Abstract
Pseudomonas aeruginosa W51D is able to grow by using branched-chain dodecylbenzene sulfonates (B-DBS) or the terpenic alcohol citronellol as a sole source of carbon. A mutant derived from this strain (W51M1) is unable to degrade citronellol but still grows on B-DBS, showing that the citronellol degradation route is not the main pathway involved in the degradation of the surfactant alkyl moiety. The structures of the main B-DBS isomers and of some intermediates were identified by gas chromatography-mass spectrometric analysis, and a possible catabolic route is proposed.
Alkylbenzene sulfonates are the most commonly used surfactants in domestic detergent formulations (8). In the United States and Europe, linear alkylbenzene sulfonates (LAS) have been used since the early 1960s, when the low rate of biodegradation of branched-chain alkylbenzene sulfonates (BAS) was recognized (2, 4, 5, 8). In some Latin American countries, BAS are currently used in different detergent formulations due to their low costs. Water pollution by BAS is a significant environmental problem in these countries.
A Pseudomonas aeruginosa strain (W51D) which is able to mineralize at least 70% of a BAS commercial mixture and completely degrade LAS has been isolated (17). This strain is resistant to high concentrations of these surfactants (17). P. aeruginosa W51D is the only reported bacterium able to mineralize BAS at a significant rate. LAS-degrading Pseudomonas strain C12B barely degrades BAS (4, 11, 19).
LAS are completely degraded in wastewater treatment plants, and different organisms participate in their mineralization, each degrading a part of the molecule. A four-member consortium was identified as responsible for LAS mineralization (9), and a larger consortium was found to be involved in mineralization in a marine environment (15). In these consortia, some members attacked the side chain, while others degraded the aromatic moiety. So far, no consortium that is able to efficiently degrade BAS has been described. The low rate of BAS biodegradation is due to the presence of highly branched alkyl groups (2, 18). Branched alkanes are generally less susceptible to biodegradation than n-alkanes and certain methyl-branched alkanes. Special 3-methyl-branched and quaternary-substituted alkyl chains can result in environmental recalcitrance (2, 7, 12–14, 18).
Some Pseudomonas strains degrade branched alkanes and alkenes. Citronellol (3,7-dimethyl-6-octen-1-ol) has been used as a model compound to study the route of degradation of branched alkenes (7, 13, 14). It has previously been reported that P. aeruginosa W51D is able to use citronellol as a sole carbon source (17). The characterization of a ctrA mutant derived from strain W51D, which has a low citronellal dehydrogenase activity (6), has also been previously reported.
The aim of this work was to contribute to the elucidation of the branched-chain dodecylbenzene sulfonate (B-DBS) degradation pathway of P. aeruginosa W51D and to evaluate the contribution of the reported route to citronellol degradation. We present evidence showing that the citronellol pathway is not the only route involved in the degradation of the surfactant alkyl lateral chain. Identification of the main B-DBS isomer structures and the direct identification of some of their degradation intermediates suggest that strain W51D completely assimilates the surfactant lateral chain prior to the aromatic ring cleavage.
Identification of the isomers present in the B-DBS mixture.
