Abstract
Vertebral Compression Fracture (VCF) occurs when the vertebral body partially collapses under the action of compressive forces. Non-traumatic VCFs can be secondary to osteoporosis fragility (benign VCFs) or tumors (malignant VCFs). The investigation of the etiology of non-traumatic VCFs is usually necessary, since treatment and prognosis are dependent on the VCF type. Currently, there has been great interest in using Convolutional Neural Networks (CNNs) for the classification of medical images because these networks allow the automatic extraction of useful features for the classification in a given problem. However, CNNs usually require large datasets that are often not available in medical applications. Besides, these networks generally do not use additional information that may be important for classification. A different approach is to classify the image based on a large number of predefined features, an approach known as radiomics. In this work, we propose a hybrid method for classifying VCFs that uses features from three different sources: i) intermediate layers of CNNs; ii) radiomics; iii) additional clinical and image histogram information. In the hybrid method proposed here, external features are inserted as additional inputs to the first dense layer of a CNN. A Genetic Algorithm is used to: i) select a subset of radiomic, clinical, and histogram features relevant to the classification of VCFs; ii) select hyper-parameters of the CNN. Experiments using different models indicate that combining information is interesting to improve the performance of the classifier. Besides, pre-trained CNNs presents better performance than CNNs trained from scratch on the classification of VCFs.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10278-022-00586-y.
Keywords: Convolutional neural network, Genetic algorithm, Computer aided diagnosis, Vertebral compression fracture
Introduction
Vertebral Compression Fracture (VCF) is a fracture related to compressive forces that cause the partial collapse of the vertebral body. The clinician and radiologist will have no doubts regarding the etiology of a VCF from high-energy trauma, given the patient’s recent history [1]. However, when the patient develops a recent and painful vertebral collapse without trauma, the diagnosis of VCFs can be challenging. VCFs are more common in the elderly population, in which osteoporosis (benign VCFs) and cancer (malignant VCFs) occur more frequently. In addition, multiple vertebral fractures can occur, and the same patient can have a simultaneous diagnosis of osteoporosis and cancer. Computed Tomography and Magnetic Resonance Images (MRIs) may help in the differential diagnosis based on morphological and signal intensity abnormalities detected in the vertebral bodies, and also in vertebral posterior elements and paravertebral soft tissues [1–3].
There is great interest in the use of Machine Learning (ML) techniques to aid medical diagnosis [4, 5]. There are several causes for the growing interest in the use of ML in computer-aided diagnosis. The cost for obtaining images has decreased over the years, causing a large increase in the number of medical images generated every year. The quality of the analysis of medical images by human radiologists is affected by fatigue, demands for speed, and experience [6]. It takes years and a high financial cost to train a qualified radiologist, and a delayed or erroneous diagnosis can be harmful to the patient.
Compared to advances in medical imaging technologies, advances in the analysis of computational medical imaging are late. However, the area has been improving recently with the help of new ML techniques. Convolutional Neural Networks (CNNs), which is a deep learning technique, have attracted a lot of attention [4, 7, 8]. The traditional ML approach for the classification of medical images is to extract characteristics using pre-defined filters [9, 10]. The definition of the best filters for a given application generally requires experience and has a great impact on the performance of the ML algorithm. The main advantage of CNNs is that these algorithms automatically discover useful representations (filters) for classification of samples in a given dataset [11]. This is possible because CNNs use input processing layers, e.g., convolutional layers, that are not employed in traditional Artificial Neural Networks (ANNs).
However, CNNs generally require large datasets that are often not available in medical applications. In many medical image analysis problems, each sample of the dataset is given by an image and not many images are available. This is the case for the dataset related to VCFs used by Pereira et al [10] and also in this work. Besides, in many of these problems, techniques for artificially increasing the dataset and transfer learning are of little help because the images have specific properties and the use of generic images is not very useful. In addition, CNNs generally do not use additional information that may be important for classification. In the case of medical images, additional information is important to aid medical diagnosis. For example, information about the patient’s age and recent trauma history is important for the classification of VCFs, as previously discussed.
In this paper, images of vertebral bodies segmented from MRIs are classified by using CNNs. Strategies are adopted to minimize the difficulties arising from the use of small datasets and for adding additional useful information to the CNN. The objective is to classify vertebral bodies without fracture, benign or malignant VCFs. Few studies in the literature have investigated the use of ML for aiding the diagnosis of VCFs using MRI. Genant et al. [12] proposed a method for classifying VCFs in conventional radiography based on the sizes of the anterior, central, and posterior portions of the vertebral bodies. In order to assist radiologists in the interpretation of images and consequently allow the early diagnosis of osteoporosis, Kasai et al. [13] developed a computerized method for detecting vertebral fractures on lateral chest radiographs. The fractured vertebral body was detected by comparing the measured vertebral height with the expected vertebral size. Ribeiro et al. [14] proposed a method for extraction and analysis of vertebral bodies for the diagnosis of compression fractures using images of lateral radiographs of the lumbar spine. Gabor filters and ANNs were applied to extract the upper and lower plateaus of each vertebral body. Ghosh et al. [15] proposed an automated method for the detection, location, and segmentation of VCFs in computed tomography images. An ensemble of five classifiers was used. Pereira et al. [9, 10, 16] proposed a method for classifying malignant VCF or benign VCF using classical ML techniques applied to MRIs. The vertebral bodies were manually segmented, and shape and texture features were extracted. Different classifiers were compared.
We propose a method based on CNNs for classifying VCFs that uses features from three different sources: i) intermediate layers of CNNs; ii) radiomics; iii) additional information from the patients and image histogram. Radiomics is related to the extraction and analysis of large amounts of predefined features from medical images [17]. Here, 106 predefined features extracted from MRIs using the pyRadiomics Library [18] are employed. In the method proposed here, features extracted from the images (radiomics, clinical, and histogram information) are inserted as additional inputs to the first dense layer of a CNN. Our hypothesis is that by using all sources of information (raw images and external features), the performance of the CNN improves when the model is used to classify VCFs. Besides, a Genetic Algorithm (GA) is used to: i) select a subset of external features (radiomics, clinical, and histogram features) relevant to the classification of VCFs; ii) find optimal hyper-parameters for the CNN.
