Abstract
A Lactococcus lactis subsp. lactis strain that can sense the bacteriocin nisin and transduce the signal into bioluminescence was constructed. By using this strain, a bioassay based on bioluminescence was developed for quantification of nisin, for detection of nisin in milk, and for identification of nisin-producing strains. As little as 0.0125 ng of nisin per ml was detected within 3 h by this bioluminescence assay. This detection limit was lower than in previously described methods.
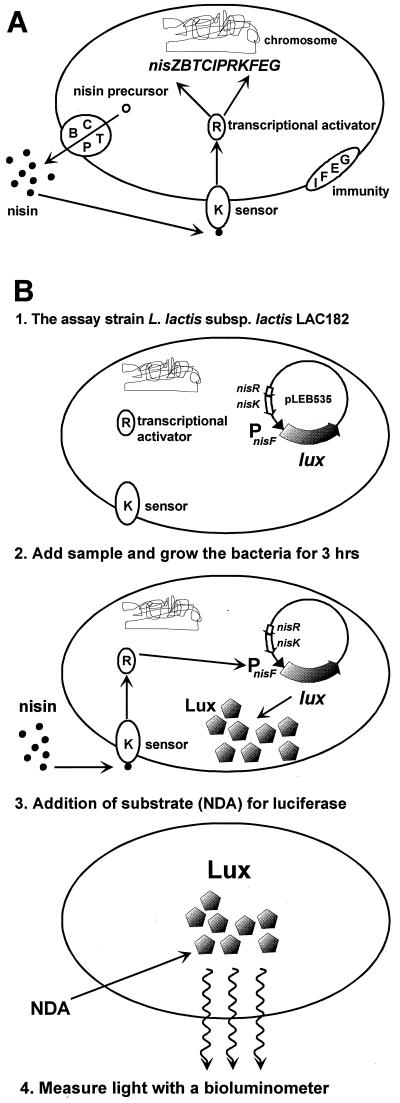
The lantibiotic nisin (17) produced by some Lactococcus lactis strains is a small antibiotic peptide that is active against several gram-positive bacteria, such as Clostridium, Bacillus, Listeria, and Staphylococcus species. Because of its nontoxicity for humans and its broad range of antimicrobial activity, it is used as a preservative in the food and dairy industries (4). Two naturally occurring nisin variants, nisin A (3) and nisin Z (16, 26), which differ in a single amino acid residue have been described. The genes encoding proteins involved in nisin biosynthesis, regulation, and self-immunity are arranged in two inducible operons, nisA/ZBTCIPRK and nisFEG (11, 19, 20, 25, 27, 28, 30, 31, 36). The genes nisR and nisK encode the response regulator (NisR) and the histidine kinase (NisK) of the two-component regulatory system (11, 20, 36), which is responsible for the autoregulation of nisin biosynthesis. Extracellular nisin acts as the environmental signal to which NisK responds. This signal is transmitted to NisR, which ultimately leads to transcriptional activation of both the nisA/Z and nisF promoters (5, 24, 27, 28) (Fig. 1A).
FIG. 1.
(A) Schematic presentation of the autoregulation of nisin biosynthesis in L. lactis subsp. lactis N8 by signal transduction. Nisin is modified and secreted by the biosynthetic machinery (BCTP). Extracellular nisin activates the histidine kinase NisK, which is autophosphorylated. NisK then phosphorylates the response regulator NisR, which activates the transcription from the promoters upstream of nisZ and nisF. The nisin immunity system (IFEG) is present to protect the nisin producer, by an unknown mechanism, from being killed by nisin. (B) Schematic presentation of the nisin bioluminescence assay based on nisin signal transduction coupled to luciferase production. The sample is added to cells of LAC182, which are grown for 3 h. If the sample contains nisin, it will activate the nisF promoter upstream of the luxHb gene in plasmid pLEB535, resulting in production of luciferase. Addition of the luciferase substrate n-decyl-aldehyde (NDA) to the cells results in emission of light, which is measured by a bioluminometer.
