Abstract
Background:
Thioredoxin Interacting Protein (TXNIP) functions as a master regulator for glucose homeostasis. Hypomethylation at the 5’-cytosine-phosphate-guanine-3’ (CpG) site cg19693031 of TXNIP has been consistently related to islet dysfunction, hyperglycemia, and type 2 diabetes. DNA methylation (DNAm) may reveal the missing mechanistic link between obesity and type 2 diabetes. We hypothesize that baseline DNAm level at TXNIP in blood may be associated with glycemic traits and their changes in response to weight-loss diet interventions.
Methods:
We included 639 adult participants with overweight or obesity, who participated in a 2-year randomized weight-loss diet intervention. Baseline blood DNAm levels were profiled by high-resolution methylC-capture sequencing. We defined the regional DNAm level of TXNIP as the average methylation level over CpGs within 500 bp of cg19693031. Generalized linear regression models were used for main analyses.
Results:
We found that higher regional DNAm at TXNIP was significantly correlated with lower fasting glucose, HbA1c, and Homeostatic Model Assessment of Insulin Resistance (HOMA-IR) at baseline (P<0.05 for all). Significant interactions were observed between dietary protein intake and DNAm on changes in insulin (P-interaction=0.007) and HOMA-IR (P-interaction=0.009) at 6 months. In participants with the highest tertile of regional DNAm at TXNIP, average protein (15%) intake was associated with a greater reduction in insulin (β: −0.14; 95% CI: −0.24, −0.03; P=0.011) and HOMA-IR (β: −0.15; 95% CI: −0.26, −0.03; P=0.014) than high protein (25%) intake, whereas no significant associations were found in those with the lower tertiles (P>0.05). The interaction was attenuated to be non-significant at 2 years, presumably related to decreasing adherence to the diet intervention.
Conclusions:
Our data indicate that higher regional DNAm level at TXNIP was significantly associated with better fasting glucose, HbA1c, and HOMA-IR; and people with higher regional DNAm levels benefited more in insulin and HOMA-IR improvement by taking the average-protein weight-loss diet.
Keywords: DNA methylation, Type 2 Diabetes, Insulin, Insulin Resistance, Weight-loss diet intervention
Introduction
Thioredoxin Interacting Protein, encoded by the TXNIP gene, functions as a master regulator for whole-body glucose homeostasis and regulates both β cell function and survival.1,2 DNA methylation (DNAm) at the cytosine-guanine nucleotide pair (CpG) controls gene expression without changing the DNA sequence and plays a pivotal role in response to environmental stressors.3 In the epigenome-wide association studies (EWAS) using peripheral blood samples, methylation of CpG cg19693031 at TXNIP has been most robustly associated with T2D.4–6 Hypomethylation of DNA at the TXNIP gene in peripheral blood, which leads to overexpression of the gene, has been consistently related to higher risks of insulin resistance,7 islet dysfunction,8,9 hyperglycemia,5 and T2D.5,10,11
Individuals with overweight and obesity have elevated risks of abnormal glucose metabolism and T2D.12,13 A variety of low-calorie weight-loss diets have shown robust effects on improving insulin sensitivity and glycemia.14–17 However, the mechanisms underlying such observations are still elusive.18 Several lines of evidence has suggested the intrinsic role for DNAm, such as that at TXNIP, in regulation gene expression on the obesity-related glucose metabolism and development of T2D.18 To date, no study has investigated whether the DNAm level at the TXNIP gene region is related to the longitudinal changes in insulin sensitivity and glucose metabolism in response to weight-loss dietary interventions. DNAm acts at the interface between the environment and genetic function, and is responsive to the modifications by environmental factors including diets. Therefore, we hypothesized that various weight-loss diets might modify the associations between baseline DNAm and the dynamic changes in glycemic traits.
The Preventing Overweight Using Novel Dietary Strategies (POUNDS Lost) trial is one of the largest and longest randomized diet interventions on weight loss to date. In the present study, we assessed the relationship of DNAm levels at the TXNIP gene region with changes in the measures of insulin sensitivity and glucose metabolism over the 2-year intervention. We particularly examined the interaction between the pre-treatment regional DNAm level at TXNIP and weight-loss diets varying in macronutrients.
