Abstract
The segmentation of the lesion region in gastroscopic images is highly important for the detection and treatment of early gastric cancer. This paper proposes a novel approach for gastric lesion segmentation by using generative adversarial training. First, a segmentation network is designed to generate accurate segmentation masks for gastric lesions. The proposed segmentation network adds residual blocks to the encoding and decoding path of U-Net. The cascaded dilated convolution is also added at the bottleneck of U-Net. The residual connection promotes information propagation, while dilated convolution integrates multi-scale context information. Meanwhile, a discriminator is used to distinguish the generated and real segmentation masks. The proposed discriminator is a Markov discriminator (Patch-GAN), which discriminates each matrix in the image. In the process of network training, the adversary training mechanism is used to iteratively optimize the generator and the discriminator until they converge at the same time. The experimental results show that the dice, accuracy, and recall are 86.6%, 91.9%, and 87.3%, respectively. These metrics are significantly better than the existing models, which proves the effectiveness of this method and can meet the needs of clinical diagnosis and treatment.
Keywords: Lesion segmentation, Deep learning, U-Net, Generative adversarial networks
Introduction
According to the latest cancer data released by the International Cancer Research Institute (IARC) in 2020, gastric cancer has become the fifth most common cancer worldwide with one million and ninety thousand cases [1]. Among the new cancer cases in China, gastric cancer is next to lung cancer and colorectal cancer, ranking third with 886,000 cases [1]. The early diagnosis of gastric cancer will contribute to decreasing incidence rate and mortality, which relies on the timely detection and correct judgment of the lesion region under endoscopy. Although it is a standard procedure, the detection accuracy of the traditional endoscope is only between 73.7 and 83.6% [2]. The reasons include mild early symptoms of gastric cancer and the heavy workload of manual analysis [3]. Therefore, researchers try to develop a method of lesion detection based on gastroscopic images to assist physicians clinically. In some recent studies, Sainju et al. [4] and Yeh et al. [5] extracted the features of gastroscope images by analyzing the color attributes. Li et al. [6, 7] used the obvious texture change characteristics of gastric cancer lesions in gastroscope images to extract texture features for lesion recognition. These methods rely on low-level feature extraction and therefore cannot learn the high-level semantic information in the image, which limits the recognition of early gastric cancer lesions.
The application of deep learning to gastroscopy image processing enables automatic annotation and extraction of lesions in images for disease recognition and diagnosis. Sakai et al. [8] proposed a high-precision automatic detection model based on the convolutional neural network (CNN) to enhance the diagnostic ability of endoscopic experts. Yoon et al. [9] used the visual geometry group network (VGG-16) [10] model to classify endoscopic images into early gastric cancer or non-early gastric cancer. Shibata et al. [11] used the mask region-based convolutional neural network (R-CNN) instance segmentation method to segment the gastroscopic images in order to obtain the boundary and label images of the gastric cancer region. Wang R et al. [12] used the fast R-CNN method to detect gastric polyps in gastroscopic images and replaced the region of interest regions (ROI) pool operation with ROI alignment operation. Cao et al. [13] proposed a feature extraction and fusion module to solve the problem that small polyps in gastroscopic images are difficult to detect from the background, and combined it with the YOLOv3 [14] network to reduce the risk of false detection and missed detection in gastroscopy. Zhu et al. [15] used ResNet to extract and classify features from gastroscopy images. The final results were used to predict the stage of gastric cancer lesions. Compared with the traditional feature extraction, the deep learning method achieves a better recognition effect, but the result is still unsatisfactory. The reason is that there are great differences in texture, lesion shape, and size among gastroscopic images. This prompted us to design a more appropriate network structure to obtain higher segmentation performance.
To develop an advanced segmentation model, we improved and expanded the U-Net [16] structure. U-Net is an enhanced model based on a full convolution neural network (FCN) [17], which can support the training model with a small amount of data and performs well in medical image segmentation. With the improvement of medical image segmentation performance requirements, many researchers have expanded the U-Net structure [18–22]. However, a common problem is the disappearance or explosion of gradients during training. In this paper, we add a residual connection [23] to the encoding and decoding parts of U-Net, which is designed to solve the problem, and further improves the convergence speed of the segmentation network. The bottleneck of U-Net, the middle part of the encoding path and decoding path, is another important structure as it mainly receives all the feature information extracted from the encoder and restores the segmented image to the original resolution through the decoder [24]. We add dilated convolution [25] at the bottleneck of U-Net to solve the problem of spatial information loss caused by continuous pooling and upsampling of the U-Net. Meanwhile, in order to capture the information of different scales in the image, we cascade multiple dilated convolution groups with different dilation rates at the bottleneck and take the results as the input of the next stage.
