Abstract
Objective
Tuft cells residing in the intestinal epithelium have diverse functions. In the small intestine, they provide protection against inflammation, combat against helminth and protist infections, and serve as entry portals for enteroviruses. In the colon, they had been implicated in tumorigenesis. Commitment of intestinal progenitor cells to the tuft cell lineage requires Rho GTPase Cell Division Cycle 42 (CDC42), a Rho GTPase that acts downstream of the Epidermal Growth Factor Receptor (EGFR) and Wnt signaling cascades, and the master transcription factor POU class 2 homeobox 3 (POU2F3). This study investigates how this pathway is regulated by the DEAD box containing RNA binding protein DDX5 in vivo.
Design
We assessed the role of DDX5 in tuft cell specification and function in control and epithelial cell-specific Ddx5 knockout mice (DDX5ΔIEC) using transcriptomic approaches.
Results
DDX5ΔIEC mice harbored a loss of intestinal tuft cell populations, modified microbial repertoire, and altered susceptibilities to ileal inflammation and colonic tumorigenesis. Mechanistically, DDX5 promotes CDC42 protein synthesis through a post-transcriptional mechanism to license tuft cell specification. Importantly, the DDX5-CDC42 axis is parallel but distinct from the known IL-13 circuit implicated in tuft cell hyperplasia, and both pathways augment Pou2f3 expression in secretory lineage progenitors. In mature tuft cells, DDX5 not only promotes integrin signaling and microbial responses, it also represses gene programs involved in membrane transport and lipid metabolism.
Conclusion
RNA binding protein DDX5 directs tuft cell specification and function to regulate microbial repertoire and disease susceptibility in the intestine.
Introduction
Intestinal epithelial cells (IEC) lining the gastrointestinal tract are essential for nutrient absorption and provide barrier protection for the host1. IECs are functionally heterogeneous. One subset called tuft cells is a major sensor of microbial challenges in the intestine. They express various unique surface molecules, including receptors that bind directly to noroviruses and receptors sensing microbial metabolites, including succinate2–4. During helminth and protist challenges, tuft cells secrete IL-25 to activate type 2 innate lymphoid cells (ILC2s). In a positive feedback circuit, activated ILC2s produces interleukin 13 (IL-13) to drive tuft cell hyperplasia and ensure robust microbial clearance2 5–9. Recent studies also revealed important roles of tuft cells in regulating inflammation and tumorigenesis in different intestine sections. In the ileum, reduction in tuft cell numbers is associated with elevated inflammation in the Crohn’s disease (CD) patients10. Succinate activation of the tuft cell circuit is found be protective against T cell mediated ileitis in a mouse model10. In the colon, tuft cells and their unique expression of the pro-survival doublecortin-like kinase 1 (DCLK1) have been implicated in tumorigenesis11 12.
IECs have diverse turnover rates ranging from 3–5 days for enterocytes to 3–6 weeks for Paneth cells13 14. They are replenished by progenitors originating from the intestinal stem cells (ISCs) residing in the crypt13 15. Progenitor differentiation to specific IEC lineages is influenced by local nutrient availability, microbial composition, and gradients of Epidermal Growth Factor (EGF), Wingless-related integration site (Wnt), Bone Morphogenetic Protein (BMP), as well as cytokines from the immune system16–18. Tuft cells in the small intestine and colon arise from the SOX4+ and ATOH1+ progenitors respectively19–21. Commitment to the tuft cell lineage requires the expression of the POU domain transcription factor, POU2F322. A recent study has implicated Cell Division Cycle 42 (CDC42) in tuft cell biogenesis23. CDC42 is a ubiquitously expressed member of the Rho GTPase family. Upon activation by EGF and Wnt signaling cascades24,23 25 26, CDC42 regulates actin cytoskeleton organization, polarity, proliferation, and migration27 28. Mutations in Cdc42 have been linked to pediatric immunodeficiency29 and severe developmental delay in patients with Takenouchi-Kosaki syndrome (TKS)30 31. Global knockout of Cdc42 in mice is embryonic lethal32. Studies using mice where Cdc42 is specifically knocked out in IECs showed that CDC42 is required for ISCs growth and survival. Biogenesis of tuft cell, but not other secretory IEC subsets, is CDC42 dependent23. However, little is known about how CDC42 contributes to tuft cell differentiation and upstream mechanisms involved in maintaining proper CDC42 expression in IECs. Here, we report that CDC42 promotes POU2F3 expression and tuft cell specification in the intestine, and this circuit is post-transcriptionally regulated by the RNA binding protein called DEAD-box helicase 5 (DDX5).
DDXs can directly bind to RNA substrates and utilize ATP hydrolysis energy to unwind RNA duplexes, facilitate RNA annealing, organize RNA-protein complex assembly,33 and promote post-transcriptional processing34. They can also partner with transcription factors to regulate gene expression35–38. Our previous study revealed that DDX5 is the highest expressed member of the DDX family in the intestine epithelium39. Overexpression of DDX5 predicts worse relapse-free survival in CRC patients40–42, and knockdown of DDX5 can significantly inhibited the growth of cancer cells in xenograft models43 44. We have recently reported that knocking out DDX5 in IECs protected against dextran sulfate sodium induced colitis as well as tumor formation in mice on a susceptible background39. Given the emerging role of intestinal tuft cells in regulating intestinal inflammation and tumorigenesis, we asked whether changes in tuft cell specification and/or function may underly the contribution of DDX5 to intestinal inflammation and tumorigenesis. Here, we report an essential role of DDX5 in maintaining CDC42-POU2F3 levels in secretory lineage progenitors to drive tuft cell specification in both small intestine and colon. In differentiated IECs, including tuft cells, DDX5 promotes anti-microbial gene programs to regulate microbial composition and disease susceptibilities in models of ileitis and tumorigenesis.
Methods
Mice
C57BL/6 wild-type (Stock No: 000664) and Villin1Cre (Stock No: 021504) mice were originally obtained from the Jackson Laboratory. Previously described Ddx5flox mice were obtained from Dr. Frances Fuller-Pace’s laboratory45. Heterozygous mice were bred to yield 6–8 week-old Ddx5+/+ Villin1Cre+ (subsequently referred to as wild-type, WTIEC) and Ddx5fl/fl Villin1Cre+ (referred to as DDX5ΔIEC) cohoused littermates for experiments. For the tumor studies, Apcflox mice were obtained from Dr. Eric Fearon’s laboratory46 to generate Apcfl/+ Ddx5+/+ Villin1Cre+ and Apcfl/+ Ddx5fl/+ Villin1Cre+ (referred as APCΔIEC DDX5WT) and Apcfl/+ Ddx5fl/fl Villin1Cre+ (APCΔIEC DDX5ΔIEC) mice. Three (3) month old wildtype and mutant mice were given either 150 mM succinate (Alfa Aesar 41983–30) or control water (containing NaCl at 300 mM to match sodium molarity with succinate treatment) for 30 days. Colons were harvested from 120–130-day old mice to assess tumor burden. Tumor measurements were determined by double-blinded analyses using ImageJ. All animal studies were approved and followed the Institutional Animal Care and Use Guidelines of the University of California San Diego. Our vivarium at UC San Diego is kept under specific pathogen free (SPF) conditions. Regular serology and PCR tests are used to monitor and ensure the absence of EDIM, MHV, MPV, MVM, TMEV, fur mites and pinworms. MNoV is normally present in our vivarium, litters born to MNoV+ parents naturally acquire the virus from their environment prior to weaning and were used for experiments between 8–12 weeks of age.
The spatial transcriptomic dataset described were performed on tissues obtained from female mice. DCLK1 immunohistochemistry experiments were performed on tissues from female mice. All other experiments reported in this study were obtained from both male and female mice in similar ratios.
Metronidazole treatment
For the antibiotic treatment studies, WTIEC and DDX5△IEC adult mice were treated with 1% sucrose alone or 1% sucrose containing 2.5 g/L of metronidazole in the drinking water for two weeks. New beddings were provided on day 7 to limit microbe reacquisition via the fecal-oral route. Mice were monitored and weighed 2–3 times per week and fecal/cecal material were collected to assess the level of protist and bacteria clearance. Pestle-homogenized fecal or cecal materials were digested at 75°C for 10min using the KAPA Express Extract kit (R&D). 1μL of the soluble fraction was used as template for qPCR quantitation.
Anti-CD3ɛ mediated model of ileitis
WTIEC and DDX5ΔIEC mice were given 300mM succinate-NaCl or equal molar of NaCl in drinking water for 2 weeks and subsequently i.p. injected 3 times with 15ug of anti-CD3ɛ per mouse every other day for 5 days47.
Histology and immunohistochemistry
Tissues harvested from the ileum and colon were fixed overnight in 10% formalin (Research Products International) at room temperature. Paraffin-embedded tissues were sectioned into 5 μm slices for Periodic Acid-Schiff (PAS) or immunohistochemical (IHC) staining (see Supplementary Table 1 for antibody information). Briefly, paraffin sections were de-paraffinized and rehydrated with Tris-buffered saline (TBST, pH 7.8 with 0.1% Tween-20) washes between each step. Sections were blocked first against endogenous peroxidases (immersed for 30 minutes in 0.3% H2O2), and then blocked against endogenous biotin using unlabeled streptavidin (Jackson ImmunoResearch, 016-000-114) and excess free biotin. Antigen retrieval was induced by heating the slide twice for 5 minutes in 10 mM sodium citrate buffer pH 6.0 (Sigma-Aldrich), followed by 20 minutes of cooling. Finally, sections were blocked against non-specific hydrophobic interactions with 1% bovine serum albumin (BSA; Biotium) in TBST. Staining was then performed with either the negative control IgG antibody or anti-DCLK1 (1:1000) antibodies (Abcam) overnight in a humid chamber at 4°C. The next day, sections were washed with TBST and then sequentially overlaid with biotinylated goat anti-rabbit (Jackson ImmunoResearch, 111-065-045) at 1:500, followed by HRP-labeled Streptavidin (Jackson ImmunoResearch, 16-030-084) at 1:500. Substrate was then overlaid with 3-amino-9-ethylcarbazole from Vector labs following manufacturer directions for 30 minutes followed by nuclear counterstain with Mayer’s hematoxylin. Images were acquired using the AT2 Aperio Scan Scope (UCSD Moores Cancer Center Histology Core). Three intestinal regions per tissue image were randomly selected for QuPath analysis. DCLK+ tuft cells were determined by QuPath Positive Cell Detection48 (minimum area = 10μm2, maximum area = 400μm2, intensity threshold = 0.4). Mucin+ goblet cells were assessed similarly (minimum area = 40μm2, maximum area = 800μm2, intensity threshold = 0.6). The average score from three regions examined in each tissue were included in the final graph.
Electron microscopy
Cardiac perfusion was performed using 5mL Ringer’s solution (Fisher Scientific, Cat. #50-980-246), followed by 5ml of fixation buffer containing 2% Paraformaldehyde/2.5% Glutaraldehyde in 0.1M Sodium Cacodylate buffer solution (BIOTREND, Cat.# 15960–01). Intestinal tissues were harvested and kept in the fixation buffer in room temperature for 3 additional hours. Fixed tissues were processed and imaged on JEOL 1400 plus at the UC San Diego Electron Microscopy Core Facility.
Intestinal epithelial and lamina propria lymphocyte harvest
Steady-state intestinal epithelial and lamina propria lymphocytes were harvested as previously described49. To isolate IECs, intestinal tissues were first incubated in 5 mM EDTA (Invitrogen) in HBSS (Gibco) containing 1 mM DTT (Invitrogen) for 20 minutes at 37°C with 200rpm agitation, and then incubated in a second wash of 5 mM EDTA in HBSS without DTT for another 20 minutes at 37°C with agitation. Single-cell suspensions released into the EDTA solutions were pooled and confirmed to contain over 85% EpCAM+ IECs as reported previously39. For the lamina propria, IEC-depleted tissues from post-EDTA washes were digested in a HBSS solution containing 10% Fetal Bovine Serum (Peak Serum), 1.0 mg/ml Collagenase D (Roche), 100 μg/ml DNase I (Sigma-Aldrich), and 50 U/ml dispase (Worthington Biochemical Corporation) at 37°C for 30–45 minutes. Lamina propria mononuclear lymphocytes were purified from the interphase of a 40:80% Percoll™ (Cytiva; formerly GE Healthcare Life Sciences) gradient.
Intestinal crypt isolation and organoid culture
Small intestinal and colonic crypts were isolated according to the manufacturer’s recommendation (STEMCELL, technical bulletin #28223). Briefly, 20cm of small intestine proximal to the stomach were harvested from 6–8 week-old WTIEC and DDX5ΔIEC littermates and cut into 2mm pieces. After 20 washes in cold PBS, tissues were resuspended in 25mL room temperature Gentle Cell Dissociation Reagent (STEMCELL, #07174) and incubated at room temperature for 15 minutes on a rocking platform at 20 rpm. The pellets enriched with intestinal crypts were resuspended in cold PBS containing 0.1% BSA. Isolated colonic crypts were embedded in Corning® Matrigel® Matrix (Corning™ 356231) and seeded onto pre-warmed, non-treated 24-well plates (CytoOne® by StarLab) and overlaid with conditioned media (STEMCELL, #6005) as described previously50. Organoids were treated with recombinant mouse IL-13 (BioLegend, 20ng/ml), NaCl (Sigma, 15μM), DMSO (Sigma), or ML141 (Cayman Chemical, 10μM). Organoid pictures were imaged using a Keyence bz-x800 microscope at 20X magnification with image stacks capturing the entire organoid volume.