The idea of combining images with other medical information is not new. For example, in the model proposed by Dimitrovski et al. [19], visual features are combined with text features to improve the classification of medical images. However, to the best of our knowledge, combining features from intermediate layers of CNNs, radiomics, clinical, and histogram information for the classification of MRIs is new. Another important contribution of the paper is the unified approach for optimization of the hyper-parameters of the CNN and feature selection. Given a dataset, finding hyper-parameters that define the architecture and influence the learning of a CNN is a very important task [20].
This paper is organized as follows. The proposed methodology is presented in “Materials and Methods”. Experimental results are presented in “Results”. In order to test our hypothesis that using all sources of information improves the performance of the CNN, the proposed system is compared in the experiments to two other approaches: i) CNN where only (raw) images are used as inputs; ii) ANN where only radiomics, clinical, and histogram features are used as inputs. Results of experiments with pre-trained CNNs and with data augmentation are also presented. The discussion of the results is presented in “Discussion”, and paper is concluded in “Conclusion”.
Materials and Methods
In the proposed method, a CNN classifies VCFs using segmented images of vertebral bodies as inputs and external features are inserted as additional inputs to the first dense layer. The CNN classifies the vertebral bodies into one of three classes: normal, benign VCF, and malignant VCF. A GA is used to select subsets of radiomics, clinical, and histogram features relevant to the classification and to optimize hyper-parameters of the CNN. Next, the proposed method is presented, as well as the dataset.
Dataset
The images dataset used comprised 61 spine MRIs from different patients diagnosed with at least one VCF at the University Hospital1. The images were acquired according to the routine clinical protocol in a Philips Achieva 1.5T MRI equipment. The images are in the median sagittal plane (central section) weighted in the T1 contrast sequence. This set of images was extracted from the PACS (Picture Archiving and Communication System) of the University Hospital. The original format of the images is DICOM (Digital Imaging and Communications in Medicine) with 16-bits/pixel (65536 gray levels). The images were later converted to TIFF (Tagged Image File Format) without compression and converted to the 8-bit/pixel scale (256 gray levels).
The images of the spines were manually segmented into rectangular windows containing one vertebral body (example) each. The dataset with segmented images was used previously in other works [10]. The anonymized spine MRI dataset is public and available for research purposes, according to previous approval by The Institutional Review Board. A total of 189 windows (examples) was obtained by the segmentation process. An example of the segmentation process is illustrated in Fig. 1. The reference standard classification of vertebral bodies was performed by a board-certified radiologist with 17 years of experience in musculoskeletal radiology at the time of the database organization. The reference standard for the classification of vertebral bodies as normal (89 examples), benign VCF (54 examples), and malignant VCF (46 examples) was based on a best valuable comparator. The best valuable comparator consisted of the reading by the musculoskeletal radiologist, as well as the review of all available medical data. These data included clinical, histologic (spinal bone biopsy), and imaging data in a follow-up of 2 years. Vertebral bodies considered normal were extracted from the lumbar spine of patients with benign VCFs.
Fig. 1.
Steps of segmentation process. The second picture shows a mask that is applied to the original image (first picture). The resulting image (third picture) is then cut to form an example of the dataset (last picture)
The dataset was split into cross-validation and test sets. The cross-validation set is composed of vertebral bodies from images of 55 patients, totaling 169 examples. The test set is composed of vertebral bodies from images of 6 patients, totaling 20 examples. The CNN is trained and tested by 10-fold cross-validation using the samples of the cross-validation set. The vertebral body images in the two sets are from different patients; one can remember that more that one image of vertebral body are extracted from the image of a patient’s spine. Separating the images into two sets was done in order to reproduce a scenario found in a hospital, where the ML-based system, trained using images of a set of patients, classifies images of a new patient.
(Raw) Images
The raw images from the central slice (2D) of the MRI for each vertebral body are used as inputs of the CNN. An example of a raw image is illustrated in the last picture of Fig. 1. CNNs have a large number of parameters to be optimized, making the use of large datasets desirable. As the number of images available in this work is small, the generation of an augmented dataset is also investigated. The Keras ImageDataGenerator Library was used to create new (raw) images for training the CNN. Transformation operations were applied to the original images, including translation, rotation, and mirroring. Flattening, elongation, and brightness changes were not used because they alter characteristics relevant to the diagnosis of VCFs. “Results” shows experiments with and without data augmentation.
Features from Clinical and Image Histogram Information
The additional features from clinical information were extracted from the patients’ electronic medical records. The information from the patients are:
Patient’s age;
Patient’s gender;
Number corresponding to the vertebral body (L1 to L5).
The image histogram is also used to extract some additional features. The histogram indicates the number of pixels in each of the gray levels of the image. The histograms corresponding to the MRI of each patient may be divergent, which can make it difficult for the classifier to learn how to generalize the classification of the images. Thus, the following information is also extracted from the central slice of the original MRIs, that is, from the 2D image:
Maximum gray level value;
Average of the gray level values;
Minimum gray level value;
Standard deviation of the gray level values.
Radiomics Features
The radiomics features were extracted from the segmented images using the pyRadiomics Library. pyRadiomics is an open source library written in Python that allows to extract radiomics features from 3D medical images [18]. Here, pyRadiomics was applied to the segmented original 3D images. All features extracted from pyRadiomics were used. They are:
First-Order Statistics: 19 features;
Shape-based (3D): 14 features;
Gray Level Co-occurrence Matrix: 23 features;
Gray Level Size Zone Matrix: 16 features;
Gray Level Run Length Matrix: 16 features;
Neighboring Gray Tone Difference Matrix: 4 features;
Gray Level Dependence Matrix: 14 features.
Hybrid Systems Based on CNN for the Diagnosis of VCFs
We propose two variations of the hybrid system. In the first hybrid system (“Hybrid System (Optimized by Hand)”), hyper-parameters of the CNN are optimized by hand running preliminary experiments. Besides, all external features (“Features from Clinical and Image Histogram Information” and “Radiomics Features”) are employed. In the second hybrid system (“Hybrid System Optimizes by GA”), a GA is used for optimizing the hyper-parameters of the CNN and for external feature selection.