Subjects and Methods
Study design and participants
The POUNDS Lost trial (clinical trial reg. no. NCT00072995, clinicaltrials.gov) is a two-year randomized clinical trial aimed to compare the effects of 4 energy-reduced diets with varying macronutrient compositions of fat, protein, and carbohydrate on weight change. The study was conducted at two sites: the Harvard T.H. Chan School of Public Health and Brigham and Women’s Hospital (BWH), Boston, MA, and the Pennington Biomedical Research Center (PBRC) of the Lousiana State University System, Baton Rouge, LA from October 2004 to December 2007. Briefly, a total of 811 participants with overweight or obestiy were randomly assigned to 1 of 4 diets; the targeted percentage of energy derived from fat, protein, and carbohydrates in the four diets were: 1) 20% fat (F), 15% protein (P), and 65% carbohydrate (C); 2) 20% F, 25% P, and 55% C; 3) 40% F, 15% P, and 45% C; and 4) 40% F, 25% P, and 35% C. The diets consist of similar foods and meet guidelines for cardiovascular health. Two diets were low-fat (20%), and the other two diets were high-fat (40%), and two diets were average-protein (15%), and the other two diets were high-protein (25%), constituting a 2-by-2 factorial design. To assess the nutritional adherence, dietary intake was assessed in a random sample of 50% of the total participants, by a review of the 5-day diet record at baseline, and by 24-hour recall during a telephone interview on 3 nonconsecutive days at 6 months and 2 years. After 2 years, 80% of the participants (n=645) had completed the trial. The primary outcome was changes in body weight after 2 years, and the secondary outcome was changes in waist circumference.16 More details of the study design and method have been described elsewhere.16 All the participants provided written informed consent. The study was approved by the human subjects committee at the Harvard School of Public Health and Brigham and Women’s Hospital, Boston, MA; the Pennington Biomedical Research Center of the Louisiana State University, Baton Rouge, LA; and a data and safety monitoring board appointed by the National Heart, Lung, and Blood Institute.
In the current analysis, we included 639 participants with complete information on regional DNAm level at TXNIP. Of the included 639 participants, 568 participants were followed up to 6 months, and 516 participants had complete data at 2 years
Measurements
Blood samples were collected fasting and stored at −80°C. Baseline peripheral blood samples were sequenced on the IlluminaNovaSeq6000 platform by a high-resolution methyl-capture sequencing (MCC-Seq) approach at Children’s Mercy Research Institute (CMRI).19 The sequenced reads were trimmed for Illumina adapters and quality (phred33 >= 20) using trimgalore v.0.4.2, a wrapper tool combining Cutadapt20 and FastQC. The trimmed reads were then aligned to the bisulfite-converted Human reference genome build 37 (GRCh37) using Illumina’s DRAGEN Bio-IT Platform v3.6 in paired end mode and the highest quality unique alignment was retained. Duplicates were flagged using Picard Tool’s MarkDuplicates v2.17.8. Methylation calls were extracted using Bismark’s bismark_methylation_extractor v0.20.0. The resulting CpG sites were further filtered using BEDTool’s subtract v2.29.2 in the following manner: 1) CpGs with total coverage <10 reads were discarded, 2) CpGs overlapping an SNP from dbSNP build 151 were removed, and 3) CpGs overlapping regions in the ENCODE Blacklist were also removed (http://hgwdev.cse.ucsc.edu/cgi-bin/hgFileUi?db=hg19&g=wgEncodeMapability). Previous evidence has shown a stronger association with T2D for regional DNAm than individual CPG site. Therefore, we extracted neighboring CpGs centering the site cg19693031 within 500bp distance (chr1:145,441,302–145,441,802). The regional DNAm level was calculated as the total number of pooled the methylated reads over the total number of pooled sequenced reads covering CpGs within this region.
Glycemic traits, including serum fasting glucose, insulin, hemoglobin A1c (HbA1c) were measured by an immunoassay with chemiluminescent detection on an Immulite analyzer (Diagnostic Products Corporation) at the Pennington Biomedical Research Center. Homeostatic assessment models were used to estimate insulin resistance (HOMA-IR) by the following equation: HOMA-IR= [fasting insulin (μU/mL)] × [fasting glucose (mg/dL)/18.01]/22.5.21 The β-cell function was indicated by HOMA-B, which is calculated as follows: [20 × fasting insulin (μU/mL)]/{[fasting glucose (mg/dL)/18.01]−3.5}.21 Body weight and waist circumference were measured in the morning before breakfast on two days at baseline, 6, 12, 18, and 24 months. Height was measured at the baseline examination. Body mass index (BMI, kg/m2) was calculated as weight (kg) divided by the square of height (m2).
Statistical analysis
The primary outcomes were changes in glycemic traits, including fasting glucose, insulin, HbA1c, HOMA-IR, and HOMA-B. Insulin, HOMA-IR, and HOMA-B were log-transformed to improve normality. F test with adjustment for age, race, and sex for continuous variables, and chi-square test for categorical variables were performed to compare the characteristics of the study participants by tertiles of regional DNAm level at TXNIP. The cross-sectional association between the baseline blood DNAm level and baseline glycemic traits were tested using generalized linear regression models, with adjustment for age, race, sex, and diet intervention groups in model 1. We further adjusted for baseline BMI in model 2. To examine the prospective association between baseline blood regional DNAm level and the changes in glycemic traits, general linear regression models were applied. In model 1, we controlled for age, race, sex, diet interventions, baseline BMI, and baseline values of respective outcome traits. In model 2, we further adjusted for 6-month weight loss, which was the period of greatest weight loss during the intervention. To test the potential interaction between the blood regional DNAm level and diets, a DNAm-diet group product term was included in the above models. We used linear mixed models, with variance component structure, to test the pre-treatment effect of blood DNAm on the trajectory of changes in the outcomes according to diet group by including a DNAm-time interaction term. Regional DNAm was analyzed continuously; for presentation purpose, we grouped it into tertiles, similar as previous publications by us and others.22–24 All statistical analyses were performed using SAS version 9.4 (SAS Institute). All-p values were 2-sided, and a p-value <0.05 was considered statistically significant.