Retroviral transduction
Authenticated Platinum-E (Plat-E) cells purchased from ATCC were grown in DMEM medium (Gibco) containing 1% L-glutamine (Gibco) and 10% fetal bovine serum (FBS; Thermo Fisher Scientific). Plat-E cells were transfected with MSCV constructs using TransIT®-LT1 Transfection Reagent (Mirus) following the manufacturer’s instructions. Virus-containing supernatant collected three days after the infection was concentrated using Retro-X concentrator (Takarabio). Virus was resuspended in organoid medium containing 8μg/ml polybrene. Dissociated organoids were resuspended in virus-containing medium for spin inoculation (600g at 32°C for 60 min) followed by a six-hour incubation at 37°C. Transduced cells were collected and seeded in Matrigel. Media was changed every 2–3 days. Organoids were analyzed by flow cytometry and/or RT-qPCR 7 days post-transduction.
Flow cytometry analysis
For analysis of tuft cells in intestinal epithelium and organoids, cells were surface stained with LIVE/DEAD Fixable Cell stain (ThermoFisher, L34957), and fluorescent conjugated antibodies against EpCAM, CD24, CD45, Siglec-F, and CD90.1 (see Supplementary Table 1 for detailed information, 1:400 in PBS) for 30 minutes. For intracellular staining of CDC42, cells were fixed/permeabilized (ThermoFisher Cat: 00-5521-00), incubated with the anti-CDC42 antibody (ThermoFisher, 1:50) for 1h in room temperature, followed by the anti-rabbit IgG (H+L) Cross-Adsorbed Secondary Antibody (Invitrogen, 1:400) for 1h in 4°C. IECs were defined as live CD4−CD8−Epcam+CD45low. Successfully transduced organoid cells were defined as CD90.1+. Proportion (%) of IECs in the tuft cell lineage were defined as Siglec-F+CD24+ as previously described51. Flow cytometry data was analyzed with FlowJo (version 10.8.1).
Western blot
Cells were lysed in a 25 mM Tris pH 8.0 (G-Bioscience, 100 mM NaCl (G-Bioscience), 0.5% NP40 (G-Bioscience) solution with protease inhibitor (Life technologies) for 30 min on ice. Samples were spun down at 14,000 × g for 15 min, and soluble protein lysates were harvested. 30–50μg protein were loaded in each lane of a SDS-PAGE gel. Blots were blocked in Odyssey Blocking buffer (Li-CoR Biosciences) and probed for CDC42. Following incubation with IRDye secondary antibody (Li-CoR® Biosciences), infrared signals on each blot were collected on the Li-CoR Odyssey CLX. The primary antibodies used in this study are listed in Supplementary Table 1.
cDNA synthesis and qPCR
Total RNA was extracted with the RNeasy Plus kit (QIAGEN) and reverse transcribed using iScript™ Select cDNA Synthesis Kit (Bio-Rad). Real time RT-PCR was performed using iTaq™ Universal SYBR® Green Supermix (Bio-Rad). For primary IECs, results were normalized to mouse Gapdh. For organoid studies, results were normalized to β-actin. Primers were designed using Primer-BLAST to span across splice junctions, resulting in PCR amplicons that span at least one intron. Primer sequences are listed in Supplementary Table 2.
RNA-seq analysis
Intestinal epithelial cell transcriptomes from WTIEC and DDX5△IEC littermates were previously reported39. Gene set enrichment analysis was carried out using the pre-ranked mode of the Gene Set Enrichment Analysis (GSEA) software with default settings52. The gene list from DEseq2 was ranked by calculating a rank score of each gene as −Log10(p-value) × sign (Log2 Fold Change), in which Fold Change is the fold change of gene expression in DDX5 △ IEC over that found in WTIEC. De novo motif enrichment in the promoter regions of differentially expressed genes (log2 fold change cutoffs of ≥ 0.5 or ≤−0.5 and p-values <0.05) was performed using the HOMER (v4.10) “findMotifs.pl” command line with the following parameters: “-nogo -start −1000 -end 500”53. All other promoters of genes expressed in those particular cells were used as background. The top 5 motifs ranked by the lowest p-value that were found in at least 3% of the differentially expressed “Target” genes were illustrated. The tumor transcriptomic sequencing data reported in this paper is available on GEO (GSE146014, reviewer access token: gbahcsuozruptyf).
For metatranscriptomic analysis of ileal associated microbial populations, reads from the DDX5ΔIEC and WTIEC IEC RNAseq dataset that were not mapped to the mouse genome were assigned with taxonomic labels using Kraken v1. The standard Kraken database encompassing annotated bacterial, archaeal, and viral genomes was used for classification of sequences with the command: “kraken --classified-out /path/to/classified_fq --unclassified-out /path/to/unclassified.fq --db $DBNAME --paired --fastq-input pair1.fa pair2.fa > /path/to/results”. A Kraken report was generated with the the command: “kraken-report --$DBNAME kraken.output”54. Differential microbial counts were assessed by DEseq2 cut off of p<0.05 with the Wald test and Log2 fold change (DDX5ΔIEC/WTIEC)>1.5 or <−1.5.
Spatial transcriptomics
Female mouse ileum samples were coiled into “Swiss rolls” and frozen immediately with chilled isopentane. The frozen tissues were embedded in Optimal Cutting Temperature (OCT) compound and cut in a pre-cooled cryostat at 20mM thickness onto two 6.5mm × 6.5mm capture areas with 5000 oligo-barcoded spots. Frozen tissue was tested for RNA quality with RIN > 7.0 (Tapestation). Slides were fixed, H&E stained, and imaged on Keyence (bz-x800) at 20x magnification. Tissues were permeabilized for 24 minutes. Reverse transcription and second strand synthesis were then performed on RNAs extracted from each tissue. cDNAs were quantified by qRT-PCR using Powerup SyBr Green master mix and the Bio-Rad system. The library integrity was confirmed by the Agilent TapeStation. Pooled libraries were sequenced on NovaSeq S4 (Illumina) using 150 base-pair paired-end dual-indexed configuration. Each sample was sequenced to a depth of 100 million. Spaceranger software (version 3.1.0) from 10X Genomics was used to process, align and summarize unique molecular identifier (UMI) counts against the mm10 human reference genome. Loupe Browser (version 5.0) was used for differential gene expression analysis and generation of visuals. The spatial transcriptomic sequencing data reported in this paper is available on GEO (GSE184564, reviewer access token: obaruukivrmpxad).
Ribosome pull-down assay
Small intestine IECs were lysed in polysome extraction buffer (10 mM HEPES, pH 7.4, 150 mM KCl, 5 mM MgCl2, 1% NP40, 2 mM dithiothreitol, 80 U/ml RNaseOUT, 100 μg/ml cycloheximide, and protease inhibitors). Cell extracts were subject to anti ribosome IP_overnight with 2 μg anti-RPL10A antibodies (Abcam) as described previously39. Level of RPL10A associated transcripts in pull-down was calculated as fraction of input for each sample.
Statistical analysis
All values are presented as mean ± standard deviation (SD). Significant differences were evaluated using GraphPad Prism 8 software (GraphPad). The Student’s t-test was used to determine significant differences between two groups with normal distribution. A two-tailed p-value of <0.05 was considered statistically significant in all experiments.
Results
DDX5 regulates tuft cell populations in the murine small intestine and colon
In our previous report, we showed that DDX5 is highly expressed in the intestinal epithelium and regulates intestinal inflammation. However, the exact mechanism remained to be elucidated. In a recent studies activation of the tuft cell circuit is also linked to intestinal inflammation10. Therefore, we asked whether DDX5 may regulate intestinal inflammation at least in part by contributing to the differentiation and function of tuft cells and/or other IEC subsets. To address this question, we performed lineage-specific Gene Set Enrichment Analysis (GSEA) on our previously reported transcriptomes of steady-state IECs obtained from two matched pairs of WTIEC and DDX5△IEC male littermates39. A loss of the tuft cell signature was found in both the small intestinal and colonic epithelia of DDX5△IEC mice (Figure 1A–B and Supplementary Figure 1A). IECs from independent pairs of male and female WTIEC and DDX5△IEC littermates were used to confirm significant reductions in the expressions of two tuft cell-specific transcripts, Pou2f3 and Dclk1. In contrast, comparable expressions of select transcripts previously shown to be enriched in other IEC subsets, including Maf (enteroendocrine cells), Muc2 (goblet cells), Lyz1 (Paneth cells), Lgr5 and Ki67 (ISCs), were found in control and DDX5ΔIEC mice (Supplementary Figure 1B). These results suggest that DDX5 may have a unique role in regulating tuft cell differentiation and/or function in the intestine.
Figure 1. Epithelial DDX5 promotes the tuft cell gene program in the intestine.
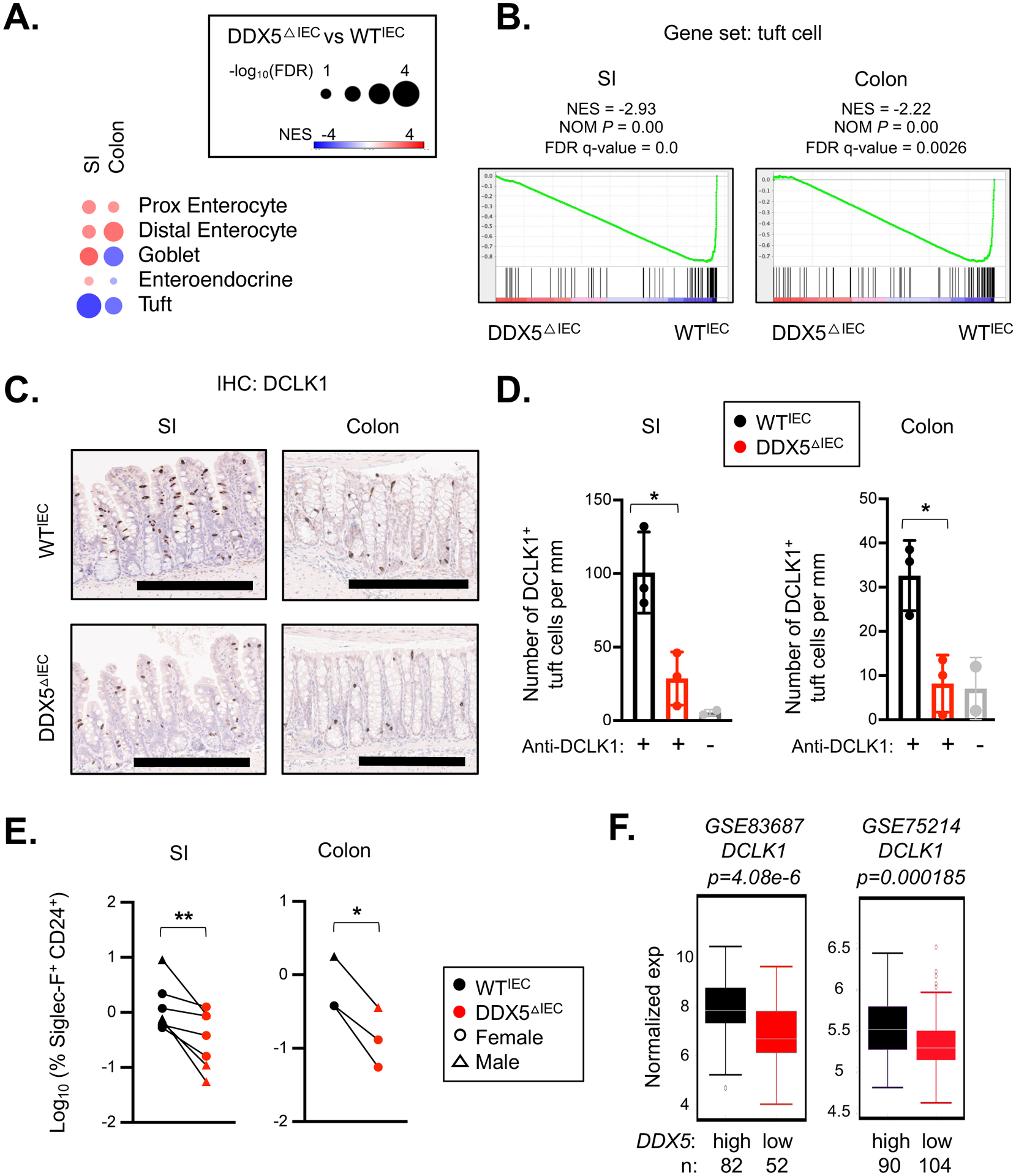
A. Heatmap summarizing the GSEA results on the indicated IEC gene subsets in the RNAseq dataset obtained from the ileal (SI) and colonic IECs of two pairs of male WTIEC and DDX5ΔIEC mice (GSE123881). NES, normalized enrichment score; FDR, false discovery rate.
B. GSEA enrichment plots of the tuft cell gene set in WTIEC and DDX5ΔIEC ileal (SI) and colonic IECs from A. NES, normalized enrichment score; NOM P, normalized nominal p-value.
C. Representative images from immunohistochemistry analysis of DCLK1+ cells in intestinal sections from female WTIEC and DDX5ΔIEC mice. Scale bar represents 300μm.