Hybrid System (Optimized by Hand)
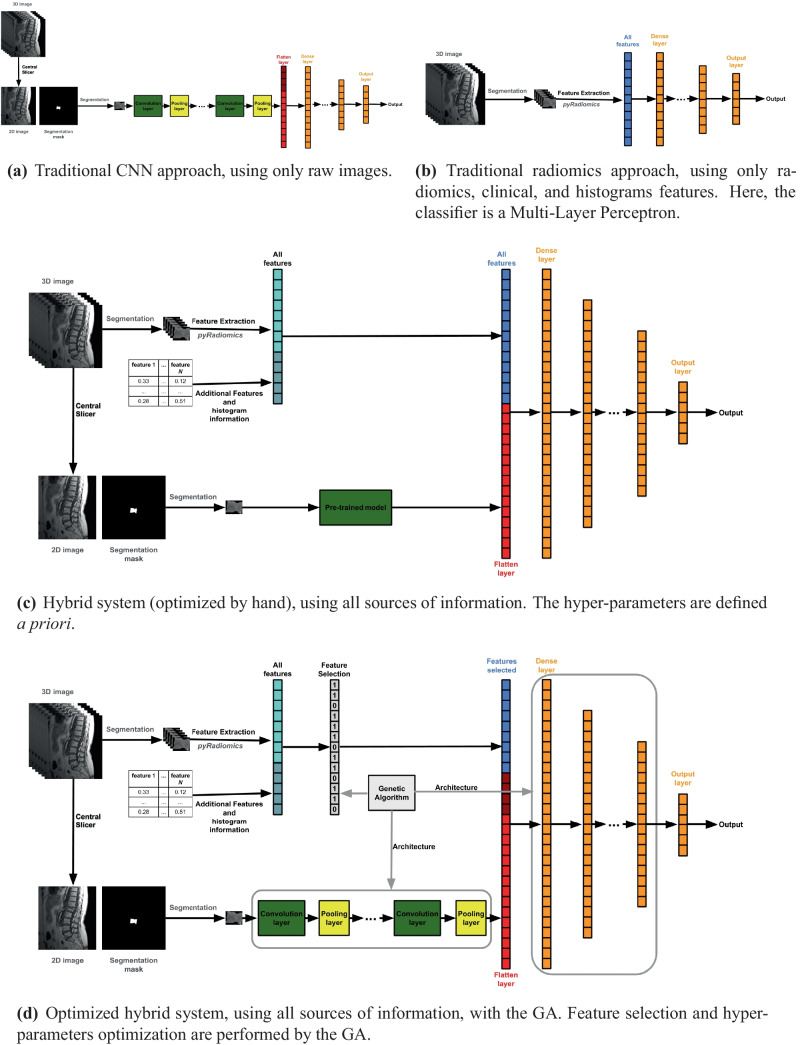
In the proposed system, three types of information are used by the CNN. The inputs of the first convolutional layer are the raw images (“(Raw) Images”), as in traditional CNNs. The inputs of the first dense layer of the CNN are given by a vector composed of radiomics (“Radiomics Features”), clinical, and histogram (“Features from Clinical and Image Histogram Information”) features, and outputs from the intermediate layers (convolutional and max-pooling layers). In practice, a CNN is built adding the external features to the first dense layer (Fig. 2c). The proposed system based on the CNN is called here hybrid system in order to distinguish it from the system based on a traditional CNN (Fig. 2a). The CNN is trained by Backpropagation. The last dense layer of the CNN is a Softmax layer. The outputs of a Softmax layer are related to the probabilities for each class in a multi-class problem. The predicted class corresponds to the class with the highest corresponding output.
Fig. 2.
Architectures of the models
In order to test the hypothesis that adding external features to the first dense layer of the CNN can improve classification, the hybrid system (Fig. 2c) is compared to two other approaches: i) a traditional CNN approach, where a CNN using only (raw) images is employed (Fig. 2a). ii) a traditional radiomics approach, where a Multi-Layer Perceptron (MLP) using only radiomics, histogram, and clinical features is employed (Fig. 2b). The approach for the MLP is similar to that one used by Pereira et al. [10], but adding histogram and clinical information to the inputs of the classifier. In the three models (Fig. 2a–c, preliminary experiments were run in order to find good hyper-parameters for the CNNs and MLP.
Pre-Trained CNNs
We also test whether using data augmentation and pre-trained CNNs can be beneficial for classifying VCFs. Here, pre-trained Keras models2 were considered. Three models were investigated in preliminary tests: Xception, InceptionV3 and VGG16. These models were chosen because they achieved some of the best performances in the ImageNet Competition3 and because they do not require too much computational power. Among the pre-trained models tested in the preliminary tests, VGG16 obtained the best performance.
We propose two pre-trained systems based on VGG16: i) The first one is based on the standard CNN model (Fig. 2a), i.e., where only the raw images are used as inputs; ii) The second one is based on the hybrid model (Fig. 2c), i.e., where the raw images are used as inputs of the first layer of the CNN, and the external information (radiomics, histogram, and clinical features) are added to the first dense layer. In both models, only the dense layers of the VGG16 are re-trained using the VCFs dataset. Both pre-trained models based on VGG16 are compared to other models in the experiments (“Results”).
Hybrid System Optimized by GA
We propose the optimization of the hyper-parameters of the CNN and the automatic feature selection by using a GA (Fig. 2d). GAs have been successfully employed in feature selection, that is an important task in ML [21, 22]. In the wrapper approach, each individual of the GA represents a subset of features; the chromosome is binary, where each element indicates the presence (1) or absence (0) of one feature. GAs have also been successfully applied for automatically designing CNN architectures for image classification [20] and for optimizing other learning hyper-parameters. The performance of a CNN is highly dependent on the settings of its hyper-parameters, and adjusting them manually is often a difficult task. When GAs are used for this task, each individual codifies in its chromosome a subset of values for the CNN’s hyper-parameters.
Here, each individual of the GA (candidate solution of the optimization problem) represents a subset of external features and hyper-parameters of the CNN4. The optimized CNNs can have up to 5 convolution layers (each one followed by a pooling layer) and up to 5 dense layers. Table 1 shows the data types and values that the elements in the chromosome of an individual of the GA can assume; a description of the meaning of each element is also presented. Each individual of the GA is evaluated by training and testing the corresponding model, i.e., the CNN with the hyper-parameters and external features defined by the individual. The cross-validation set is used for training and testing the CNNs in this step. The set is split in two, one for training (80%) and another for testing (20%). The evaluation (fitness) of the individual is the balanced accuracy for the test examples. Figure 3 shows the simplified flowchart for the GA.
