Abstract
Unlocking the potential of personalized medicine in point-of-care settings requires a new generation of biomarker and proteomic assays. Ideally, assays could inexpensively perform hundreds of quantitative protein measurements in parallel at the bedsides of patients. This goal greatly exceeds current capabilities. Furthermore, biomarker assays are often challenging to translate from benchtop to clinic due to difficulties achieving and assessing the necessary selectivity, sensitivity, and reproducibility. To address these challenges, we developed an efficient (<5 min), robust (comparatively lower CVs), and inexpensive (decreasing reagent use and cost by >70%) immunoassay method. Specifically, the immunoblot membrane is dotted with the sample and then developed in a vortex fluidic device (VFD) reactor. All assay steps – blocking, binding, and washing – leverage the unique thin-film microfluidics of the VFD. The approach can accelerate direct, indirect, and sandwich immunoblot assays. The applications demonstrated include assays relevant to both the laboratory and the clinic.
Keywords: immunoassay, vortex fluidics, diagnostics
Graphical Abstract
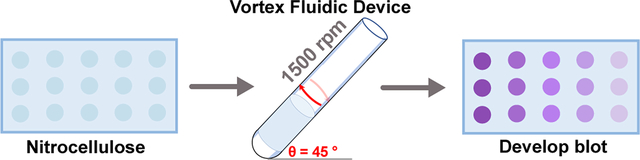
The VFD-accelerated immunoblot assay (VAIA) improves conventional processing time from a hours-long process to <5 minutes. Here, VAIA is performed with three major immunoassay formats with purified proteins and diluted biofluids.
The promises of personalized medicine require efficient, inexpensive testing for the presence and concentration of biomarkers. Low-cost diagnostics for broad deployment of precision medicine also represent a health justice issue, as high-tech medical devices often neglect resource-limited areas.[1] The extreme disparity between technologically lagging and advanced settings directly impacts disease mortality and morbidity, particularly for infectious diseases.[2] Thus, a clear need exists for a simple, cost-effective platform technology to advance precision medicine worldwide.
Point-of-care (PoC) tests have revolutionized diagnostics and patient care. For example, the rapid Strep A test has reduced unnecessary antibiotic treatments with clear benefits to public health.[3] Similarly, PoC influenza tests can allow early antiviral intervention, if conducted within 72 hours post-symptom onset.[4,5] The pregnancy test for chorionic gonadotropin has changed women’s reproductive health and has emerged as both the most common at-home and PoC diagnostic test.[6] Despite these successful examples, a clear gap exists between the thousands of evidence-based biomarkers reported and their validation in the clinic.[7] Thus, technology allowing PoC biomarker validation and widespread deployment is required to close this gap.
Already used extensively for biomarker-based tests, immunoblot assays (IAs) offer a low-tech, but highly effective disease diagnostic. For example, an IA was developed as a cost-effective tool for detection of Dengue, a rampant ailment in countries lacking medical infrastructure for more complicated testing.[8] Similarly, IAs have been developed for the diagnosis of myofibrillar myopathies.[9] Most prominently, ßan IA is used in concert with an enzyme-linked immunosorbent assay (ELISA) to diagnose human immunodeficiency virus.[10,11]
However, IAs typically incur high costs and have complex protocols and low sensitivity.[12,13] Despite these limitations, IAs in laboratories are often used to optimize conditions for the more experimentally demanding and time-consuming Western blot. A conventional IA typically requires >2 hr and consumes significant amounts of expensive reagents (e.g., primary and secondary antibodies).[14] Kurien et al. previously described a shortened, >40 min IA protocol reliant on processing the blot with reagents pre-warmed to 37 °C, which suggests thermally driving equilibration is one approach to accelerating IAs.[15] An alternative, especially for temperature sensitive applications, the Vitrozm Zoom Blot Plate, a single-use apparatus, can perform a multiplexed IA in 60 min.[16]
Eliminating the IA’s background is key to improving its sensitivity. Wu et al. determined that inefficient washing is the main contributor to high background in IAs.[17] We envisioned applying the mechanical energy of a vortex fluidic device (VFD) to provide stringent washes, accelerated equilibration, and decrease IA background. Previously, the VFD has been used to drive protein purification and tethering[18], recovery of DNA from formalin-preserved tissue[19], protein folding[20], and embedding active enzyme in xerogels.[21] Here, we report using the VFD to improve IA sensitivity and reduce processing time to <5 min.