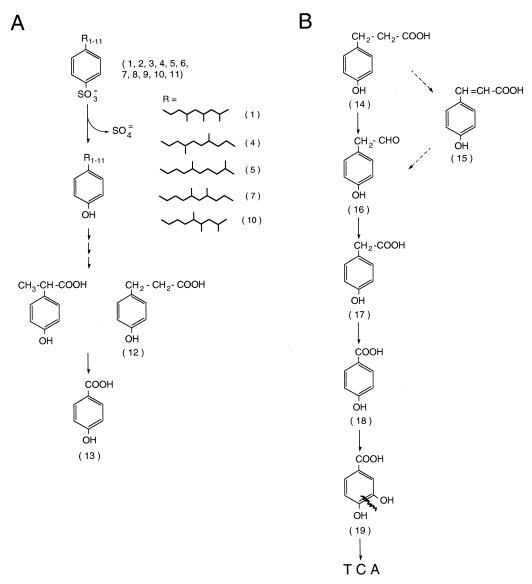
The substrate used for P. aeruginosa W51D degradation studies, B-DBS, was purified by high-performance liquid chromatography (HPLC) from a commercial BAS preparation as a single peak on a semipreparative (250- by 22-mm) Econosil C18 column (Alltech Associates Inc.) by treatment with 60:40 H2O-acetonitrile (ACN) for 10 min, followed by a linear gradient reaching pure ACN in 5 min; this solvent was kept for an additional 2 min. The flow rate used was 5 ml/min, and the elution profile was monitored as described above. The B-DBS mixture obtained from HPLC was desulfonated by reflux treatment in the presence of phosphoric acid as previously described (18) and was injected onto a gas chromatograph coupled with mass spectrometry (MS). The desulfonated B-DBS mixture showed the existence of multiple isomers. The gas chromatography-MS (GC-MS) analysis of 11 of the branched-chain isomers showed that all of them had a molecular weight (M+, molecular ion) of 246 m/z (Table 1). The mass spectrum analysis showed an important ion peak of 91 m/z, indicating the C6H5CH2+ ion. The electron impact fractionation also showed a branched nature of the alkyl moiety. Upon electron impact, saturated hydrocarbons fragment preferentially at the branching points; the positive charge remains on the more highly substituted carbon atom, and elimination of the longest carbon chain is favored. The absence of an (M-15)+ ion in the mass spectrum of the alkylbenzenes is not surprising, although there are many methyl groups present. The methyl radical is the least stable of the alkyl radicals and will not be eliminated readily if other fragmentations are facile. The analytical techniques used in this work do not allow the determination of the positions of both the methyl groups and phenyl moiety. The suggested structures of the alkyl chains of the five most abundant B-DBS isomers are shown in Fig. 1.
TABLE 1.
GC-MS data for a purified and desulfonated B-DBS mixture
| Compound | Area (%)a | RT (min)b | m/z of major ion peaks (%)c |
|---|---|---|---|
| 1 | 13.6 | 15.0 | 246(M+) (5), 204 (1), 203 (10), 190 (1), 189 (10), 175 (3), 162 (2), 161 (17), 159 (1), 148 (2), 147 (20), 133 (5), 131 (2), 120 (1), 119 (15), 114 (5), 113 (6), 112 (1), 106 (12), 105 (100), 103 (2), 92 (3), 91 (35), 79 (2), 78 (1), 77 (2), 71 (3) |
| 2 | 2.48 | 15.4 | 246(M+) (17), 175 (19), 161 (22), 147 (16), 133 (9), 119 (15), 117 (7), 106 (2), 105 (36), 104 (9), 92 (9), 91 (100), 71 (10) |
| 3 | 1.34 | 15.7 | 246(M+) (9), 133 (15), 120 (15), 119 (100), 118 (6), 106 (5), 105 (7), 91 (21), 71 (2) |
| 4 | 13.92 | 16.5 | 246(M+) (5), 217 (4), 175 (1), 147 (3), 134 (1), 133 (15), 120 (12), 119 (100), 118 (7), 117 (3), 115 (2), 106 (3), 105 (20), 104 (1), 103 (2), 92 (2), 91 (13), 79 (2), 77 (2), 71 (1) |
| 5 | 12.34 | 16.9 | 246(M+) (5), 133 (2), 120 (11), 119 (100), 118 (6), 117 (1), 105 (3), 103 (2), 92 (1), 91 (13), 79 (2), 77 (2), 71 (1) |
| 6 | 7.93 | 17.4 | 246(M+) (4), 147 (4), 133 (5), 120 (10), 119 (100), 118 (6), 117 (1), 106 (2), 105 (12), 103 (2), 92 (2), 91 (17), 79 (3), 77 (2), 71 (1) |
| 7 | 17.87 | 18.1 | 246(M+) (5), 217 (3), 189 (2), 175 (3), 161 (4), 147 (1), 134 (1), 133 (8), 120 (10), 119 (100), 118 (6), 117 (2), 115 (2), 106 (2), 105 (23), 103 (2), 92 (2), 91 (24), 79 (3), 78 (1), 77 (2), 71 (2 |