Most of the methods developed for quantification of nisin are based on its inhibitory activity to a test organism (8). The agar diffusion bioassay (14, 35) is still the most widely used. Several parameters affect the accuracy and sensitivity of this method (37). In addition, equal concentrations of nisins A and Z produce inhibition zones of different sizes due to the better diffusion properties of nisin Z (7). Immunological detection methods have also been developed. The sandwich-type enzyme-linked immunosorbent assay for nisin A, based on sheep polyclonal antibodies and developed by Falahee et al. (13), has a detection limit of 0.5 ng/ml for pure nisin and 0.2 μg/ml for nisin analyzed from spiked cheese. Other methods for immunodetection of nisin involving competitive direct enzyme-linked immunosorbent assays with polyclonal (32) and monoclonal (33) antibodies from mice have been developed. These assays showed nisin detection limits of 5 to 10 and 10 ng/ml, respectively. A rabbit antiserum against nisin Z was used to develop an immunodot method for detection of nisin, with a detection limit of 3 ng/ml for pure nisin Z and 0.155 μg/ml for nisin analyzed from milk and whey (2). All the immunoassays described are considerably more sensitive than the agar diffusion assay. Still, methods based on antibodies against nisin are not totally reliable due to possible cross-reactions with related compounds (12). The gusA gene has been placed under control of the nisin promoters for studies of transcriptional activity (5, 6, 24). Strains with the nisA or nisF promoter fused to gusA could be induced by small amounts of nisin to produce β-glucuronidase activity in a linear dose-response relationship. The specificity of nisin recognition was high, because the other tested bacteriocins, including subtilin, which is the closest structural homolog of nisin, were not functional inducers (24). However, even if these strains could be used to develop a nisin quantification assay, the assay method requires breakage of the cells and takes several hours to perform, making this approach unfavorable. In this communication, we present a fast, simple, and sensitive bioassay based on the autoinducibility of the nisin promoter PnisF and bioluminescence derived from bacterial luciferase genes fused to the nisin promoter (Fig. 1B). This assay detects nisin in milk, water and supernatants from nisin-producing strains.
Construction of the indicator strain.
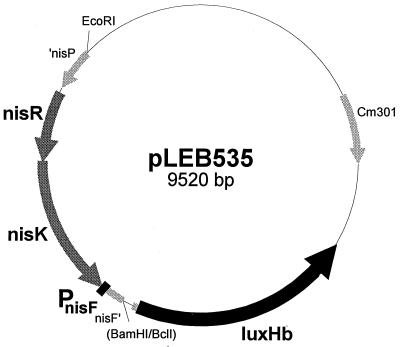
A genomic fragment including the intact nisR and nisK genes and the promoter of nisF originating from the nisin Z-producing strain L. lactis subsp. lactis N8 was cut from plasmid pLEB190 (20) as an EcoRI-BamHI fragment. This fragment was used to replace the EcoRI-BclI fragment containing the P45 promoter in plasmid pTCluxHb (21). In the resulting plasmid, pLEB535 (Fig. 2), the bioluminescence genes luxAB from Xenorhabdus luminescens (named luxHb in plasmid pTCluxHb) are placed under control of the PnisF promoter. This construct was electroporated (18) into the non-nisin producer L. lactis MG1614 (15) and plated at 30°C on M17 (34) plates containing 0.5% (wt/vol) glucose and 0.5% (wt/vol) sucrose (M17GS) and chloramphenicol (5 μg/ml). The resulting indicator strain was named LAC182.
FIG. 2.
Physical map of the nisin bioluminescence assay plasmid pLEB535.
Nisin bioluminescence assay.