Table 1.
List of CNN’s hyper-parameters and features optimized by the GA
| Parameter | Values | Type | Description |
|---|---|---|---|
| activation_cnn | [’relu’, ’tanh’] | string | Activation function to be used in the convolutional layers. |
| activation_dense | [’relu’, ’tanh’] | string | Activation function to be used in dense layers. |
| dropout | [0.0, 0.1, 0.2, 0.3, 0.4] | float | Dropout rate to be used in the layer. |
| epochs | [30, 45, 60, 90, 120, 150, 200] | integer | Number of iterations on which the model should be trained. |
| filters | [4, 5, 6, 7, 8, 9] | integer | Integer that defines the the number of filters in convolution layers. The number of filters is defined by the function: , where i is the value of the element (second column). |
| kernel_size | [2, 3, 4, 5, 6] | integer | Integer specifying the height and width of the 2D convolution window. The same value is used for both dimensions. |
| learning_rate | [0.0001, 0.001, 0.01, 0.05, 0.1] | float | Learning rate. |
| optimizer | [’adam’, ’sgd’] | string | Optimizer used for training the model. |
| padding | [’valid’, ’same’] | string | “valid” means no padding. “same” results in padding evenly to the left/right or up/down of the input such that the output has the same height/width dimension as the input. |
| batchNormalization | [’true’, ’false’] | Boolean | True: normalizes the activations of the previous layer, that is, applies a transformation that keeps the average activation close to 0 and the standard deviation of activation close to 1. False: does nothing. |
| max_pool | [’true’, ’false’] | Boolean | True: Applies the max-pooling operation. False: do nothing. |
| pool_size | [2, 3, 4, 5] | integer | Reducing factor (vertical, horizontal). The same window length is used for both dimensions. |
| scaler | [’standard’, ’minmax’] | string | Scaling method for the data. |
| units | [20, 50, 80, 100, 150, 200, 250, 500] | integer | Number of neurons in the dense layer. |
| feature | [’true’, ’false’] | Boolean | The corresponding feature is selected (True) or not (False). There is an element of the chromosome for each feature. |
| activate | [’true’, ’false’] | Boolean | The layer is added (True) to the model or not (False). |
Fig. 3.
GA used for feature selection and optimization of CNN’s hyper-parameters
The chromosomes of the GA are composed of elements with different data types (Table 1). In this way, different types of mutations are implemented. In the sequence, each type of mutation is presented.
Bit Flip Mutation The bit flip mutation is applied when the element of the chromosome is a Boolean. When bit flip mutation occurs, the bit is inverted.
Window Mutation The window mutation is applied for elements with integer or real data types. The window mutation consists of randomly choosing a value in a window with up to 4 neighbors. Table 2 shows examples of window mutation.
Table 2.
Examples of window mutation
| Position | Value | Possible values | Window values | Value after mutation |
|---|---|---|---|---|
| dropout | 0.1 | [0.0, 0.1, 0.2, 0.3, 0.4] | [0.0, 0.2, 0.3] | 0.2 |
| dropout | 0.2 | [0.0, 0.1, 0.2, 0.3, 0.4] | [0.0, 0.1, 0.3, 0.4] | 0.4 |
| epochs | 60 | [30, 45, 60, 90, 120, 150, 200] | [30, 45, 90, 120] | 90 |
| epochs | 30 | [30, 45, 60, 90, 120, 150, 200] | [45, 60] | 60 |
| epochs | 200 | [30, 45, 60, 90, 120, 150, 200] | [120, 150] | 120 |
Nominal Mutation: The nominal mutation is applied for elements with categorical data types. Because there is no order in this case, the element is uniformly randomly chosen. Table 3 shows examples of nominal mutation.
Table 3.
Examples of nominal mutation
| Position | Value | Possible values | Value after mutation |
|---|---|---|---|
| optimizer | ’adam’ | [’adam’, ’sgd’] | ’sgd’ |
| padding | ’same’ | [’valid’, ’same’] | ’valid’ |
Results
In this section, the experimental results of the proposed hybrid systems are presented. All the codes used in this work were developed in Python using the Keras API with the TensorFlow backend for implementing the ANNs5. In total, 6 models are compared:
MLP, using only radiomics, histogram, and clinical features (Fig. 2b);
Manually optimized CNN, using only (raw) images (Fig. 2a);
Manually optimized CNN, using only (raw) images, with data augmentation (same architecture showed in Fig. 2a, but with data augmentation);
Pre-trained CNN (“Pre-Trained CNNS”), using only (raw) images;
Hybrid model using pre-trained CNN (“Pre-Trained CNNS”), using all sources of information (Fig. 2c);
Optimized hybrid model (Fig. 2d), using all sources of information.
The model parameters were adjusted by hand interactively in initial experiments (not shown here)6. The architecture and parameters of the best model found in each approach during the cross-validation were selected, and a final training was carried out using the entire training set and evaluating the model in the test set. For the optimized hybrid model, the GA was run 5 times with different pseudo-random seeds. The population size was 30, and the number of generations is 50. Two-point crossover was employed. The crossover rate was 0.6, while the mutation rate was 0.02. Elitism was used, where the 2 best individuals of each generation are automatically selected to the next generation. Tournament selection was used to select the other individuals; the pool size in tournament selection was 3. The results are presented in the following. The results of the cross-validation and the test sets are shown considering the best model obtained in each approach7.
Model Using Only Radiomics, Histogram, and Clinical Features
MLP
The average balanced accuracy obtained by the best MLP model in the cross-validation set was 73.43%, while the best result for the test set was 58.8%. One can remember that the cross-validation and test sets are composed of vertebral bodies of different patients. Table 4 presents the results of additional metrics used to evaluate the classifier for the test set.
Table 4.
