Abstract
Multiple sclerosis (MS) is a kind of central nervous system (CNS) autoimmune disease, which mainly damages nerves, the brain, and the spinal cord. Recently, several clinical cases reported the relativity between Coronavirus Disease 2019 (COVID-19) and the development of MS, but the mechanism of how COVID-19 affects the occurrence of MS was still not clear. It is bioinformatics technology that we use to explore the potential association at the gene level. The genetic information related to the two diseases was collected from the DisGNET platform for functional protein network analysis and used STRING to identify the complete gene set. The protein–protein interaction (PPI) network was analyzed by STRING. Finally, in the GEO database, we selected peripheral blood mononuclear cell (PBMC) RNA sequencing data (GSE164805, GSE21942) from COVID-19 patients and MS patients to verify the potential cross mechanism between the two diseases. The similar gene set of immune or inflammation existed between the patients with COVID-19 and ones with MS, including L2RA, IFNG, IL1B, NLRP3, and TNF. Interaction network analysis among proteins revealed that IL1B, P2RX7, IFNB1, IFNB1, TNF, and CASP1 enhanced the network connectivity between the combined gene set of COVID-19 and MS associated with NOD-like receptor (NLR) signaling. The involvement of NLR signaling in both diseases was further confirmed by comparing peripheral blood monocyte samples from COVID-19 and MS patients. Activation of NLR signaling was found in both COVID-19 and MS. The PBMC samples analyses also indicated the involvement of the NLR signaling pathway. Taken together, our data analyses revealed that the NLR signaling pathway might play a critical role in the COVID-19-related MS.
Keywords: COVID-19, Multiple sclerosis, SARS-CoV-2
Introduction
By November 8, 2021, there were 249 million people who infected COVID-19 all over the world, and over 5 million died in this infection. Mainly disseminating by respiratory infection, the pathogen SARS-CoV-2 enters the host cells by binding the cell surface functional receptor ACE2, leading to symptoms such as fever and dry cough (Shi et al. 2021). Risk factors affecting its prognosis include age, smoking, diabetes, cardiovascular, and cerebrovascular diseases (Hu et al. 2020). Besides common respiratory symptoms, some patients with COVID-19 had also discovered various nervous system symptoms in both central and peripheral neurological diseases, such as ischemic stroke, encephalitis, and delineation (Meppiel et al. 2020). As well as the SARS-CoV-2-induced inflammatory response, its immune response and the direct injury of the virus were considered as the possible mechanisms of how the nervous system symptoms came into being (Jha et al. 2021).
MS is an autoimmune inflammatory disease of the CNS. Their pathological characteristics are mainly local or diffuse inflammation, demyelination, and neuronal damage to the optic nerve, brain, and spinal cord. Few past articles about COVID-19 mentioned MS (Palao et al. 2020; Fragoso et al. 2021; Naser Moghadasi 2021). Many studies of COVID-19-related MS focused on its treatment, prevention management, and vaccine. However, with further researches about the COVID-19-related nervous system symptoms, SARS-CoV-2 infection was found to possibly cause the occurrence and development of MS (Barzegar et al. 2021). In addition, an Italian control study told us that the crowd of patients with MS has a great deal heavier clinical symptoms when they suffered COVID-19 infection (Sormani et al. 2022). Because of the lack of diagnostic biomarkers, MS typing mainly depends on clinical evaluation and identification, including relapse remission (RRMS), secondary progression (SPMS), primary progression (PPMS), and progressive relapse (PRMS). Approximately 85% of patients were initially RRMS with pathology characterized by myelin lesions (demyelination) with T cell and microglia infiltration causing (Kuhlmann et al. 2002). The most common clinical manifestations of MS patients include limb weakness, paraesthesia, decreased vision, and ataxia. Environmental and genetic factors and viral infection are the common influencing factors of MS. Epstein–Barr Virus (EBV), human herpes virus 6 (HHV-6), and the human endogenous retroviruses (HERVs) are the pathogens associated with MS. EBV plays a major role in MS, with enhanced human immunoreactivity after EBV infection, EB virus-specific T lymphocytes gradually fail or lack as the duration of the disease increases. The persistence of viral infection leads to sustained immune-inflammatory damage in the central and peripheral systems (Zivadinov et al. 2016; Pender et al. 2017; Serafini et al. 2019). In addition to the above mechanisms, the INF-1, the TH-17 axis, and the inflammasome pathways are also involved in the MS pathological process (Luo et al. 2020).