| 8 | 1.75 | 18.4 | 246(M+) (9), 203 (12), 187 (15), 174 (8), 173 (48), 159 (12), 147 (28), 133 (17), 131 (27), 129 (8), 128 (8), 120 (6), 119 (39), 118 (8), 117 (14), 115 (10), 106 (22), 105 (100), 91 (47), 85 (10), 79 (7), 71 (14) |
| 9 | 2.21 | 18.7 | 246(M+) (4), 217 (6), 134 (3), 133 (26), 131 (2), 120 (11), 119 (100), 118 (4), 117 (5), 106 (4), 105 (35), 92 (3), 91 (28), 79 (4), 71 (3) |
| 10 | 16.97 | 19.5 | 246(M+) (5), 173 (1), 161 (3), 159 (4), 147 (1), 131 (1), 120 (11), 119 (100), 118 (7), 117 (3), 115 (1), 106 (3), 105 (22), 103 (2), 92 (1), 91 (18), 79 (3), 77 (2), 71 (4) |
| 11 | 2.96 | 19.9 | 246(M+) (4), 174 (2), 173 (15), 161 (2), 133 (2), 131 (4), 120 (12), 119 (100), 118 (5), 117 (4), 115 (2), 106 (2), 105 (11), 103 (2), 91 (20), 79 (2), 77 (2) |
Based on the total ion current response of the MS detector.
RT, retention time.
Ions with more than 1% abundance are shown. The ion abundance percentages are shown in parentheses.
FIG. 1.
Schematic representation of the proposed degradation route of B-DBS (A) and 4-hydroxyphenylpropionate (B) by P. aeruginosa W51D. The numbers in parentheses correspond to the compound numbers in Tables 1 and 2, where the MS results are given. The structures of the alkyl lateral chains of the most abundant B-DBS isomers are shown as R groups. TCA, tricarboxylic acid cycle.
Participation of the citronellol pathway in B-DBS degradation.
Pseudomonas degradation of citronellol (12) is the only enzymatic pathway characterized in bacteria for the assimilation of methyl-ramified alkanes so far. The isolation and characterization of a P. aeruginosa W51D mutant (W51M1) unable to degrade citronellol due to the reduced activity of the enzyme citronellal dehydrogenase have previously been described (6). To determine whether the citronellol pathway was used by strain W51D to degrade the B-DBS alkyl chain, we studied the ability of mutant W51M1 to assimilate the surfactants. We found that even though mutant W51M1 is unable to use citronellol as a carbon source, it is still able to grow on M9 plus B-DBS (0.2% [wt/vol]). Quantitative determination of B-DBS degradation by strain W51M1 was obtained from HPLC analysis of culture supernatants, in which the extent of B-DBS consumed by this mutant on M9 plus glucose (0.2% [wt/vol]) plus B-DBS was estimated after 48 h of growth. The consumption by strain W51M1 was reduced by 40% compared to that by the wild-type strain W51D. Even though the total amount of surfactant consumed on this medium was reduced, we could not detect differences in the relative proportion of the accumulated intermediates. These results show that the enzymes involved in citronellol degradation could also be involved in B-DBS degradation, presumably at the first steps of the catabolism, but it is apparent that this pathway is not the main route of B-DBS degradation by strain W51D.
It has been reported that the wild-type strain PAO1 (supplied by Bruce Holloway) is able to grow with citronellol as the sole source of carbon (6); however, this strain is unable to use B-DBS as a carbon source. These results suggest that the capability of strain W51D to degrade B-DBS is mainly due to the presence of a degradation route of the B-DBS lateral chain that is different from the citronellol pathway, which is not present in strain PAO1.
Analysis of the W51D pathway for B-DBS degradation.