The VFD houses a rapidly rotating tube tilted at the optimum angle of 45° (Fig. 1A). At low rotational speeds (>3000 rpm) the mechanoenergy of the dynamic thin film forces liquid across, in and over the membrane via shear stress and “tornado-like” topological fluid flows. The resultant high mass transfer throughout the membrane allows for IAs to be performed rapidly inside a VFD’s quartz reactor. Briefly, antigens were dotted on a nitrocellulose membrane that was sandwiched between two sheets of filter paper. The paper assembly was rolled into a cylinder and placed concentrically within the VFD reactor. All blocking, binding, and washing fluids were subsequently added and discarded between each assay step. The membrane was removed from the VFD, and the colorimetric substrate added.
Figure 1.
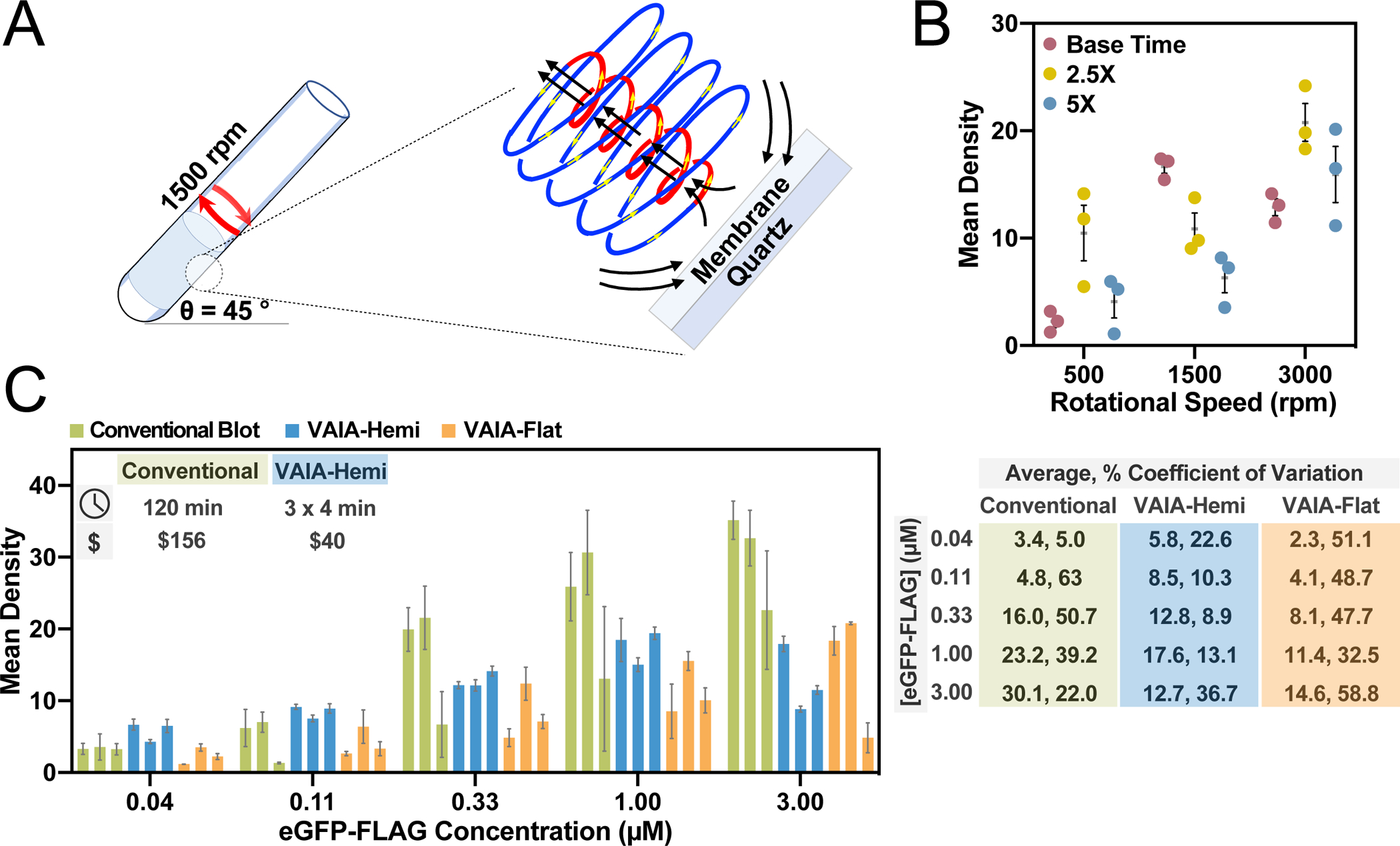
Optimization of the VAIA method. A) Nitrocellulose membranes dotted with target antigens are concentrically placed in a VFD reactor housing a quartz tube (20 mm OD, 17.5 mm ID, 18.5 cm in length). The VFD reactor tube is tilted at 45° and rotated at 1500 rpm throughout the assay steps. At this speed the shear stress is dominated by a spinning top topological fluid flow, as depicted. Membranes are removed from the VFD reactor, and the assay dots are visualized through addition of a colorimetric reagent. B) An indirect phage IA was used to optimize VAIA rotational speed and assay time. Systematic screening of 9 combinations of these parameters revealed 1500 rpm and a 4.5 min assay time (Base Time) yielded the highest signal-to-noise ratios and lowest error. Error bars represent SEM (n = 3). C) A direct eGFP-FLAG detection IA was used to compare quantitation with VAIA to the conventional IA method. VAIA was faster, more robust, more sensitive, and less costly than the conventional method. The fluid flow was examined by performing VAIA with either hemispherical-bottom VFD reactors or flat-bottom VFD reactors. Error bars represent SEM for each group of dots on each immunoblot (n = 3). Percent coefficients of variation are based upon independent replicates (n = 3).
