Abstract
Prevotella species in the human gut microbiome are primarily comprised of Prevotella copri, and its diversity and function were recently investigated in detail. Much less is known about other Prevotella species in the human gut. Here, we examined the composition of Prevotella species in human guts by mapping publicly available gut metagenomes to a dereplicated set of metagenome-assembled genomes (MAGs) representing Prevotella lineages found in human guts. In most human cohorts, P. copri is the most relatively abundant species (e.g. up to 14.3% relative abundance in Tangshan, China). However, more than half of the metagenome reads in several cohorts mapped to Prevotella MAGs representing P. stercorea and several other species sister to P. stercorea and P. copri. Analyses of genes encoded in these genomes indicated that P. stercorea and related lineages lacked many hemicellulose degrading enzymes and were thus less likely to metabolise hemicelluloses compared with P. copri and copri-related lineages. Instead, P. stercorea genomes possess several carbohydrate esterases that may be involved in releasing ester modifications from carbohydrates to facilitate their degradation. These findings reveal unexplored Prevotella diversity in the human gut and indicate possible niche partitions among these related species.
Subject terms: Microbial ecology, Microbiome
Introduction
Several Prevotella species inhabit the human gut, among which Prevotella copri is the most common with estimates indicating ~ 40% prevalence in the wider human population and relative abundances that exceed 50% in some individuals1. This species is more prevalent in non-Western populations likely due to its association with high fibre low fat diets2–4, and is often linked with desirable health measures such as reduced visceral fat and improved glucose metabolism5,6. However, there are conflicting reports that implicate P. copri in adverse conditions such as insulin resistance7, hypertension8 and persistent gut inflammation9. Recently, Tett and colleagues demonstrated that the existing P. copri lineage is comprised of at least four distinct species-level lineages each sharing less than 95% average nucleotide identity with one another1. Furthermore, the use of polysaccharides among P. copri isolates can markedly vary indicating distinct metabolic patterns within this lineage10. These inconsistencies between traditional taxonomic designations and species boundaries coupled with strain-specific metabolic diversity and dietary preferences could be reasons behind the conflicting observations related to P. copri and human health11, and more broadly underscores the need to characterise microbial diversity in the human microbiome to understand its function.
Other than P. copri, the human gut hosts several other Prevotella species such as P. stercorea12, P. rectalis13, P. histicola14 and some only known by genomes recovered from human gut metagenomes15–18. Of these, P. stercorea is likely the second most prevalent and relatively abundant species in the healthy human gut after P. copri based on healthy human gut metagenomes19 while much less is known about the distribution of other Prevotella species. For example, P. rectalis13 and P. ihumii20 were recently isolated from individuals with no apparent disease and to our knowledge have not been implicated in any health conditions. Furthermore, DNA-based gut microbiome analyses often mention unclassified Prevotella but fall short of pinpointing exact species21,22 likely due to uncharacterised diversity in this lineage.
Since P. copri are important members of the human gut microbiome due to their roles in human health as well as putative associations with several adverse conditions, we hypothesised that uncharacterised Prevotella species could also play important functions in the human gut as they are likely to share similar traits with P. copri. As such, an assessment of overall Prevotella diversity and distribution in the human gut will greatly facilitate understanding of their roles in health. Here, we examined the genomes of Prevotella species recovered from human gut metagenomes and show that the most prevalent and relatively abundant species are primarily related to P. copri and P. stercorea, and these species possess distinct gene repertoires likely reflecting adaptations to metabolic niches. Specifically, P. stercorea and related species lack several genes for xylan metabolism but have carbohydrate esterases that suggest they work cooperatively with P. copri in metabolising dietary fibre.