The indicator strain LAC182 was grown in M17GS plus chloramphenicol (5 μg/ml) without aeration until an optical density at 600 nm (OD600) of 0.5 was reached. Glycerol (87%) was added to one-fifth of the volume, and the bacteria were stored at −70°C before use. For nisin detection, these precultured cells of LAC182 were diluted 1:100 in M17GS plus 0.1% Tween 80. Nisin (Sigma) standards were made in 0.1% Tween 80. The solution was prepared with distilled water acidified to pH 2.5 with HCl (hereafter referred to as 0.1% Tween 80). Volumes between 5 and 70 μl of the nisin dilutions were mixed with 1 ml of diluted indicator bacteria, which were then grown for 3 h at 30°C without aeration. The 1-ml bacterial sample was added to 40 μl of n-decyl-aldehyde (1% [vol/vol]) in absolute ethanol in a cuvette, and the luciferase activity was determined immediately as relative light units with the Bio-Orbit 1253 luminometer connected to a computer using Lucyfer version 1.1β software (Teemu Teeri, University of Helsinki, Finland). Growth was measured as OD600 by using an UltrospecII spectrophotometer (Pharmacia LKB).
Background and nisin-induced luciferase activity.
Luciferase background activity was detectable 1 h after initiation of growth and increased slowly for the next 4 h. Between 5 and 6 h after initiation of growth (OD600 of 0.3 to 0.5), the background luciferase activity increased rapidly. After 6 h, before the bacteria reached the stationary phase, the activity dropped suddenly, so that by 8 h the activity was the same as at the beginning of the growth. The luciferase activity in induced (0.01 IU of nisin/ml, final assay concentration) bacteria showed a steady increase from the beginning of growth, reaching its maximum level at 6 h, after which it dropped. After 3 h of growth, before an OD600 of 0.1 was reached, the ratio of induced luciferase activity to uninduced was highest; therefore, this time point was chosen for the nisin bioluminescence assay. However, an increase in luciferase activity of induced cells compared with the control could already be detected after 1 h of growth, showing that this bioassay could be used to determine rapidly whether a sample contains nisin (data not shown).
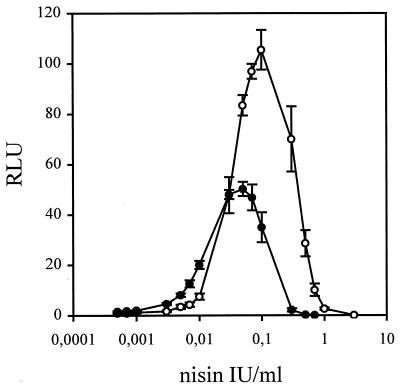
When luciferase activity was induced with different amounts of nisin, the presence of Tween 80 proved to be crucial. Nisin is an amphipathic molecule with a high tendency to adhere to various surfaces. This may cause a large loss of nisin, especially when low concentrations are being used. The aggregation and adsorption of nisin can be prevented by addition of Tween 80 (22). Without Tween 80, the lowest concentration of nisin that could be detected as an increase in luciferase activity compared with the control was 0.01 IU/ml. When 0.1% Tween 80 was included in the diluent, the detection limit decreased to 0.0005 IU/ml, corresponding to 0.0125 ng/ml. This indicates that 95% of the nisin was lost during dilution without Tween 80. In both cases, the luciferase activity increased linearly with nisin concentration until enough nisin was present to exert its inhibitory effect on the indicator bacteria. In the presence of 0.1% Tween 80, the highest luciferase activity was reached at a nisin concentration of 0.03 to 0.07 IU/ml (0.75 to 1.75 ng/ml) (Fig. 3). After the 3-h growth period chosen for the bioassay, the maximum luciferase activity of nisin-induced cells was 40 to 50 times higher than the activity of the control cells. Induction with nisin concentrations higher than 0.3 IU/ml (7.5 ng/ml) gave lower luciferase activity values than those of the control, due to inhibition of the indicator bacteria by nisin.
FIG. 3.
Luciferase activity recorded as relative light units (RLU) of L. lactis subsp. lactis LAC182 cells induced for 3 h with different amounts of nisin diluted in 0.1% Tween 80 (●) or milk (○) with or without 0.1% Tween 80. The nisin concentrations are given as final assay concentrations. The result shown is the mean and standard deviation from five experiments.