The initial optimization of the VFD-accelerated IA (VAIA) was performed with a previously described, robust assay.[22] The dotted HSA antigen was captured by addition of a small quantity (2 mL of 1 nM phage) of a phage-displayed HSA binding ligand and visualized by colorimetric activity of a peroxidase-conjugated, phage-specific antibody (2 mL of 1:10,000 diluted antibody in PBS) (Fig. 1A). The VFD conditions, including rotational speed and time, were subject to optimization. Ultimately, 1500 rpm and 4.5 min total of blocking (1 min), phage binding (2 min), antibody binding (1 min), and washing (2 × 15 s) steps yielded the greatest signal-to-noise ratios and the lowest levels of non-specific background binding (Fig. 1B).
Next, a direct assay further demonstrated the generality of the technique and revealed the effects of fluid flow on VAIA (Fig. 1C). The model protein, enhanced green fluorescent protein with a C-terminal FLAG-tag (eGFP-FLAG)[23], was detected with an anti-eGFP peroxidase-conjugated antibody. The assay was performed in either a hemispherical quartz VFD tube (VAIA-Hemi) or a flat-bottom quartz VFD tube (VAIA-Flat). At the optimal speed, the curved hemispherical base of the tube and the curved wall of the tube are expected to create a Coriolis fluid flow impacting the inner surface of the tube. Overall, the fluid flow afforded by the VAIA-Hemi resulted in more sensitivity and lower variability, where the rapid processing arises from the Coriolis fluid flow inducing high mass transfer into and out of the membrane. The cost per IA ($, inset in Fig. 1C) considers the decreased reagent and consumables cost for the VFD with approximate costs shown in 2022 USD.