Methods
Obtaining Prevotella genomes from public repositories and gut metagenome data sets
This study was approved by the Joint Chinese University of Hong Kong New Territories East Cluster Clinical Research Ethics Committee (reference number 2016.707 and 2016.407) and was performed in accordance with relevant guidelines, regulations, and the Declaration of Helsinki. Written informed consent was obtained from all participants. Microbial genomes were downloaded from publicly available sources including the Genome Taxonomy Database (GTDB)23 R202 which contains RefSeq genomes and from several studies of human microbiome diversity15,17,18. In addition, we obtained Prevotella genomes from two previous studies examining gut microbiome diversity in Hong Kong24 and Yunnan25. For these two studies, metagenome sequences were quality filtered using Trimmomatic26 v0.39 to remove adapter sequences and low-quality regions. Quality filtered reads were then de novo assembled using MEGAHIT27 v1.2.9. Metagenome-assembled genomes (MAGs) were reconstructed from each metagenome assembly by mapping quality filtered reads to their respective assemblies using BWA MEM28 v0.7.17 and then using these coverage profiles as input into MetaBAT29 v2.10.2, v2.12.1 and MaxBin30 v2.2.7. The resulting MAGs were dereplicated using DAS Tool31 and checked for completeness and contamination using CheckM32 (Parks et al., 2015) v1.1.3. Taxonomy was inferred using GTDB-Tk33 v1.4.1 with the R95 database.
Inferring Prevotella phylogeny
In addition to completeness and contamination estimates from CheckM and taxonomies from GTDB-Tk, all MAGs were scanned using Kraken234 v2.1.1 with the StandardPlusPF precompiled database (v20210517) to assess the fraction of each genome consistent with taxonomy inferred from GTDB-Tk to avoid including chimeric genome assemblies. Genomes that scored > 90% completeness and < 5% contamination based on CheckM, inferred as Prevotella by GTDB-Tk and had > 70% of their genomes classified as Prevotella by Kraken2 were selected for phylogenetic inference and genome annotation. Identical genomes were dereplicated based on a threshold of 99% average nucleotide identity (ANI) using CoverM v0.6.0 (https://github.com/wwood/CoverM). The genomes were also dereplicated at 95% ANI to obtain a set of unique species. Bootstrapped phylogenetic tree (1000 bootstraps) was inferred using IQ-TREE235 v2.1.2 based on a concatenated alignment of 120 phylogenetically informative single copy bacterial marker genes generated by GTDB-Tk for both the set of 99% and 95% ANI genomes.
Estimating Prevotella prevalence and relative abundance in diverse human populations
To estimate the prevalence and abundance of Prevotella in the human population, we downloaded publicly available human gut metagenomes (Table S1) from countries including Austria36, Denmark, Spain37, France38, Sweden39, China8,40, Japan41, El Salvador, Peru42, Fiji43, Israel44, Mongolia45, Tanzania46,47 and USA48,49. These data were quality filtered using Trimmomatic v0.39 and then mapped to the species dereplicated set of Prevotella genomes (95% ANI) using CoverM v0.6.0 which produces relative abundance estimates of each genome in each cohort based on coverage and number of mapped reads (see https://github.com/wwood/CoverM/#calculation-methods). Prevalence was calculated as the number of metagenomes in which Prevotella was detected divided by the total number of metagenomes in each cohort. For surveys with case–control cohorts (e.g. colorectal cancer in Japan41), only non-disease subjects were included in this study.
Annotating genes
We used Panaroo50 v1.2.8 to determine gene orthologues shared among the Prevotella genomes. Orthologues were assigned Kyoto Encyclopedia of Genes and Genomes (KEGG) Orthologs (KOs) and Carbohydrate active enzymes (CAZy) families by searching against a database of Hidden Markov Models implemented in EnrichM (https://github.com/geronimp/enrichM) v0.5.0. Distribution of KOs and CAZy families among the MAGs were visualised using non-metric multidimensional scaling (NMDS) ordinations on Jaccard distances using the Vegan package51 v2.5.7 in R v4.0.3.
Results
Phylogenetic diversity of human gut associated Prevotella species
To assess the extent of Prevotella diversity in the human gut, we first downloaded publicly available collections of MAGs derived from human gut metagenomes15,17,18. Additionally, we obtained MAGs from gut metagenome surveys of Hong Kong24 and Yunnan populations25 as we were interested in the distribution of Prevotella in these locales. From these data sets, we selected MAGs classified as members of the Prevotella genus based on GTDB taxonomy. After quality filtering and dereplication at 99% ANI to remove identical genomes, 3943 MAGs remained of which more than 60% were comprised of five highly represented species including P. copri (1579 genomes), P. sp003447235 (416 genomes), P. sp000434975 (185 genomes), P. stercorea (174 genomes) and P. sp002265625 (164 genomes) (Tables S2 and Table S3).