D. DCLK1+ tuft cell counts on ileal (SI) and colonic sections from 3 pairs of female WTIEC (black, n=3) and DDX5ΔIEC (red, n=3) mice analyzed according to the automated workflow described in Supplementary Figure 2A. Mice were fasted overnight to minimize the impact of varying food intake prior to tissue harvest. Each dot represents the result from one mouse. No primary antibody controls are shown in grey. * p<0.05 (unpaired t-test).
E. Summary of flow cytometry results of WTIEC and DDX5ΔIEC ileal (SI) and colonic epithelium. IECs were defined as live CD4−CD8−Epcam+CD45low. Proportion (%) of IECs in the tuft cell lineage defined as Siglec-F+CD24+ are graphed. See Supplementary Figure 3A for gating strategy. Each dot represents the result from one mouse. * p<0.05, ** p<0.01 (paired t-test).
F. Human DCLK1 expression in colonic biopsies with DDX5 expression below (low) or above (high) the mean of each respective dataset (GSE8368755 and GSE7521456).
Next, we investigated whether the loss of tuft cell gene signature in DDX5ΔIEC IECs may be due to a loss of the intestinal tuft cell population. Indeed, immunohistochemistry analysis revealed a significant reduction of DCLK1+ tuft cells in the DDX5△IEC small intestine and colon (Figure 1C–D, workflow described in Supplementary Figure 2A). In contrast, Periodic acid-Schiff staining (PAS) of the small intestine and colon sections from control and DDX5△IEC mice showed comparable crypt density and goblet cell numbers (Supplementary Figure 2B–D). Consistent with the histology results, flow cytometry analysis of the small intestine and colonic IECs (defined as CD45−EpCAM+) also confirmed a significant reduction of tuft cells (defined as Siglec-F+CD24+, similar to a previous report51) in the DDX5△IEC IECs from male and female mice (Figure 1E, gating strategy detailed in Supplementary Figure 3A). These results demonstrate that DDX5 promotes tuft cell biogenesis in the murine small intestine and colon. In two human colonic IEC transcriptomic datasets previously reported55 56, elevated DCLK1 expression was also associated with higher DDX5 expression (Figure 1F), suggesting regulation of the tuft cell gene program by DDX5 is likely evolutionarily conserved.
Impaired expression of the tuft cell commitment factor, Pou2f3, in DDX5△IEC IEC secretory lineage progenitors
In the small intestine, ATOH1 and SOX4 expressing IECs are progenitors of the secretory lineages, including tuft cells, goblet cells, Paneth cells, and enteroendocrine cells20 57 58. Next, we asked whether DDX5 contribute to the generation of these progenitor populations and/or their ability to express factors required for the generation of each secretory lineage, including Pou2f3-Gfi1b for tuft cells, Spdef for Goblet cells, Sox9 for Paneth cells, and Neurod1 for enteroendocrine cells. Spatial transcriptomics (10X Visium) analysis of ileal sections revealed similar number of Atoh1high and Sox4high spots were present in the WTIEC and DDX5△IEC tissues (Figure 2A). As expected, Pou2f3 transcripts were more abundantly found in Sox4hi spots than Atoh1hi spots in the wildtype ileum. However, its expression was greatly reduced in both Sox4high and Atoh1hi spots from the DDX5△IEC ileum (Figure 2B). In contrast, Atoh1high spots from the DDX5△IEC ileum showed higher levels of Neurod1, but similar levels of Gfl1, Spdef, and Sox9 compared to Atoh1high spots from WTIEC ileum (Supplementary Figure 4A). Similar differential gene expression analysis on Lgr5hi spots, but revealed only a relatively minor footprint of DDX5 in the ISCs (Supplementary Figure 4B and Supplementary Table 3), consistent with our earlier observation from our bulk IEC analysis (Supplementary Figure 1B). These results suggest that DDX5 may regulate tuft cell biogenesis by promoting Pou2f3 expression in the secretory lineage progenitors.
Figure 2. Spatial transcriptomic analysis revealed altered secretory lineage progenitor signature in the DDX5ΔIEC mice.
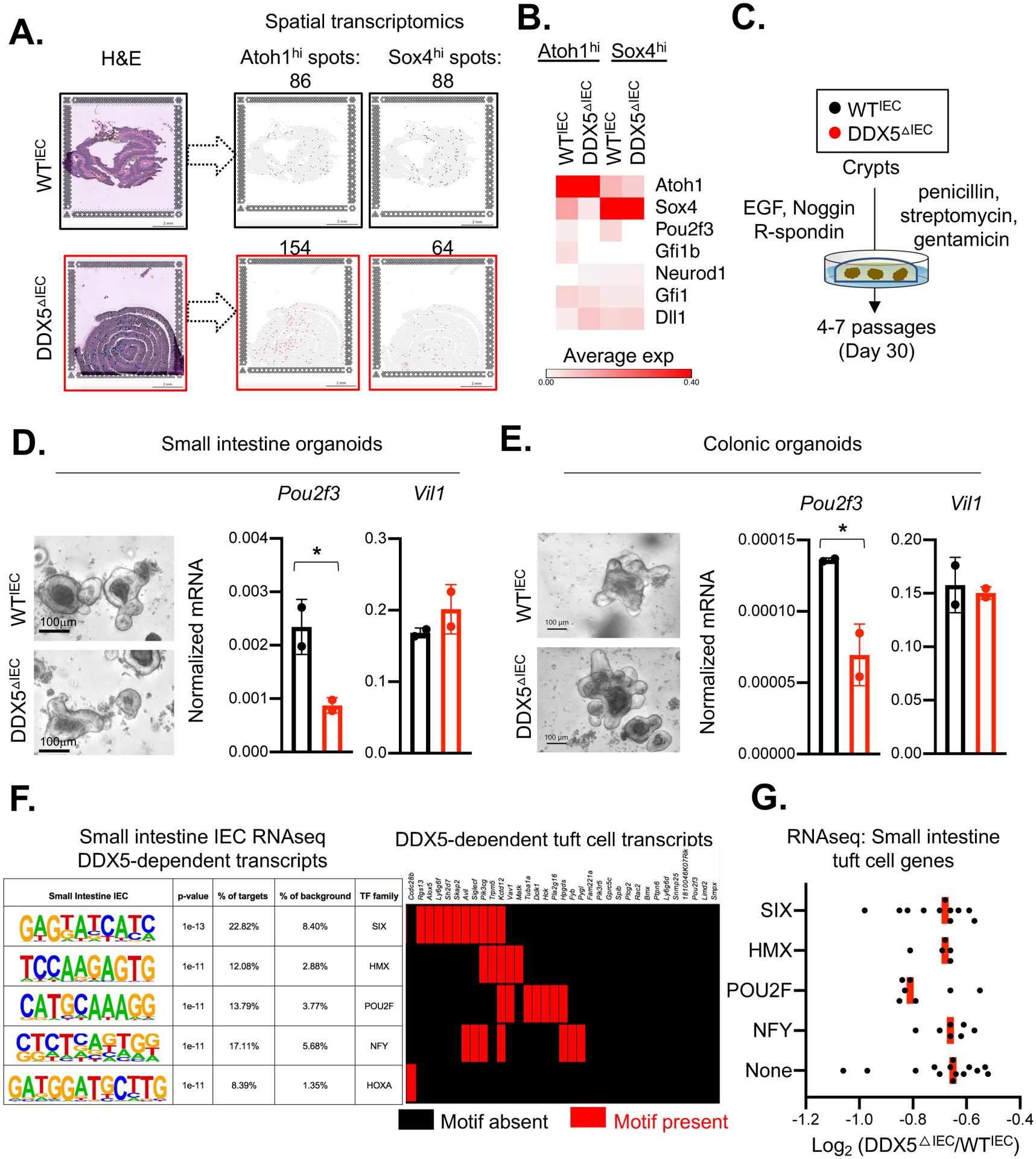
A. Left: H&E staining of the small intestinal swiss rolls from one pair of female WTIEC and DDX5ΔIEC littermates used for the spatial transcriptomic analysis (Visium, 10X Genomic). Right: Loupe browser displaying Atoh1high or Sox4high spots on each tissue section. Scale bar: 2mm.
B. Heatmap indicating the average expression of IEC lineage-specifying genes in Atoh1high or Sox4high spots indicated in A.
C. Schematic of establishing intestinal crypt organoid cultures.
D. Left: Representative bright field images of organoids established from small intestinal crypts of WTIEC and DDX5ΔIEC mice following the protocol outlined in50. Scale bar, 100μm. Right: Normalized mRNA expressions of Pou2f3 and Vil1 from WTIEC (n=2) and DDX5ΔIEC (n=2) small intestinal organoids cultured over 4–7 passages. Each dot represents the result from an independent organoid experiment. Results were averaged from two independent biological experiments. * p<0.05 (unpaired t-test).
E. Left: Representative bright field images of organoids established from colonic crypts of WTIEC and DDX5ΔIEC mice following the protocol outlined in50. Scale bar, 100μm. Right: Normalized mRNA expressions of Pou2f3 and Vil1 from WTIEC and DDX5ΔIEC colonic organoids cultured over 4–7 passages. Each dot represents the result from an independent organoid experiment. Results were averaged from two independent biological experiments. * p<0.05 (unpaired t-test).
F. Left: HOMER de novo analysis identified the top 5 motifs present at the promoters (defined as −1 kb to +500 bp from the transcription start site) of all ileal IEC DDX5-dependent genes identified in Figure 1A39. Promoters from all other IEC expressed genes were used as background. Right: The presence (red) or absence (black) of the motifs indicated on the left at the tuft cell DDX5-dependent gene promoters.
G. Average log2 fold change of the transcript abundance of tuft cell-expressed genes harboring SIX, HMX, POU2F, NFY motifs, or None of the above motifs at their promoters in ileal IECs from two independent pairs of WTIEC and DDX5ΔIEC littermates. Each dot represents one gene.
Consistent with findings from the spatial transcriptomic studies, small intestine crypts enriched with ISC and progenitors isolated from DDX5ΔIEC mice had lower Pou2f3 mRNA abundance compared to those found in crypts obtained from their WTIEC littermates (Supplementary Figure 4C). In addition, DDX5ΔIEC small intestinal crypts cultured ex vivo over 4–7 passages in the presence of the Wnt pathway agonist R-spondin (Rspo), epidermal growth factor (EGF), the BMP inhibitor Noggin (as previously described16), penicillin, streptomycin, and gentamicin developed into organoids and maintained lower Pou2f3 expression compared to those derived from WTIEC mice (Figure 2C–D). Similar observations were found in DDX5ΔIEC colonic organoids (Figure 2E). No significant change in organoid viability and size were found between cultures obtained from control or DDX5ΔIEC crypts (Supplementary Figure 5A). Furthermore, tuft cell-specific genes harboring a promoter POU2F motif had the greatest loss in the expression in the DDX5ΔIEC tissue (Figure 2F–G). Together, these results demonstrate an epithelial cell-intrinsic role of DDX5 in maintaining Pou2f3 expression in vivo and ex vivo.
DDX5 is dispensable for IL-13 driven tuft cell hyperplasia
Differentiation of IEC subsets, including tuft cells, is heavily influenced by the local microbial composition and immune cytokine profiles. In the presence of protozoan Trichomonas muris, for example, the small intestinal tuft cell population expands rapidly in an IL-13-dependent manner3 4. Therefore, we asked whether the loss of tuft cell numbers in the DDX5ΔIEC intestine may be explained by a reduction in local IL-13 level, difference in Trichomonas abundance, and/or expressions of IL-13 receptors in the ISC and progenitors. At the RNA level, we found a similar abundance of transcripts encoding Il13 expressed by immune cells in the lamina propria of cohoused WTIEC and DDX5ΔIEC littermates (Supplementary Figure 6A). Furthermore, cohoused WTIEC and DDX5ΔIEC littermates acquired similar levels of Trichomonas species from their parents prior to weaning (Supplementary Figure 6B).
To assess whether the Trichomonas species present in our mouse colony were implicated in the tuft cell specification, we treated cohoused WTIEC and DDX5ΔIEC littermates with metronidazole in drinking water for 14 days to deplete majority of the Trichomonas species together with other metronidazole sensitive bacterial species (Supplementary Figure 6B). Interestingly, WTIEC mice treated with metronidazole had no significant change in the proportion of tuft cells in the small intestine and colon as measured by flow cytometry (defined as Siglec-F+CD24+). Likewise, the proportion of tuft cells and epithelial Pou2f3 transcript levels in the metronidazole-treated DDX5△IEC mice remained lower than levels found in WTIEC mice under similar treatment (Supplementary Figure 6C). These results suggest that the metronidazole-sensitive Trichomonas and bacteria species present in our mouse colony were not involved in the establishment of tuft cell in the WTIEC and DDX5ΔIEC mice. It remained to be investigated whether specific metronidazole-insensitive microbe(s) and/or metabolites present in our colony contribute to the DDX5-dependent generation of tuft cell we observed.
Furthermore, we found no defect in the ability of DDX5ΔIEC ISC and secretory lineage progenitors to express genes encoding receptors for IL-13 (Supplementary Figure 7A). Therefore, we hypothesized that DDX5 is likely dispensable for tuft cell hyperplasia in response to IL-13. To test this possibility, WTIEC and DDX5ΔIEC small intestinal and colonic organoids were treated with recombinant IL-13 for three days. At the RNA level, both WTIEC and DDX5ΔIEC organoids upregulated Pou2f3 to a similar extent, resulting in a ≥2 fold increase in tuft cell numbers compared to vehicle-treated organoids (Supplementary Figure 7B–D). Together, these results suggest that DDX5 is dispensable for type 2 immune cytokine-driven tuft cell hyperplasia.