Results for the MLP
| Precision | Recall | F1-score | Support | Specificity | Sensitivity | |
|---|---|---|---|---|---|---|
| Benign | 0.33 | 0.17 | 0.22 | 6 | 0.85 | 0.16 |
| Malignant | 0.75 | 0.60 | 0.67 | 5 | 0.93 | 0.6 |
| Normal | 0.69 | 1.00 | 0.82 | 9 | 0.63 | 1.0 |
Models Using Only (Raw) Images
Manually Optimized CNN
The average balanced accuracy obtained by the best model found using only (raw) images as inputs, i.e., when traditional CNNs are employed, was 67.8% on the cross-validation set, while the best result for the test set was 77.3%. Table 5 presents the results of additional metrics used to evaluate the classifier for the test set.
Table 5.
Results for the CNN that uses only (raw) images as inputs
| Precision | Recall | F1-score | Support | Specificity | Sensitivity | |
|---|---|---|---|---|---|---|
| Benign | 0.75 | 0.50 | 0.60 | 6 | 0.91 | 0.5 |
| Malignant | 0.67 | 0.80 | 0.73 | 5 | 0.87 | 0.8 |
| Normal | 0.90 | 1.00 | 0.95 | 9 | 0.91 | 1.0 |
Manually Optimized CNN Using Data Augmentation
In this experiment, data augmentation was employed for artificially increasing the number of images in the cross-validation dataset. The CNN uses here only (raw) images as inputs. The average balanced accuracy obtained in the cross-validation set was 67.4%, while the best result for the test set was again 75.5%. Table 6 presents the results of additional metrics used to evaluate the classifier for the test set.
Table 6.
Results for the CNN that uses only (raw) images as inputs and data augmentation
| Precision | Recall | F1-score | Support | Specificity | Sensitivity | |
|---|---|---|---|---|---|---|
| Benign | 0.8 | 0.50 | 0.73 | 6 | 0.92 | 0.666 |
| Malignant | 0.75 | 0.60 | 0.67 | 5 | 0.92 | 0.6 |
| Normal | 0.82 | 1.00 | 0.90 | 9 | 0.84 | 1.0 |
Pre-Trained CNN
The average balanced accuracy obtained by the VGG16 using only (raw) images as inputs was 71.9% in the cross-validation set, while the best result for the test set was 82.96%. Table 7 presents the results of additional metrics used to evaluate the classifier for the test set.
Table 7.
Results for the VGG16 that uses only (raw) images as inputs
| Precision | Recall | F1-score | Support | Specificity | Sensitivity | |
|---|---|---|---|---|---|---|
| Benign | 0.75 | 1.00 | 0.86 | 6 | 0.85 | 1.0 |
| Malignant | 0.75 | 0.60 | 0.67 | 5 | 0.93 | 0.6 |
| Normal | 1.00 | 0.89 | 0.94 | 9 | 1.00 | 0.88 |
Proposed Hybrid Models (Using All Sources of Information)
Hybrid Model Using Pre-Trained CNN
The average balanced accuracy obtained by the hybrid model with the pre-trained VGG16 in the cross-validation set was 71.53%, while the balanced accuracy for the test set was 87.77%. Table 8 presents the results of additional metrics used to evaluate the classifier for the test set.
Table 8.
Results for the pre-trained hybrid model
| Precision | Recall | F1-score | Support | Specificity | Sensitivity | |
|---|---|---|---|---|---|---|
| Benign | 0.83 | 0.83 | 0.83 | 6 | 0.93 | 0.83 |
| Malignant | 0.80 | 0.80 | 0.80 | 5 | 0.93 | 0.80 |
| Normal | 1.00 | 1.00 | 1.00 | 9 | 1.00 | 1.00 |
Hybrid Model Optimized by the Genetic Algorithm
Table 9 shows the results obtained by the best individual from each of the runs of the GA. The best model for the cross-validation set (Run 3) selected 50 out of a total of 113 external features8. In the following, the detailed results for the test set obtained by the best model (Run 3) are presented.
Table 9.
Balanced accuracy for the best individual of each of the 5 executions of the GA for the cross-validation and test sets
| Run | Cross-validation set | Test set |
|---|---|---|
| 1 | 0.757 | 0.644 |
| 2 | 0.787 | 0.729 |
| 3 | 0.792 | 0.700 |
| 4 | 0.748 | 0.644 |
| 5 | 0.770 | 0.755 |
| avg | 0.770 | 0.694 |
| std | 0.018 | 0.049 |
The average balanced accuracy obtained in the cross-validation set was 79.25%, while the balanced accuracy obtained in the test set was 70.00%. Table 10 presents the results of additional metrics used to evaluate the classifier for the test set.
Table 10.
Results for the hybrid model optimized by the GA
| Precision | Recall | F1-score | Support | Specificity | Sensitivity | |
|---|---|---|---|---|---|---|
| Benign | 0.6 | 0.50 | 0.55 | 6 | 0.85 | 0.5 |
| Malignant | 1.00 | 0.60 | 0.75 | 5 | 1.00 | 0.6 |
| Normal | 0.75 | 1.00 | 0.86 | 9 | 0.72 | 1.00 |
Comparison of the Models with Different Sources of Information
Table 11 summarizes results obtained by all models. Table 12 shows the accuracy results for each fold of the cross-validation set, considering 10-fold stratified cross-validation. After verifying that normality and variance criteria were satisfied for these results, the Student’s t-test with a 5% significance level was applied. The results of the CNN manually optimized were statistically compared to the results of the other models. No significant difference was observed between the CNN models with and without data augmentation, both using only (raw) images as inputs. The results for the optimized hybrid model were better, with significant difference, than the results for the CNN that uses only (raw) images. Table 13 shows the model with better results for sensitivity and specificity considering each class for the test set.
Table 11.
Summary of the results for all models. The best results are in bold
| Metric | Set | MLP | CNN manually optimized | CNN manually optimized with data augmentation | Pre-trained CNN | Hybrid model using pre-trained CNN | Hybrid model optimized by the GA |
|---|---|---|---|---|---|---|---|
| Balanced accuracy (Cross-validation) | cross-validation | 73.4% | 67.8% | 67.4% | 71.9% | 71.5% | 79.2% |
| Balanced accuracy | Test | 58.8% | 77.3% | 75.5% | 82.9% | 87.7% | 70.0% |
| F1-Score | Test | 57% | 76% | 76.6% | 76.6% | 87.6% | 70% |
| Specificity | Test | 80.3% | 89.6% | 89.3% | 92.6% | 95.3% | 85.6% |
| Sensitivity | Test | 58.6% | 76.6% | 75.3% | 82.6% | 87.6% | 70% |
| Macro-average ROC | Test | 95% | 94% | 90% | 97% | 98% | 92% |
Table 12.