Results
Baseline characteristics of the study participants according to the tertile categories of the regional DNAm level at TXNIP are shown in Table 1. Women were more likely to have a higher level of DNAm at TXNIP, presumably due to lower BMI and better glycemic profile among women. There were no significant differences in age, race, body weight, waist circumference, BMI, blood pressure, or lipids profiles across the tertile categories of DNAm at TXNIP. Participants with a higher level of regional DNAm were more likely to have a lower level of fasting glucose (P=0.01) and HbA1c (P=0.003) at baseline. Dietary intakes of macronutrients were similar across the tertiles.
Table 1.
Baseline characteristics of study population according to the tertiles of the blood DNAm level at TXNIP.
| Tertiles of DNAm level at TXNIP | p | |||
|---|---|---|---|---|
| T1 | T2 | T3 | ||
| Age, years | 52.2 (9.5) | 50.2 (8.7) | 50.1 (9.4) | .08 |
| White | 177 (83.9) | 172 (81.0) | 161 (74.9) | .32 |
| Female | 104 (49.3) | 135 (63.4) | 152 (70.7) | <.001 |
| Weight, kg | 95.1 (15.6) | 93 (15.9) | 92.8 (15.1) | .60 |
| BMI, kg/m2 | 32.8 (3.8) | 32.7 (3.9) | 32.8 (4.0) | .81 |
| Waist circumference, cm | 105.8 (13.0) | 103.1 (13.3) | 102.7 (12.9) | .79 |
| SBP, mmHg | 121.3 (13.7) | 119 (13.3) | 118.7 (13.9) | .78 |
| DBP, mmHg | 76.2 (10.0) | 75.4 (9.1) | 75.1 (9.4) | .88 |
| LDL-c, mg/dL | 125.7 (34.4) | 125.3 (31.7) | 128.1 (31.4) | .76 |
| HDL-c, mg/dL | 47.4 (14.7) | 47.7 (12.8) | 50.8 (14.5) | .16 |
| Total cholesterol, mg/dL | 203.2 (39.6) | 201 (35.8) | 204.1 (35.8) | .55 |
| Glucose, mg/dL | 94.4 (14.4) | 91.4 (10.0) | 89.7 (9.9) | .01 |
| Insulin, μU/mL | 11.5 [9.3] | 9.9 [7.1] | 10.0 [7.9] | .47 |
| HbA1c, % | 5.4 (0.4) | 5.4 (0.4) | 5.3 (0.3) | .003 |
| HOMA-IR | 2.7 [2.4] | 2.2 [1.9] | 2.2 [1.8] | .26 |
| HOMA-B | 139.3 [102.9] | 127.3 [91.1] | 131.5 [101.1] | .62 |
| Diet group | ||||
| High-fat diet | 120 (56.9) | 110 (51.6) | 102 (47.4) | .15 |
| High-protein diet | 110 (52.1) | 98 (46.0) | 111 (51.6) | .37 |
| Dietary intake per day | ||||
| Energy, kcal | 2004.4 (582.3) | 1943.1 (515.0) | 1915.9 (548.8) | .87 |
| Fat, % of energy | 44.5 (8.4) | 45 (7.2) | 44.7 (7.3) | .50 |
| Protein, % of energy | 36.7 (6.6) | 36.5 (5.6) | 37.3 (6.0) | .97 |
| Carbohydrate, % of energy | 18.2 (3.2) | 18.1 (3.0) | 18.2 (3.7) | .83 |
Data are mean (SD), median [IQR], or N (%).
P values were calculated by χ2 test for categorical variables and an F test after adjusting for age, sex, and race for continuous variables.
BMI: body mass index; SBP: systolic blood pressure; DBP: diastolic blood pressure; LDL-c: low lipoprotein cholesterol; HDL-c: high lipoprotein cholesterol; HOMA-IR: homeostatic model assessment of insulin resistance; HOMA-B: homeostatic model assessment of β-cell function.
The nutrient intake and biomarkers of adherence are shown in Supplementary Table 1. Consistent with the entire study population in POUNDS Lost, the reported dietary intake and biomarkers of adherence confirmed that participants modified their macronutrient intake in the direction of the intervention. Participants assigned to the high-protein diet reported higher dietary protein intake and lower carbohydrate intake than those assigned to the average-protein diet at 6 months; the adherence generally declined for both groups from 6 months to 2 years.
We observed significant associations between baseline blood regional DNAm level at TXNIP and baseline levels of fasting glucose, HbA1c, and HOMA-IR after adjustment for age, race, sex, diet intervention groups, and baseline BMI (Figure 1, Table 2, model 2). Higher regional DNAm at TXNIP was associated with lower fasting glucose (P<0.001), insulin (P<0.001), and HOMA-IR (P=0.03) at baseline. Compared to the lowest tertile group (lower methylation), participants in the highest tertile (higher methylation) had 3.4 mg/dL lower fasting glucose, 0.13 % lower HbA1c, and 0.10 lower HOMA-IR level (Figure 1). When analyzing the association between regional DNAm level at TXNIP and changes in glycemic traits in participants from all the diet groups combined, we found no significant associations between regional DNAm level at TXNIP and changes in glycemic traits, including fasting glucose, insulin, HbA1c, HOMA-IR, or HOMA-B.