The multifunctional eGFP-FLAG fusion protein provided a useful model to demonstrate a multitude of classic IA methods. The FLAG-tag was used as the antigen in both indirect (4.5 min) and direct (3.25 min) IA formats (Fig. 2A). Both formats delivered dose-dependent responses and different binding patterns. Specifically, the direct FLAG IA has a higher apparent binding affinity (EC50 = 0.231) and an apparent hook effect at higher concentrations.[24,25] In contrast, the indirect FLAG immunoblot has a lower apparent binding affinity (EC50 = 0.358) and does not appear to have a hook effect, even at the highest concentration. Thus, the observed hook effect in the previous assay may simply be an artifact of the larger error for data points at the assay’s higher concentrations. These apparent variations indicate that VAIA can analyze a variety of typical IA binding patterns.
Figure 2.
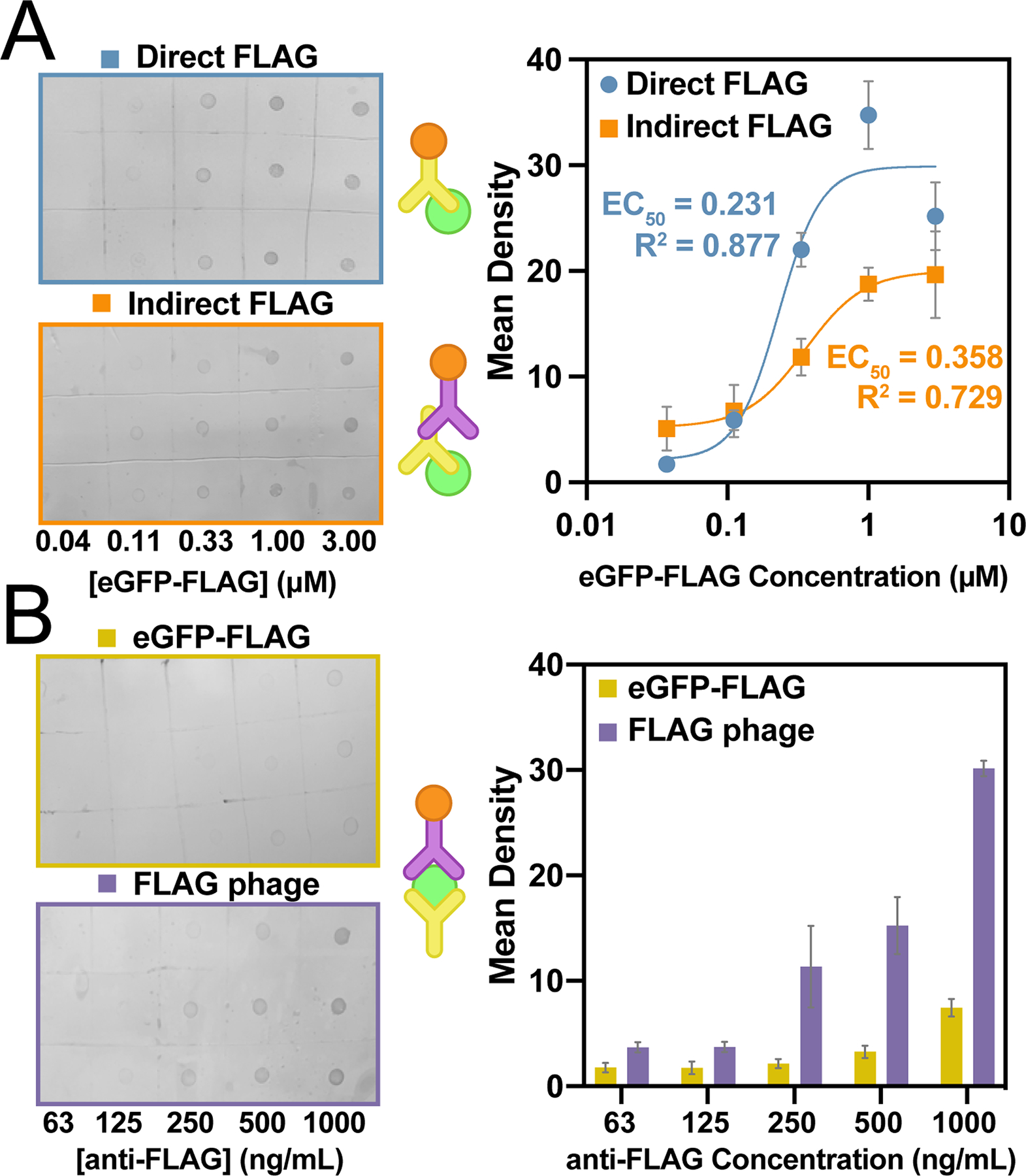
Generalization of VAIA to three common immunoassay formats. A) A direct and indirect anti-FLAG VAIA allowed dose-dependent quantification of eGFP-FLAG. The different binding modes cause the data to vary in both sensitivity and saturation limit. Data were fit with a four-parameter logistical curve fit. EC50 and R2 values are as indicated on the graph. B) Anti-FLAG antibodies captured either FLAG phage or eGFP-FLAG for sandwich VAIAs. Detection of FLAG phage was more sensitive, but both methods resulted in useful signal. A schematic for each assay format is provided (middle). Error bars represent SEM (n = 3).