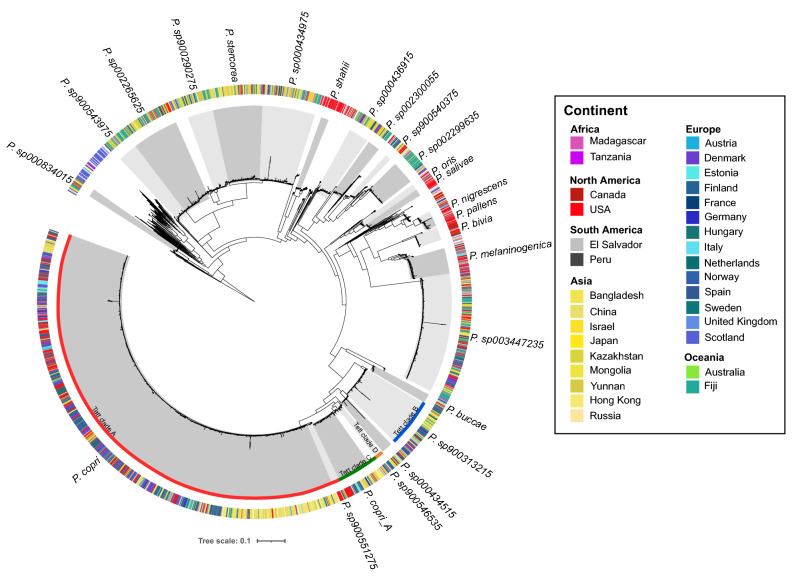
To establish phylogenetic relationships among the 3943 MAGs representing Prevotella species detected in the guts of the wider human population, we downloaded an additional 222 NCBI RefSeq Prevotella genomes held in the GTDB database and incorporated these with the set of 3943 MAGs. Phylogenetic trees were then inferred based on a concatenated alignment of 120 bacterial single copy marker genes to produce a bootstrapped consensus tree (Fig. 1). Within the lineage consisting of P. copri species, MAGs from the Asian continent were phylogenetically closer to one another compared with those from Western geographies (USA, Canada, Europe) possibly reflecting adaptations to local populations. These patterns were also observed in P. stercorea in which 128 out of 174 genomes were derived from Asian populations. Conversely, species such as P. pallens (24 of 33), P. salivae (18 of 23), and P. bivia (41 of 47) were largely represented by MAGs from Canada and USA. As for MAGs derived from gut metagenomes representing rural populations (e.g. Tanzania, Fiji, El Salvador), they were primarily classified as several not yet formally characterised species including sp002299635, sp002265625, sp000434975 and sp900543975. These observations indicate that numerous Prevotella species are found in human guts (at least 25 when considering those represented by > 20 MAGs for confidence; Table S2), many of which have not been formally characterised especially those from rural populations.
Figure 1.
Phylogenetic tree representing phylogenetic relationships among Prevotella isolates and metagenome-assembled genomes (MAGs) recovered from human gut metagenomes. Five Paraprevotella genomes were used as outgroup to root the tree. MAGs included in this tree were downloaded from publicly available repositories15,17,18 and from healthy human gut metagenome surveys of Hong Kong24 and Yunnan populations25. Genomes were dereplicated at 99% average nucleotide identity (ANI) and a concatenated amino acid alignment consisting of 120 phylogenetically informative marker genes (from GTDB) was used to infer phylogenetic trees using IQTREE2. Taxonomy labels from GTDB are shown for species represented by at least 20 genomes (i.e., lower odds of spurious misassemblies or genome binning) (see Table S2 for genome counts); species boundaries are indicated by the alternating grey shades. Note that the GTDB-based taxonomy used here is not comparable to the four P. copri clades A–D from Tett et al.1- these four clades from Tett et al.1 are indicated separately in the figure. The outer colour strip indicates source country/continent of the respective genomes shaded by continent.