DDX5 promotes Cdc42 translation to induce tuft cell biogenesis
Next, we asked whether DDX5 promotes Pou2f3 expression and drives tuft cell biogenesis by directly binding to Pou2f3 mRNA and/or transcripts encoding its upstream regulators. To this end, we examined the DDX5-associated RNA interactome identified by the enhanced cross-linked immunoprecipitation (eCLIPseq) experiment on IECs derived from the steady-state WT small intestine39. To our surprise, we did not identify any DDX5 footprint on the Pou2f3 transcript, suggesting that DDX5 likely regulate Pou2f3 expression indirectly. Of the eleven known molecules previously implicated in Pou2f3 expression and tuft cell biogenesis (Gfi1b, Mtor, Rptor, Stat6, Ogt, Tas1r3, Sox4, Hes1, Dll1, Atoh1, and Cdc42), only the Cdc42 transcripts were bound by DDX5 (Figure 3A). CDC42 is a GTP-binding protein previously implicated in ISC survival, growth, as well as tuft cell biogenesis23. In the absence of DDX5, IECs had reduced CDC42 protein abundance (Figure 3B). Spatial transcriptomics analysis revealed that Cdc42 is ubiquitously expressed across all IEC subsets, and most abundantly found in Lgr5hi ISCs and Sox4hi progenitors (Figure 3C). Murine retrovirus carrying a CDC42 (isoform 1, V1) expression construct partially rescued CDC42 protein levels in DDX5ΔIEC small intestinal organoids (Supplementary figure 8A–B) and restored the Siglec-F+CD24+ tuft cell population to levels found in WTIEC cultures (Figure 3D–E). These results demonstrate that DDX5 promotes tuft cell biogenesis by regulating CDC42.
Figure 3. DDX5 regulates Pou2f3 expression and tuft cell specification by promoting Cdc42 translation.
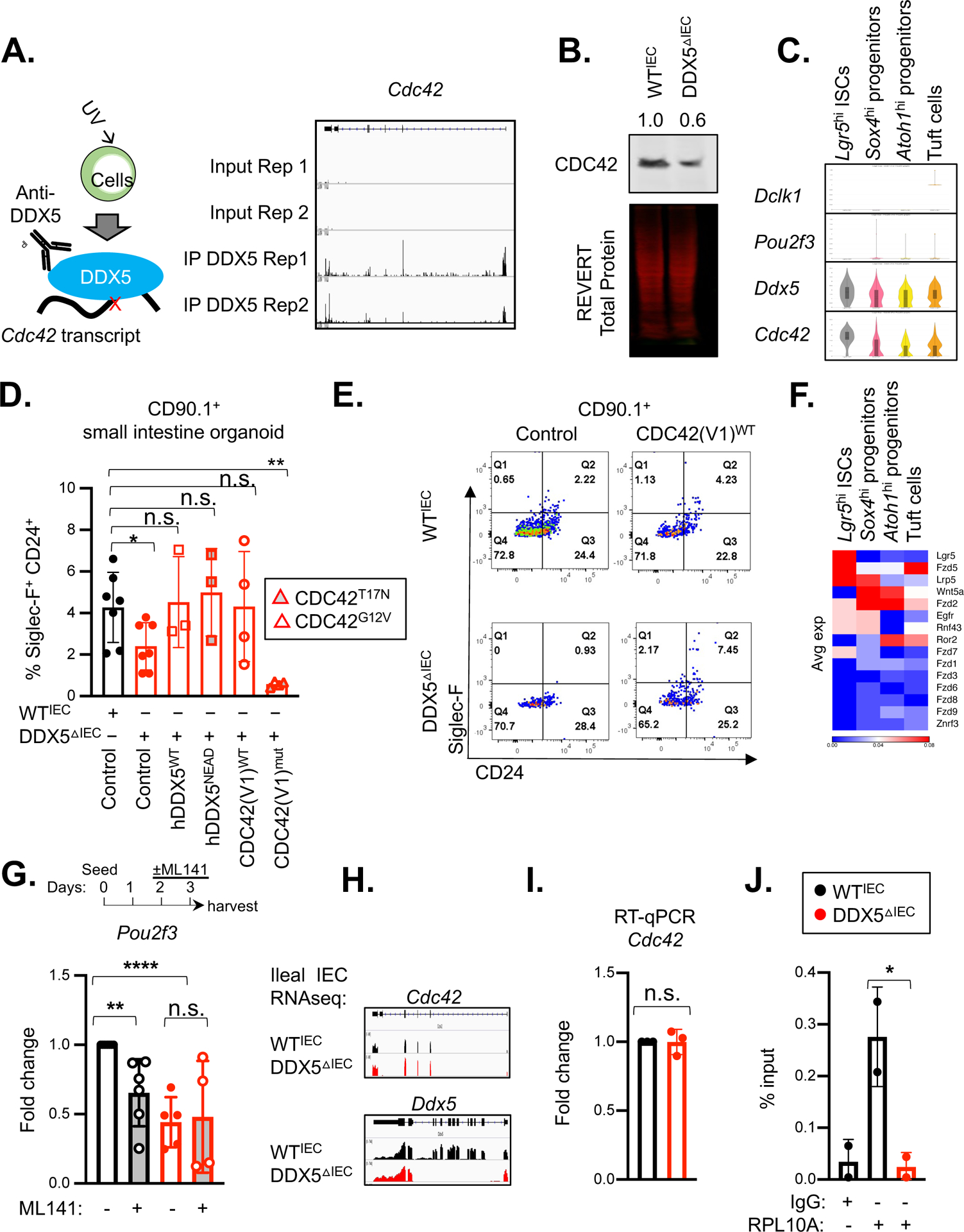
A. Left: Schematic representation of the enhanced CLIP (eCLIPseq) experiment. Right: Integrative Genomics Viewer (IGV) browser view of DDX5 eCLIPseq signals at the Cdc42 loci from two independent experiments.
B. Representative western blot for CDC42 in whole cell lysates of small intestinal IECs from WTIEC and DDX5ΔIEC mice. Experiments were repeated three times using independent biological samples with similar results.
C. Violin plots displaying expression of Dclk1, Pou2f3, Ddx5, and Cdc42 in small intestinal stem cells (ISC, Lgr5hi), Atoh1hi and Sox4hi secretory lineage progenitors, or tuft cells from the spatial transcriptomics dataset described in Figure 2A.
D. Summary of the % of SiglecF+CD24+ tuft cells in WTIEC and DDX5ΔIEC small intestinal organoids 7 days post-transduction with retrovirus (marked by CD90.1+) carrying the indicated expression vectors. Each dot represents the result from an independent organoid experiment. n.s. not significant, * p<0.05, ** p<0.01 (unpaired t-test).
E. Representative flow cytometry plots from D.
F. Heatmap indicating the average expression of known signaling molecules upstream of CDC42 activation in ISCs, progenitors, and tuft cells from Figure 2A.
G. Fold change of Pou2f3 mRNA in WTIEC (n=6) or DDX5ΔIEC (n=5) small intestine organoids cultured in the presence of CDC42 inhibitor ML141 (10μM) for 24 hours relative to WTIEC cells treated with vehicle control (DMSO). Each dot represents the result from one mouse. n.s. not significant, ** p<0.01, **** p<0.001 (unpaired t-test).
H. IGV browser view of WTIEC and DDX5ΔIEC ileal IEC RNAseq-derived sample reads mapped to the Ddx5 and Cdc42 loci (GSE123881).
I. Fold change of ileal Cdc42 mRNA in DDX5ΔIEC mice (n=3) relative to values found in WTIEC mice (n=3). Each dot represents the result from one mouse. n.s. not significant (unpaired t-test).
J. Ribosome RPL10A enrichment from Cdc42 mRNA in small intestinal IECs. Results shown represent the average of two independent experiments. Each dot represents the result from one mouse. * p<0.05 (unpaired t-test).
EGF and R-spondin in the organoid culture media engage EGFR and Wnt signaling pathways known to drive CDC42 activity. At the RNA level, small intestinal Sox4hi progenitors abundantly expressed Egfr, Fzd2, and Lrp5 (Figure 3F). Therefore, we asked whether CDC42 GTPase activity may be implicated in the generation of tuft cells in the organoid culture system. Consistent with this possibility, DDX5ΔIEC small intestinal organoids transduced with the dominant negative CDC42T17N or CDC42G12V expression constructs failed to restore the Siglec-F+CD24+ tuft cell population (Figure 3D), suggesting that the GTPase activity of CDC42 is required for DDX5-mediated tuft cell biogenesis. To bypass the essential function of CDC42 in ISC reported previously23 59, established WT organoids were passaged, seeded on fresh Matrigel, and allowed to differentiate normally for 48hrs and then subjected to CDC42 inhibition by ML141. In this assay, acute ML141 treatment did not result in measurable difference in organoid survival or growth (Supplementary Figure 9A). We also did not detect significant change in the abundance of Klf4, Lyz1, and Lgr5 transcripts in ML141 treated organoids (Supplementary Figure 9B). However, Pou2f3 expression was significantly diminished in WT organoids transiently treated with ML141 for 24hrs (Figure 3G). Intriguingly, ML141 treated wildtype small intestinal and colonic organoids remained responsive to IL-13 by expanding their tuft cell population to a similar extent as control treated cultures (Supplementary Figure 9C–D). Together, these results confirm that the DDX5-CDC42 axis is dispensable for IL-13-induced tuft cell expansion ex vivo.
Mechanistically, DDX5 did not alter IEC Cdc42 transcript abundance (Figure 3HI). Ribosome capture assay revealed that optimal translation machinery engagement on Cdc42 mRNA transcripts in small intestinal IECs was DDX5-dependent (Figure 3J). Intriguingly, retrovirus carrying wildtype DDX5 or the helicase dead DDX5DEAD, expression constructs were able to rescue the Siglec-F+CD24+ tuft cell population in DDX5ΔIEC small intestinal organoids to levels found in WTIEC cultures (Figure 3D). This suggests that DDX5 helicase activity is dispensable for promoting Cdc42 ribosomal engagement. Together, results from our genetic, genomic, and pharmacological experiments demonstrate that epithelial DDX5 binds Cdc42 transcripts and promotes CDC42 protein translation in a helicase activity-independent manner to control tuft cell biogenesis.
DDX5 negatively regulates tuft cell lipid and protein metabolic programs
Despite a significant loss of tuft cells in the DDX5△IEC intestine, both immunohistochemistry and flow cytometry analyses demonstrated a small population of tuft cells can be generated in the absence of DDX5 (Figure 1C–E). Next, we tested whether DDX5△IEC tuft cells may be morphological and/or functional distinct from those with intact DDX5 expression. Electron microscopy revealed similar brush-like microvilli structures characteristic of elongated tuft cells in both WTIEC and DDX5△IEC small intestine (Figure 4A), suggesting that DDX5 does not influence tuft cell morphology.
Figure 4. DDX5-dependent tuft cell gene programs in the small intestine.
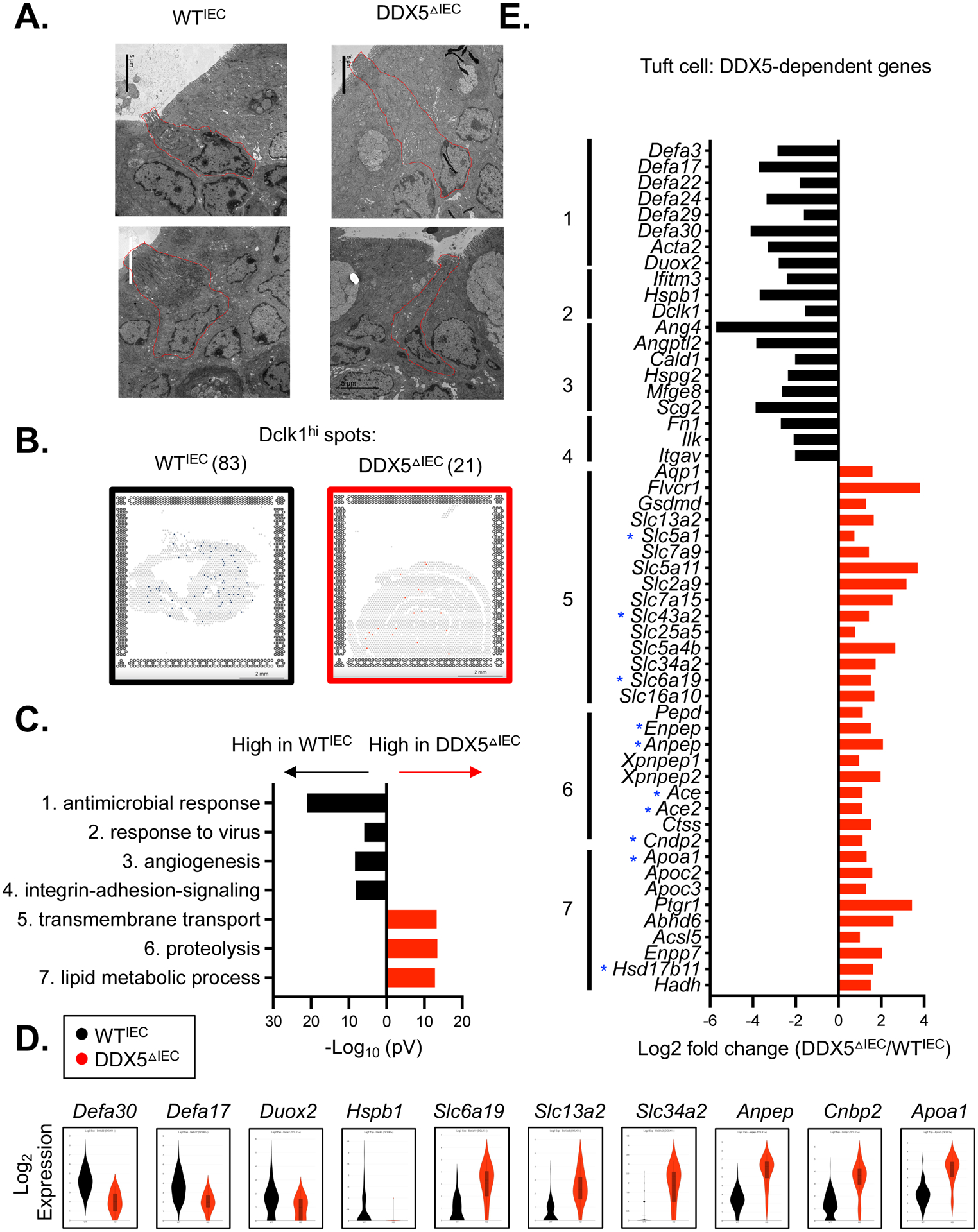
A. Representative electron microscopy images of small intestinal tuft cells from WTIEC and DDX5ΔIEC mice. Scale bar: 5μm.