Another common IA, the sandwich-format, features an antigen entrapped between two noncompetitive antibodies. VAIA enabled two different sandwich-format assays to be performed with each requiring <5 min (Fig. 2B). An anti-FLAG antibody was dotted on the membrane and captured either eGFP-FLAG or FLAG-tagged M13 bacteriophage (FLAG phage). The eGFP-FLAG fusion was sandwiched with anti-eGFP-HRP; the FLAG phage was sandwiched with anti-M13-HRP. Both immunoblots demonstrated dose-dependent binding; however, the FLAG phage signal was significantly more intense and sensitive. Taken as a whole, the wide variety of IA formats demonstrate the generality and robustness of VAIA.
The previously described assays were all performed with a commercial blocking agent, ChonBlock (CB). However, researchers typically block with solutions of bovine serum albumin (BSA), non-fat milk (NFM), or high concentrations of the non-ionic detergent Tween-20. Therefore, the eGFP-FLAG direct detection with anti-eGFP-HRP was repeated with these more common blocking conditions (Fig. S1). All four blocking conditions resulted in a robust signal; however, 5% CB had the best signal-to-noise and therefore, the best sensitivity. The experiments illustrate the adaptability of the VAIA platform for application to a variety of IA conditions and reagents.
VAIA also works well for potential clinical applications. In clinical samples, endogenous proteins can be challenging to detect due to the complex composition of the biofluid, including interfering substances. Human serum albumin (HSA) levels in the body are a biomarker for malnutrition, cirrhosis, and kidney disease.[26,27] VAIA’s ability to detect HSA was first shown using a direct assay with an HSA reference standard (Fig. 3A). This assay with reference standard was built upon by detection of endogenous HSA in diluted plasma, sera, urine, and blood from human patients validated testing of biofluids with VAIA (Fig. 3B). As shown through comparison with the reference standard assay, HSA levels were the highest in blood, followed by plasma, then sera. In urine, the measured HSA levels were undetectable (i.e., comparable to the negative control). This result is predictable, as urine from healthy donors should be relatively HSA-free.[28]
Figure 3.