In recent years, the adversarial training idea of generative adversarial networks (GAN) [26] has been introduced to semantic segmentation tasks. Some studies have shown that the application of generative adversarial networks to medical image segmentation can obtain better results [27–31]. Inspired by the above researchers, the paper optimizes the proposed segmentation model by adversarial training. We use the improved segmentation network based on U-Net to replace the generator in GAN. Besides, we use Patch-GAN [32] to replace the discriminator in GAN. The discriminator is used as a loss function to improve the similarity between the segmentation result and the ground truth. In the process of confrontation training, the discriminator takes the segmentation result as the input and the original gastroscopic image as the condition constraint. Through adversarial training, the segmentation results are constantly close to the real results, thereby improving the accuracy of the segmentation. In general, this work focuses on the following points.
This paper proposes a model to segment the lesions in gastroscopic images by using the confrontation training method of GAN.
The method designs a new segmentation network, which adds a residual connection to the encoding and decoding part of U-Net, while introduces cascaded dilated convolutions at the bottleneck of U-Net.
We use the improved segmentation network as the generator in the GAN model and set the discriminator in GAN as Patch-GAN structure.
Finally, the work evaluates and presents the segmentation results. Experimental results show that the model can accurately segment the lesion area in the gastroscopic image, and the segmentation result is better than other models.
Methods
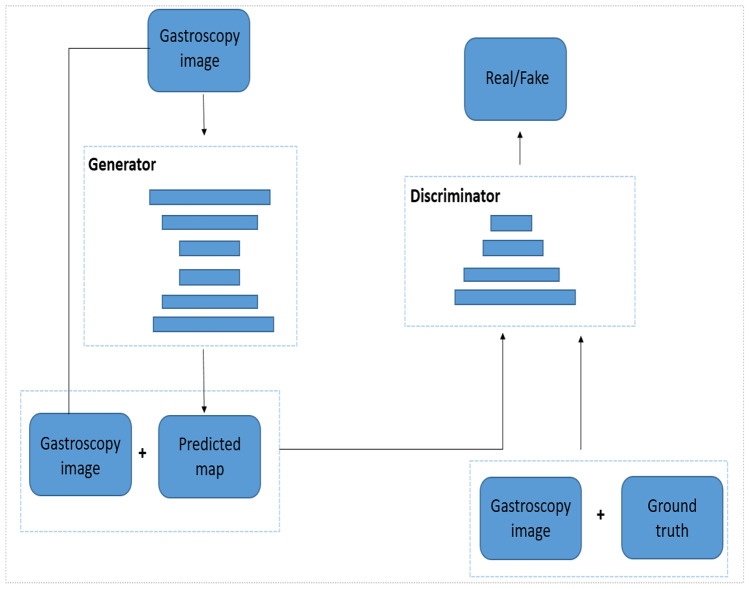
The overall framework of the proposed model is shown in Fig. 1. The proposed model consists of a generator and a discriminator. The generator is a segmentation network based on U-Net. We input the gastroscopic image into the segmentation network, and the output is the segmentation result of the lesion area. The discriminator is a Patch-GAN model that receives two sets of inputs: one is a four-channel tensor of the original gastroscopic image and the segmentation result image of the generator; the other is a four-channel tensor of the manual label and the original gastroscopic image. The output of the discriminator is a matrix. Each element in the matrix represents the true or false judgment of a patch size receptive field in the original gastroscopic image.
Fig. 1.
The overall framework of the proposed model
Generator
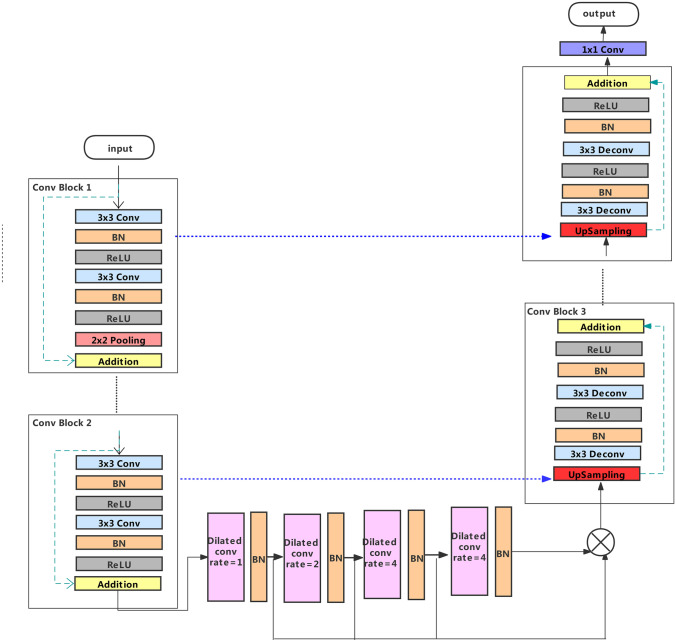
In the U-Net network, the function of the encoding path is to obtain context information. The decoding path is used for accurate positioning, and the jump connection can integrate all levels of information. To make the model have better nonlinear expression ability, the encoder path on the left is composed of five groups of convolution units. The structure of the first four groups of convolution units is the same. Each convolution unit uses two convolutional layers with kernel size and a stride of 1 for feature extraction. After each convolution layer, there are a batch normalization layer (BN) and correction rectified linear unit (ReLU) activation functions. After that, a max-pooling layer with filter size is used to halve the size of the feature map. In this work, the residual connection is added to the convolution unit. We add the input feature map and the final output feature map of each convolution unit to get the combination of different layer features. Adding the input features to the output features can reuse the features and eliminate the gradient disappearance to a certain extent. In the fifth convolution unit, we remove the max-pooling layer to keep the size of the feature map unchanged, so that the corresponding layers of the encoder and decoder subnets can achieve skip-connection of the same scale.