B. Loupe browser screenshots displaying quantified Dclk1hi spots on the small intestinal sections from WTIEC and DDX5ΔIEC mice described in Figure 2A. Scale bar: 2mm.
C. Gene ontology analysis revealed the top 7 pathways downregulated (black) or upregulated (red) in DDX5ΔIEC Dclk1hi cells from B.
D. Violin plots displaying expressions of DDX5-dependent genes—with or without DDX5-associated transcripts—involved in the antimicrobial response (Defa3 and Defa17), viral response (Duox2 and Hspb1), transport (Slc6a19), proteolysis (Anpep and Cnbp2), or lipid metabolism (Apoa1) in WTIEC and DDX5ΔIEC Dclk1hi cells from C.
E. Log fold changes (DDX5ΔIEC/ WTIEC) in the expression of tuft cell DDX5-dependent genes participating in pathways identified in C. Small intestinal IEC DDX5-associated transcripts identified in eCLIP analysis are indicated with a blue star (GSE124023).
Previous reports clustered the small intestinal tuft cells into two main subsets: one having higher expression of genes related to neuronal development (type 1), and the other (type 2) enriched with immune-related genes, such as the Protein Tyrosine Phosphatase Receptor Type C gene encoding the pan-immune cell marker CD451 60. Of the few DDX5△IEC tuft cells present in the small intestine, their proportion of CD45 low- and mid-subpopulations were similar to those found among WTIEC tuft cells (Supplementary Figure 10A). GSEA of the small intestinal IEC transcriptomes39 also confirmed that both type 1 and 2 programs were significantly impaired in the DDX5△IEC epithelium (Supplementary Figure 10B).
To fully elucidate IEC subset-specific DDX5-dependent gene programs, we performed differential gene analysis on the spatial transcriptomic datasets obtained from the WTIEC and DDX5△IEC small intestinal sections discussed in Figure 2A. We uncovered 17.5% of the DDX5-induced transcripts and 3.8% of the DDX5-repressed genes were shared among tuft, enteroendocrine, goblet, and Paneth cells (Supplementary Figure 11A–B and Supplementary Table 4). The numbers of DDX5-regulated genes in goblet and enteroendocrine cells is lower than those found in tuft and Paneth cells. Notably, DDX5△IEC small intestinal tuft cells had reduced expression of genes involved in microbial responses (Figure 4B–D), including a loss of Cd300lf transcripts encoding a surface molecule used by the Murine norovirus (MNoV) as a key docking site for host entry2 (Supplementary Figure 12A). As a result, DDX5ΔIEC mice raised by MNoV+ parents had significantly fewer MNoV transcripts in their small intestinal epithelium compared to their cohoused wildtype littermates (Supplementary Figure 12A). In addition, meta-transcriptomic analyses of the WTIEC and DDX5△IEC intestinal epithelium transcriptomes revealed significantly lower mucosal-associated reads mapping to viral and bacterial genomes from the DDX5ΔIEC small intestine and colon, respectively (Supplementary Figure 12B–C). In particular, species of Helicobacters and Prevotella were significantly reduced in the DDX5ΔIEC colon (Supplementary Figure 12D and Supplementary Table 5).
While none of the DDX5-dependent genes involved in microbial responses harbor a DDX5 eCLIPseq footprint, ten of the genes involved in transmembrane transport and protein and lipid metabolism that were upregulated in DDX5△IEC tuft cells were direct targets of DDX5 (indicated with a blue star on Figure 4E). These results reveal a novel role of DDX5 as a repressor of transmembrane transport and protein and lipid metabolic programs in tuft cells of the small intestine. Together, these results extend the role of DDX5 in shaping IEC subset gene programs and modulating the microbial community in the intestine.
Succinate-induced tuft cell hyperplasia protects against ileitis and restores colon tumorigenic potential in DDX5△IEC mice
Small intestinal tuft cells have been recently reported to provide protection against T-lymphocyte mediated ileitis upon anti-CD3ɛ challenge10. Therefore, we hypothesized that the reduced tuft cell numbers in the small intestine of DDX5ΔIEC mice may result in enhanced susceptibility to ileitis. Consistent with our hypothesis, 50% of the DDX5ΔIEC mice challenged in this model succumbed to the disease by day 18 (Figure 5A). Of the ones that survived, mononuclear immune cells from their ileal lamina propria had elevated transcripts encoding the inflammatory cytokine TNF (Figure 5B). Pre-administration of succinate, a microbial-derived metabolite known to promote tuft cell hyperplasia in an IL-13 dependent manner4 6, in drinking water was able to partially restore Dclk1 expression in the small intestine epithelium of DDX5ΔIEC mice (Figure 5C), similar to previous reports3 10, dampened Tnf expression, and significantly protected them against mortality from the anti-CD3ɛ challenge (Figure 5A–B).
Figure 5. Succinate-induced tuft cell hyperplasia protects DDX5ΔIEC mice against exacerbated ileitis and restores tumorigenic potential in the DDX5ΔIEC colon.
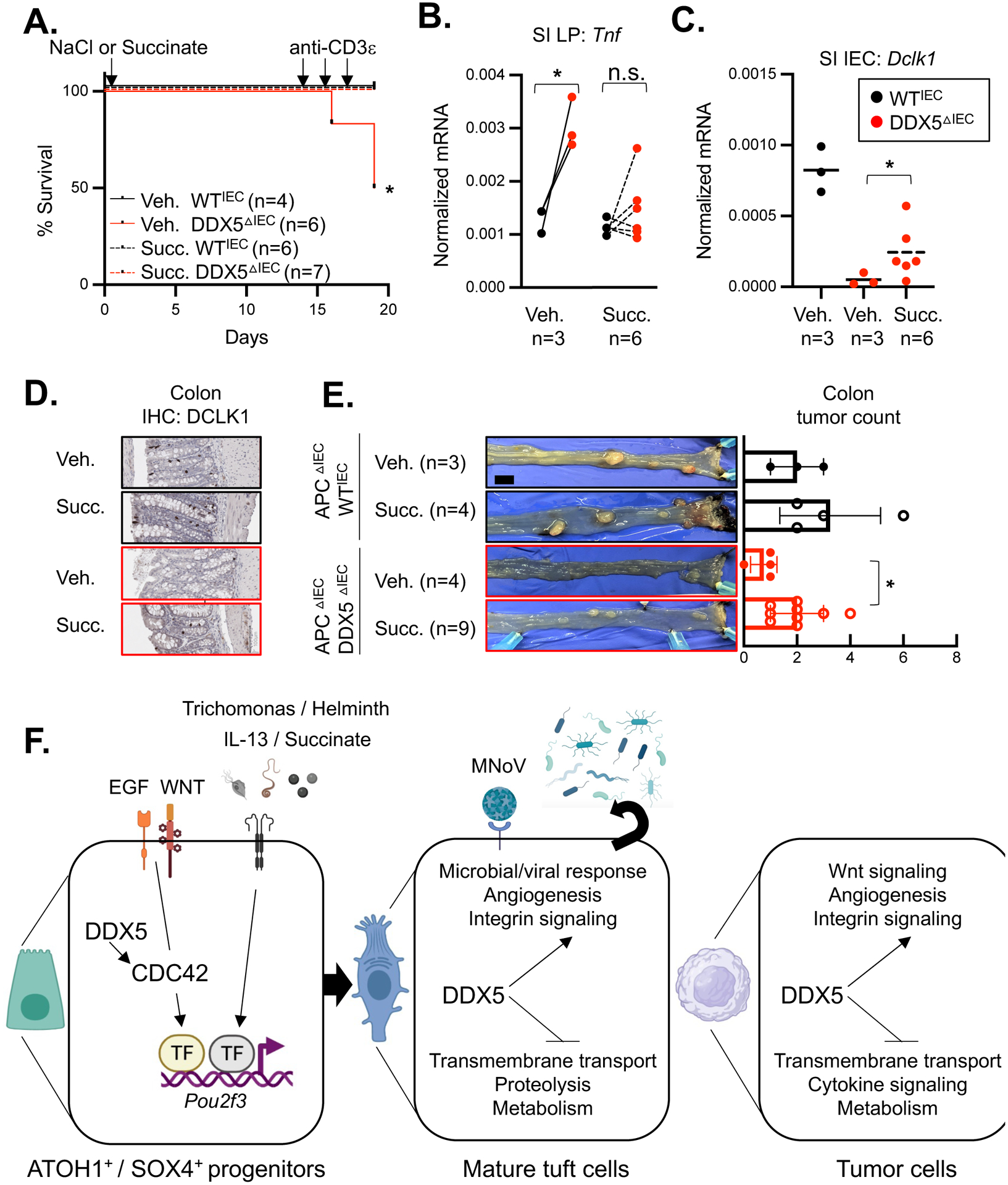
A. Survival plot of mice treated with succinate-NaCl (Succ., WTIEC, n=6; DDX5ΔIEC, n=7) or equal molar of NaCl (Veh., WTIEC, n=4; DDX5ΔIEC, n=6) in drinking water for 2 weeks and subsequently challenged by by 15μg of anti-CD3ɛ per mouse for 5 days. * p<0.05 (Mantel-Cox test).
B. Normalized mRNA expression of Tnf in small intestinal lamina propria (LP) mononuclear cells isolated from mice from A on day 19. Each dot represents the result of one mouse. n.s. not significant, * p<0.05 (paired t-test).
C. Normalized mRNA expression of Dclk1 in the small intestinal IECs isolated from mice from A on day 19. One dot represents the result of one mouse. * p<0.05 (unpaired t-test).
D. Representative images of DCLK1 staining in colon from mice treated with NaCl (Veh.) or succinate-NaCl in drinking water for 30 days. Scale bar represents 100μm.
E. Left: Representative images of colonic tumors from 120 days old APCΔIEC DDX5WT and APCΔIEC DDX5ΔIEC mice treated with succinate-NaCl or equal molar of NaCl in drinking water on day 90 for 30 days. Right: Summary of colonic tumor counts from treated APCΔIEC DDX5WT and APCΔIEC DDX5ΔIEC mice. Each dot represents the result from one mouse. * p<0.05 (unpaired t-test). Scale bar represents 1cm.
F. Working model. Left: DDX5-CDC42-POU2f3 axis regulating progenitor commitment toward the tuft cell lineage. Right: DDX5-dependent gene programs in differentiated tuft cells.
In the colon, multiple IEC subsets, including DCLK1+ tuft cells, have been reported to harbor tumorigenic potential11 12. We recently reported that DDX5ΔIEC mice on the intestinal tumor-susceptible ApcΔIEC background46 harbored fewer intestinal tumors39. Transcriptomic analysis of control and DDX5 deficient tumors revealed pathways as well as a subset of DDX5-dependent genes that were similarly regulated in tuft cells (Supplementary Figure 13A–C and Supplementary Table 6). Therefore, we tested whether succinate supplementation can also promote colonic tuft cell programs, including Dclk1 expression, and restore the tumorigenic potential of the APCΔIECDDX5ΔIEC mice. Immunohistochemistry analysis confirmed partial restoration of the DCLK1+ population in succinate treated DDX5ΔIEC mice (Figure 5D). In the tumor susceptible background APCΔIEC, succinate treated DDX5ΔIEC mice had higher colonic tumor counts than those treated with vehicle control alone (Figure 5E). Altogether, these results highlight the critical roles of epithelial DDX5 in protecting against ileal inflammation yet contributing to colonic tumorigenesis (modeled in Figure 5F).