Figure 1.
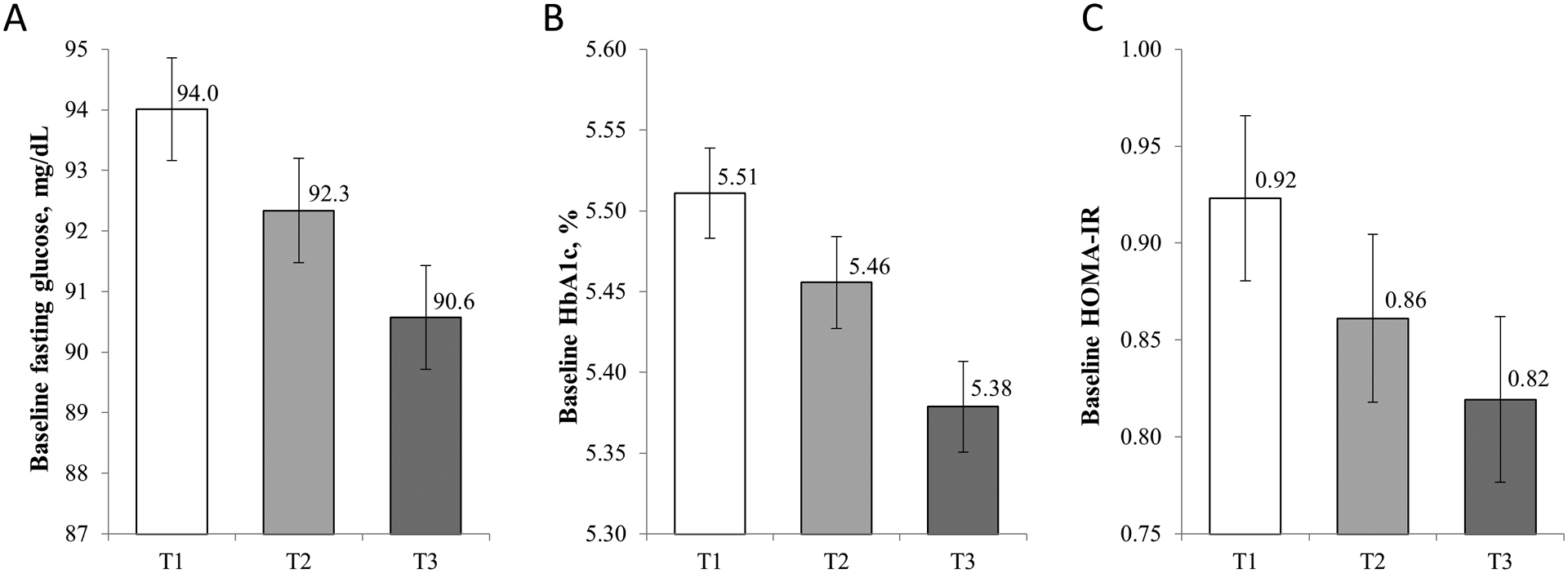
Association of blood regional DNAm level at TXNIP with baseline fasting glucose, HbA1c, and HOMA-IR.,
Model was adjusted for age, race, sex, diet intervention groups, and baseline body mass index.
Insulin, HOMA-IR was log transformed before analyses. HbA1c: hemoglobin A1c; HOMA-IR: homeostatic model assessment of insulin resistance. Panel A: fasting glucose, panel B: HbA1c, panel C: HOMA-IR.
Table 2.
Association of blood DNA methylation level at TXNIP with baseline glycemic traits.
| Model 1 | Model 2 | ||||
|---|---|---|---|---|---|
| β (SE) | p | β (SE) | p | ||
| Fasting glucose, mg/dL | −8.70 (2.62) | <0.001 | −8.74 (2.59) | <.001 | |
| Fasting insulin, μU/mL | −0.19 (0.13) | 0.15 | −0.19 (0.12) | .10 | |
| HbA1c, % | −0.32 (0.09) | <0.001 | −0.32 (0.09) | <.001 | |
| HOMA-IR | −0.28 (0.14) | 0.05 | −0.28 (0.13) | .03 | |
| HOMA-B | 0.07 (0.13) | 0.60 | 0.06 (0.12) | .61 | |
Model 1 was adjusted for age, race, sex, diet intervention groups.
Model 2: model 1 + baseline body mass index.