Balanced accuracy obtained for each fold of 10-fold stratified cross-validation to separate the Benign x Malignant x Normal classes. The letter S indicates that the differences are statistically significant, considering the test t, when the corresponding model is compared with the reference model. The symbols + and -, respectively, mean that average results obtained by the model were better or worse than the average results of the reference model
| Fold | MLP | CNN manually optimized | CNN manually optimized with data augmentation | Pre-trained CNN | Hybrid model with pre-trained CNN | Hybrid model optimized by the GA |
|---|---|---|---|---|---|---|
| 1 | 0.666 | 0.640 | 0.656 | 0.944 | 0.833 | 0.833 |
| 2 | 0.411 | 0.697 | 0.669 | 0.705 | 0.588 | 0.705 |
| 3 | 0.882 | 0.676 | 0.686 | 0.647 | 0.764 | 0.882 |
| 4 | 0.941 | 0.650 | 0.728 | 0.647 | 0.882 | 0.764 |
| 5 | 0.764 | 0.698 | 0.703 | 0.941 | 0.823 | 0.823 |
| 6 | 0.823 | 0.705 | 0.628 | 0.470 | 0.823 | 0.882 |
| 7 | 0.529 | 0.626 | 0.686 | 0.882 | 0.529 | 0.823 |
| 8 | 0.823 | 0.698 | 0.644 | 0.705 | 0.470 | 0.647 |
| 9 | 0.812 | 0.681 | 0.680 | 0.750 | 0.812 | 0.750 |
| 10 | 0.687 | 0.662 | 0.662 | 0.500 | 0.625 | 0.812 |
| avg | 0.734 (+) | 0.678 | 0.674 (-) | 0.719 (+) | 0.715 (+) | 0.792 (S+) |
| std | 0.163 | 0.029 | 0.017 | 0.166 | 0.147 | 0.075 |
Table 13.
Models with better results for sensitivity and specificity for each class
| Specificity | Sensitivity | |
|---|---|---|
| Benign | Hybrid model with pre-trained CNN (0.93) | Pre-trained CNN (1.0) |
| Malignant | Hybrid model optimized by the GA (1.0) | CNN manually optimized, Hybrid model with pre-trained CNN (0.8) |
| Normal | Pre-trained CNN Hybrid model with pre-trained CNN (1.0) | MLP, CNN manually optimized, CNN manually optimized with data augmentation, Hybrid model with pre-trained CNN, Hybrid model optimized by the GA (1.0) |
Discussion
Some observations can be made about the results presented in “Results”.
Impact of Data Augmentation
First, CNN’s accuracy with data augmentation (“Manually Optimized CNN Using Data Augmentation”) was slightly worse than CNN’s accuracy without data augmentation (“Manually Optimized CNN”). In the experiments for both models, only (raw) images were used as inputs. The model without data augmentation obtained better accuracy for the cross-validation and the test sets (Table 11). However, the CNN with data augmentation was slightly better for the F1-Score. In data augmentation, artificial examples were generated by modifying the original images using pre-defined transformations, e.g., rotation. The addition of the artificial examples did not significantly improve the generalization of the CNN model. This can be explained because the original images have similar properties, e.g., orientation of the vertebral bodies in the images is similar. In this way, the transformations used to augment the dataset do not add relevant information to help generalize the classification made by the CNN. In addition, the average time to run the model with data augmentation is longer: 6 min for the model without data augmentation against 13 min for the model with data augmentation.
Impact of Using Pre-Trained Architectures
The pre-trained models (“Pre-Trained CNNS”) obtained better results for the test set than the respective models without pre-training. When the hybrid models with and without pre-training are compared (last two columns of Table 11), the hybrid pre-trained CNN obtained better results for all measures when the test set is considered. When the CNN manually optimized is compared to the pre-trained CNN (4 and 6 columns of Table 11), the results for the cross-validation set are also better. The dataset originally used to train the pre-trained CNN (VGG16) is composed of a large set of different types of images. The VGG16 discovered useful filters for classifying images by using a large generic dataset. When re-trained with the VCF dataset here, these features showed to be useful for classifying the vertebral bodies. Besides, pre-training a CNN requires much less computational effort than training the same CNN from scratch. Finally, the VCF dataset used here is small, which impacts in the generalization power of the ANNs trained from scratch.
Impact of Using all Sources of Information
Table 11 shows that adding radiomics, clinical, and histogram features to the first dense layer improves the performance of the CNN for the cross-validation set. The pre-trained CNN trained with (raw) images (6 column) obtained slight better results for the cross-validation set than the proposed hybrid model using pre-trained CNN (7 column). However, the hybrid model proposed using pre-trained CNN obtained better results for the test set. The hybrid model using the pre-trained CNN also obtained better results for the test set compared to the MLP (3 column) and the manually optimized CNN (4 column). The MLP uses only external features, while the standard CNN uses only (raw) images.
It is important to observe that the radiomics features were extracted from 3D MRI using the pyRadiomics library. As a consequence, the features vector may contain relevant features present only in the 3D images. In addition, the vector contains shape and texture features similar to the characteristics analyzed by the radiologist when classifying benign and malignant VCFs. Clinical and histogram information can be useful too. The pre-trained CNN obtained an accuracy of 75% for the benign and malignant classes (Table 7), while the hybrid model obtained an accuracy of 83% for the benign class and 80% for the malignant class (Table 8). Thus, the traditional CNN, which uses only (raw) images as input, obtained good overall results, being able to detect the presence of VCFs, i.e., differentiating normal vertebral bodies from fractured bodies. However, it performed worse when differentiating malignant VCFs from benign VCFs compared to the hybrid model using all sources of information.
Impact of Using GA to Select Features and CNN’s Hyper-Parameters
The hybrid model optimized by the GA (“Hybrid Model Optimized by the Genetic Algorithm”) obtained the better balanced accuracy for the cross-validation set (Table 11). Besides, there is statistically difference from the results obtained by the CNN optimized manually (Table 12). The results show that the GA can find good hyper-parameters and select relevant features for obtaining good CNNs.