To measure the induction in response to nisin A compared with nisin Z, we analyzed supernatants from the nisin A producer L. lactis ATCC 11454 (American Type Culture Collection, Rockville, Md.) and the nisin Z producer L. lactis N8 (16). The same amounts of nisins A and Z, as estimated from the MIC (7) determined in the agar diffusion assay (27), gave approximately the same response in the bioluminescence assay (data not shown). However, nisin A resulted in an approximately 25% higher maximum luciferase activity than nisin Z did. This indicates that in this assay nisin A is either a more efficient inducer or a less efficient killer than nisin Z.
To detect possible cross-reactions of other bacteriocins, we tested the bioassay on supernatants from the subtilin producer Bacillus subtilis ATCC 6633 (American Type Culture Collection) the carnocin producer Carnobacterium piscicola LMG233 (Valio Ltd., Helsinki, Finland), and the sakacin A producer Lactobacillus sake Lb706(pSAK27) (1). None of the tested bacteriocins, including subtilin, the structurally closest homolog of nisin, gave a positive response in the bioluminescence assay (data not shown).
Detection of nisin in milk.
The applicability of the bioluminescence assay to the detection of trace levels of nisin in food samples was tested by using milk (homogenized and pasteurized, 1.5% fat). Nisin was diluted in milk with or without 0.1% Tween 80, and different amounts of nisin in 70 μl of milk were mixed with the indicator bacteria; otherwise the samples were processed as described above. We observed that no nisin was lost when milk was used as the diluent. The background luciferase activity of the indicator bacteria was not affected by the presence of milk; instead, larger amounts of nisin could be added without inhibition of the bacteria. The sensitivity was slightly reduced in the presence of milk. The lowest concentration in each experiment that clearly resulted in an increase in luciferase activity was 0.003 IU/ml (0.075 ng/ml) (Fig. 3).
Identification of nisin-producing strains by the nisin bioluminescence assay.
Screening programs aimed at finding new bacteriocins result in the isolation of bacteria that produce antimicrobial substances. It is of value to rapidly and easily identify previously described bacteriocins in order to be able to concentrate efforts on novel ones. Because nisin is a broad-spectrum bacteriocin and is produced by many strains of L. lactis, it has been repeatedly detected. Therefore, a rapid and simple identification test for nisin is needed.
We conducted a test in a blind manner on strains provided by Quest Ltd. (Bussum, The Netherlands). Seven randomized samples were shipped in duplicate as supernatants and stored at 8°C for 14 days before being tested. The samples were diluted 1:100 and 1:10,000 in 0.1% Tween 80. For nisin detection, 5 μl of the supernatants and the dilutions were mixed with indicator bacteria and processed as described above. The nisin producers were identified correctly among the unknown strains (Table 1). Higher luciferase activity than in the control was obtained from the nisin producers only, including the unknown test microorganism. The undiluted supernatant from the Lactobacillus strain reduced the luciferase activity, as did three of the supernatants containing nisin. Diluting the Lactobacillus supernatant did not cause induction, which proves that the inhibitory substance is not nisin.
TABLE 1.
Identification of nisin-producing strains by the nisin bioluminescence assay
| Strain | Luciferase activitya at:
|
Nisin producer | ||
|---|---|---|---|---|
| 1:1 | 1:100 | 1:10,000 | ||
| Lactobacillus sp. | 0.2 | 1.0 | 1.0 | − |
| Pediococcus sp. | 1.0 | 0.9 | 1.1 | − |
| Lactococcus sp. | 0.9 | 1.0 | 1.0 | − |
| Nisin Z producer | 60.5 | 1.7 | 1.1 | + |
| Nisin A producer | <0.1 | 16.2 | 1.2 | + |
| Variant of nisin A | <0.1 | 15.4 | 1.0 | + |
| Unknown test microorganism | <0.1 | 17.6 | 1.1 | + |
The values indicate the ratio of the mean activities of the diluted duplicate supernatants compared with that of the uninduced indicator strain LAC182.