Insulin, HOMA-IR, and HOMA-B were log-transformed before analyses. HbA1c: hemoglobin A1c; HOMA-IR: homeostatic model assessment of insulin resistance; HOMA-B: homeostatic model assessment of β-cell function
We then analyzed the associations according to the diet intervention groups. We observed significant interactions between the dietary protein intake and regional DNAm on changes in insulin (P-interaction=0.007) and HOMA-IR (P-interaction=0.009) at 6 months after multivariable adjustment (Figure 2). In the highest tertile of regional DNAm at TXNIP, average-protein intake was associated with a greater reduction in insulin (β: −0.14; 95% CI: −0.24, −0.03; P=0.011) and HOMA-IR (β: −0.15; 95% CI: −0.26, −0.03; P=0.014), whereas no significant associations were found in lower tertiles (P>0.05). The interaction was attenuated to be non-significant at 2 years, presumably related to less adherence to the intervention. Dietary fat/carbohydrate intake did not interact with regional DNAm in relation to the glycemic changes.
Figure 2.
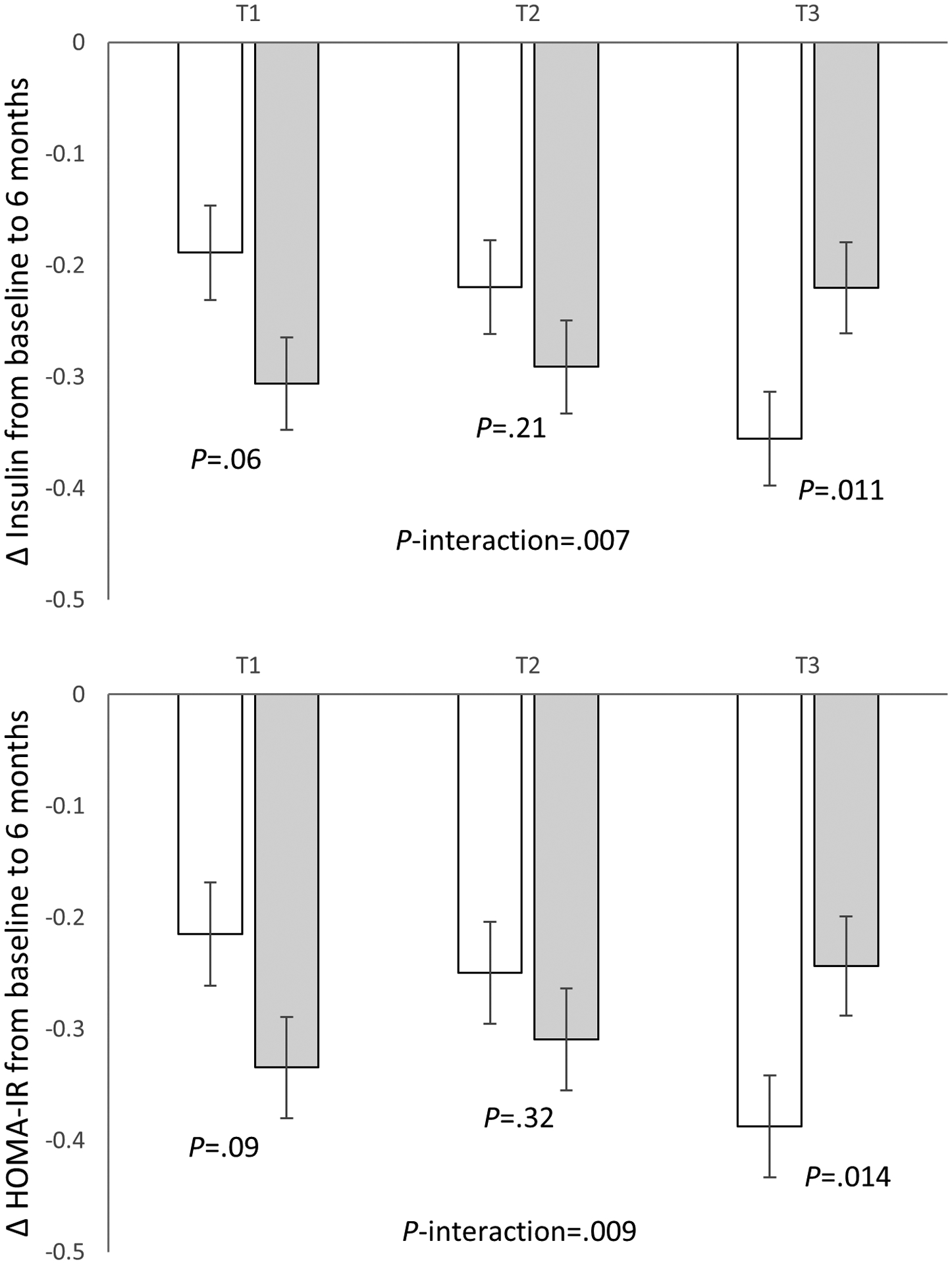
Changes in fasting insulin and HOMA-IR from baseline to 6 months according to tertiles of regional DNAm level around TXNIP in average- and high-protein diet groups.
Data are means ± SE values after adjustment for age, race, sex, body mass index, log transferred baseline values of respective outcome traits, and weight loss from baseline to 6 months. T1: lowest tertile; T3: highest tertile.