The latest COVID-19-related studies found that SARS-CoV-2 can elicit drastic inflammatory responses and release a large number of cytokines, leading to tissue damage and organ failure. COVID-19 has been implicated in immune diseases, and can cause multi-system inflammatory syndrome (MIS-C) in children, while the autoantibody profiles indicate the involvement of multiple autoantibodies (Consiglio et al. 2020). After anatomizing the brains of COVID-19 corpses, the researchers discovered the facts of inflammatory damage in the nervous system. A most recent review evaluates the possible link between COVID-19 and MS systematically, indicating that immune/inflammation is their common pathophysiological basis (Bellucci et al. 2021). Many cases of COVID-19 combined with demyelination have been reported continuously, and yet the pathophysiology is unclear. Therefore in this bioinformatics study, the potential genetic link of the two diseases was described to further elucidate the pathogenesis of COVID-19-associated MS.
Methods
COVID-19-and MS-related gene collection
DisGNET is a multi-functional platform that has included a large number of variations related to human diseases and genes. The related genetic information of these two diseases was obtained from DisGeNET (COVID-19, Data code: C000657245, version 5, Total Number of PMIDs ≥ 7; MS, Data code: C0026769, gda Score ≥ 0.3). These genes were used for functional protein network analysis using STRING to identify the complete gene set.
PPI of COVID-19 and MS-associated genes
We analyzed the protein–protein interaction (PPI) network of these COVID-19 and MS-associated genes by STRING. The minimum interaction score requirement was set with the maximum credibility, 0.900, and hiding all disconnected nodes. The thickness of the Network link represented the strength of the data support. Active interaction sources chose 'Co-occurrence', 'Co-expression', 'Databases' and 'Experiments'. We use the Markov Cluster Algorithm (MCL), a convenient and scalable-based clustering algorithm, and set MCL Inflation Parameter = 3. The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway was analyzed by STRING to find the potential cross mechanism between the two diseases.
Transcriptome analysis
The COVID-19 transcriptomic data were obtained from the GSE164805 dataset (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE164805) in the GEO database, and we selected RNA sequencing data from peripheral blood mononuclear cell (PBMC) samples from 4 of these COVID-19 patients and four healthy controls. Similarly, the MS transcriptomic data were obtained from the GSE21942 dataset (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi) in the GEO database, where 12 patients with MS and 15 healthy controls were selected for the peripheral blood mononuclear cell (PBMC) samples. The STRING is used to enrich the two disease-related differences in KEGG function, and the standard screening criteria is P Adjust value < 0.05, Q Value < 0.05. ComplexheatMap is used to draw the HeatMap of COVID-19 and MS-related differential genes, respectively.
Results
PPI networks and KEGG for COVID-19 and MS-related genes
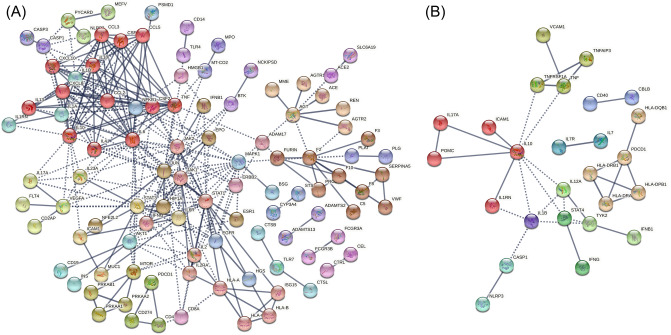
The lists of COVID-19 and MS-related genes were firstly screened for pathways for these two diseases. COVID-19-related genes (Data code: C000657245, version 5, Total Number Of PMIDs ≥ 7) and MS (Data code: C0026769, gda Score ≥ 0.3) were filtered out in DisGeNET, and the connecting genes of the two diseases were identified (Table 1). A PPI analysis of the gene list using STRING showed a high number of connections to IL-10 in both networks, and the number of nodes in IL-10 was 17 in COVID-19 and 9 in MS, respectively (Fig. 1). The KEGG functional enrichment of the two groups of genes is mainly distributed in the direction of immune inflammation (Table 2), including "Cytokine-cytokine receptor interaction", "NLR signaling pathway", "NF-kappa B signaling pathway" and "IL-17 signaling pathway" (FDR < 0.0001).
Table 1.