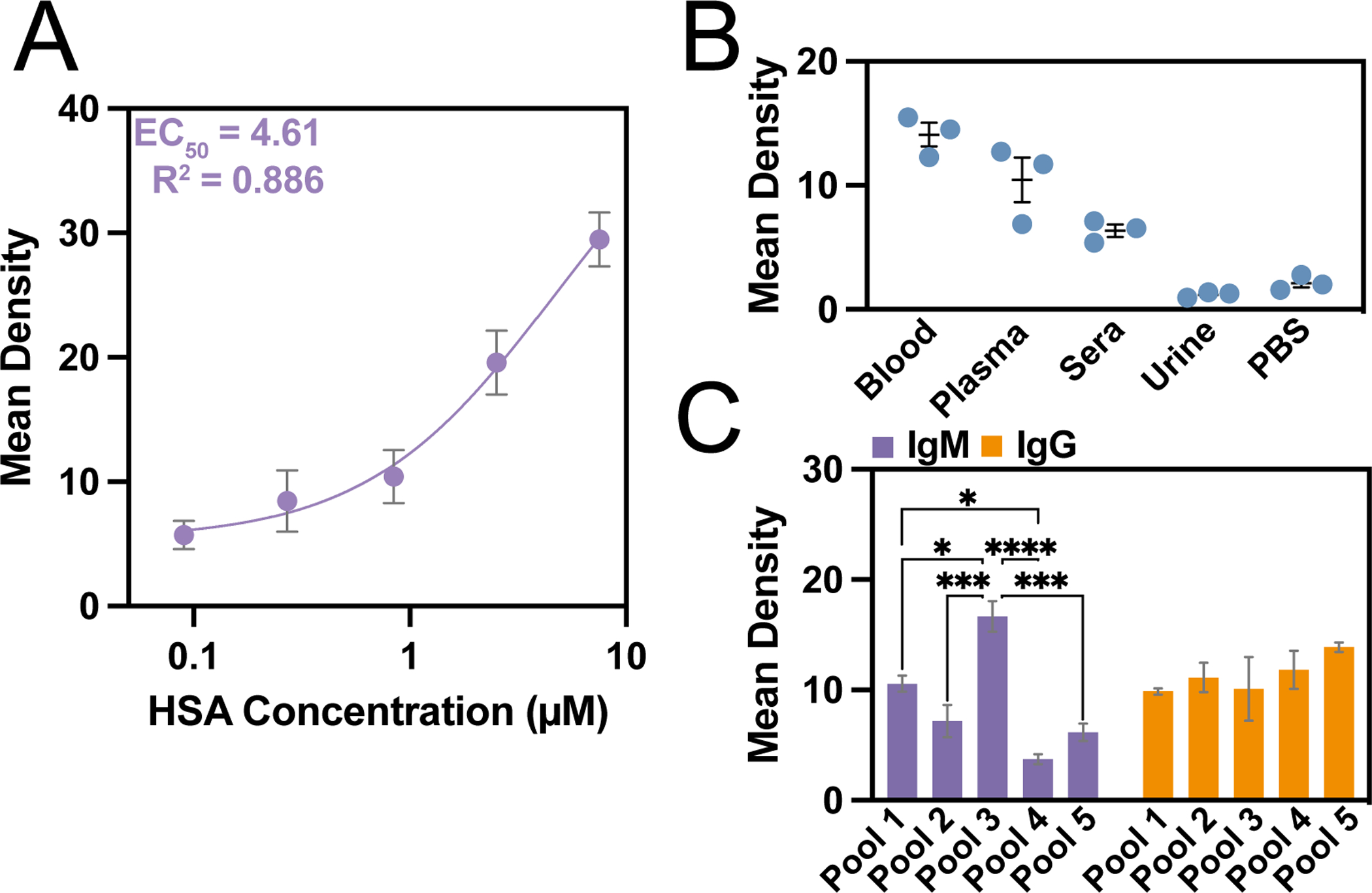
Clinical potential of VAIAs with biofluids. A) A direct VAIA with reference standard HSA provided a standard curve for measuring biomarker concentrations. Data were fit with a three-parameter logistical curve fit with the indicated EC50 and R2 values. B) An indirect VIAIA for HSA in plasma, sera, urine, blood, and PBS demonstrated VAIA applicability to a variety of biofluids. C) IgM and IgG levels in pooled plasma were assayed with VAIA. Error bars represent SEM (n = 3). ANOVA with Tukey’s multiple comparisons yields p-values of *<0.05, ***<0.001, ****<0.0001.