We further evaluated the trajectory of changes in insulin and HOMA-IR over 2 years in average- and high-protein group according to regional DNAm level at TXNIP (Supplementary Figure 1). In the highest tertile of regional DNAm at TXNIP, we found significant interactions between the dietary protein and intervention time on changes in insulin and HOMA-IR (P-interaction=0.024, and 0.038, respectively). Participants with the highest tertile group of DNAm level at TXNIP in the average-protein group showed a greater reduction in insulin and HOMA-IR at 6 months, compared to those in the high-protein group. Such difference converged to be similar from 6 months to 2 years, presumably related to less adherence to the diet intervention. While among participants with the two lower tertiles of DNAm, similar trajectories of insulin or HOMA-IR were observed between the average- and high-protein group (P-interaction>0.05).
Discussion
In this 2-year weight-loss dietary intervention trial, we found that higher regional DNAm level at TXNIP was significantly associated with lower fasting glucose, HbA1c, and HOMA-IR among participants who were overweight or obese. Moreover, we found that dietary protein intakes significantly modified the relation between regional DNAm level at TXNIP and changes in insulin and HOMA-IR at 6 months. Such effect modification became non-significant at 2 years.
Our findings on the associations of the regional DNAm level at TXNIP with glucose and HbA1c at baseline were directionally consistent with previous findings,5–7,9,25,26 which confirms the validity of analyzing the regional DNAm level at TXNIP in our study population. Previous investigations combining human physiology, genome-wide expression profiling, and cellular studies have shown that TXNIP was an independent determinant of glucose uptake in the human body.2 It has also been demonstrated that TXNIP expression in human enhanced β cell apoptosis.2,8 Our findings, together with evidence from these previous studies, support the functional link between TXNIP and glucose metabolism in humans.27
Interestingly, we found that the pre-treatment DNAm level significantly interacted with dietary protein intake on changes in insulin and HOMA-IR at 6 months, independent of weight loss. Participants with the highest tertile of regional DNAm at TXNIP benefited more in insulin and HOMA-IR improvement by consuming an average-protein weight-loss diet. It is well known that dietary intakes have indisputable impact on DNAm.28 For example, very low protein diets or malnutrition during gestation has been reported to result in both hypomethylation and hypermethylation at specific genes in offspring.29–32 However, the underlying mechanisms of the interaction between DNAm at TXNIP and dietary protein intake on changes in insulin and HOMA-IR are still elusive. TXNIP is a critical component of pancreatic β-cell biology, nutrient sensing, energy metabolism, and regulation of cellular redox states.11,33 Previous studies in animal models reported that TXNIP downregulation protected against obesity-induced diabetes by preventing β-cell apoptosis and preserving β-cell mass.8 Downregulation of mediobasal hypothalamic TXNIP expression was also reported to prevent diet-induced obesity and insulin resistance.34 The observed interaction was attenuated and became non-significant at 2 years, along with the diminished adherence to macronutrient goals and weight regain from 6 months to 2 years, similar to other long-term weight-loss dietary intervention trials.16,35
To the best of our knowledge, this is the first study to examine the regional DNAm level at TXNIP on changes in glycemic traits in response to weight-loss dietary interventions. Our study has several strengths. First, the high-resolution MCC-seq approach comprehensively captured the DNAm profiles in the target region. Second, repeated measures of glucose metabolism in POUNDS Lost allowed us to assess the relations between DNAm and longitudinal changes in these markers. We also acknowledge several limitations. First, we only measured DNAm levels in peripheral blood; DNAm levels in diabetes-related tissues, such as pancreatic islets, liver, or adipose tissue, were not available. However, DNAm levels in blood at TXNIP have been consistently related to glycemic measures and T2D, and reflect the methylation changes in the relevant tissues in previous studies.11,36,37 Chambers et al reported a significant correlation between DNAm in peripheral blood and DNAm in the liver at the TXNIP locus.11 Hypomethylation at the TXNIP locus in pancreatic islets and skeletal muscle was also found to be directionally consistent with DNAm in blood.37,38 Second, 80% of our study population was white. Whether such findings are applicable to other ethnic groups need to be further investigated. Third, we only measured the DNAm at baseline and were unable to investigate whether the epigenetic changes responded to weight-loss dietary interventions.
In conclusion, we found that the higher blood DNAm level at TXNIP was significantly related to lower fasting glucose, HbA1c, and HOMA-IR; and participants with higher DNAm levels benefited more in insulin and HOMA-IR improvement by taking the average-protein weight-loss diet, which was independent of weight loss. Our findings, together with previous evidence, highlight DNAm at TXNIP as a potential intervention target for obesity management.
Supplementary Material
Acknowledgments
The authors thank all the POUNDS Lost participants for their dedication and contribution to the research.
Funding sources
The study was supported by grants from the National Heart, Lung, and Blood Institute (HL071981, HL034594, HL126024), the National Institute of Diabetes and Digestive and Kidney Diseases (DK115679, DK091718, DK100383), the Fogarty International Center (TW010790), and Tulane Research Centers of Excellence Awards. Xiang Li was the recipient of the American Heart Association Predoctoral Fellowship Award (19PRE34380036).