The features selected by the GA can be analyzed and later used in clinical practice. The age feature (from clinical information) was selected in all runs of the GA. The vertebral body feature was selected in 3 out of 5 runs, while the sex feature was selected in only 1 run. Regarding the features extracted from the image histogram, max was selected in 4 runs, while mean, min, and STD were selected in 3 runs. Therefore, the additional features proved to be relevant to the problem studied here. Regarding gray level intensity features, firstorder RobustMeanAbsoluteDeviation was selected in all runs. Two shape features (shape Compactness1 and shape Maximum2DDiameterRow), three gray intensity features (firstorder Energy, firstorder InterquartileRange and firstorder Skewness), and seven texture features ( gldm DependenceVariance, glrlm ShortRunEmphasis, glrlm RunEntropy, glszm ZoneEntropy, glszm LargeAreaEmphasis, glszm HighGrayLevelZoneEmphasis and glszm LargeAreaLowGrayLevelEmphasis) were selected in 4 runs9.
Despite the better results for the cross-validation set, the optimized hybrid model obtained worse results for the test set when compared to the hybrid model using pre-trained CNNs. As the dataset used in this work is small, it can lead to a difficulty in generalizing the model, that is, the best attributes found by the GA in the training set may not represent the best attributes for the test set. Therefore, the better performance of the hybrid model using pre-trained CNNs in the test set is explained by the use of a much larger CNN and the use of filters discovered when the model was trained in a large set containing different types of images (“Impact of Using Pre-Trained Architectures”). However, deeper CNNs demands more time and computational and energy power for operation and training. In this way, an advantage of using the GA is that smaller CNNs, with similar or better performance than larger CNNs optimized by hand, can be obtained by the optimization process.
From Tables 11 and 13, one can observe that the pre-trained CNN obtained the best sensitivity for the benign class, while the hybrid model with pre-trained CNN obtained the best specificity. For the malignant class, the model with best specificity was the hybrid model optimized by the GA, while the best sensitivity was obtained by the CNN optimized manually and the hybrid model using pre-trained CNN. Regarding the normal class, the models with best specificity were the pre-trained CNN and the hybrid models. The best sensitivity for the normal class was obtained by the MLP, manually optimized CNNs, and hybrid models.
Conclusion
In this work, we propose adding radiomics, clinical, and histogram information to the first dense layer of a CNN used for classifying VCFs in MRIs. The input of the first layer of the CNN is the (raw) image. Two hybrid models are proposed: i) one where pre-trained CNNs are used; ii) one where GAs are used to select radiomics, clinical, and histogram features and optimize the CNN’s hyper-parameters.
The hybrid model optimized by the GA obtained better accuracy results for the cross-validation set than: i) a model in the standard radiomics approach, where an MLP using only radiomics, clinical, and histogram features is used; ii) a model in the standard CNN approach, where a CNN using only raw images are used as inputs; iii) a CNN approach (same as ii) with data augmentation; iv) a pre-trained CNN, where a CNN using only raw images are used as inputs. By using the GA, smaller CNNs were obtained. Besides, features selected by the GA can be important in order to find relevant characteristics in the classification of VCFs in clinical practice.
Regarding the test set, the best results were obtained by the hybrid model with the pre-trained CNN. Using pre-trained CNNs allowed to use larger CNNs. Besides, the use of filters discovered when the model was trained in a large set containing different types of images was useful because the VCF dataset is small. The experimental results indicate that the hybrid models are promising. The strategy of combining several sources of information is similar to that one adopted by a specialist in the clinical practice. The methodology proposed in this work is generic and can be applied to other problems related to the analysis of medical images.
The system proposed here is limited to using images of isolated vertebral bodies as input, while a radiologist uses the entire image, allowing her/him to compare the vertebral body with other vertebral bodies of the same image, as well as reviewing all available medical data. Another limitation is due to the fact that models that use radiomics information need 3D images (MRIs) to extract the radiomics features using the pyRadiomics library. The use of information from neighboring vertebral bodies to classify a given sample, as well as using the maximum amount of information from the patient’s medical record, is a relevant future work. Another way to combine information from different sources is to use sets of trained classifiers with different information. Other strategies can also be used to combine radiographic resources, clinical information and (raw) images. Finally, using GAs to optimize the pre-trained model is another relevant future work.
Supplementary Information
Below is the link to the electronic supplementary material.
Author Contributions
R. Del-Lama contributed to conceptualization, methodology, software, validation, investigation, writing (original draft, review, editing) and formal analysis. R. Candido, N. Chiari-Correia contributed to resources, software, data curation and writing (review and editing). M. Nogueira-Barbosa, P. Azevedo-Marques contributed to resources, data curation, validation and writing (review and editing). R. Tinós contributed to conceptualization, methodology, supervision, formal analysis, validation and writing (original draft, review and editing).
Funding
This work was partially supported by São Paulo Research Foundation - FAPESP (under grants #2019/01219-2, #2019/07665-4, and, #2021/09720-2) and National Council for Scientific and Technological Development - CNPq (under grant 305755/2018-8).
Data Availability
The anonymized spine MRI dataset is public and available for research purposes.
Code Availability
The source codes for the proposed hybrid systems are publicly available on GitHub (https://github.com/rafaelsdellama/nnga).
Declarations
The anonymized spine MRI dataset is public and available for research purposes. Approval was obtained from the ethics committee of the University Hospital. The procedures used in this study adhere to the tenets of the Declaration of Helsinki.
Ethics Approval
Approval (Number: 1.951.052) was obtained from the ethics committee of the Ribeirão Preto Medical School, University of São Paulo (CAAE: 64951317.0.0000.5440). The procedures used in this study adhere to the tenets of the Declaration of Helsinki. The anonymized spine MRI dataset is public and available for research purposes. The dataset with segmented images was used previously in other works [9, 10, 16].
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Conflict of Interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Footnotes
Hospital das Clínicas, Ribeirão Preto Medical School, University of São Paulo.
The ImageNet project is a large database of images, with more than 14 million images of more than 20,000 categories.
Figure S1 of the supplementary materials file shows how the hyper-parameters of the CNN and external features are codified in the chromosome of an individual of the GA.
The source codes for the proposed hybrid systems are freely available on GitHub (https://github.com/rafaelsdellama/nnga).