The bottleneck is the key part of U-Net. In this paper, four dilated convolution layers are added in the middle of the bottleneck. These convolutions make the receptive field control arbitrarily without reducing the image resolution. To prevent the “grid effect” in the dilated convolution, we use the dilation rate of 1 in the first layer. The dilation rate of the second layer is increased by 2 times based on the first layer, while the dilation rate of the third and fourth layers is increased by 2 times based on the second layer. In order to capture multi-scale information in gastroscopic images, we cascade four dilated convolutions with different dilation rates, and use the final result as the input of the next stage.
The decoding path of the proposed segmentation network consists of four groups of convolution units. Similar to the encoding path, the unit contains two convolution layers. After the convolution layer is a BN and a ReLU. However, the decoding path needs to upsample the feature map from a lower level before each unit to restore the image size, and it needs to connect with the feature map from the corresponding encoding path. After the last stage of the decoding path, a convolution layer with filter size and a sigmoid activation function is used to map the multi-channel features into the desired segmentation. The general structure of the generator is shown in Fig. 2.
Fig. 2.
Schematic of generator
Discriminator
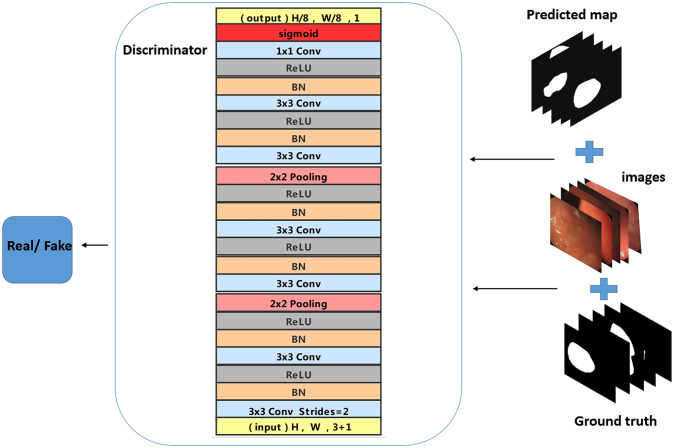
Considering the influence of different parts of gastroscopic images on the judgment of lesion area, the proposed discriminant network is a Patch-GAN. The discriminator in GAN maps the input to a probability value of the true sample, while Patch-GAN maps the input image to a matrix C of N × N. The value of represents the probability that each patch is a true sample, and the mean value of is the final result of the discriminator. We can trace a certain position in the original image from the feature matrix C. The discriminator makes every part of the original image influence the final output.
In this work, Patch-GAN is composed of three groups of convolution units. The first two groups of convolution units include two convolutional layers with kernel size and stride of 1 for feature extraction. After each convolution layer, there are BN, ReLU, and a max-pooling layer. We remove the max-pooling layer in the last convolution. After three convolution units, there is a convolution layer with kernel size and a sigmoid activation layer. Patch-GAN outputs a matrix with the size of The mean value of the elements in the matrix is the final judgment result. The structure of the discriminator is shown in Fig. 3.
Fig. 3.