Uncharacterised Prevotella species are prevalent and relatively abundant in the human gut
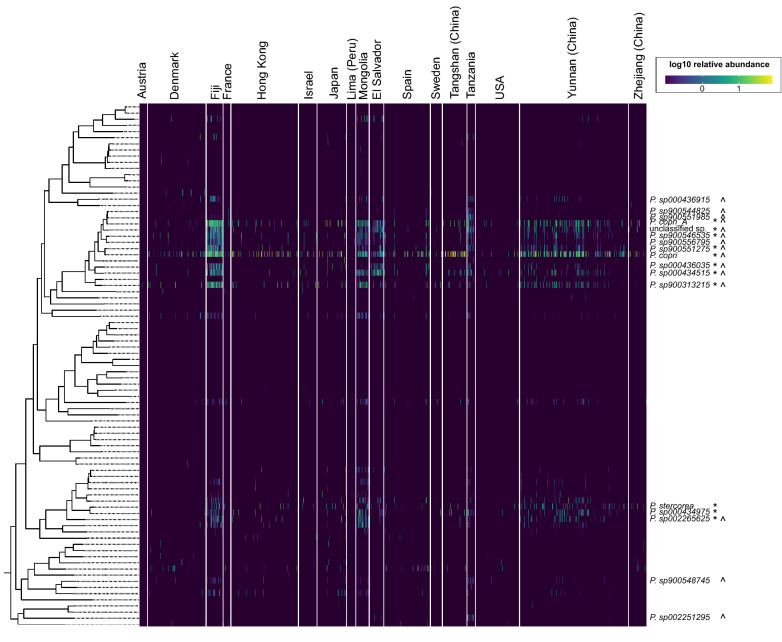
Since many of these Prevotella MAGs are directly derived from human gut metagenomes and may not be identified using conventional microbial community profiling tools, we sought to estimate their prevalence and relative abundances in the human population by mapping metagenome reads. In total, we downloaded 4095 human gut metagenome data sets from non-disease cohorts representing populations from China, Japan, USA, Austria, Denmark, France, Spain, Israel, Sweden, El Salvador, Peru, Fiji, Mongolia, and Tanzania, and mapped quality filtered reads to the set of Prevotella MAGs dereplicated at the species level (95% ANI). As expected, total Prevotella relative abundance was highest in Fiji, a rural population data set (average 36.1% relative abundance), followed by Tangshan (China; 20.5%) and Mongolia (19.6%) (Fig. 2) (Table 1). They were also more widespread in rural communities such as Fiji (97.8% prevalence), El Salvador (94.7%) and Mongolia (91.8%) compared with urban cohorts in which prevalence ranged between 11 and 58% (Table 1). Among the Prevotella species identified above, P. copri was the most prevalent (average 30.7%) and relatively abundant overall (average 3.3%) (Table 2, Table S4). Other prevalent and relatively abundant species were primarily from two distinct lineages, one more closely related to P. copri and the second to P. stercorea (hereafter referred to as copri and stercorea groups) (Fig. 2) (Table S4). Collectively, these species other than P. copri account for an average 4.3% relative abundance which is comparable to the 3.2% for P. copri (Table S5). Like P. copri, most of them are more prevalent and relatively abundant in rural populations such as Fiji, Mongolia and El Salvador, possibly indicating a similar association with non-Western lifestyles and/or high fibre diets. In addition, we detected a few lineages at lower prevalence and relative abundances that are possibly more restricted in their distribution such as P. sp001275135 and P. sp003447235 (Table S4). For example, P. sp001275135 has an average prevalence of only 3.7% across all data sets included in this study, but its prevalence in France and Spain is 14.8% and 17.5%, respectively. Taken together, these observations indicate that the most common Prevotella species in the human gut are primarily comprised of P. copri, P. stercorea and several closely related lineages, although some species with more limited distributions may be prevalent in certain biogeographies.
Figure 2.
Estimated Prevotella relative abundances across several human gut metagenome data sets. Phylogenetic tree indicates relationships of the Prevotella genomes dereplicated at the species level (> 95% average nucleotide identity). Only genomes with reads mapped are included in this figure. Taxonomy labels are shown for species with at least 1% relative abundance (indicated with *) or at least 50% prevalence (indicated with ^) in any one data set. See Table S4 for relative abundance and prevalence values.
Table 1.