The architecture and parameters of the best model found in each approach are available in Tables S14–S18 of the supplementary materials. The average time for each model is available in Table S13. The description of the hardware used to perform this work is found in Chapter 3 of the supplementary material.
ROC Curves, precision results, and confusion matrices are presented in the supplementary materials (Tables S1–S12 and Figs. S2–S7).
The features selected in each run of the GA are presented in Tables S19 and S20 of the supplementary materials.
The description of all features extracted by pyRadiomics is available at: https://pyradiomics.readthedocs.io/en/latest/features.html.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.J. Tehranzadeh and C. Tao, “Advances in mr imaging of vertebral collapse,” in Seminars in Ultrasound, CT and MRI, 25(6), 440–460, Elsevier (2004). [DOI] [PubMed]
- 2.Jung H-S, Jee W-H, McCauley TR, et al. Discrimination of metastatic from acute osteoporotic compression spinal fractures with mr imaging1. Radiographics. 2003;23(1):179–187. doi: 10.1148/rg.231025043. [DOI] [PubMed] [Google Scholar]
- 3.Cuenod CA, Laredo J-D, Chevret S, et al. Acute vertebral collapse due to osteoporosis or malignancy: Appearance on unenhanced and gadolinium-enhanced mr images. Radiology. 1996;199(2):541–549. doi: 10.1148/radiology.199.2.8668809. [DOI] [PubMed] [Google Scholar]
- 4.Shen D, Wu G, Suk H-I. Deep learning in medical image analysis. Annual review of biomedical engineering. 2017;19:221–248. doi: 10.1146/annurev-bioeng-071516-044442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan H-P, Hadjiiski LM, Samala RK. Computer-aided diagnosis in the era of deep learning. Medical Physics. 2020;47(5):e218–e227. doi: 10.1002/mp.13764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ker J, Wang L, Rao J, et al. Deep learning applications in medical image analysis. Ieee Access. 2017;6:9375–9389. doi: 10.1109/ACCESS.2017.2788044. [DOI] [Google Scholar]
- 7.Greenspan H, Van Ginneken B, Summers RM. Guest editorial deep learning in medical imaging: Overview and future promise of an exciting new technique. IEEE Transactions on Medical Imaging. 2016;35(5):1153–1159. doi: 10.1109/TMI.2016.2553401. [DOI] [Google Scholar]
- 8.Litjens G, Kooi T, Bejnordi BE, et al. A survey on deep learning in medical image analysis. Medical image analysis. 2017;42:60–88. doi: 10.1016/j.media.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 9.L. Frighetto-Pereira, R. Menezes-Reis, G. A. Metzner, et al., “Classification of vertebral compression fractures in magnetic resonance images using shape analysis,” in 2015 E-Health and Bioengineering Conference (EHB), 1–4, IEEE (2015). [DOI] [PubMed]
- 10.Frighetto-Pereira L, Rangayyan RM, Metzner GA, et al. Shape, texture and statistical features for classification of benign and malignant vertebral compression fractures in magnetic resonance images. Computers in Biology and Medicine. 2016;73:147–156. doi: 10.1016/j.compbiomed.2016.04.006. [DOI] [PubMed] [Google Scholar]
- 11.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521(7553):436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 12.Genant HK, Wu CY, Van Kuijk C, et al. Vertebral fracture assessment using a semiquantitative technique. Journal of bone and mineral research. 1993;8(9):1137–1148. doi: 10.1002/jbmr.5650080915. [DOI] [PubMed] [Google Scholar]
- 13.Kasai S, Li F, Shiraishi J, et al. Computerized detection of vertebral compression fractures on lateral chest radiographs: Preliminary results with a tool for early detection of osteoporosis. Medical Physics. 2006;33(12):4664–4674. doi: 10.1118/1.2364053. [DOI] [PubMed] [Google Scholar]
- 14.E. Ribeiro, M. H. Nogueira-Barbosa, R. Rangayyan, et al., “Detection of vertebral compression fractures in lateral lumbar x-ray images,” in XXIII Congresso Brasileiro em Engenharia Biomédica (CBEB), 1–4 (2012).
- 15.S. Ghosh, A. Raja’S, V. Chaudhary, et al., “Automatic lumbar vertebra segmentation from clinical ct for wedge compression fracture diagnosis,” in Medical Imaging 2011: Computer-Aided Diagnosis, 7963, 796303, International Society for Optics and Photonics (2011).
- 16.P. M. Azevedo-Marques, H. Spagnoli, L. Frighetto-Pereira, et al., “Classification of vertebral compression fractures in magnetic resonance images using spectral and fractal analysis,” in 2015 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), 723–726, IEEE (2015). [DOI] [PubMed]
- 17.Kumar V, Gu Y, Basu S, et al. Radiomics: the process and the challenges. Magnetic resonance imaging. 2012;30(9):1234–1248. doi: 10.1016/j.mri.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Griethuysen JJ, Fedorov A, Parmar C, et al. Computational radiomics system to decode the radiographic phenotype. Cancer research. 2017;77(21):e104–e107. doi: 10.1158/0008-5472.CAN-17-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dimitrovski I, Kocev D, Kitanovski I, et al. Improved medical image modality classification using a combination of visual and textual features. Computerized Medical Imaging and Graphics. 2015;39:14–26. doi: 10.1016/j.compmedimag.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 20.Sun Y, Xue B, Zhang M, et al. Automatically designing cnn architectures using the genetic algorithm for image classification. IEEE Transactions on Cybernetics. 2020;50(9):3840–3854. doi: 10.1109/TCYB.2020.2983860. [DOI] [PubMed] [Google Scholar]
- 21.Xue B, Zhang M, Browne WN, et al. A survey on evolutionary computation approaches to feature selection. IEEE Transactions on Evolutionary Computation. 2015;20(4):606–626. doi: 10.1109/TEVC.2015.2504420. [DOI] [Google Scholar]
- 22.Tinós R. Artificial neural network based crossover for evolutionary algorithms. Applied Soft Computing. 2020;95:106512. doi: 10.1016/j.asoc.2020.106512. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The anonymized spine MRI dataset is public and available for research purposes.
The source codes for the proposed hybrid systems are publicly available on GitHub (https://github.com/rafaelsdellama/nnga).