Structure of discriminator. The input of the discriminator is the data that the original gastroscopic image and the real label are spliced together or the data that the gastroscopic image and the generated prediction label are spliced together. The function of the original gastroscopic image is to judge the input conditions. The discriminator tries to distinguish real data from false data
Loss Function
The proposed model is a conditional GAN (cGAN) [33], so the input is no longer limited by random noise. The loss of the original GAN is a mutation function, which cannot indicate the quality of network training. In this work, the loss of cGAN is used to train the segmentation network. The original gastroscopic image is input into the discriminator network as an additional condition of the model. and represent the original gastroscopic image data and the lesion label image data, respectively. The mapping of the generator G is expressed as follows: →, and the mapping of the discriminator D is expressed as {, }→ 0 and 1 indicate whether x is the lesion segmentation image generated by the generator or a manually labeled lesion segmentation image, respectively. N represents the size of the output matrix of Patch-GAN, where . The formula is as follows:
| 1 |
In this work, the generator and discriminator have been trained alternately. The parameters of the generator are fixed when training the discriminator. The discriminator tries to judge the real segmentation map as true and the segmentation map generated by the generator as false. Therefore, the parameters of the discriminator are updated to maximize D (,) and minimize D (, G ()). When training the generator, the parameters of the discriminator are fixed. In order to prevent the discriminator from making correct judgments, the parameters of the generator are updated to maximize D (, G ()). Therefore, the definition of the objective function is shown in Eq. (2).
| 2 |
The segmentation of lesions in gastroscopic images can be regarded as a pixel-level binary classification problem, so the selection of loss function can be regarded as the selection of loss function in a binary classification task. The binary cross-entropy loss function is commonly used in a binary classification task. In this work, the binary cross-entropy loss function is used to measure the distance of the lesion area between the real label and the predicted label, as shown in Eq. (3), where G() represents the predicted label and represents the real label.
| 3 |
The final loss function is obtained by adding the generated confrontation loss and partition loss, as shown in Eq. (4), where γ is the weight coefficient.
| 4 |
Dataset and Evaluation Metrics
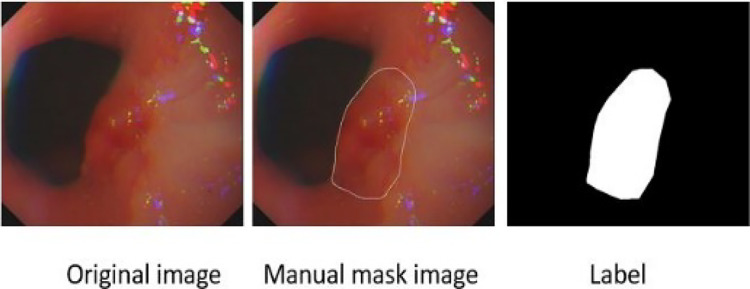
The gastroscopic image dataset used in this work is from the digestive endoscopy center of General Hospital of the People’s Liberation Army. Some poor-quality images are removed in the experiment. We finally select 630 pairs of original gastroscopic images and corresponding lesion annotation images. We use Labelme to further process the dataset, and transform the lesion annotation map into a real label, as shown in Fig. 4.
Fig. 4.
Illustration of the dataset
The format of gastroscopic images in the dataset is JPG. We randomly select 100 images for testing, and the rest 530 images are used for network training. Considering that in the real scene, different endoscopic procedures for color calibration and camera movement lead to differences between gastroscopic images, we need to preprocess the images. In this paper, each channel of the image is processed by zero-mean. The normalized data can speed up the convergence speed of training and improve the segmentation accuracy. The image size for this dataset is inconsistent, so we resize its images to . In addition, we enhanced the training data in the experiment to prevent over-fitting. The training data is expanded by flipping the image horizontally or vertically, rotating at any angle from 0 to 360°, and randomly enhancing the image chroma between 0.8 and 1.2. The final training data have 9360 images. We randomly select 5% images from the enhanced training data to form the validation dataset, and the number of images in the remaining training data is 8892.
We use Tensorflow as the backend Keras framework to build the environment, and use GTX3080 GPU to train discriminator and generator alternately. A total of 150 epochs are set in the experiment. The Adam optimizer was used to update the network weights and bias on a mini-batch size of 4, using a fixed learning rate of . We set the weight coefficient in Eq. (4) to 10. We evaluate the segmentation performance of the proposed method on 100 test images by seven metrics, namely dice, accuracy, recall, specificity, IOU, PR curve, and ROC curve, respectively. Evaluation metrics are defined as follows:
| 5 |
| 6 |
| 7 |
| 8 |
| 9 |
where TP, TN, FP, and FN show the true positive, true negative, false positive, and false negative samples, respectively.
Experimental Results and Analysis
We study the effect of adding a residual connection, dilated convolution, and GAN based on U-Net on the final segmentation results. Table 1 shows the comparison of segmentation results of ablation experiments.
Table 1.