Disease-associated genes retrieved from DisGeNET
| Diseases | Disease-associated genes |
|---|---|
| COVID-19 | ACE2, CRP, S, IL6, ORF1ab, ACE, REN, TNF, TMPRSS2, CD4, CD8A, AGT, ALB, IL1B, GPT, F2, IL10, LOC102724971, LOC102723407, INS, IFNG, N, CXCL8, AMH, IL1A, IFNA1, FURIN, CSF2, DPP4, TNNI3, E, F3, IL2, GOT1, CRX, IFNB1, NFKB1, AGTR1, RPGR, PDB1, IL6R, PLAT, CALCA, CCL2, IL17A, SERPINA13P, HLA-C, LSAMP, LAMP3, ABO, HBA1, CENPJ, IL2RA, NELFCD, MB, CXCL10, VWF, CTSL, NLRP3, ORF8, CTRL, CTSB, IL4, SPECC1, MAS1, ZFYVE9, MS4A1, PDCD1, RTN1, RTN4, BSG, MTOR, NCKIPSD, AHI1, SOAT1, VEGFA, PLG, KRT20, F8, JAK1, SERPINA5, CD19, IFNA2, PSMD1, STAT3, TTR, KNG1, NPPB, EMSLR, AGTR2, CSF3, IL7, IL18, PROS1, LINC01672, ESR1, NFE2L2, PSS, ORF3a, C5, EEF1A2, ERBB2, G6PD, GGT1, NCAM1, ADAM17, TLR4, GGTLC5P, GGTLC3, GGT2, GGTLC4P, FCGR3A, FCGR3B, HSPA5, IL1RN, INSRR, JAK2, CCL3, CCL5, TLR3, KLK4, SH2D3C, BTK, VPS51, ABCB1, WDTC1, PGR-AS1, C3, CD14, F10, HIF1A, HLA-A, SERPINF2, SLC5A2, ADAMTS2, TLR7, STS, CYP3A4, FLT4, HMGB1, LTF, EPCAM, COX2, PRF1, MAPK1, PKD2L1, SLC33A1, ADAMTS13, IGKV2D-29, PYCARD, ASZ1, M, ORF6, ORF7a, AR, CASP1, CDSN, CEL, HLA-B, IGHE, IL13, LCT, MME, MUC1, OCA2, PRKAA2, STAT1, B3GALNT1, RHOD, SCPEP1, FUZ, SCAI, ORF10, MS2, AKT1, C5AR1, CASP3, CD48, CD68, EGFR, DMTN, EPHA3, EPO, ICAM1, JUN, MBL2, MEFV, MPO, PRKAA1, PRKAB1, NECTIN1, SELP, TFPI, APOL1, RNMT, HGS, ISG15, CABIN1, CD2AP, CD274, IL23A, CDCA7L, OTOR, IFIH1, SLTM, OMA1, SLC6A19 |
| MS | HLA-DRB1, IL2RA, IL7R, TNFRSF1A, CLEC16A, CD40`8, TYK2, KIF1B, HLA-DRA, CBLB, STAT4, CD6, TNFAIP3, TNFSF14, IFNB1, IL17A, IFNG, APOE, IL10, VDR, IL1B, HLA-DQB1, HLA-DPB1, IL7, NLRP3, IL1RN, ICAM1, P2RX7, IRF8, CNR1, GC, CASP1, SLC11A1, POMC, CLDN11, SELE, VCAM1, PDCD1, NECTIN2, IL12A, RBPJ, MCAM, TNF, KCNJ10, BCHE |
Genes associated with both COVID-19 and MS are bolded
COVID-19 coronavirus disease 2019, MS Multiple sclerosis
Fig. 1.
Functional protein–protein association networks of (A) COVID-19 and (B) MS-related genes were analysed by STRING. Genes associated with the two disease entities (disease id: C000657245, version 5, N.PMIDs ≥ 7; MS, disease id: C0026769, Gene-Disease Association Score ≥ 0.3) were retrieved from DisGeNET, acknowledge platform for disease genomics. Only highly confident interactions derived from “Experiments”, “Databases”, “Co-expression” and “Co-occurrence” with the minimum required interaction score of 0.900 were shown. The Markov cluster (MLC) algorithm with an inflation parameter of 3 was used for network clustering
Table2.