Alphabetical List of Abbreviations
- BMI
body mass index
- DNAm
DNA methylation
- HbA1c
hemoglobin A1c
- HOMA-B
homeostatic model assessment of β-cell function
- HOMA-IR
homeostatic model assessment of insulin resistance
- T2D
type 2 diabetes
- TXNIP
Thioredoxin Interacting Protein
Footnotes
Competing Interests:
All authors declare no conflict of interest.
References:
- 1.Yoshihara E TXNIP/TBP-2: A Master Regulator for Glucose Homeostasis. Antioxidants. 2020; 9. doi: 10.3390/antiox9080765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parikh H, Carlsson E, Chutkow WA, Johansson LE, Storgaard H, Poulsen P et al. TXNIP Regulates Peripheral Glucose Metabolism in Humans. PLOS Med 2007; 4: e158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moore LD, Le T, Fan G. DNA Methylation and Its Basic Function. Neuropsychopharmacology 2013; 38: 23–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walaszczyk E, Luijten M, Spijkerman AMW, Bonder MJ, Lutgers HL, Snieder H et al. DNA methylation markers associated with type 2 diabetes, fasting glucose and HbA1c levels: a systematic review and replication in a case–control sample of the Lifelines study. Diabetologia 2018; 61: 354–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Florath I, Butterbach K, Heiss J, Bewerunge-Hudler M, Zhang Y, Schoettker B et al. Type 2 diabetes and leucocyte DNA methylation: an epigenome-wide association study in over 1,500 older adults. Diabetologia 2016; 59: 130–138. [DOI] [PubMed] [Google Scholar]
- 6.Meeks KAC, Henneman P, Venema A, Addo J, Bahendeka S, Burr T et al. Epigenome-wide association study in whole blood on type 2 diabetes among sub-Saharan African individuals: findings from the RODAM study. Int J Epidemiol 2019; 48: 58–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hidalgo B, Irvin MR, Sha J, Zhi D, Aslibekyan S, Absher D et al. Epigenome-Wide Association Study of Fasting Measures of Glucose, Insulin, and HOMA-IR in the Genetics of Lipid Lowering Drugs and Diet Network Study. Diabetes 2014; 63: 801 LP–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Minn AH, Hafele C, Shalev A. Thioredoxin-interacting protein is stimulated by glucose through a carbohydrate response element and induces β-cell apoptosis. Endocrinology 2005; 146: 2397–2405. [DOI] [PubMed] [Google Scholar]
- 9.Volkmar M, Dedeurwaerder S, Cunha DA, Ndlovu MN, Defrance M, Deplus R et al. DNA methylation profiling identifies epigenetic dysregulation in pancreatic islets from type 2 diabetic patients. EMBO J 2012; 31: 1405–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cardona A, Day FR, Perry JRB, Loh M, Chu AY, Lehne B et al. Epigenome-Wide Association Study of Incident Type 2 Diabetes in a British Population: EPIC-Norfolk Study. Diabetes 2019; 68: 2315–2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chambers JC, Loh M, Lehne B, Drong A, Kriebel J, Motta V et al. Epigenome-wide association of DNA methylation markers in peripheral blood from Indian Asians and Europeans with incident type 2 diabetes: A nested case-control study. Lancet Diabetes Endocrinol 2015; 3: 526–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Must A, Spadano J, Coakley EH, Field AE, Colditz G, Dietz WH. The Disease Burden Associated With Overweight and Obesity. JAMA 1999; 282: 1523–1529. [DOI] [PubMed] [Google Scholar]
- 13.Colditz GA, Willett WC, Rotnitzky A, Manson JE. Weight Gain as a Risk Factor for Clinical Diabetes Mellitus in Women. Ann Intern Med 1995; 122: 481–486. [DOI] [PubMed] [Google Scholar]
- 14.Group DPP (DPP) R. The Diabetes Prevention Program (DPP): description of lifestyle intervention. Diabetes Care 2002; 25: 2165–2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lindström J, Louheranta A, Mannelin M, Rastas M, Salminen V, Eriksson J et al. The Finnish Diabetes Prevention Study (DPS). Diabetes Care 2003; 26: 3230 LP–3236. [DOI] [PubMed] [Google Scholar]
- 16.Sacks FM, Bray GA, Carey VJ, Smith SR, Ryan DH, Anton SD et al. Comparison of weight-loss diets with different compositions of fat, protein, and carbohydrates. N Engl J Med 2009; 360: 859–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pan X-R, Li G-W, Hu Y-H, Wang J-X, Yang W-Y, An Z-X et al. Effects of Diet and Exercise in Preventing NIDDM in People With Impaired Glucose Tolerance: The Da Qing IGT and Diabetes Study. Diabetes Care 1997; 20: 537 LP–544. [DOI] [PubMed] [Google Scholar]
- 18.Dobosz AM, Dziewulska A, Dobrzyń A. Spotlight on epigenetics as a missing link between obesity and type 2 diabetes. Postepy Biochem 2018; 64: 157–165. [DOI] [PubMed] [Google Scholar]
- 19.Allum F, Shao X, Guénard F, Simon M-M, Busche S, Caron M et al. Characterization of functional methylomes by next-generation capture sequencing identifies novel disease-associated variants. Nat Commun 2015; 6: 7211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin M Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal; Vol 17, No 1 Next Gener Seq Data Anal - 1014806/ej171200 2011.http://journal.embnet.org/index.php/embnetjournal/article/view/200. [Google Scholar]