Comparison of ablation experiment results
| Method | Res | Dila | GAN | Dice | Accuracy | Recall | Specificity | IOU | PR | ROC |
|---|---|---|---|---|---|---|---|---|---|---|
| Endoscopist | - | - | - | - | 0.836 | - | - | - | - | - |
| U-Net | - | - | - | 0.776 | 0.872 | 0.751 | 0.849 | 0.636 | 0.842 | 0.896 |
| U-GAN | - | - | Y | 0.848 | 0.908 | 0.858 | 0.929 | 0.736 | 0.909 | 0.944 |
| DU-GAN | Y | - | Y | 0.861 | 0.915 | 0.872 | 0.934 | 0.755 | 0.935 | 0.968 |
| RU-GAN | - | Y | Y | 0.852 | 0.911 | 0.862 | 0.931 | 0.742 | 0.925 | 0.963 |
| Ours | Y | Y | Y | 0.866 | 0.919 | 0.873 | 0.939 | 0.763 | 0.931 | 0.965 |
Bold characters indicate the best performance. “Y” indicates that the method is added, and “-” indicates that the method is not added, “Res” is the residual connection, and “Dila” is the dilated convolution
In this paper, U-Net is used as the baseline. Its segmentation results are the most unsatisfactory, with dice and accuracy of 77.6% and 87.2%. The U-GAN in Table 1 uses U-Net as the generator in GAN, and it can be seen that the result has been greatly improved. Dice reaches 84.8%, an increase of 7.2% over the baseline network. The DU-GAN is an improvement on U-GAN. Its generator is a segmentation network with dilated convolution introduced into U-Net. The dice is 8.5% higher than the baseline network. Correspondingly, the RU-GAN adds a residual connection to U-Net encoding and decoding based on U-GAN, and the dice increases by 7.6% compared with the baseline network. For the proposed method, we replace the generator in GAN with a network that integrates both dilated convolution and residual connection into the baseline. The dice of the proposed method is 9% higher than the baseline network and 1.8% higher than the U-GAN. The accuracy of the endoscopist labeling is only 83.6%, while our proposed model achieves 91.9%. This proves the effectiveness of the proposed method. It further shows that GAN adversarial training can produce better segmentation results. The introduction of residual connection and dilated convolution into the model can also further improve the segmentation performance. We show the segmentation results of the above networks in Fig. 5.
Fig. 5.
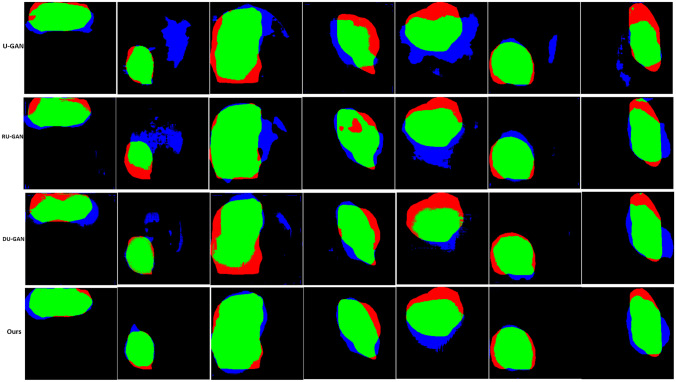
Segmentation result of gastroscopic images. Input is the original endoscopic image; label is the real label image; columns (a), (b), and (c) are the lesion segmentation images obtained by U-GAN, DU-GAN, and RU-GAN methods in Table 1; and (d) is the segmentation image of the proposed model in this paper. We further extract the contours from the segmentation results of columns (a), (b), (c), and (d). In addition, we label the gastroscopic images to obtain columns (a1), (b1), (c1), and (d1), respectively
It can be seen from Fig. 5 that the segmentation result of column d is closest to the real label image. Once again, the model we proposed can produce higher quality segmentation image. The images in column a are segmentation maps that use U-Net as the generator, and the results are relatively poor. The images in column b and column c are segmentation maps with residual connection and dilation convolution on U-Net, respectively. Compared with column a, the results are closer to the real label. However, it can be seen from the segmentation image that the edge of the images in column c with residual connections is fuzzy. This indicates that adding dilated convolution with different dilation rates in the bottleneck of U-Net can help the image to obtain better context information and prevent the loss of edge features. The lesions mark by the blue line are the result of manual labeling, while the segmentation result of the model marks by the green line. From the drawn contours, it can be seen more clearly that the proposed model is more accurate in the segmentation of the lesions.
Figure 6 analyzes the performance of the model from the perspective of false positive and false negative. It can be seen that the error rate of the proposed method is the lowest.
Fig. 6.