Several diseases can be diagnosed through the assessment of immunoglobulin levels in biofluids.[29–31] Unfortunately, the current state-of-the-art immunoglobulin assays are lengthy and complicated. Here, pooled plasma further characterized the clinical potential of VAIA (Fig. 3C). Interestingly, immunoglobulin G (IgG) levels were consistent amongst the pooled plasma (no significance by ANOVA), and immunoglobulin M (IgM) levels varied drastically from one another. In summary, the strong wash conditions of VAIA can overcome interfering substances during IAs with clinical samples.
The approach described here could find use in many chemical processes requiring the interactions of solid and liquid states. In diagnostics, for example, molecular recognition often requires molecules in liquid-phase to bind to a target affixed to solid support.
The VFD-driven rapid equilibration could accelerate equilibration of otherwise slow binding events. Furthermore, the >10-fold acceleration, combined with decreased costs, and simplified execution suggests the work reported here could advance IAs in academic, industrial, and clinical spaces.
We conclude by noting that VAIA has the potential to satisfy the requirements for bringing proteomic assays to PoC settings. VAIA is robust, rapid, and technically simple to execute, unlike conventional IAs. This conclusion was verified by an independent operator who replicated the Materials and Methods. Additionally, the large number of assay formats in this report demonstrates VAIA’s adaptability to a variety of established immunoassay formats. The approach can be readily scaled to examine hundreds to potentially thousands of proteins in one assay using a <5 min, inexpensive sandwich format assay. Most importantly, the data are unambiguous and digitizable with a cell phone camera. Therefore, VAIA could address the gap between development and implementation of biomarker-based precision medicine.
Supplementary Material
Acknowledgements
We gratefully acknowledge the patients who donated samples to make this work possible (research approved by the UCI IRB as projects HS#2012-8716 and HS#2014-1758). This work was supported by the UCI COVID-19 Basic, Translational and Clinical Research Fund (CRAFT), the Allergan Foundation, and UCOP Emergency COVID-19 Research Seed Funding. E.C.S. was supported by a Dissertation Fellowship from the UCI Department of Chemistry. S.R.S. was supported by a Public Impact Fellowship from the UCI Graduate Division. A.A.G. and A.M.S. thank the Minority Access to Research Careers Program, funded by the NIH (GM-69337). We thank Arjun S. Pamidi for coining the acronym “VAIA”.
Footnotes
Supporting information for this article is found in an additional submitted document.
Competing Interest Statement
The authors declare the following competing financial interests: E.C.S., C.L.R., and G.A.W. are pursuing a patent for VAIA. G.A.W. has a financial interest in PhageTech Inc., a diagnostics company, which was not involved in this study. The terms of this arrangement have been reviewed and approved by the University of California, Irvine in accordance with its conflict-of-interest policies.
References
- [1].Howitt P, Darzi A, Yang GZ, Ashrafian H, Atun R, Barlow J, Blakemore A, Bull AMJ, Car J, Conteh L, Cooke GS, Ford N, Gregson SAJ, Kerr K, King D, Kulendran M, Malkin RA, Majeed A, Matlin S, Merrifield R, Penfold HA, Reid SD, Smith PC, Stevens MM, Templeton MR, Vincent C, Wilson E, Lancet 2012, 380, 507–535. [DOI] [PubMed] [Google Scholar]
- [2].Gavazzi G, Herrmann F, Krause KH, Clin. Infect. Dis 2004, 39, 83–91. [DOI] [PubMed] [Google Scholar]