Discussion
Tuft cells in the small intestine and colon arise from the SOX4+ and ATOH1+ progenitors respectively19–21. Commitment to the tuft cell lineage requires the expression of the POU domain transcription factor, POU2F322. Induction of Pou2f3 is best characterized in response to IL-13 and IL-4 stimulation during helminth and protist challenges2 5–9. However, the molecular mechanisms that control Pou2f3 expression in the absence of IL-13 and IL-4 stimulation remain elusive. Here, we report that CDC42 and its GTPase activity is essential for Pou2f3 expression. Consistent with our findings, epithelial specific knockout of CDC42 resulted in a loss of intestinal tuft cells, but no change to other secretory IEC subsets23. In the small intestine, we show that Cdc42 is the abundantly expressed in ISCs and secretory lineage progenitor cells. Optimal translation of the Cdc42 transcript rely on the RNA binding protein DDX5 in a helicase-activity independent manner. Knocking out DDX5 specifically in IECs results in reduced CDC42 protein abundance and loss of tuft cell populations in both the small intestine and colon. Ex vivo, retroviral expression of wildtype CDC42 rescued the tuft cell biogenesis defect in small intestinal DDX5ΔIEC organoids.
Surprisingly, the DDX5-CDC42 axis is dispensable for tuft cell hyperplasia in response to IL-13. WT organoids cells treated with the CDC42 inhibitor and DDX5ΔIEC organoids remain capable of responding to IL-13, upregulating Pou2f3 expression, and inducing tuft cell expansion. Future studies will be needed to investigate whether these parallel DDX5-CDC42 and IL-13-induced pathways recruit shared or distinct transcription factors and/or machineries to drive Pou2f3 expression in the secretory lineage progenitors. Compared to the secretory lineage progenitors, DDX5 has a limited transcription footprint on ISCs. We also did not find abnormalities in growth and survival in the DDX5ΔIEC crypts containing ISCs and progenitors, unlike those observed in cells from the CDC42ΔIEC mice23. We speculate that this likely suggest that the remaining CDC42 levels in the DDX5ΔIEC epithelium is sufficient to maintain ISC growth and survival and/or CDC42 expression in ISC may be DDX5-independent. Future studies that genetically ablate Ddx5 in specific IEC progenitor and/or IEC subsets will be needed to more definitively address whether the contribution of DDX5 to tuft cell differentiation and function may be cell-autonomous and/or indirect.
In the small intestine, tuft cells protect against ileal inflammation in a mouse model and a loss of tuft cells is identified in Crohn’s disease patients10. As expected, DDX5ΔIEC mice with reduced intestine tuft cell biogenesis were more susceptible to disease in a model of ileitis. Administration of succinate partially restored tuft cell numbers in DDX5ΔIEC mice and allowed for greater resilience during ileitis. Compared to the small intestine, the colon epithelium harbored fewer tuft cells and their physiologic and pathologic functions are not well understood. Results from this study suggest that the DDX5-CDC42 axis is also implicated in colonic tuft cell biogenesis. In contrast to the anti-inflammatory role of DDX5 in the small intestine IECs, we previously reported that DDX5 in the colonic IECs promotes inflammation in a DSS-induced model of colitis39. Regional specific roles of DDX5 in intestine inflammation may be related to distinct microbe-host interactions and/or distinct gene programs regulated by DDX5 in colonic IECs. Future single-cell transcriptomic studies will be needed to fully elucidate the mechanisms underlying these specificities. Development of additional IEC-subset specific knockout models will be needed for definitively attributing specific DDX5 regulated programs to the various phenotypes observed here.
In summary, this study uncovered an epithelial cell intrinsic pathway involving the RNA binding protein DDX5 and its ability to regulate the CDC42-POU2F3 axis critical for tuft cell biogenesis that is parallel but distinct from the IL-13 induced hyperplasia responses and can serve as new molecular targets for the restoration of mucosal homeostasis in disease settings.
Supplementary Material
Supplementary Figure 1: IEC-subset specific gene expression in the ileal and colonic epithelium.
A. Expression of genes determined by RNAseq (DESeq2) analysis of the ileal and colonic IECs from two pairs of WTIEC and DDX5ΔIEC mice as described in Figure 1A. Proximal enterocytes (Prox Ent), distal enterocytes (Dist Ent); Paneth cells (P), enteroendocytes (E.E). Black: log2 fold changes (DDX5ΔIEC/WTIEC) > 0.5; blue: log fold changes (DDX5ΔIEC/WTIEC) < −0.5.
B. Subset-specific gene expression analysis by RT-qPCR of ileal (left) and colonic (right) IECs from WTIEC (n=5) and DDX5ΔIEC mice (n=5). Each dot represents the result from one mouse pair.
Supplementary Figure 2: Intestinal crypt density and goblet cell numbers are DDX5-independent.
A. DCLK1+ cell quantitation workflow in QuPath48.
B. Representative Periodic acid-Schiff (PAS) staining of WTIEC and DDX5ΔIEC small intestines. Scale bar represents 100μm.
C. WTIEC (n=3) and DDX5ΔIEC (n=3) mice Ileal and colonic crypt densities. Each dot represents the result from one mouse. n.s. not significant (unpaired t-test).
D. WTIEC (n=3) and DDX5ΔIEC (n=3) mice ileal and colonic goblet cell counts. Each dot represents the result from one mouse. n.s. not significant (unpaired t-test).
Supplementary Figure 3: Tuft cell gating strategy.
A. Small intestines were incubated in 5 mM EDTA containing HBSS for 20 minutes at 37°C for the isolation of IECs. SSC: side scatter (an index for cell granularity). Tuft cells were defined as live EpCAM+CD4−CD8β−CD45low-midSiglec-F+CD24+ 4.
FSC-A: area under forward scatter (an index for cell size).
Supplementary Figure 4: DDX5 dependency of IEC subset defining genes in the intestinal ISC and secretory lineage progenitors.
A. Violin plots of expression from the indicated genes in Atoh1hi or Sox4hi cells from the spatial transcriptomic analysis from figure 2A.
B. Venn diagrams of distinct and overlapping DDX5-dependent genes (p<0.05) among ISC and the secretory lineage progenitors.
C. Normalized RNA expressions of Pou2f3 and Vil1 in small intestinal crypts isolated from two pairs of WTIEC and DDX5ΔIEC mice. Each dot represents the result from one mouse. ** p<0.01 (unpaired t-test).
Supplementary Figure 5: DDX5 is not involved in intestinal organoid survival and growth.
A. Viability and size of small intestinal organoids (46 WTIEC from 5 random fields; 62 DDX5ΔIEC organoids from 7 random fields) and colonic organoids (10 WTIEC from 5 random fields; 11 DDX5ΔIEC organoids from 5 random fields) two days after re-seeding of established cultures. Images were acquired under a 20X magnification.
Supplementary figure 6. Similar type 2 cytokines and trichomonas levels in WTIEC and DDX5ΔIEC mice.
A. RNA expressions of Il4, Il13, and Gata3 normalized to Gapdh in intestinal lamina propria lymphocytes from WTIEC (SI n=4; colon n=3) and DDX5ΔIEC (SI n=4; colon n=3) mice.
B. Levels of Trichomonas (left) and bacterial (right) genomes normalized to host genome detected in fecal or cecal contents. Each dot represents the result from one mouse. n.s. not significant, **** p<0.001 (t-test). Metro: metronidazole administered by drinking water.
C. Proportion of tuft cells and normalized RNA expression of Pou2f3 in the intestines of WTIEC and DDX5ΔIEC treated with or without metronidazole. Results shown are the average and standard deviation of two independent experiments. Each dot represents result from one mouse. * p<0.05 (t-test).
Supplementary Figure 7. IL-13 driven tuft cell hyperplasia is DDX5-independent.
A. Heatmap indicating the average expression of genes encoding receptors for IL-13 and IL-4 in ISCs, progenitors, and tuft cells from Figure 2A.
B. Normalized RNA expression of Pou2f3 in small intestine or colon organoids stimulated with IL-13 (20ng/ml) or vehicle (NaCl at 15μM) for 48hrs. Results shown are the average and standard deviation of two independent experiments. n.s. not significant, * p<0.05 (paired t-test).
C. Representative flow cytometry plots of SiglecF+CD24+ tuft cell populations from WTIEC and DDX5ΔIEC small intestinal and colonic organoids stimulated with IL-13 (20ng/ml) or vehicle (NaCl at 15μM) for 72hrs.
D. Summary of fold changes in the proportions of tuft cells in intestinal organoids stimulated with IL-13 compared to vehicle. Each dot represents the result from one independent experiment.
Supplementary Figure 8. Restoring CDC42 protein levels in DDX5ΔIEC small intestinal organoids by retroviral transduction.
A. Representative retrovirus transduction efficiency in small intestinal organoids was assessed using flow cytometry to measure surface expression of CD90.1. Experiments were repeated three times using independent biological samples with similar results.
B. Left: Representative CDC42 protein abundance in transduced small intestinal organoids as assessed by flow cytometry. Right: Summary of CDC42hi small intestine organoid IECs from three independent experiments. n.s. not significant, * p<0.05 (paired t-test).
Supplementary Figure 9. Transient inhibition of CDC42 activity did not alter wildtype intestinal organoid viability, growth, or response to IL-13 stimulation.
A. Viability and size of WTIEC small intestinal organoids 24hrs post treatment (DMSO, n=553 from 4 random fields; ML141, n=551 from 4 random fields) and 72hrs post treatment (DMSO, n=259 from 2 random fields; ML141, n=267 from 2 random fields). **** p<0.001, n.s. not significant (unpaired, t-test).
B. Normalized RNA expressions of Lyz1, Klf4 and Lgr5 in WTIEC small intestinal organoids cultured in the presence or absence of CDC42 inhibitor ML141 (10μM) for 24hrs. Each dot represents the result from one mouse. n.s. not significant (unpaired, t-test).
C. Representative flow cytometry plots of WTIEC small intestinal or colonic organoids cultured for 72hrs in the presence or absence of recombinant IL-13 (20ng/ml) and/or CDC42 inhibitor ML141 (10μM).
D. Summary of fold changes in tuft cell frequencies among WTIEC small intestinal and colonic organoids upon treatment with DMSO or ML141 inhibitor from C. Tuft cell lineage was defined as Siglec-F+CD24+ IECs4. Each dot represents the result from one independent experiment.
Supplementary Figure 10. DDX5 promotes both type 1 and type 2 tuft cell programs in the small intestine.
A. Left: Histogram of CD45 expression on gated ileal tuft cells (CD4−CD8−Epcam+Siglec-F+CD24+). Right: Proportion (%) of CD45low or CD45mid cells within the tuft cell lineage. Results from four pairs of WTIEC and DDX5ΔIEC cohoused littermates were graphed. n.s. not significant (unpaired t-test). B. GSEA of Type 1 or Type 2 tuft cell gene sets in ileal IEC transcriptomes from WTIEC and DDX5ΔIEC mice. NES, normalized enrichment score; NOM P, normalized p-value.
Supplementary Figure 11. DDX5-dependent and -independent genes among small intestinal IEC lineages.
A. Violin plots displaying expressions of IEC-lineage marker genes among IEC lineages assessed from the spatial transcriptomic dataset described in Figure 2A.
B. Venn diagrams of distinct and overlapping downregulated and upregulated DDX5-dependent genes (p<0.05) among small intestine IEC lineages.
Supplementary Figure 12. DDX5-dependent viral and bacterial populations in the murine intestine.
A. Left: Normalized RNAseq read counts of Cd300lf and Dclk1 in two pairs of ileal IECs from cohoused WTIEC and DDX5ΔIEC littermates. Right: Normalized MNoV RNA abundance from five pairs of ileal IECs from cohoused WTIEC and DDX5ΔIEC littermates. * p<0.05 (unpaired t-test).
B. Schematic of meta-transcriptomic analysis of ileal IEC RNAseq data from two pairs of WTIEC and DDX5ΔIEC mice.
C. Normalized transcript counts assigned to viral, bacterial and archaeal genomes by meta-transcriptomic analysis as illustrated in B. * p<0.05 (unpaired t-test).
D. Significantly altered microbial species in the DDX5ΔIEC colons from C (DESeq2). p<0.05, Log2 fold change (DDX5ΔIEC/WTIEC)>1.5 or <−1.5.
Supplementary Figure 13. DDX5-dependent programs in colonic tumors.
A. Volcano plot depicting DDX5-dependent transcripts in murine colonic tumors. One dot represents one gene (black dot, pV<0.05).
B. Gene ontology analysis revealed the top 7 pathways downregulated (black) or upregulated (red) in DDX5ΔIEC colonic tumor cells from A.
C. Heatmap of select DDX5-dependent transcripts and their expression in control or DDX5 deficient colonic tumors.
What is already known on this subject?
A subset of intestinal epithelial cells known as tuft cells orchestrates mucosal defense against helminths and parasites, contributes to epithelial repair, and reduces ileal inflammation, but also serves as a viral reservoir and harbors tumor stem cell properties.
Tuft cell specification requires Rho GTPase Cell Division Cycle 42 (CDC42) and the master transcription factor POU class 2 homeobox 3 (POU2F3). However, little is known about how this pathway is regulated in vivo.
RNA helicase DDX5 promotes tumorigenesis in the small intestine and colon. However, little is known about its role in normal IEC differentiation and function.
What are the new findings?
In the small intestine, knocking out DDX5 specifically in intestinal epithelial cells results in a dramatic reduction of intestinal tuft cell numbers and exacerbation of ileal inflammation,
In the colon, knocking out DDX5 specifically in intestinal epithelial cells results in alteration of microbial composition and protection against colonic tumorigenesis.
Mechanistically, DDX5 binds to and drives the translation of Cdc42 transcripts to maintain proper CDC42 level in the secretory progenitor cells required for Pou2f3 expression and tuft cell specification.