Comparison of results of segmentation of lesions in gastroscopic images. Green is the overlap, red is false negative, and blue is false positive
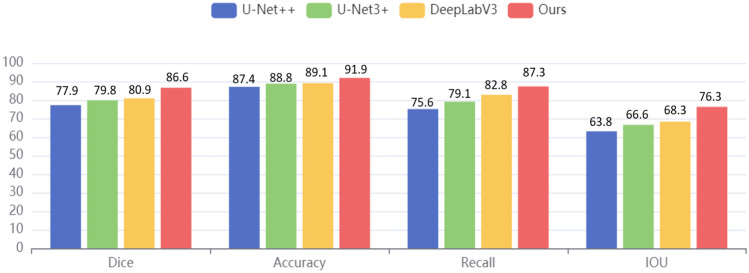
In this work, the classical segmentation models U-Net + + [34], U-Net3 + [35], and DeepLabV3 [24] are selected as the contrast models. The same gastroscopic dataset is used in all the contrast experiments. The segmentation results are shown in Fig. 7. The bar illustration was used to show the comparisons. As seen in Fig. 7, our dice, accuracy, recall, and IOU scores are 86.6%, 91.9%, 87.3%, and 76.3%, respectively. Compared with Deeplabv3, U-Net + + , and U-Net3 + , the dice of the proposed method is increased by 5.7%, 8.7%, and 6.8%, respectively, and the accuracy is also increased by 2.8%, 4.5%, and 3.1%. The data in Fig. 7 clearly show that the proposed model has the best segmentation performance among all the above segmentation models, which is enough to reflect the superiority of our model in the segmentation of lesions in gastroscopic images.
Fig. 7.
Compared with other classical segmentation models
Discussion
We analyze the segmentation results of the proposed model. The residual blocks are added to the model to alleviate the problem of gradient disappearance and improve the training effect. As the segmentation results show, the addition of the residual blocks increases dice, accuracy, and sensitivity, but the boundaries of the segmented image are blurred. Considering that the continuous pooling and upsampling of the U-net causes the loss of spatial resolution, we add dilated convolutions with different dilation rates to the bottleneck of U-Net. This enables the segmented images to obtain more context information and retrieve more edge features. The improved segmentation network is used as the generator of GAN. The segmentation network learns the high-order consistency between pixels through adversarial training with the discriminator of GAN so that the segmentation result is constantly close to the ground truth. The efficiency of the proposed model is validated by experimental results obtained on gastroscopic images. Performance indicators show that our model achieves a very satisfactory 86.6% dice, 91.9% accuracy, and 93.9% specificity in the segmentation of the lesion region.
Recall index indicates that the overlap between the lesions in the segmentation map of the proposed method and the manually labeled lesions is 87.3%. It shows that the edge detail recognition of the lesions in the segmented images is slightly different from the real label, but the segmentation images of our model cover the main lesions. Accuracy indicates how many pixels are correctly predicted. In this work, the total accuracy of predicting the lesion area and non-lesion area in gastroscopic images reached 91.9%, while the accuracy of endoscopist labeling was only 73.7 to 83.6%. This shows that the model can provide some help to doctors in early cancer diagnosis, which undoubtedly brings hope to patients with gastric cancer. This work not only saves the lives of patients but also alleviates the problem of the supply and demand balance of medical resources, which has important research significance.
On the other hand, the labeling of lesions in medical images is easily affected by the subjective visual judgment of the endoscopist, which easily leads to over-segmentation or under-segmentation of ground truth. In the pixel-by-pixel recognition task, the fuzzy boundary labeling of the lesion leads to the poor learning ability of the model in the edge region, which is also the reason for the relatively low IOU. In future studies, we consider the average of the training dataset labeled by multiple endoscopists as our final training dataset, which may improve the segmentation results.
Conclusion
In this paper, we develop a novel gastric lesion segmentation model based on GAN, using the improved segmentation network of U-Net as the generator and Patch-GAN as the discriminator, which effectively segments the lesions from the gastroscopic images with high accuracy. We add the residual connections in the encoder and decoder of U-Net to improve the robustness of the network. The residual blocks also solve the problem of gradients disappearing and exploding during the training process. By cascading the dilated convolutions with different dilation rates at the bottleneck of U-Net, more contextual information can be obtained and the segmentation boundary can be refined. The discriminant mechanism in the parallel cGAN is used to provide the trainable loss function, which makes the segmentation network get full training. The experimental results show that the proposed model has a clearer segmentation boundary after adding cascaded dilated convolutions, which achieves a reduced loss of accuracy while keeping the receptive field unchanged. In addition, the lower false negatives and false positives also proved the effectiveness of our discriminator setting of Patch-GAN. Compared with previous medical image segmentation models, the proposed model has better performance in dice, accuracy, recall, and IOU. The segmentation result of the model is closer to the manual segmentation standard for lesions in gastroscopic images. This work can reduce the burden on clinicians, save them time and physical labor, and improve the accuracy of lesion detection in gastroscopic images. Considering that we only use the gastroscopic images for experiments at present, the proposed model can be applied to other medical image segmentation tasks in future work.