Significantly enriched ‘KEGG pathways’ in COVID-19- and MS-associated gene networks
| Diseases | KEGG pathways of disease-associated gene networks |
|---|---|
| COVID-19 | Cytokine-cytokine receptor interaction, JAK-STAT signaling pathway, Toll-like receptor signaling pathway, Th17 cell differentiation, Hematopoietic cell lineage, PI3K-Akt signaling pathway, NOD-like receptor signaling pathway, Complement and coagulation cascades, IL-17 signaling pathway, Natural killer cell mediated cytotoxicity, Necroptosis, HIF-1 signaling pathway, C-type lectin receptor signaling pathway, TNF signaling pathway, Osteoclast differentiation, T cell receptor signaling pathway, Cytosolic DNA-sensing pathway, Th1 and Th2 cell differentiation, Renin-angiotensin system, Viral protein interaction with cytokine and cytokine receptor, EGFR tyrosine kinase inhibitor resistance, Antigen processing and presentation, Phagosome, Insulin resistance, Adipocytokine signaling pathway, RIG-I-like receptor signaling pathway, Cell adhesion molecules, Fc epsilon RI signaling pathway, Prolactin signaling pathway, FoxO signaling pathway, Autophagy—animal, NF-kappa B signaling pathway, MAPK signaling pathway, Phospholipase D signaling pathway, Chemokine signaling pathway, Cellular senescence, Apoptosis, B cell receptor signaling pathway |
| MS | Cytokine-cytokine receptor interaction, Cell adhesion molecules, Necroptosis, Th1 and Th2 cell differentiation, NF-kappa B signaling pathway, JAK-STAT signaling pathway, TNF signaling pathway, Hematopoietic cell lineage, Th17 cell differentiation, C-type lectin receptor signaling pathway, NOD-like receptor signaling pathway, Osteoclast differentiation, IL-17 signaling pathway, Intestinal immune network for IgA production, Toll-like receptor signaling pathway, T cell receptor signaling pathway |
KEGG pathways are ranked according to the false discovery rate (all < 0.0001) in an ascending order. Pathways under the category of ‘Human Diseases’ were omitted. Pathways associated with both COVID-19 and MS are bolded
COVID-19 coronavirus disease 2019, MS Multiple sclerosis
United PPI network related to COVID-19-and MS-related genes
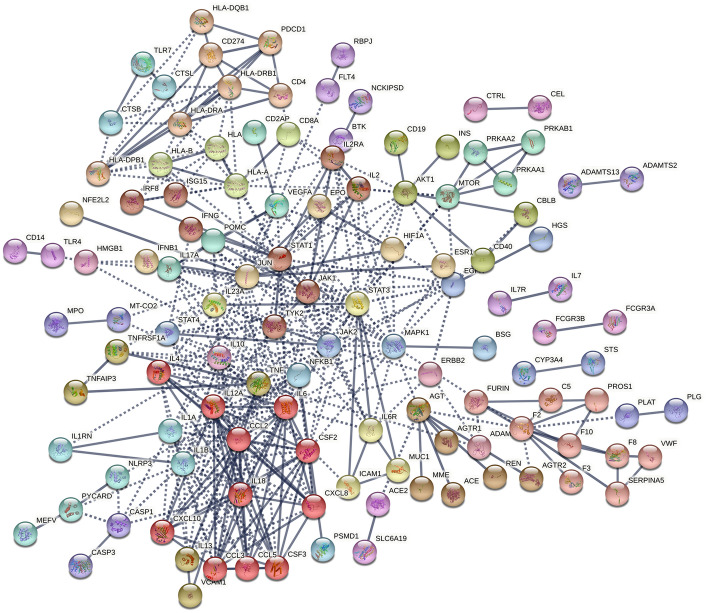
PPI analysis after integration of COVID-19-related genes and MS-related genes was used to analyze potential associations between the two diseases. The integrated gene networks were better integrated than the previous. The previously unconnected clusters were reconnected into the primary network (e.g., the clusters CBLB/CD40 and HLA-DPB1/HLA-DQB1/HLA-DRA/HLA-DRB1/PDCD1 in MS, and the clusters CTSB/CTSL/TLR7 in COVID-19) (Fig. 2). Moreover, the integrated networks have more average number of edges than those of both diseases, in which the gene interactions get tighter. The effect of COVID-19 on the PPI network of MS-related genes was further clarified by calculating the number of connected genes for 46 MS-related genes in the MS, COVID-19, and COVID-19-MS combinations. There is more of the number of 46 MS-related genes in the combined gene network (Fig. 3) than the gene networks of COVID-19 and MS. Plotting the Heatmap, ComplexHeatmap is used to cluster the number of 46 MS-associated gene connections. There are two main modes in Heatmap, namely enhancement (IL12A, TYK2, STAT4, HLA A-DRA, HLA-DRB1, HLA-DPB1) and accumulation (IL2RA, CASP1, IL17A, IFNB1, NLRP3, IL1RN, IFNG, and ICAM1) effects (Fig. 4). Enrichment analysis of these 14 MS-associated genes affected by COVID-19 indicated that the NLR signaling pathway is involved in the COVID-19-associated MS.
Fig. 2.
STRING analysis of combined COVID-19/MS gene sets. Interaction map shows better integration of COVID-19-and MS-associated clusters
Fig. 3.