What is the foreseeable clinical impact?
Our findings reveal an epithelial cell intrinsic (DDX5-CDC42-POU2F3) pathway in promoting intestinal tuft cell specification. Modulation of this pathway may have therapeutic potentials for combating ileal inflammation and colonic cancers.
Acknowledgements
We thank Frances Fuller-Pace for sharing the DDX5 conditional mutant mice previously described45. We thank Jakob von Moltke at the University of Washington, Seattle, Elina Zuniga, Stephen Hedrick, and Karen Sykes at the University of California, San Diego, for critical reading of our manuscript.
T.L., N.A., Y.L., J.E.H., and W.J.M.H. are partially funded by the Edward Mallinckrodt, Jr. Foundation and the National Institutes of Health (R01-GM124494 to W.J.M.H). P.G and S.D. were funded by the National Institutes for Health (NIH) (R01-AI141630 to P.G., DK107585 to S.D. and UG3TR003355, UG3TR002968 and R01-AI55696 to P.G. and S.D.). P.G. and S.D. were also supported by the Leona M. and Harry B. Helmsley Charitable Trust. RNAseq was conducted at the IGM Genomics Center, University of California San Diego. We thank the UC San Diego Electron Microscopy Core, HUMANOID Core, and the Moores Cancer Center Histology Core supported by the National Cancer Institute (CCSG P30-CA23100).
Footnotes
Conflict of Interest Statement
The authors declare no competing financial interests.
References
- 1.Haber AL, Biton M, Rogel N, et al. A single-cell survey of the small intestinal epithelium. Nature 2017;551(7680):333–39. doi: 10.1038/nature24489 [published Online First: 2017/11/17] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wilen CB, Lee S, Hsieh LL, et al. Tropism for tuft cells determines immune promotion of norovirus pathogenesis. Science 2018;360(6385):204–08. doi: 10.1126/science.aar3799 [published Online First: 2018/04/14] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lei W, Ren W, Ohmoto M, et al. Activation of intestinal tuft cell-expressed Sucnr1 triggers type 2 immunity in the mouse small intestine. Proc Natl Acad Sci U S A 2018;115(21):5552–57. doi: 10.1073/pnas.1720758115 [published Online First: 2018/05/08] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nadjsombati MS, McGinty JW, Lyons-Cohen MR, et al. Detection of Succinate by Intestinal Tuft Cells Triggers a Type 2 Innate Immune Circuit. Immunity 2018;49(1):33–41 e7. doi: 10.1016/j.immuni.2018.06.016 [published Online First: 2018/07/19] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banerjee A, McKinley ET, von Moltke J, et al. Interpreting heterogeneity in intestinal tuft cell structure and function. J Clin Invest 2018;128(5):1711–19. doi: 10.1172/JCI120330 [published Online First: 2018/05/02] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Howitt MR, Lavoie S, Michaud M, et al. Tuft cells, taste-chemosensory cells, orchestrate parasite type 2 immunity in the gut. Science 2016;351(6279):1329–33. doi: 10.1126/science.aaf1648 [published Online First: 2016/02/06] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hogan SP, Seidu L, Blanchard C, et al. Resistin-like molecule beta regulates innate colonic function: barrier integrity and inflammation susceptibility. J Allergy Clin Immunol 2006;118(1):257–68. doi: 10.1016/j.jaci.2006.04.039 [published Online First: 2006/07/04] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerbe F, Sidot E, Smyth DJ, et al. Intestinal epithelial tuft cells initiate type 2 mucosal immunity to helminth parasites. Nature 2016;529(7585):226–30. doi: 10.1038/nature16527 [published Online First: 2016/01/15] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.von Moltke J, Ji M, Liang HE, et al. Tuft-cell-derived IL-25 regulates an intestinal ILC2-epithelial response circuit. Nature 2016;529(7585):221–5. doi: 10.1038/nature16161 [published Online First: 2015/12/18] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Banerjee A, Herring CA, Chen B, et al. Succinate Produced by Intestinal Microbes Promotes Specification of Tuft Cells to Suppress Ileal Inflammation. Gastroenterology 2020;159(6):2101–15.e5. doi: 10.1053/j.gastro.2020.08.029 [published Online First: 2020/08/24] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chandrakesan P, Weygant N, May R, et al. DCLK1 facilitates intestinal tumor growth via enhancing pluripotency and epithelial mesenchymal transition. Oncotarget 2014;5(19):9269–80. doi: 10.18632/oncotarget.2393 [published Online First: 2014/09/12] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Westphalen CB, Quante M, Wang TC. Functional implication of Dclk1 and Dclk1-expressing cells in cancer. Small GTPases 2017;8(3):164–71. doi: 10.1080/21541248.2016.1208792 [published Online First: 2016/07/28] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barker N Adult intestinal stem cells: critical drivers of epithelial homeostasis and regeneration. Nat Rev Mol Cell Biol 2014;15(1):19–33. doi: 10.1038/nrm3721 [published Online First: 2013/12/12] [DOI] [PubMed] [Google Scholar]
- 14.Gassler N Paneth cells in intestinal physiology and pathophysiology. World J Gastrointest Pathophysiol 2017;8(4):150–60. doi: 10.4291/wjgp.v8.i4.150 [published Online First: 2017/12/01] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clevers H The intestinal crypt, a prototype stem cell compartment. Cell 2013;154(2):274–84. doi: 10.1016/j.cell.2013.07.004 [published Online First: 2013/07/23] [DOI] [PubMed] [Google Scholar]
- 16.Yamada K, Sato K, Morishita S, et al. Establishment of a primary culture method for mouse intestinal epithelial cells by organ culture of fetal small intestine. Biosci33 Biotechnol Biochem 2009;73(8):1849–55. doi: 10.1271/bbb.90246 [published Online First: 2009/08/08] [DOI] [PubMed] [Google Scholar]
- 17.Park JH, Kotani T, Konno T, et al. Promotion of Intestinal Epithelial Cell Turnover by Commensal Bacteria: Role of Short-Chain Fatty Acids. PLoS One 2016;11(5):e0156334. doi: 10.1371/journal.pone.0156334 [published Online First: 2016/05/28] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hooper LV, Gordon JI. Commensal host-bacterial relationships in the gut. Science 2001;292(5519):1115–8. doi: 10.1126/science.1058709 [published Online First: 2001/05/16] [DOI] [PubMed] [Google Scholar]
- 19.Gracz AD, Samsa LA, Fordham MJ, et al. Sox4 Promotes Atoh1-Independent Intestinal Secretory Differentiation Toward Tuft and Enteroendocrine Fates. Gastroenterology 2018;155(5):1508–23 e10. doi: 10.1053/j.gastro.2018.07.023 [published Online First: 2018/07/29] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gerbe F, van Es JH, Makrini L, et al. Distinct ATOH1 and Neurog3 requirements define tuft cells as a new secretory cell type in the intestinal epithelium. J Cell Biol 2011;192(5):767–80. doi: 10.1083/jcb.201010127 [published Online First: 2011/03/09] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Banerjee A, Herring CA, Chen B, et al. Succinate Produced by Intestinal Microbes Promotes Specification of Tuft Cells to Suppress Ileal Inflammation. Gastroenterology 2020;159(6):2101–15 e5. doi: 10.1053/j.gastro.2020.08.029 [published Online First: 2020/08/24] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsumoto I, Ohmoto M, Narukawa M, et al. Skn-1a (Pou2f3) specifies taste receptor cell lineage. Nat Neurosci 2011;14(6):685–7. doi: 10.1038/nn.2820 [published Online First: 2011/05/17] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang X, Bandyopadhyay S, Araujo LP, et al. Elevating EGFR-MAPK program by a nonconventional Cdc42 enhances intestinal epithelial survival and regeneration. JCI Insight 2020;5(16) doi: 10.1172/jci.insight.135923 [published Online First: 2020/07/21] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tu S, Wu WJ, Wang J, et al. Epidermal growth factor-dependent regulation of Cdc42 is mediated by the Src tyrosine kinase. J Biol Chem 2003;278(49):49293–300. doi: 10.1074/jbc.M307021200 [published Online First: 2003/09/25] [DOI] [PubMed] [Google Scholar]
- 25.Wang X-Y, Gan M-X, Li Y, et al. Cdc42 induces EGF receptor protein accumulation and promotes EGF receptor nuclear transport and cellular transformation. FEBS Letters 2015;589(2):255–62. doi: 10.1016/j.febslet.2014.11.049 [DOI] [PubMed] [Google Scholar]
- 26.Schlessinger K, Hall A, Tolwinski N. Wnt signaling pathways meet Rho GTPases. Genes Dev 2009;23(3):265–77. doi: 10.1101/gad.1760809 [published Online First: 2009/02/11] [DOI] [PubMed] [Google Scholar]
- 27.Melendez J, Liu M, Sampson L, et al. Cdc42 Coordinates Proliferation, Polarity, Migration, and Differentiation of Small Intestinal Epithelial Cells in Mice. Gastroenterology 2013;145(4):808–19. doi: 10.1053/j.gastro.2013.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang L, Yang L, Debidda M, et al. Cdc42 GTPase-activating protein deficiency promotes genomic instability and premature aging-like phenotypes. Proceedings of the National Academy of Sciences 2007;104(4):1248. doi: 10.1073/pnas.0609149104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Szczawinska-Poplonyk A, Ploski R, Bernatowska E, et al. A Novel CDC42 Mutation in an 11-Year Old Child Manifesting as Syndromic Immunodeficiency, Autoinflammation, Hemophagocytic Lymphohistiocytosis, and Malignancy: A Case Report. Frontiers in Immunology 2020;11:318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Motokawa M, Watanabe S, Nakatomi A, et al. A hot-spot mutation in CDC42 (p.Tyr64Cys) and novel phenotypes in the third patient with Takenouchi-Kosaki syndrome. Journal of Human Genetics 2018;63(3):387–90. doi: 10.1038/s10038-017-0396-5 [DOI] [PubMed] [Google Scholar]
- 31.Bucciol G, Pillay B, Casas-Martin J, et al. Systemic Inflammation and Myelofibrosis in a Patient withTakenouchi-Kosaki Syndrome due to CDC42 Tyr64CysMutation. Journal of Clinical Immunology 2020;40(4):567–70. doi: 10.1007/s10875-020-00742-5 [DOI] [PubMed] [Google Scholar]
- 32.Chen F, Ma L, Parrini MC, et al. Cdc42 is required for PIP(2)-induced actin polymerization and early development but not for cell viability. Curr Biol 2000;10(13):758–65. doi: 10.1016/s0960-9822(00)00571-6 [published Online First: 2000/07/19] [DOI] [PubMed] [Google Scholar]
- 33.Linder P, Jankowsky E. From unwinding to clamping - the DEAD box RNA helicase family. Nat Rev Mol Cell Biol 2011;12(8):505–16. doi: 10.1038/nrm3154 [DOI] [PubMed] [Google Scholar]
- 34.Lee YJ, Wang Q, Rio DC. Coordinate regulation of alternative pre-mRNA splicing events by the human RNA chaperone proteins hnRNPA1 and DDX5. Genes Dev 2018;32(15–16):1060–74. doi: 10.1101/gad.316034.118 [published Online First: 2018/07/26] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Giraud G, Terrone S, Bourgeois CF. Functions of DEAD box RNA helicases DDX5 and DDX17 in chromatin organization and transcriptional regulation. BMB Rep 2018. [published Online First: 2018/10/09] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Endoh H, Maruyama K, Masuhiro Y, et al. Purification and identification of p68 RNA helicase acting as a transcriptional coactivator specific for the activation function 1 of human estrogen receptor alpha. Mol Cell Biol 1999;19(8):5363–72. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Fuller-Pace FV. The DEAD box proteins DDX5 (p68) and DDX17 (p72): multi-tasking transcriptional regulators. Biochim Biophys Acta 2013;1829(8):756–63. doi: 10.1016/j.bbagrm.2013.03.004 [published Online First: 2013/03/26] [DOI] [PubMed] [Google Scholar]
- 38.Jensen ED, Niu L, Caretti G, et al. p68 (Ddx5) interacts with Runx2 and regulates osteoblast differentiation. Journal of cellular biochemistry 2008;103(5):1438–51. doi: 10.1002/jcb.21526 [published Online First: 2007/10/26] [DOI] [PubMed] [Google Scholar]
- 39.Abbasi N, Long T, Li Y, et al. DDX5 promotes oncogene C3 and FABP1 expressions and drives intestinal inflammation and tumorigenesis. Life Sci Alliance 2020;3(10) doi: 10.26508/lsa.202000772 [published Online First: 2020/08/21] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Causevic M, Hislop RG, Kernohan NM, et al. Overexpression and poly-ubiquitylation of the DEAD-box RNA helicase p68 in colorectal tumours. Oncogene 2001;20(53):7734–43. doi: 10.1038/sj.onc.1204976 [DOI] [PubMed] [Google Scholar]
- 41.Lee H, Flaherty P, Ji HP. Systematic genomic identification of colorectal cancer genes delineating advanced from early clinical stage and metastasis. BMC Med Genomics 2013;6:54–54. doi: 10.1186/1755-8794-6-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cristescu R, Lee J, Nebozhyn M, et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nature medicine 2015;21(5):449–56. doi: 10.1038/nm.3850 [published Online First: 2015/04/22] [DOI] [PubMed] [Google Scholar]