To obtain information on the main W51D route for B-DBS degradation, we determined the structure of the degradation intermediates by analyzing the 72-h culture supernatants of W51D cells grown on M9 plus B-DBS by GC-MS. These supernatants were acidified with HCl to pH 3 to eliminate the extracellular matrix of the biofilm formed in this medium. The acidified supernatant was centrifuged, the supernatant was extracted three times with ethylacetate, the organic fraction was concentrated by evaporation, and the extracted product was dried with N2. The dried product was resuspended in methanol at a final concentration of 1 mg/ml. Samples were silylated prior to GC-MS analysis with ClSi(CH3)3 (Sigma) according to the manufacturer’s instructions. One microliter of the compound solutions separated by HPLC or directly silylated after being concentrated was injected onto a gas chromatograph coupled with an MS detector (Hewlett-Packard HP6890 GC-MS system). Table 2 shows the MS data of these identified metabolites.
TABLE 2.
GC-MS data for products formed by P. aeruginosa W51D grown in the compound indicated
| Compound source and no. | m/z of major ion peaks (%)a | Suggested structure (% identity)b |
|---|---|---|
| B-DBS | ||
| 12 | 310(M+) (8), 295 (4), 281 (10), 267 (4), 208 (6), 207 (24), 193 (12), 192 (40), 191 (4), 181 (4), 180 (11), 179 (64), 177 (12), 163 (5), 149 (5), 147 (12), 136 (4), 133 (8), 131 (5), 129 (4), 117 (6), 116 (4), 105 (8), 95 (15), 93 (32), 79 (16), 77 (15), 76 (5), 75 (42), 74 (10), 73 (100) | Hydrocinnamic acid, p-[(TMS)oxy]-, TMS estere (97) |
| 13 | 282(M+) (8), 268 (9), 267 (36), 224 (7), 223 (36), 217 (6), 207 (5), 204 (5), 195 (4), 194 (7), 193 (38), 190 (6), 179 (4), 151 (4), 149 (6), 147 (8), 135 (6), 133 (5), 126 (14), 105 (4), 95 (8), 94 (4), 93 (8), 91 (6), 76 (4), 75 (16), 74 (10), 73 (100) | Benzoic acid, 4-[(TMS)oxy]-, TMS ester (97) |
| 3(4-hydroxyphenyl-propionic) acid | ||
| 14 | 310(M+) (20), 311 (4), 295 (5), 194 (4), 193 (12), 192 (70), 181 (5), 180 (18), 179 (100), 177 (22), 163 (6), 149 (4), 140 (4), 75 (24), 74 (6), 73 (58) | Hydrocinnamic acid, p-[(TMS)oxy]-, TMS ester (99) |
| 15 | 308(M+) (40), 309 (9), 310 (4), 295 (5), 194 (12), 293 (52), 250 (8), 249 (36), 233 (4), 221 (4), 220 (16), 219 (72), 203 (8), 192 (5), 191 (5), 179 (16), 175 (8), 147 (8), 135 (5), 115 (10), 102 (4), 89 (4), 76 (4), 75 (34), 74 (10), 73 (100) | Cinnamic acid, p-[(TMS)oxy], TMS ester (99) |
| 16 | 208(M+) (24), 195 (5), 194 (18), 193 (100), 151 (14), 149 (4), 147 (4), 135 (6), 133 (5), 123 (4), 93 (8), 91 (9), 89 (14), 77 (6), 75 (13), 74 (6), 73 (33) | Benzene acetaldehyde, 4-[(TMS)oxy]c |
| 17 | 252(M+) (28), 207 (4), 193 (7), 192 (8), 180 (15), 179 (100), 177 (12), 164 (11), 163 (18), 149 (8), 135 (7), 133 (9), 131 (11), 107 (8), 91 (9), 89 (22), 78 (8), 75 (15), 74 (7), 73 (88) | 4-Hydroxypehnylacetic acid ethyl ester TMS (92) |
| 18 | 282(M+) (14), 269 (5), 268 (12), 267 (56), 263 (6), 262 (28), 225 (4), 224 (8), 223 (45), 207 (6), 194 (8), 193 (4), 179 (4), 151 (4), 149 (5), 147 (8), 135 (6), 133 (5), 130 (5), 126 (14), 117 (4), 103 (4), 95 (8), 93 (24), 91 (5), 89 (5), 77 (4), 75 (17), 74 (8), 73 (100) | Benzoic acid, 4-[(TMS)oxy]-, TMS ester (97) |
| 19 | 370(M+) (8), 355 (16), 343 (10), 342 (12), 341 (38), 327 (6), 325 (8), 302 (6), 297 (12), 283 (8), 267 (17), 255 (6), 254 (8), 253 (27), 239 (14), 238 (24), 237 (100), 221 (7), 208 (6), 191 (10), 179 (9), 165 (8), 164 (14), 163 (70), 149 (7), 147 (10), 135 (7), 133 (10), 119 (6), 117 (6), 105 (6), 93 (6), 89 (30), 73 (62) | Benzoic acid 3,4-[(TMS)-dioxy]-, TMS esterc |
Ions with more than 4% abundance are shown. The ion abundance percentages are shown in parentheses.