In conclusion, the nisin bioluminescence assay can be used to quantitate nisin, to detect nisin in milk, and to identify strains that produce either nisin A or nisin Z. The overall response range (given as the final assay concentration) in the bioassay is 0.0005 to 0.3 IU/ml, corresponding to 0.0125 to 7.5 ng/ml. In milk, the detection range is 0.003 to 1 IU/ml (0.075 to 25 ng/ml), meaning that undiluted milk samples containing 0.04 to 1,000 IU/ml (0.001 to 25 μg/ml) can be assayed without inhibition of the indicator strain. The sensitivity is thus higher than that of the traditional agar diffusion assay and most of the immunoassays described previously (2, 13, 32, 33). Previously described methods for detection of nisin in milk include pretreatment of the milk samples. Still, the sensitivity remained quite low compared with the detection limit for pure nisin (2, 29). The major advantages of the above-described bioluminescence assay for detection of nisin in milk are that it is much less complicated and far more sensitive. The fact that large dilutions are required in some cases before the detection is possible is an advantage in instances where interfering substances might be present. To test undiluted samples containing more than 300 IU of nisin/ml, a more resistant indicator strain is needed. Since the nisin A promoter was inducible by nisin in a linear dose-response relationship in several heterologous hosts (10, 23), transfer of the system into another species could provide a way of overcoming the present limitations. The traditional agar diffusion assay measures the inhibitory activity of nisin, while this bioluminescence assay measures the inducing activity. These two activities do not necessarily correlate for a given nisin molecule (9, 24). A bioassay based on the induction capacity is therefore an important complement when engineered nisin molecules are being characterized. The simplicity of our method makes it an ideal system for automation of nisin quantification. Because an increase in the luciferase activity is detected after 1 h, it could be used as an on-line method of detection of nisin concentrations in fermentation experiments.
Acknowledgments
Tiina Immonen is acknowledged for critical reading of the manuscript.
Financial support was provided by Academy of Finland.
REFERENCES
- 1.Axelsson L, Holck A. The genes involved in production of and immunity to sakacin A, a bacteriocin from Lactobacillus sake Lb706. J Bacteriol. 1995;177:2125–2137. doi: 10.1128/jb.177.8.2125-2137.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bouksaim M, Fliss I, Meghrous J, Simard R, Lacroix C. Immunodot detection of nisin Z in milk and whey using enhanced chemiluminescence. J Appl Microbiol. 1998;84:176–184. doi: 10.1046/j.1365-2672.1998.00315.x. [DOI] [PubMed] [Google Scholar]
- 3.Buchman G W, Banerjee S, Hansen J N. Structure, expression, and evolution of a gene encoding the precursor of nisin, a small protein antibiotic. J Biol Chem. 1988;263:16260–16266. [PubMed] [Google Scholar]
- 4.Delves-Broughton J, Blackburn P, Evans R J, Hugenholz J. Applications of the bacteriocin, nisin. Antonie Leeuwenhoek. 1996;69:193–202. doi: 10.1007/BF00399424. [DOI] [PubMed] [Google Scholar]