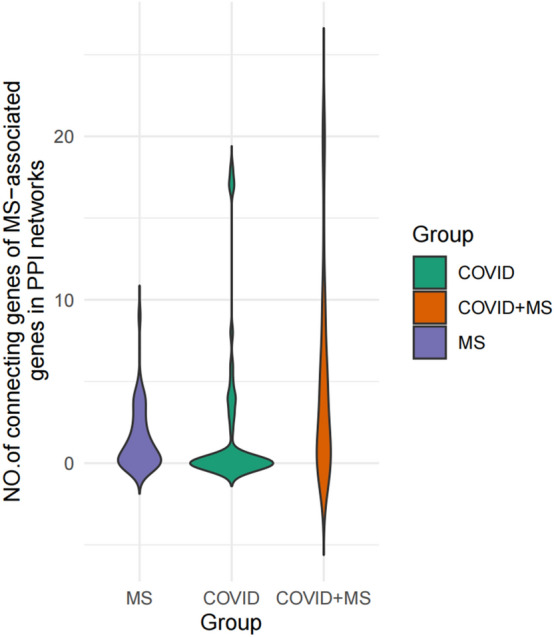
GBS-associated genes showed higher connectivity in the combined network as compared to two disease entities alone
Fig. 4.
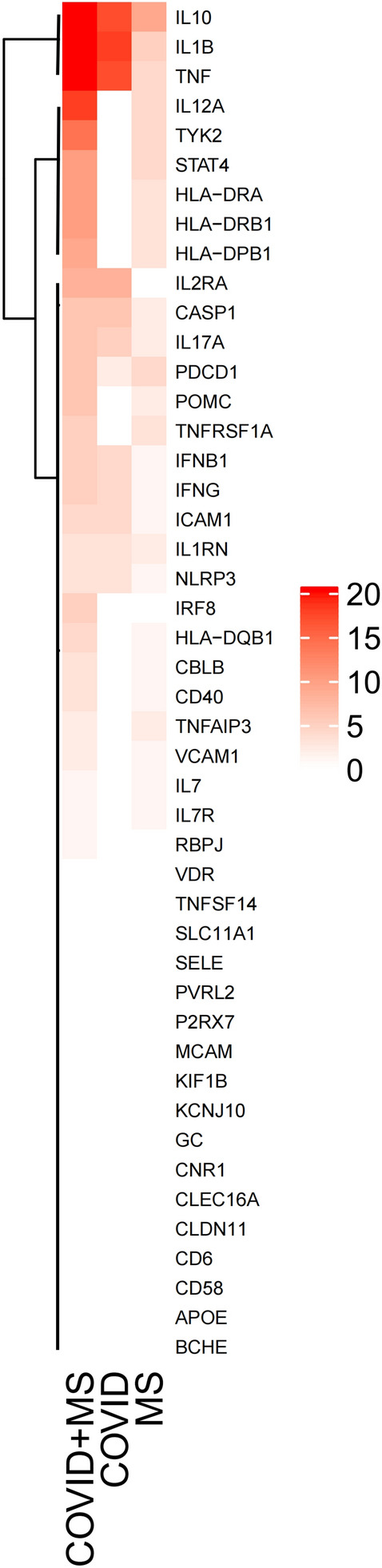
Heap map visualizes the additive and potentiating effects of COVID-19-associated genes on the network connectivity of MS-associated genes
PMBC difference gene by COVID-19 and MS patients
To validate our data, we selected RNA sequencing datasets from PBMC samples from COVID-19 and MS patients from the public database for differential gene analysis, KEGG functional enrichment, and heatmap drawing. The selection criteria for differential genes were: adjusted P value < 0.05 and the absolute value of log2 fold change > 1. A total of 11,594 differential genes (5,392 upregulated and 6,202 downregulated) were found in COVID-19 patients, mainly enriched in immune/inflammation-related pathways such as "NF-kappa B signaling pathway", "TNF signaling pathway", and "NLR signaling pathway" (adjusted P value < 0.05, Q value < 0.05). 593 different genes (268 upregulated and 325 downregulated) were analyzed in data concentration in MS patients. A KEGG enrichment analysis was performed on these differential genes, and the pathways were mainly enriched in immune/inflammatory pathways, such as "B cell receptor signaling pathway", "NLR signaling pathway", and "Th17 cell differentiation" (adjusted P value < 0.05, Q value < 0.05)(Table 3). There was a total of 106 differential genes between COVID-19 and MS (54 upregulated and 52 downregulated), which were significantly enriched in the NLR signaling pathway: consistent with NLR involvement in the COVID-19-associated MS.
Table 3.