- 43.Du C, Li D-q, Li N, et al. DDX5 promotes gastric cancer cell proliferation in vitro and in vivo through mTOR signaling pathway. Scientific Reports 2017;7(1):42876. doi: 10.1038/srep42876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ma Z, Feng J, Guo Y, et al. Knockdown of DDX5 Inhibits the Proliferation and Tumorigenesis in Esophageal Cancer. Oncology research 2017;25(6):887–95. doi: [published Online First: 2017/03/01] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nicol SM, Bray SE, Derek Black H, et al. The RNA helicase p68 (DDX5) is selectively required for the induction of p53-dependent p21 expression and cell-cycle arrest after DNA damage. Oncogene 2012. doi: 10.1038/onc.2012.426 [published Online First: 2012/09/19] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grivennikov SI, Wang K, Mucida D, et al. Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth. Nature 2012;491(7423):254–8. doi: 10.1038/nature11465 [published Online First: 2012/10/05] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miura N, Yamamoto M, Fukutake M, et al. Anti-CD3 induces bi-phasic apoptosis in murine intestinal epithelial cells: possible involvement of the Fas/Fas ligand system in different T cell compartments. International Immunology 2005;17(5):513–22. doi: 10.1093/intimm/dxh231 [DOI] [PubMed] [Google Scholar]
- 48.Bankhead P, Loughrey MB, Fernandez JA, et al. QuPath: Open source software for digital pathology image analysis. Sci Rep 2017;7(1):16878. doi: 10.1038/s41598-017-17204-5 [published Online First: 2017/12/06] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sano T, Huang W, Hall JA, et al. An IL-23R/IL-22 Circuit Regulates Epithelial Serum Amyloid A to Promote Local Effector Th17 Responses. Cell 2015;163(2):381–93. doi: 10.1016/j.cell.2015.08.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miyoshi H, Stappenbeck TS. In vitro expansion and genetic modification of gastrointestinal stem cells in spheroid culture. Nature protocols 2013;8(12):2471–82. doi: 10.1038/nprot.2013.153 [published Online First: 11/14] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McGinty JW, Ting HA, Billipp TE, et al. Tuft-Cell-Derived Leukotrienes Drive Rapid Anti-helminth Immunity in the Small Intestine but Are Dispensable for Anti-protist Immunity. Immunity 2020;52(3):528–41 e7. doi: 10.1016/j.immuni.2020.02.005 [published Online First: 2020/03/12] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102(43):15545–50. doi: 10.1073/pnas.0506580102 [published Online First: 2005/10/04] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Heinz S, Benner C, Spann N, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 2010;38(4):576–89. doi: 10.1016/j.molcel.2010.05.004 [published Online First: 2010/06/02] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wood DE, Salzberg SL. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biology 2014;15(3):R46. doi: 10.1186/gb-2014-15-3-r46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Peters LA, Perrigoue J, Mortha A, et al. A functional genomics predictive network model identifies regulators of inflammatory bowel disease. Nat Genet 2017;49(10):1437–49. doi: 10.1038/ng.3947 [published Online First: 2017/09/12] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vancamelbeke M, Vanuytsel T, Farre R, et al. Genetic and Transcriptomic Bases of Intestinal Epithelial Barrier Dysfunction in Inflammatory Bowel Disease. Inflamm Bowel Dis 2017;23(10):1718–29. doi: 10.1097/MIB.0000000000001246 [published Online First: 2017/09/09] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Herring CA, Banerjee A, McKinley ET, et al. Unsupervised Trajectory Analysis of Single-Cell RNA-Seq and Imaging Data Reveals Alternative Tuft Cell Origins in the Gut. Cell Syst 2018;6(1):37–51 e9. doi: 10.1016/j.cels.2017.10.012 [published Online First: 2017/11/21] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gracz AD, Samsa LA, Fordham MJ, et al. Sox4 Promotes Atoh1-Independent Intestinal Secretory Differentiation Toward Tuft and Enteroendocrine Fates. Gastroenterology 2018;155(5):1508–23.e10. doi: 10.1053/j.gastro.2018.07.023 [published Online First: 2018/07/29] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sakamori R, Das S, Yu S, et al. Cdc42 and Rab8a are critical for intestinal stem cell division, survival, and differentiation in mice. J Clin Invest 2012;122(3):1052–65. doi: 10.1172/JCI60282 [published Online First: 2012/02/23] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McKinley ET, Sui Y, Al-Kofahi Y, et al. Optimized multiplex immunofluorescence single-cell analysis reveals tuft cell heterogeneity. JCI Insight 2017;2(11) doi: 10.1172/jci.insight.93487 [published Online First: 2017/06/02] [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: IEC-subset specific gene expression in the ileal and colonic epithelium.
A. Expression of genes determined by RNAseq (DESeq2) analysis of the ileal and colonic IECs from two pairs of WTIEC and DDX5ΔIEC mice as described in Figure 1A. Proximal enterocytes (Prox Ent), distal enterocytes (Dist Ent); Paneth cells (P), enteroendocytes (E.E). Black: log2 fold changes (DDX5ΔIEC/WTIEC) > 0.5; blue: log fold changes (DDX5ΔIEC/WTIEC) < −0.5.
B. Subset-specific gene expression analysis by RT-qPCR of ileal (left) and colonic (right) IECs from WTIEC (n=5) and DDX5ΔIEC mice (n=5). Each dot represents the result from one mouse pair.
Supplementary Figure 2: Intestinal crypt density and goblet cell numbers are DDX5-independent.
A. DCLK1+ cell quantitation workflow in QuPath48.
B. Representative Periodic acid-Schiff (PAS) staining of WTIEC and DDX5ΔIEC small intestines. Scale bar represents 100μm.
C. WTIEC (n=3) and DDX5ΔIEC (n=3) mice Ileal and colonic crypt densities. Each dot represents the result from one mouse. n.s. not significant (unpaired t-test).
D. WTIEC (n=3) and DDX5ΔIEC (n=3) mice ileal and colonic goblet cell counts. Each dot represents the result from one mouse. n.s. not significant (unpaired t-test).
Supplementary Figure 3: Tuft cell gating strategy.
A. Small intestines were incubated in 5 mM EDTA containing HBSS for 20 minutes at 37°C for the isolation of IECs. SSC: side scatter (an index for cell granularity). Tuft cells were defined as live EpCAM+CD4−CD8β−CD45low-midSiglec-F+CD24+ 4.
FSC-A: area under forward scatter (an index for cell size).
Supplementary Figure 4: DDX5 dependency of IEC subset defining genes in the intestinal ISC and secretory lineage progenitors.
A. Violin plots of expression from the indicated genes in Atoh1hi or Sox4hi cells from the spatial transcriptomic analysis from figure 2A.
B. Venn diagrams of distinct and overlapping DDX5-dependent genes (p<0.05) among ISC and the secretory lineage progenitors.
C. Normalized RNA expressions of Pou2f3 and Vil1 in small intestinal crypts isolated from two pairs of WTIEC and DDX5ΔIEC mice. Each dot represents the result from one mouse. ** p<0.01 (unpaired t-test).
Supplementary Figure 5: DDX5 is not involved in intestinal organoid survival and growth.
A. Viability and size of small intestinal organoids (46 WTIEC from 5 random fields; 62 DDX5ΔIEC organoids from 7 random fields) and colonic organoids (10 WTIEC from 5 random fields; 11 DDX5ΔIEC organoids from 5 random fields) two days after re-seeding of established cultures. Images were acquired under a 20X magnification.
Supplementary figure 6. Similar type 2 cytokines and trichomonas levels in WTIEC and DDX5ΔIEC mice.
A. RNA expressions of Il4, Il13, and Gata3 normalized to Gapdh in intestinal lamina propria lymphocytes from WTIEC (SI n=4; colon n=3) and DDX5ΔIEC (SI n=4; colon n=3) mice.
B. Levels of Trichomonas (left) and bacterial (right) genomes normalized to host genome detected in fecal or cecal contents. Each dot represents the result from one mouse. n.s. not significant, **** p<0.001 (t-test). Metro: metronidazole administered by drinking water.
C. Proportion of tuft cells and normalized RNA expression of Pou2f3 in the intestines of WTIEC and DDX5ΔIEC treated with or without metronidazole. Results shown are the average and standard deviation of two independent experiments. Each dot represents result from one mouse. * p<0.05 (t-test).
Supplementary Figure 7. IL-13 driven tuft cell hyperplasia is DDX5-independent.
A. Heatmap indicating the average expression of genes encoding receptors for IL-13 and IL-4 in ISCs, progenitors, and tuft cells from Figure 2A.
B. Normalized RNA expression of Pou2f3 in small intestine or colon organoids stimulated with IL-13 (20ng/ml) or vehicle (NaCl at 15μM) for 48hrs. Results shown are the average and standard deviation of two independent experiments. n.s. not significant, * p<0.05 (paired t-test).
C. Representative flow cytometry plots of SiglecF+CD24+ tuft cell populations from WTIEC and DDX5ΔIEC small intestinal and colonic organoids stimulated with IL-13 (20ng/ml) or vehicle (NaCl at 15μM) for 72hrs.
D. Summary of fold changes in the proportions of tuft cells in intestinal organoids stimulated with IL-13 compared to vehicle. Each dot represents the result from one independent experiment.
Supplementary Figure 8. Restoring CDC42 protein levels in DDX5ΔIEC small intestinal organoids by retroviral transduction.
A. Representative retrovirus transduction efficiency in small intestinal organoids was assessed using flow cytometry to measure surface expression of CD90.1. Experiments were repeated three times using independent biological samples with similar results.
B. Left: Representative CDC42 protein abundance in transduced small intestinal organoids as assessed by flow cytometry. Right: Summary of CDC42hi small intestine organoid IECs from three independent experiments. n.s. not significant, * p<0.05 (paired t-test).
Supplementary Figure 9. Transient inhibition of CDC42 activity did not alter wildtype intestinal organoid viability, growth, or response to IL-13 stimulation.
A. Viability and size of WTIEC small intestinal organoids 24hrs post treatment (DMSO, n=553 from 4 random fields; ML141, n=551 from 4 random fields) and 72hrs post treatment (DMSO, n=259 from 2 random fields; ML141, n=267 from 2 random fields). **** p<0.001, n.s. not significant (unpaired, t-test).
B. Normalized RNA expressions of Lyz1, Klf4 and Lgr5 in WTIEC small intestinal organoids cultured in the presence or absence of CDC42 inhibitor ML141 (10μM) for 24hrs. Each dot represents the result from one mouse. n.s. not significant (unpaired, t-test).
C. Representative flow cytometry plots of WTIEC small intestinal or colonic organoids cultured for 72hrs in the presence or absence of recombinant IL-13 (20ng/ml) and/or CDC42 inhibitor ML141 (10μM).
D. Summary of fold changes in tuft cell frequencies among WTIEC small intestinal and colonic organoids upon treatment with DMSO or ML141 inhibitor from C. Tuft cell lineage was defined as Siglec-F+CD24+ IECs4. Each dot represents the result from one independent experiment.
Supplementary Figure 10. DDX5 promotes both type 1 and type 2 tuft cell programs in the small intestine.
A. Left: Histogram of CD45 expression on gated ileal tuft cells (CD4−CD8−Epcam+Siglec-F+CD24+). Right: Proportion (%) of CD45low or CD45mid cells within the tuft cell lineage. Results from four pairs of WTIEC and DDX5ΔIEC cohoused littermates were graphed. n.s. not significant (unpaired t-test). B. GSEA of Type 1 or Type 2 tuft cell gene sets in ileal IEC transcriptomes from WTIEC and DDX5ΔIEC mice. NES, normalized enrichment score; NOM P, normalized p-value.
Supplementary Figure 11. DDX5-dependent and -independent genes among small intestinal IEC lineages.
A. Violin plots displaying expressions of IEC-lineage marker genes among IEC lineages assessed from the spatial transcriptomic dataset described in Figure 2A.
B. Venn diagrams of distinct and overlapping downregulated and upregulated DDX5-dependent genes (p<0.05) among small intestine IEC lineages.
Supplementary Figure 12. DDX5-dependent viral and bacterial populations in the murine intestine.
A. Left: Normalized RNAseq read counts of Cd300lf and Dclk1 in two pairs of ileal IECs from cohoused WTIEC and DDX5ΔIEC littermates. Right: Normalized MNoV RNA abundance from five pairs of ileal IECs from cohoused WTIEC and DDX5ΔIEC littermates. * p<0.05 (unpaired t-test).
B. Schematic of meta-transcriptomic analysis of ileal IEC RNAseq data from two pairs of WTIEC and DDX5ΔIEC mice.
C. Normalized transcript counts assigned to viral, bacterial and archaeal genomes by meta-transcriptomic analysis as illustrated in B. * p<0.05 (unpaired t-test).
D. Significantly altered microbial species in the DDX5ΔIEC colons from C (DESeq2). p<0.05, Log2 fold change (DDX5ΔIEC/WTIEC)>1.5 or <−1.5.
Supplementary Figure 13. DDX5-dependent programs in colonic tumors.
A. Volcano plot depicting DDX5-dependent transcripts in murine colonic tumors. One dot represents one gene (black dot, pV<0.05).
B. Gene ontology analysis revealed the top 7 pathways downregulated (black) or upregulated (red) in DDX5ΔIEC colonic tumor cells from A.
C. Heatmap of select DDX5-dependent transcripts and their expression in control or DDX5 deficient colonic tumors.