- 21.Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia 1985; 28: 412–419. [DOI] [PubMed] [Google Scholar]
- 22.Zhou T, Sun D, Heianza Y, Li X, Champagne CM, LeBoff MS et al. Genetically determined vitamin D levels and change in bone density during a weight-loss diet intervention: the Preventing Overweight Using Novel Dietary Strategies (POUNDS Lost) Trial. Am J Clin Nutr 2018; 108: 1129–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yaskolka Meir A, Keller M, Müller L, Bernhart SH, Tsaban G, Zelicha H et al. Effects of lifestyle interventions on epigenetic signatures of liver fat: Central randomized controlled trial. Liver Int 2021; 41: 2101–2111. [DOI] [PubMed] [Google Scholar]
- 24.Yamazaki M, Yamada H, Munetsuna E, Maeda K, Ando Y, Mizuno G et al. DNA methylation level of the gene encoding thioredoxin-interacting protein in peripheral blood cells is associated with metabolic syndrome in the Japanese general population. Endocr J 2021. doi: 10.1507/endocrj.EJ21-0339. [DOI] [PubMed] [Google Scholar]
- 25.Soriano-Tárraga C, Jiménez-Conde J, Giralt-Steinhauer E, Mola-Caminal M, Vivanco-Hidalgo RM, Ois A et al. Epigenome-wide association study identifies TXNIP gene associated with type 2 diabetes mellitus and sustained hyperglycemia. Hum Mol Genet 2016; 25: 609–619. [DOI] [PubMed] [Google Scholar]
- 26.Dayeh T, Volkov P, Salö S, Hall E, Nilsson E, Olsson AH et al. Genome-wide DNA methylation analysis of human pancreatic islets from type 2 diabetic and non-diabetic donors identifies candidate genes that influence insulin secretion. PLoS Genet 2014; 10: e1004160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Juvinao-Quintero DL, Marioni RE, Ochoa-Rosales C, Russ TC, Deary IJ, van Meurs JBJ et al. DNA methylation of blood cells is associated with prevalent type 2 diabetes in a meta-analysis of four European cohorts. Clin Epigenetics 2021; 13: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kadayifci FZ, Zheng S, Pan Y-X. Molecular Mechanisms Underlying the Link between Diet and DNA Methylation. Int J Mol Sci 2018; 19: 4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Straten EME, Bloks VW, Huijkman NCA, Baller JFW, Meer H van, Lütjohann D et al. The liver X-receptor gene promoter is hypermethylated in a mouse model of prenatal protein restriction. Am J Physiol Integr Comp Physiol 2010; 298: R275–R282. [DOI] [PubMed] [Google Scholar]
- 30.Heijmans BT, Tobi EW, Stein AD, Putter H, Blauw GJ, Susser ES et al. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci 2008; 105: 17046–17049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burdge GC, Slater-Jefferies J, Torrens C, Phillips ES, Hanson MA, Lillycrop KA. Dietary protein restriction of pregnant rats in the F0 generation induces altered methylation of hepatic gene promoters in the adult male offspring in the F1 and F2 generations. Br J Nutr 2007; 97: 435–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tobi EW, Lumey LH, Talens RP, Kremer D, Putter H, Stein AD et al. DNA methylation differences after exposure to prenatal famine are common and timing-and sex-specific. Hum Mol Genet 2009; 18: 4046–4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cha-Molstad H, Saxena G, Chen J, Shalev A. Glucose-stimulated expression of Txnip is mediated by carbohydrate response element-binding protein, p300, and histone H4 acetylation in pancreatic beta cells. J Biol Chem 2009; 284: 16898–16905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blouet C, Schwartz GJ. Nutrient-Sensing Hypothalamic TXNIP Links Nutrient Excess to Energy Imbalance in Mice. J Neurosci 2011; 31: 6019 LP–6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnston BC, Kanters S, Bandayrel K, Wu P, Naji F, Siemieniuk RA et al. Comparison of weight loss among named diet programs in overweight and obese adults: a meta-analysis. JAMA 2014; 312: 923–933. [DOI] [PubMed] [Google Scholar]
- 36.Bacos K, Gillberg L, Volkov P, Olsson AH, Hansen T, Pedersen O et al. Blood-based biomarkers of age-associated epigenetic changes in human islets associate with insulin secretion and diabetes. Nat Commun 2016; 7: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dayeh T, Tuomi T, Almgren P, Perfilyev A, Jansson P-A, de Mello VD et al. DNA methylation of loci within ABCG1 and PHOSPHO1 in blood DNA is associated with future type 2 diabetes risk. Epigenetics 2016; 11: 482–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walaszczyk E, Luijten M, Spijkerman AMW, Bonder MJ, Lutgers HL, Snieder H et al. DNA methylation markers associated with type 2 diabetes, fasting glucose and HbA1c levels: a systematic review and replication in a case–control sample of the Lifelines study. Diabetologia 2018; 61: 354–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