Significantly enriched ‘KEGG pathways’ with differentially expressed genes in COVID-19 and MS peripheral blood mononuclear cells (PBMCs)
| Diseases | KEGG pathways of disease-associated differentially expressed genes in PBMCs |
|---|---|
| COVID-19 | NF-kappa B signaling pathway, TNF signaling pathway, Lysosome, NOD-like receptor signaling pathway |
| MS | Hematopoietic cell lineage, B cell receptor signaling pathway, Apoptosis, NOD-like receptor signaling pathway, Th17 cell differentiation, p53 signaling pathway |
Pathways under the category of ‘Human Diseases’ were omitted. Pathways associated with both COVID-19 and MS are bolded
COVID-19 coronavirus disease 2019, MS Multiple sclerosis
Discussion
It is still not clear whether the COVID-19-associated neurological symptoms are due to direct viral invasion or immune/inflammation-mediated damage. SARS-CoV-2 was found to be neuropathic and neurotoxic by binding the angiotensin-converting enzyme-2 (ACE-2) receptor that is mainly distributed in various organs such as the nervous system (Miller and Arnold 2019), and SARS-CoV-2 can directly damage the nervous system. What’s more, SARS-CoV-2 through the blood pathway and the neuronal pathway can also directly invade the nervous system (Wu et al. 2020). A case of SARS-CoV-2-associated meningitis was firstly reported by Moriguchi T, et al. The presence of SARS-CoV-2 virus in the cerebrospinal fluid (CSF) by detecting SARS-CoV-2 RNA, further demonstrating that SARS-CoV-2 may cause direct damage to the nervous system (Moriguchi et al. 2020). In addition, viral infection has been shown to induce an inflammatory response and activate myelinated myelin-specific T cells, which can accelerate an early or delayed virus-induced demyelination process. SARS-CoV-2 elicited a more severe self-aggressive immunoinflammatory response compared to viruses in other respiratory ways (Subbarao and Mahanty 2020). Factually, it was found that the pathogenesis of COVID-19-related demyelination is mainly a dramatic immune response initiated by inflammatory cytokines (IL-1β, IL-6) through COVID-19-related encephalopathy (Bodro et al. 2020). The genes related to ACE-2 and their access were not found in this study about the bioinformatics analysis for MS and COVID-19, but our functional analysis showed that immune/inflammation is closely related to these two diseases. The genes related to COVID-19 and MS, as well as the differential genes of PBMC samples, were significantly enriched in the NLR signaling pathway, which provides new clues for COVID-19-related MS.
MS is an immune-inflammatory disease of the CNS, which usually involves the optic nerve, brain, and spinal cord, and its pathological basis is axonal inflammatory demyelination. It is characterized by a unique oligoclonal band, IL-1β, in CSF (Stangel et al. 2013). Promoting inflammatory factors that can cross the blood–brain barrier are discovered in the CSF of the patients with COVID-19, such as IL-1β, IL-2, IL-4, IL-6, IL-8, IL-10, TNF-α, and IFN-γ. They can affect macrophages, microglia, and astrocytes and mediate major cells of CNS innate immunity leading to cytokine storm (Desforges et al. 2019; Han et al. 2020). The past articles about the pathogenesis of MS show that cytokines like IL-1β, IL-6, and IL-10 are also involved in the pathophysiology of MS.
The nod-like receptor (NLR) pathway is responsible for detecting various pathogens and producing an innate immune response, driving the activation of NFkB and MAPK, cytokine production and apoptosis. In addition, NLR of some pattern recognition receptors, such as NLRP1 and NLRP3, oligomerize to form multiprotein complexes, resulting in the activation of Caspase-1 (Zhao et al. 2020), and then the emergence of IL-1B and IL-18 inflammatory factors induced cell death. In MS, the NLRP 3 inflammasome is significantly involved in the chronic inflammation caused by microglia, thus leading to neurodegeneration. Inflammasomes may affect IL-1B and IFN-1 to promote MS progression (Guarda et al. 2011; Malhotra et al. 2020), and gain-of-function variants in the NLRP3 gene have been associated with the severity and progression of MS (Soares et al. 2019). In the COVID-19-related reports, NLRP3 inflammasomes overactivated in cytokine storms (Rodrigues et al., 2021; Ratajczak et al. 2021). Both the above results and our bioinformatics analysis demonstrate that the NLR signaling pathway may be an important intersection between COVID-19 and MS and may provide new ideas and potential targets for treating COVID-19-related MS and prevention of MS recurrence in patients with COVID-19.
There are still some limitations in our study, for instance, the most important one of which is the genes and pathways we found need proving in animal models related to COVID-19 and MS. The pathogenesis of COVID-19-related MS, for example, can be proved in the COVID-19 mouse model with experimental autoimmune neuritis (EAN). The animal model improvement is observed through treating to intervene in the production of NLRP3 inflammation, which may provide a new direction for the treatment of COVID-19-related MS.