Identification was based on an instrument library match. TMS, trimethylsilyl.
Tentative assignment of structure.
Considering the structure of the identified compounds, it is possible to suggest the partial pathway for B-DBS degradation shown in Fig. 1. The first step in B-DBS degradation seems to be the desulfonation of the benzene ring and its concomitant hydroxylation. This proposition was initially made based on the finding that all the degradation intermediates detected were hydroxylated derivatives (Table 2) and no sulfonated molecules were found. To further confirm this result, we treated the ethylacetate extract from the cell-free culture supernatant with diazomethane to form the methylsulfonate derivatives and then analyzed the products by GC-MS. In accordance with our original observation, only the sulfonated B-DBS substrate was detected and no other sulfonated intermediate was found. Desulfonation with the concomitant hydroxylation of the aromatic ring has been reported as the first step in LAS degradation by Pseudomonas putida strains (9, 21).
The GC-MS analysis of the culture supernatants of W51D cells grown on M9 plus B-DBS showed the production of 4-hydroxypropionate and 4-hydroxybenzoate (Table 2). These aromatic compounds, as well as phenylacetate, are readily degraded by strain W51D (all supplied as a carbon source to M9 medium at a concentration of 5 mM), suggesting that they could be B-DBS degradation intermediates. The identification of 4-hydroxybenzoate as a possible B-DBS degradation intermediate suggests that strain W51D completely oxidizes the B-DBS lateral chain prior to the aromatic ring cleavage.
The oxidation of the branched lateral B-DBS chain could proceed by the removal of C-1 units through α-oxidation, as reported for LAS degradation (3, 18), in conjunction with classical β-oxidation, or by a modified β-oxidation in which two carbons are cleaved from the main hydrocarbon chain together with a methyl ramification, as has been reported to occur in the degradation of methyl-branched alkanes by an unclassified gram-positive bacterium (12).
W51D pathway for the degradation of 4-hydroxyphenylpropionate.
Considering that 4-hydroxyphenylpropionate is a B-DBS catabolic intermediate, we studied the W51D route for the degradation of this compound as a way to analyze further B-DBS catabolism by strain W51D. The proposed route of W51D degradation of 4-hydroxyphenylpropionate (Fig. 1) was elucidated by using this compound as a carbon source and by identifying the intermediates (Table 2) by GC-MS analysis, as described above. The degradation intermediates were purified by HPLC as follows: 50 μl of the concentrated supernatant or of the supernatant of strain W51D grown for 48 h on 4-hydroxyphenylpropionate was injected on a Nova-Pak C18 3.9- by 150-mm reverse-phase column (Waters), and the elution solvent was H2O-ACN-acetic acid 75/4/1, vol/vol/vol for 3 min, followed by a linear gradient to reach 100% ACN in 6 min. The flux used was 1 ml/min, and the elution was monitored at 254 nm with a Hewlett-Packard 1050 diode array UV detector. We found, in accordance with the results obtained by studying B-DBS degradation, that the propionate moiety of this molecule was completely oxidized and degraded to 4-hydroxybenzoate prior to the aromatic ring cleavage. This is unusual, since most bacteria cleave the aromatic ring of short-chain alkylbenzenes without previous oxidation of the alkyl chain (16).