- [3].Bin Hendi S, Malik ZA, Khamis AH, Al-Najjar FYA, BMC Pediatr. 2021, 21, 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Balish A, Garten R, Klimov A, Villanueva J, Influenza Other Respi. Viruses 2013, 7, 491–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Uyeki TM, Pediatr. Infect. Dis. J 2003, 22, 164–177. [DOI] [PubMed] [Google Scholar]
- [6].Gnoth C, Johnson S, Geburtshilfe Frauenheilkd. 2014, 74, 661–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Selleck MJ, Senthil M, Wall NR, Biomark. Insights 2017, 12, 1177271917715236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Falconar AKI, Romero-Vivas CME, Virol. J 2013, 10, 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Marini M, Guglielmi V, Faulkner G, Piffer S, Tomelleri G, Vattemi G, Electrophoresis 2015, 36, 3097–3100. [DOI] [PubMed] [Google Scholar]
- [10].Esteban JI, Tai CC, Kay JWD, Shih JWK, Bodner AJ, Alter HJ, Lancet 1985, 326, 1083–1086. [DOI] [PubMed] [Google Scholar]
- [11].CDC Laboratory Procedure Manual, HIV Western Blot Confirmatory Test, 2013. [Google Scholar]
- [12].Montagnese F, Babačić H, Eichhorn P, Schoser B, J. Neurol 2019, 266, 1358–1366. [DOI] [PubMed] [Google Scholar]
- [13].Harrell J, Rubio XB, Nielson C, Hsu S, Motaparthi K, Clin. Dermatol 2019, 37, 692–712. [DOI] [PubMed] [Google Scholar]
- [14].Stott DI, Immunol J. Methods 1989, 119, 153–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Kurien BT, Danda D, Bachmann M, Scofield RH, Methods Mol. Biol 2015, 1312, 449–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Mai J, Method and Apparatus for Biomolecule Analysis, 2017, US 9,568,404. [Google Scholar]
- [17].Wu M, Stockley PG, Martin WJ, Electrophoresis 2002, 23, 2373–2376. [DOI] [PubMed] [Google Scholar]
- [18].Britton J, Dyer RP, Majumdar S, Raston CL, Weiss GA, Angew. Chemie - Int. Ed 2017, 56, 2296–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Totoiu CA, Phillips JM, Reese AT, Majumdar S, Girguis PR, Raston CL, Weiss GA, PLoS One 2020, 15, DOI 10.1371/journal.pone.0225807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Britton J, Smith JN, Raston CL, Weiss GA, Methods Mol. Biol 2017, 1586, 211–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Luo X, Mohammed Al-Antaki AH, Igder A, Stubbs KA, Su P, Zhang W, Weiss GA, Raston CL, ACS Appl. Mater. Interfaces 2020, 12, 51999–52007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ogata AF, Edgar JM, Majumdar S, Briggs JS, Patterson SV, Tan MX, Kudlacek ST, Schneider CA, Weiss GA, Penner RM, Anal. Chem 2017, 89, 1373–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Sen SR, Sanders EC, Gabriel KN, Miller BM, Isoda HM, Salcedo GS, Garrido JE, Dyer RP, Nakajima R, Jain A, Caldaruse A-M, Santos AM, Bhuvan K, Tifrea DF, Ricks-Oddie JL, Felgner PL, Edwards RA, Majumdar S, Weiss GA, mSphere 2021, 6, e00203–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Ross GMS, Filippini D, Nielen MWF, Salentijn GIJ, Anal. Chem 2020, 92, 15587–15595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Miles LEM, Lipschitz DA, Bieber CP, Cook JD, Anal. Biochem 1974, 61, 209–224. [DOI] [PubMed] [Google Scholar]
- [26].Lee S, Sung DB, Kang S, Parameswaran S, Choi JH, Lee JS, Han MS, Sensors 2019, 19, 5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Doumas BT, Peters T, Clin. Chim. Acta 1997, 258, 3–20. [DOI] [PubMed] [Google Scholar]
- [28].Choi S, Choi EY, Kim DJ, Kim JH, Kim TS, Oh SW, Clin. Chim. Acta 2004, 339, 147–156. [DOI] [PubMed] [Google Scholar]
- [29].Strassburg CP, Best Pract. Res. Clin. Gastroenterol 2010, 24, 667–682. [DOI] [PubMed] [Google Scholar]
- [30].Phillips AC, Carroll D, Drayson MT, Batty GD, Epidemiol J. Community Health 2015, 69, 129–135. [DOI] [PubMed] [Google Scholar]
- [31].Zhang H, Li P, Wu D, Xu D, Hou Y, Wang Q, Li M, Li Y, Zeng X, Zhang F, Shi Q, Medicine (Baltimore). 2015, 94, e387. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