Distribution of total Prevotella in the human gut.
| Data set | Relative abundance ± standard deviation (%) | Prevalence (%) | Number of metagenomes |
|---|---|---|---|
| Austria | 0.09 ± 0.35 | 11.11 | 63 |
| Denmark | 2.42 ± 6.19 | 41.35 | 474 |
| Fiji | 36.08 ± 17.64 | 97.83 | 138 |
| France | 2.26 ± 5.99 | 52.46 | 61 |
| Hong Kong | 6.16 ± 13.21 | 33.27 | 547 |
| Israel | 5.82 ± 10.34 | 58.00 | 150 |
| Japan | 8.15 ± 14.75 | 41.95 | 236 |
| Lima (Peru) | 0.37 ± 1.56 | 19.48 | 77 |
| Mongolia | 19.61 ± 16.05 | 91.82 | 110 |
| El Salvador | 10.24 ± 9.70 | 94.74 | 114 |
| Spain | 3.38 ± 9.69 | 39.68 | 378 |
| Sweden | 2.58 ± 9.87 | 37.00 | 100 |
| Tangshan | 20.48 ± 24.84 | 56.63 | 196 |
| Tanzania | 7.36 ± 8.85 | 79.10 | 67 |
| USA | 1.12 ± 4.85 | 17.60 | 358 |
| Yunnan rural | 8.22 ± 12.32 | 69.32 | 427 |
| Yunnan urban | 8.70 ± 13.67 | 56.39 | 454 |
| Zhejiang | 4.87 ± 10.66 | 37.93 | 145 |
Table 2.
Most relatively abundant and prevalent Prevotella species in the human gut.
| Species (GTDB taxonomy) | Average relative abundance ± standard deviation (%) | Average prevalence (%) |
|---|---|---|
| Prevotella copri | 3.27 ± 3.62 | 30.73 ± 26.43 |
| Prevotella copri_ A | 1.10 ± 1.59 | 20.11 ± 25.05 |
| Prevotella sp000434515 | 0.59 ± 0.82 | 16.89 ± 25.17 |
| Prevotella sp900313215 | 0.58 ± 0.92 | 14.44 ± 20.81 |
| Unclassified Prevotella | 0.37 ± 0.61 | 15.53 ± 25.16 |
| Prevotella sp000434975 | 0.28 ± 0.43 | 10.22 ± 14.77 |
| Prevotella stercorea | 0.25 ± 0.29 | 8.59 ± 8.51 |
| Prevotella sp900546535 | 0.25 ± 0.36 | 14.18 ± 22.96 |
| Prevotella sp002265625 | 0.20 ± 0.38 | 10.48 ± 18.11 |
| Prevotella sp900551275 | 0.19 ± 0.36 | 14.47 ± 23.20 |
| Prevotella sp000436035 | 0.16 ± 0.37 | 8.05 ± 16.36 |
| Prevotella sp900290275 | 0.11 ± 0.16 | 7.31 ± 13.58 |
| Prevotella sp900556795 | 0.10 ± 0.21 | 14.75 ± 25.02 |
| Prevotella sp000436915 | 0.09 ± 0.15 | 8.83 ± 16.88 |
| Prevotella sp900543975 | 0.09 ± 0.21 | 7.43 ± 15.23 |
Human gut Prevotella species occupy distinct functional niches
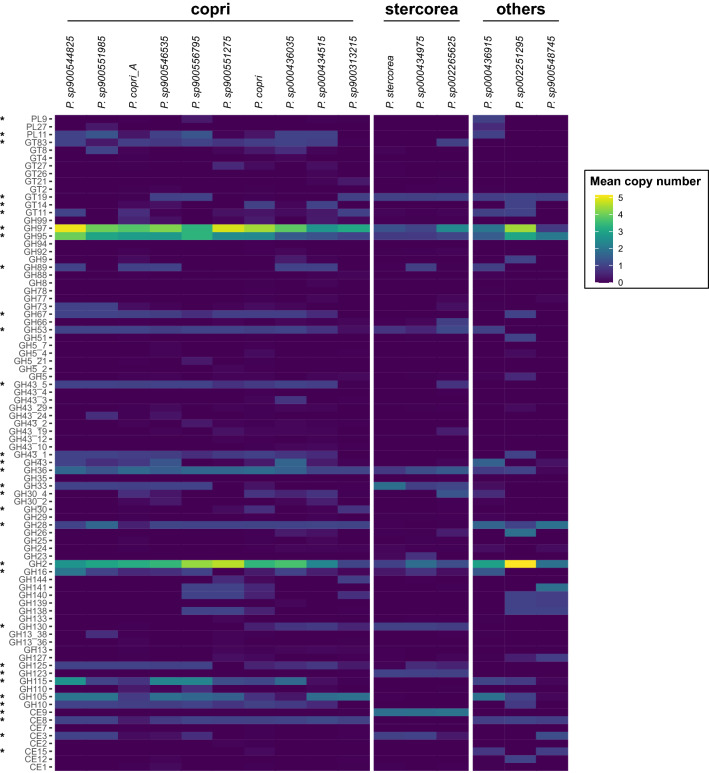
Prevotella copri in the human gut are associated with high fibre non-Western diets as they possess extensive repertoires of carbohydrate active enzymes that allow this species to metabolise complex polysaccharides. We thus hypothesised that other gut Prevotella species are also adapted to carbohydrate-related metabolism although their exact gene content likely differs compared with P. copri, thus allowing them to utilise distinct fractions of dietary fibre. To identify differences in gene content, we first predicted protein coding genes in each genome and then compared predicted genes to the KEGG Orthology (KO) database to infer metabolic potential. NMDS ordinations of KO counts indicated that genomes of prevalent and relatively abundant species belonging to the two copri and stercorea groups identified above significantly differed in their gene content (Fig. S1), and this