It has been reported that cinnamic acid is accumulated as a nonmetabolized by-product of the degradation of long-chain alkylbenzenes (16). We detected the production of 4-hydroxycinnamic acid when strain W51D was grown on M9 plus 4-hydroxyphenylpropionate (Table 2). Our data are insufficient to conclude whether this compound is an intermediate or a by-product of this catabolic route. However, the oxidation of the cinnamic acid lateral chain by W51D, as judged by the detected degradation intermediates of cells grown on M9 plus glucose plus cinnamic acid (5 mM) for 48 h, is consistent with 4-hydroxycinnamic acid being a 4-hydroxyphenylpropionate degradation intermediate, since the same degradation products were detected in both cases (Fig. 1). The oxidation of the cinnamic acid lateral chain by W51D is similar to styrene lateral chain oxidation by Pseudomonas sp. strain Y2 (20).
Quantitative data were obtained from the GC analysis by using the flame ionization detection response. After 48 h of W51D growth on M9 plus 4-hydroxyphenylpropionate, the product/substrate ratios (according to the areas of the peaks in the chromatograms) for the different compounds detected were as follows: for 4-hydroxycinnamic acid (isomer A), 0.442; for 4-hydroxycinnamic acid (isomer B), 0.218; for 4-hydroxybenzene acetaldehyde, 0.007; for 4-hydroxyphenylacetate, 0.003; for 4-hydroxybenzoate, 0.035; and for 3,4-dihydroxybenzoate, 0.004.
In conclusion, our studies show, so far, that P. aeruginosa W51D degrades the surfactant B-DBS by using in part the citronellol pathway but that most of the surfactant degradation seems to be carried out by a new catabolic pathway. The data obtained suggest that strain W51D desulfonates B-DBS prior to alkyl chain oxidation. Desulfonation is followed by a complete oxidation of the alkyl moiety before the aromatic ring is cleaved. To our knowledge, there has been no report of a similar pathway involved in the degradation of alkylbenzenes. Therefore, whatever route strain W51D uses for B-DBS degradation, its characterization will reveal a novel catabolic pathway, and its complete elucidation is a matter of great importance which remains to be further analyzed.
Acknowledgments
The portion of this research done in Spain was founded in part by BIO-CT97-0641. Jesús Campos-García held a CONACyT scholarship during the development of this work and received support from Programa de Apoyo al Posgrado PADEP (UACPyP/UNAM), project no. 030506.
REFERENCES
- 1.Abril M-A, Michan C, Timmis K N, Ramos J L. Regulator and enzyme specificities of the TOL plasmid-encoded upper pathway for degradation of aromatic hydrocarbons and expansion of the substrate range of the pathway. J Bacteriol. 1989;171:6782–6790. doi: 10.1128/jb.171.12.6782-6790.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexander M. Nonbiodegradable and other recalcitrant molecules. Biotechnol Bioeng. 1973;15:611–647. [Google Scholar]
- 3.Baggi G, Catelani D, Galli E, Treccani V. Microbial degradation of phenylalkanes. 2-Phenylbutane, 3-phenylpentane, 3-phenyldodecane, and 4-phenylheptane. Biochem J. 1972;126:1091–1097. doi: 10.1042/bj1261091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cain R B. Biodegradation of anionic surfactants. Biochem Soc Trans. 1987;15(Suppl.):7–22. [PubMed] [Google Scholar]
- 5.Cain R B. Biodegradation of detergents. Curr Opin Biotechnol. 1994;5:266–274. [Google Scholar]
- 6.Campos-García, J., and G. Soberón-Chávez. Cloning and characterization of the Pseudomonas aeruginosa W51D ctrA gene, involved in citronellol degradation and bacterial chemotaxis. Submitted for publication.
