy of culture-dependent methods for describing microbial community structure. Appl Environ Microbiol. 1993;59:1520–1525. doi: 10.1128/aem.59.5.1520-1525.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wallner G, Amann R, Beisker W. Optimizing fluorescent in situ-hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry. 1993;14:136–143. doi: 10.1002/cyto.990140205. [DOI] [PubMed] [Google Scholar]
- 65.Weinbauer M G, Suttle C A. Comparison of epifluorescence and transmission electron microscopy for counting viruses in natural marine waters. Aquat Microb Ecol. 1997;13:225–232. [Google Scholar]
- 66.Weiss P, Schweitzer B, Amann R, Simon M. Identification in situ and dynamics of bacteria on limnetic organic aggregates (lake snow) Appl Environ Microbiol. 1996;62:1998–2005. doi: 10.1128/aem.62.6.1998-2005.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wright T D, Vergin K L, Boyd P W, Giovannoni S J. A novel delta-subdivision proteobacterial lineage from the lower ocean surface layer. Appl Environ Microbiol. 1997;63:1441–1448. doi: 10.1128/aem.63.4.1441-1448.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zavarzin G A, Stackebrandt E, Murray R G. A correlation of phylogenetic diversity in the Proteobacteria with the influences of ecological forces. Can J Microbiol. 1991;37:1–6. doi: 10.1139/m91-001. [DOI] [PubMed] [Google Scholar]
- 69.Zobell C E, Upham H C. A list of marine bacteria including descriptions of sixty new species. Bull Scripps Inst Oceanogr. 1944;5:239–292. [Google Scholar]