- 5.de Ruyter P G G A, Kuipers O P, Beerthuyzen M M, van Alen-Boerrigter I, de Vos W M. Functional analysis of promoters in nisin gene cluster of Lactococcus lactis. J Bacteriol. 1996;178:3434–3439. doi: 10.1128/jb.178.12.3434-3439.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Ruyter P G G A, Kuipers O P, de Vos W M. Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl Environ Microbiol. 1996;62:3662–3667. doi: 10.1128/aem.62.10.3662-3667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Vos W M, Mulders J W M, Siezen R J, Hugenholtz J, Kuipers O P. Properties of nisin Z and distribution of its gene, nisZ, in Lactococcus lactis. Appl Environ Microbiol. 1993;59:213–218. doi: 10.1128/aem.59.1.213-218.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Vuyst L, Vandamme E J. Nisin, a lantibiotic produced by Lactococcus lactis subsp. lactis: properties, biosynthesis, fermentation and applications. In: de Vuyst L, Vandamme E J, editors. Bacteriocins of lactic acid bacteria. Oxford, United Kingdom: Chapman & Hall, The Alden Press; 1994. pp. 152–199. [Google Scholar]
- 9.Dodd H M, Horn N, Chan W C, Giffard C J, Bycroft B W, Roberts G C K, Gasson M J. Molecular analysis of the regulation of nisin immunity. Microbiology. 1996;142:2385–2392. doi: 10.1099/00221287-142-9-2385. [DOI] [PubMed] [Google Scholar]
- 10.Eichenbaum Z, Federle M J, Marra D, de Vos W M, Kuipers O P, Kleerebezem M, Scott J R. Use of the lactococcal nisA promoter to regulate gene expression in gram-positive bacteria: comparison of induction level and promoter strength. Appl Environ Microbiol. 1998;64:2763–2769. doi: 10.1128/aem.64.8.2763-2769.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Engelke G, Gutowski-Eckel Z, Kiesau P, Siegers K, Hammelmann M, Entian K-D. Regulation of nisin biosynthesis and immunity in Lactococcus lactis 6F3. Appl Environ Microbiol. 1994;60:814–825. doi: 10.1128/aem.60.3.814-825.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Falahee M B, Adams M R. Cross-reactivity of bacteriocins from lactic acid bacteria and lantibiotics in a nisin bioassay and ELISA. Lett Appl Microbiol. 1992;15:214–216. doi: 10.1111/j.1472-765x.1992.tb00766.x. [DOI] [PubMed] [Google Scholar]
- 13.Falahee M B, Adams M R, Dale J W, Morris B A. An enzyme immunoassay for nisin. Int J Food Sci Technol. 1990;25:590–595. [Google Scholar]
- 14.Fowler G G, Jarvis B, Tramer J. The assay of nisin in foods. Soc Appl Bacteriol Tech Ser. 1975;8:91–105. [Google Scholar]
- 15.Gasson M J. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J Bacteriol. 1983;154:1–9. doi: 10.1128/jb.154.1.1-9.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Graeffe T, Rintala H, Paulin L, Saris P. A natural nisin variant. In: Jung G, Sahl H-G, editors. Nisin and novel lantibiotics. Leiden, The Netherlands: ESCOM Science Publishers B.V.; 1991. pp. 260–268. [Google Scholar]
- 17.Gross E, Morell J L. The structure of nisin. J Am Chem Soc. 1971;93:4634–4635. doi: 10.1021/ja00747a073. [DOI] [PubMed] [Google Scholar]
- 18.Holo H, Nes I F. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl Environ Microbiol. 1989;55:3119–3123. doi: 10.1128/aem.55.12.3119-3123.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Immonen T, Saris P E J. Characterization of the nisFEG operon of the nisin Z producing Lactococcus lactis subsp. lactis N8 strain. DNA Sequence. 1998;9:263–274. doi: 10.3109/10425179809008466. [DOI] [PubMed] [Google Scholar]
- 20.Immonen T, Ye S, Ra R, Qiao M, Paulin L, Saris P E J. The codon usage of the nisZ operon in Lactococcus lactis N8 suggests a non-lactococcal origin of the conjugative nisin-sucrose transposon. DNA Sequence. 1995;5:203–218. doi: 10.3109/10425179509030968. [DOI] [PubMed] [Google Scholar]
- 21.Jacobs M F, Tynkkynen S, Sibakov M. Highly bioluminescent Streptococcus thermophilus strain for the detection of dairy-relevant antibiotics in milk. Appl Microbiol Biotechnol. 1995;44:405–412. doi: 10.1007/BF00169936. [DOI] [PubMed] [Google Scholar]
- 22.Joosten H M L J, Nuñez M. Adsorption of nisin and enterocin 4 to polypropylene and glass surfaces and its prevention by Tween 80. Lett Appl Microbiol. 1995;21:389–392. [Google Scholar]