Acknowledgements
The data was provided by the digestive endoscopy center of General Hospital of People’s Liberation Army.
Funding
This work was supported by the National Key R&D Program of China (2017YFB0403801) and the Natural National Science Foundation of China (NSFC) (61835015).
Declarations
Ethics Approval
This article does not contain any data, or other information from studies or experimentation, with the involvement of human or animal subjects.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Conflict of Interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yaru Sun and Yunqi Li contributed equally to this work and should be considered co-first authors.
Contributor Information
Dongzhi He, Email: victor@bjut.edu.cn.
Zhiqiang Wang, Email: wzq301@263.net.
References
- 1.Latest global cancer data: Cancer burden rises to 19.3 million new cases and 10.0 million cancer deaths in 2020. Available at https://www.iarc.who.int/news-events. Accessed 26 January 2021
- 2.Cheng J, Xi W, Yang A, Jiang Q, and Fang W: Model to identify early-stage gastric cancers with a deep invasion of submucosa based on endoscopy and endoscopic ultrasonography findings. J Surgical Endoscopy 32(2), 2018 [DOI] [PubMed]
- 3.Cui ZH, Zhang QY, Zhao L, Sun X, Lei Y. Application of intelligent target detection technology based on a gastroscopic image in early gastric cancer screening. J China Digital Medicine. 2021;16(02):7–1. [Google Scholar]
- 4.Sainju S, Bui FM, Wahid KA. Automated bleeding detection in capsule endoscopy videos using statistical features and region growing. J Journal of medical systems. 2014;38(4):1–11. doi: 10.1007/s10916-014-0025-1. [DOI] [PubMed] [Google Scholar]
- 5.Yeh JY, Wu TH, Tsai WJ. Bleeding and ulcer detection using wireless capsule endoscopy images. J Journal of Software Engineering and Applications. 2014;7(5):422. doi: 10.4236/jsea.2014.75039. [DOI] [Google Scholar]
- 6.Li Y, He Z, Ye X, Han K. Spatial-temporal graph convolutional networks for skeleton-based dynamic hand gesture recognition. J EURASIP Journal on Image and Video Processing. 2019;2019(1):1–7. doi: 10.1186/s13640-018-0395-2. [DOI] [Google Scholar]
- 7.Li B, Meng MQH. Computer-aided detection of bleeding regions for capsule endoscopy images. J IEEE Transactions on biomedical engineering. 2009;56(4):1032–1039. doi: 10.1109/TBME.2008.2010526. [DOI] [PubMed] [Google Scholar]
- 8.Sakai Y, Takemoto S, Hori K: Automatic detection of early gastric cancer in endoscopic images using a transferring convolutional neural network. 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), pp 4138–4141, 2018 [DOI] [PubMed]
- 9.Yoon HJ, Kim S, Kim JH, Keum JS, Noh SH. A lesion-based convolutional neural network improves endoscopic detection and depth prediction of early gastric cancer. J Journal of clinical medicine. 2019;8(9):1310. doi: 10.3390/jcm8091310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simonyan K, Zisserman A: Very Deep Convolutional Networks for Large-Scale Image Recognition. J Computer Science, 2014
- 11.Shibata T, Teramoto A, Yamada H, Ohmiya N, Fujita H. Automated detection and segmentation of early gastric cancer from endoscopic images using mask R-CNN. J Applied Sciences. 2020;10(11):3842. doi: 10.3390/app10113842. [DOI] [Google Scholar]
- 12.Wang R, Zhang W, Nie W, Yu Y: Gastric Polyps Detection by Improved Faster CNN. Proceedings of the 2019 8th International Conference on Computing and Pattern Recognition 128–133, 2019
- 13.Cao C, Wang R, Yu Y, Zhang H, Yu Y, and Sun C: Gastric polyp detection in gastroscopic images using deep neural network. PloS one 16(4), 2021 [DOI] [PMC free article] [PubMed]
- 14.Redmon J, Farhadi A: YOLOv3: An Incremental Improvement. Eprint Arxiv, arXiv: 1804.02767 v1, 2018
- 15.Zhu Y, Wang QC, Xu MD. Application of convolutional neural network in the diagnosis of the invasion depth of gastric cancer based on conventional endoscopy. J Gastrointestinal endoscopy. 2019;89(4):806–815. doi: 10.1016/j.gie.2018.11.011. [DOI] [PubMed] [Google Scholar]