- 7.Fall R R, Brown J L, Schaeffer T L. Enzyme recruitment allows the biodegradation of recalcitrant branched hydrocarbons by Pseudomonas citronellolis. Appl Environ Microbiol. 1979;38:715–722. doi: 10.1128/aem.38.4.715-722.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greek B F. Sales of detergents growing despite recession. Chem Eng News. 1991;69:25–52. [Google Scholar]
- 9.Jiménez L, Breen A, Thomas N, Federle T W, Sayler G S. Mineralization of linear alkylbenzene sulfonate by a four-member aerobic bacterial consortium. Appl Environ Microbiol. 1991;57:1566–1569. doi: 10.1128/aem.57.5.1566-1569.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kertesz M A, Kölbener P, Stockinger H, Beil S, Cook A M. Desulfonation of linear alkylbenzenesulfonate surfactants and related compounds by bacteria. Appl Environ Microbiol. 1994;60:2296–2303. doi: 10.1128/aem.60.7.2296-2303.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Payne W J, Feisal V E. Bacterial utilization of dodecyl sulfate and dodecyl benzene sulfonate. Appl Microbiol. 1963;11:339–344. doi: 10.1128/am.11.4.339-344.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pirnik M P. Microbial oxidation of methyl branched alkanes. Crit Rev Microbiol. 1977;5:413–422. doi: 10.3109/10408417709102812. [DOI] [PubMed] [Google Scholar]
- 13.Rontani J-F, Gilewicz M J, Michotey V D, Zheng T L, Bonin P C, Bertrand J-C. Aerobic and anaerobic metabolism of 6,10,14-trimethylpentadecan-2-one by a denitrifying bacterium isolated from marine sediments. Appl Environ Microbiol. 1997;63:636–643. doi: 10.1128/aem.63.2.636-643.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schaeffer T L, Cantwell S G, Brown J L, Watt D S, Fall R R. Microbial growth on hydrocarbons: terminal branching inhibits biodegradation. Appl Environ Microbiol. 1979;38:742–746. doi: 10.1128/aem.38.4.742-746.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sigoillot J-C, Nguyen M-H. Complete oxidation of linear alkylbenzene sulfonate by bacterial communities selected from coastal seawater. Appl Environ Microbiol. 1992;58:1308–1312. doi: 10.1128/aem.58.4.1308-1312.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith M R. The biodegradation of aromatic hydrocarbons by bacteria. Biodegradation. 1990;1:191–206. doi: 10.1007/BF00058836. [DOI] [PubMed] [Google Scholar]
- 17.Soberón-Chávez G, Haïdour A, Ramos J L, Campos J, Ortigoza J. Selection and preliminary characterization of a Pseudomonas aeruginosa strain mineralizing some isomers in a branched-chain dodecylbenzene sulfonate mixture. World J Microbiol Biotechnol. 1996;12:367–362. doi: 10.1007/BF00340213. [DOI] [PubMed] [Google Scholar]
- 18.Swisher R D. Biodegradation of ABS in relation to chemical structure. J Water Pollut Control Fed. 1963;35:877–892. [Google Scholar]
- 19.Thomas O R T, White G F. Immobilization of the surfactant-degrading bacterium Pseudomonas C12B in polyacrylamide gel. III. Biodegradation specificity for raw surfactants and industrial wastes. Enzyme Microb Technol. 1991;13:338–343. doi: 10.1016/0141-0229(90)90010-n. [DOI] [PubMed] [Google Scholar]
- 20.Velasco A, Alonso S, García J L, Perera J, Díaz E. Genetic and functional analysis of the styrene catabolic cluster of Pseudomonas sp. strain Y2. J Bacteriol. 1998;180:1063–1071. doi: 10.1128/jb.180.5.1063-1071.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Willetts A J, Cain R B. Microbial metabolism of alkylbenzenesulphonates. Bacterial metabolism of undecylbenzene-p-sulphonate. Biochem J. 1972;129:389–402. doi: 10.1042/bj1290389. [DOI] [PMC free article] [PubMed] [Google Scholar]