- 23.Kleerebezem M, Beerthuyzen M M, Vaughan E E, de Vos W M, Kuipers O P. Controlled gene expression systems for lactic acid bacteria: transferable nisin-inducible expression cassettes for Lactococcus, Leuconostoc, and Lactobacillus spp. Appl Environ Microbiol. 1997;63:4581–4584. doi: 10.1128/aem.63.11.4581-4584.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuipers O P, Beerthuyzen M M, de Ruyter P G G A, Luesink E J, de Vos W M. Autoregulation of nisin biosynthesis in Lactococcus lactis by signal transduction. J Biol Chem. 1995;270:27299–27304. doi: 10.1074/jbc.270.45.27299. [DOI] [PubMed] [Google Scholar]
- 25.Kuipers O P, Beerthuyzen M M, Siezen R J, de Vos W M. Characterization of the nisin gene cluster nisABTCIPR of Lactococcus lactis. Requirement of expression of the nisA and nisI genes for development of immunity. Eur J Biochem. 1993;216:281–291. doi: 10.1111/j.1432-1033.1993.tb18143.x. [DOI] [PubMed] [Google Scholar]
- 26.Mulders J W M, Boerrigter I J, Rollema H S, Siezen R J, de Vos W M. Identification and characterization of the lantibiotic nisin Z, a natural nisin variant. Eur J Biochem. 1991;201:581–584. doi: 10.1111/j.1432-1033.1991.tb16317.x. [DOI] [PubMed] [Google Scholar]
- 27.Qiao M, Ye S, Koponen O, Ra R, Usabiaga M, Immonen T, Saris P E J. Regulation of the nisin operons in Lactococcus lactis N8. J Appl Bacteriol. 1996;80:626–634. doi: 10.1111/j.1365-2672.1996.tb03267.x. [DOI] [PubMed] [Google Scholar]
- 28.Ra S R, Qiao M, Immonen T, Pujana I, Saris P E J. Genes responsible for nisin synthesis, regulation and immunity form a regulon of two operons and are induced by nisin in Lactococcus lactis N8. Microbiology. 1996;142:1281–1288. doi: 10.1099/13500872-142-5-1281. [DOI] [PubMed] [Google Scholar]
- 29.Rossano R, Del Fiore A, D’Elia A, Pesole G, Parente E, Riccio P. New procedure for the determination of nisin in milk. Biotechnol Tech. 1998;12:783–786. [Google Scholar]
- 30.Siegers K, Entian K-D. Genes involved in immunity to the lantibiotic nisin produced by Lactoccoccus lactis 6F3. Appl Environ Microbiol. 1995;61:1082–1089. doi: 10.1128/aem.61.3.1082-1089.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steen M T, Chung Y J, Hansen J N. Characterization of the nisin gene as part of a polycistronic operon in the chromosome of Lactococcus lactis ATCC11454. Appl Environ Microbiol. 1991;57:1181–1188. doi: 10.1128/aem.57.4.1181-1188.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suárez A M, Rodríguez J M, Hernández P E, Azcona-Olivera J I. Generation of polyclonal antibodies against nisin: immunization strategies and immunoassay development. Appl Environ Microbiol. 1996;62:2117–2121. doi: 10.1128/aem.62.6.2117-2121.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suárez A M, Rodríguez J M, Morales P, Hernández P E, Azcona-Olivera J I. Development of monoclonal antibodies to the lantibiotic nisin A. J Agric Food Chem. 1996;44:2936–2940. [Google Scholar]
- 34.Terzaghi B E, Sandine W E. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol. 1975;29:807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tramer J, Fowler J J. Estimation of nisin in foods. J Sci Food Agric. 1964;15:522–528. [Google Scholar]
- 36.van der Meer J R, Polman J, Beerthuyzen M M, Siezen R J, Kuipers O P, de Vos W M. Characterization of the Lactococcus lactis nisin A operon genes nisP, encoding a subtilisin-like serine protease involved in precursor processing, and nisR, encoding a regulatory protein involved in nisin biosynthesis. J Bacteriol. 1993;175:2578–2588. doi: 10.1128/jb.175.9.2578-2588.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wolf C E, Gibbons W R. Improved method for quantification of the bacteriocin nisin. J Appl Bacteriol. 1996;80:453–457. doi: 10.1111/j.1365-2672.1996.tb03242.x. [DOI] [PubMed] [Google Scholar]