- 16.Ronneberger O, Fischer P, Brox T: U-net: Convolutional networks for biomedical image segmentation, in International Conference on Medical Image Computing and Computer-Assisted Intervention, Cham pp. 234–241, 2015
- 17.Long J, Shelhamer E, Darrell T. Fully Convolutional Networks for Semantic Segmentation. IEEE Transactions on Pattern Analysis and Machine Intelligence. 2015;39(4):640–651. doi: 10.1109/TPAMI.2016.2572683. [DOI] [PubMed] [Google Scholar]
- 18.Oktay O, Schlemper J, and Folgoc LL, Lee M, Heinrich M, Misawa K, Mori K, Mcdonagh S, Hammerla NY, and Kainz B: Attention U-Net: Learning Where to Look for the Pancreas. arXiv: Computer Vision and Pattern Recognition, 2018
- 19.Chen W, Zhang Y, He J, Qiao Y, Chen Y, Shi H, and Tang X: Prostate Segmentation using 2D Bridged U-net. arXiv: 1807.04459, 2018
- 20.Liu Z, Song YQ, Sheng VS, Wang L, Jiang R, Zhang X, and Yuan D: Liver CT sequence segmentation based with improved U-Net and graph cut. Expert Systems with Applications 126(JUL.): 54–63, 2019
- 21.Moradi, S, Ghelich-Oghli M, Alizadehasl A, Shiri I, Oveisi N, and Oveisi M: MFP-U-Net: A novel deep learning based approach for left ventricle segmentation in echocardiography. Physica Medica 58–69,2019 [DOI] [PubMed]
- 22.Lin TY, Dollar P, Girshick R, He K, Hariharan B, and Belongie S: Feature Pyramid Networks for Object Detection. Computer Vision and Pattern Recognition 936–944, 2017
- 23.He KM, Zhang XY, Ren SQ, Sun J: Deep Residual Learning for Image Recognition. IEEE Conference on Computer Vision & Pattern Recognition. IEEE Computer Society, 2015
- 24.Yin XH, Wang YC, Li DY. Review of medical image segmentation technology based on improved u-net structure. J Journal of software. 2021;32(02):519–550. [Google Scholar]
- 25.Chen LC, Papandreou G, Schroff F, Adam H: Rethinking Atrous Convolution for Semantic Image Segmentation. arXiv: 1706. 05587v3, 2017
- 26.Goodfellow IJ, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, Courville A, Bengio Y. Generative Adversarial Networks. Advances in Neural Information Processing Systems. 2014;3:2672–2680. [Google Scholar]
- 27.Moeskops P, Veta M, Lafarge MW, Eppenhof KA, Pluim JPW: Adversarial Training and Dilated Convolutions for Brain MRI Segmentation. International Workshop on Deep Learning in Medical Image Analysis International Workshop on Multimodal Learning for Clinical Decision Support, DLMIA 2017
- 28.Xue Y, Xu T, Zhang H, Long LR, and Huang X. SegAN: Adversarial network with multi-scale L1 loss for medical image segmentation. Neuroinformatics 16-(3–4):383–392, 2018 [DOI] [PubMed]
- 29.Rezaei M, Yang H, Harmuth K, and Meinel C: Conditional Generative Adversarial Refinement Networks for Unbalanced Medical Image Semantic Segmentation. 2019 IEEE Winter Conference on Applications of Computer Vision (WACV). IEEE, 2019
- 30.Liu SP, Hong JM, Liang JP, Jia XP, OuYang J, Yin J. Semi-supervised conditional generation antagonism network for medical image segmentation. J Journal of software. 2020;31(08):310–324. [Google Scholar]
- 31.Gao YH, Huang R, Yang YW, Zhang J, Shao KN, Tao CJ, Chen YY, Metaxas DN, Li HS, and Chen M: FocusNetv2: Imbalanced large and small organ segmentation with adversarial shape constraint for head and neck CT images. J Medical Image Analysis 67, 2021 [DOI] [PubMed]
- 32.Isola P, Zhu JY, Zhou T, Efros AA: Image-to-Image Translation with Conditional Adversarial Networks. IEEE Conference on Computer Vision & Pattern Recognition. IEEE, 2016
- 33.Mirza M, Osindero S: Conditional Generative Adversarial Nets. J Computer Science 2672–2680, 2014
- 34.Zhou Z, Siddiquee M, Tajbakhsh N, Liang J:UNet++: A Nested U-Net Architecture for Medical Image Segmentation. 4th Deep Learning in Medical Image Analysis (DLMIA) Workshop, 2018 [DOI] [PMC free article] [PubMed]
- 35.Huang H, Lin L, Tong R, Hu H, and Wu J: U-Net 3+: A Full-Scale Connected U-Net for Medical Image Segmentation. ICASSP 2020 - 2020 IEEE International Conference on Acoustics, Speech, and Signal Processing (ICASSP). IEEE, 2020 [DOI] [PMC free article] [PubMed]