Conclusion
We analyze the COVID-19 and MS-related genes, PPI and KEGG in this research, and the NLR signaling pathway is considered as a key pathway for COVID-19-related MS. Once again, we verify the NLR signaling pathway participated in two diseases through the transcription group analysis of PBMC. We lastly clarify that the NLR signaling pathway plays a key role in COVID-19-associated MS in our study.
Author contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by Dongtai Zhang, Zhenyang Yu, Yiwen Jiang and Dan Zhu. The first draft of the manuscript was written by Dong Qiu and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Funding
Heilongjiang Postdoctoral Science Foundation, 21042190093, Dan Zhu.
Data availability
Data will be available upon request.
Declarations
Competing interests
The authors have no financial or proprietary interests in any material discussed in this article.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Dong Qiu, Email: 776956774@qq.com.
Dan Zhu, Email: 154696464@qq.com.
References
- Barzegar M, Vaheb S, Mirmosayyeb O, et al. Can coronavirus disease 2019 (COVID-19) trigger exacerbation of multiple sclerosis? A retrospective study. Mult Scler Relat Disord. 2021;52:102947. doi: 10.1016/j.msard.2021.102947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellucci G, Rinaldi V, Buscarinu MC, et al. Multiple sclerosis and SARS-CoV-2: has the interplay started? Front Immunol. 2021;12:1–14. doi: 10.3389/fimmu.2021.755333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodro M, Compta Y, Llansó L, et al. Increased CSF levels of IL-1β, IL-6, and ACE in SARS-CoV-2-associated encephalitis. Neurol Neuroimmunol Neuroinflammation. 2020;7:1–6. doi: 10.1212/NXI.0000000000000821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consiglio CR, Cotugno N, Sardh F, et al. The immunology of multisystem inflammatory syndrome in children with COVID-19. Medrxiv. 2020;14:3159. doi: 10.1101/2020.07.08.20148353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desforges M, Le Coupanec A, Dubeau P, et al. Human coronaviruses and other respiratory viruses: underestimated opportunistic pathogens of the central nervous system? Viruses. 2019;12:1–28. doi: 10.3390/v12010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragoso YD, Pacheco FAS, Silveira GL, et al. COVID-19 in a temporal relation to the onset of multiple sclerosis. Mult Scler Relat Disord. 2021;50:102863. doi: 10.1016/j.msard.2021.102863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarda G, Braun M, Staehli F, et al. Type I interferon inhibits interleukin-1 production and inflammasome activation. Immunity. 2011;34:213–223. doi: 10.1016/j.immuni.2011.02.006. [DOI] [PubMed] [Google Scholar]
- Han H, Ma Q, Li C, et al. Profiling serum cytokines in COVID-19 patients reveals IL-6 and IL-10 are disease severity predictors. Emerg Microbes Infect. 2020;9:1123–1130. doi: 10.1080/22221751.2020.1770129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L, Chen S, Fu Y, et al. Risk factors associated with clinical outcomes in 323 coronavirus disease 2019 (COVID-19) hospitalized patients in Wuhan, China. Clin Infect Dis : Off Publ Infect Dis Soc Am. 2020;71:2089–2098. doi: 10.1093/cid/ciaa539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha NK, Ojha S, Jha SK, et al. Evidence of coronavirus (CoV) pathogenesis and emerging pathogen SARS-CoV-2 in the nervous system: a review on neurological impairments and manifestations. J Mol Neurosci. 2021;71:2192–2209. doi: 10.1007/s12031-020-01767-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlmann T, Lingfeld G, Bitsch A, et al. Acute axonal damage in multiple sclerosis is most extensive in early disease stages and decreases over time. Brain. 2002;125:2202–2212. doi: 10.1093/brain/awf235. [DOI] [PubMed] [Google Scholar]
- Luo D, Wang J, Zhang X, et al. Identification and functional analysis of specific MS risk miRNAs and their target genes. Mult Scler Relat Disord. 2020;41:1–8. doi: 10.1016/j.msard.2020.102044. [DOI] [PubMed] [Google Scholar]
- Malhotra S, Costa C, Eixarch H, et al. NLRP3 inflammasome as prognostic factor and therapeutic target in primary progressive multiple sclerosis patients. Brain. 2020;143:1414–1430. doi: 10.1093/brain/awaa084. [DOI] [PubMed] [Google Scholar]
- Meppiel E, Peiffer-Smadja N, Maury A, et al. (2020) Since January 2020 Elsevier has created a COVID-19 resource centre with free information in English and Mandarin on the novel coronavirus COVID-19. The COVID-19 resource centre is hosted on Elsevier Connect, the company’s public news and information website. Elsevier hereby grants permission to make all its COVID-19-related research that is available on the COVID-19 resource centre - including this research content - immediately available in PubMed Central and other publicly funded repositories , such as the WHO COVID database with rights for unrestricted research re-use and analyses in any form or by any means with acknowledgement of the original source. These permissions are granted for free by Elsevier for as long as the COVID-19 resource centre remains active. Neurologic manifestations associated with COVID-19: a multicentre registry
- Miller AJ, Arnold AC. The renin–angiotensin system in cardiovascular autonomic control: recent developments and clinical implications. Clin Auton Res. 2019;29:231–243. doi: 10.1007/s10286-018-0572-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriguchi T, Harii N, Goto J, et al. A first case of meningitis/encephalitis associated with SARS-Coronavirus-2. Int J Infect Dis. 2020;94:55–58. doi: 10.1016/j.ijid.2020.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naser Moghadasi A. A 31 year-old female patient with concurrent clinical onset of multiple sclerosis and COVID-19: Possible role of SARS-CoV-2 in the pathogenesis of multiple sclerosis. Autoimmun Rev. 2021;20:102803. doi: 10.1016/j.autrev.2021.102803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palao M, Fernández-Díaz E, Gracia-Gil J, et al. Multiple sclerosis following SARS-CoV-2 infection. Mult Scler Relat Disord. 2020;45:102377. doi: 10.1016/j.msard.2020.102377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pender MP, Csurhes PA, Burrows JM, Burrows SR. Defective T-cell control of Epstein-Barr virus infection in multiple sclerosis. Clin Transl Immunol. 2017;6:e126–e217. doi: 10.1038/cti.2016.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratajczak MZ, Bujko K, Ciechanowicz A, et al. SARS-CoV-2 entry receptor ACE2 is expressed on very small CD45− precursors of hematopoietic and endothelial cells and in response to virus spike protein activates the Nlrp3 inflammasome. Stem Cell Rev Reports. 2021;17:266–277. doi: 10.1007/s12015-020-10010-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues TS, de Sá KSG, Ishimoto AY, et al. Inflammasomes are activated in response to SARS-cov-2 infection and are associated with COVID-19 severity in patients. J Exp Med. 2021;218:e20201707. doi: 10.1084/JEM.20201707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serafini B, Rosicarelli B, Veroni C, et al. Epstein-Barr virus-specific CD8 T cells selectively infiltrate the brain in multiple sclerosis and interact locally with virus-infected cells: clue for a virus-driven immunopathological mechanism. J Virol. 2019;93:1–21. doi: 10.1128/jvi.00980-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi JC, Yu ZJ, He GQ, et al. Epidemiological features of 105 patients infected with the COVID-19. J Natl Med Assoc. 2021;113:212–217. doi: 10.1016/j.jnma.2020.09.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares JL, Oliveira EM, Pontillo A. Variants in NLRP3 and NLRC4 inflammasome associate with susceptibility and severity of multiple sclerosis. Mult Scler Relat Disord. 2019;29:26–34. doi: 10.1016/j.msard.2019.01.023. [DOI] [PubMed] [Google Scholar]
- Sormani MP, Schiavetti I, Carmisciano L, et al. COVID-19 severity in multiple sclerosis. Neurol - Neuroimmunol Neuroinflammation. 2022;9:e1105. doi: 10.1212/nxi.0000000000001105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stangel M, Fredrikson S, Meinl E, et al. The utility of cerebrospinal fluid analysis in patients with multiple sclerosis. Nat Rev Neurol. 2013;9:267–276. doi: 10.1038/nrneurol.2013.41. [DOI] [PubMed] [Google Scholar]
- Subbarao K, Mahanty S. Respiratory virus infections: understanding COVID-19. Immunity. 2020;52:905–909. doi: 10.1016/j.immuni.2020.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Xu X, Chen Z, et al. Nervous system involvement after infection with COVID-19 and other coronaviruses. Brain Behav Immun. 2020;87:18–22. doi: 10.1016/j.bbi.2020.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao N, Li C, cui, Di B, Xu L li, Recent advances in the NEK7-licensed NLRP3 inflammasome activation: mechanisms, role in diseases and related inhibitors. J Autoimmun. 2020 doi: 10.1016/j.jaut.2020.102515. [DOI] [PubMed] [Google Scholar]
- Zivadinov R, Cerza N, Hagemeier J, et al. Humoral response to EBV is associated with cortical atrophy and lesion burden in patients with MS. Neurol - Neuroimmunol Neuroinflammation. 2016;3:e190. doi: 10.1212/nxi.0000000000000190. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be available upon request.