could be attributed to several pectin and hemicellulolytic enzymes detected in the copri group but missing from stercorea (Table S6). Hemicellulose is a polysaccharide polymer in plant cell walls and typically consists of sugars such as xylose, arabinose, mannose, galactose and rhamnose. The stercorea group either lacks or has fewer copies of genes such as xylanase (K01181), xylosidase (K01198), xylose symporters (K08138), mannosidase (K01218), pectinesterase (K01051), rhamnosidase (K05989) and endoglucanase (K01179) suggesting that stercorea species have reduced ability to utilise hemicelluloses compared with copri-related species. Instead, stercorea species may participate in breaking down oligosaccharides and glycoproteins based on detection of genes such as N-acetylglucosamine-6-phosphate deacetylase (K01443) and endo-beta-N-acetylglucosaminidase (K01227). In addition, stercorea species possess components of the glycine cleavage system and histidine utilisation system (hut) which could indicate a more extensive amino acid metabolism compared with the copri group. These carbohydrate utilisation patterns are also reflected in CAZy profiles indicating the stercorea group as non-hemicellulose degraders. CAZy families involved in hemicellulose degradation such as CE8 (pectin methylesterase), GH10 (β-1,4-xylanase), GH28 (polygalacturonase), GH30 (endo-xylanase), GH43_1 (β-xylosidase, a-l-arabinofuranosidase), GH67 (α-glucuronidase), GH115 (xylan α-1,2-glucuronidase) and GH141 (α-l-fucosidase, xylanase) were absent in nearly all stercorea group genomes, whereas the presence of CE3 (acetyl xylan esterase) and CE9 (N-acetylglucosamine 6-phosphate deacetylase) suggest a possible role in assisting with removal of ester modifications to facilitate carbohydrate degradation52 by P. copri or other gut bacteria with the necessary gene repertoire (Table S7) (Fig. 3). The presence of GH33 (sialidases) and GH123 (N-acetylgalactosaminidases) further suggest that the stercorea species could metabolise sialic acids derived from dietary (e.g. glycolylneuraminic acid in red meat53) and/or human glycans (e.g. acetylneuraminic acid) and are likely to utilise these substrates in addition to or instead of dietary fibre. Other Prevotella species such as sp001275135 prevalent in France and Spain possess hemicellulolytic CAZy families akin to copri, whereas sp002299635 and sp003447235 are more similar with stercorea in that they lack hemicellulolytic families. These similarities in CAZy content among species are supported by an NMDS of total CAZy counts showing two broad types of CAZy profiles in human gut Prevotella, one representing the copri group and the other the stercorea group with reduced capacity to utilise hemicelluloses (Fig. S2).
Figure 3.
Distribution of carbohydrate active enzyme (CAZy) families in human gut Prevotella (average copies per genome). Only genomes with > 1% relative abundance or > 50% prevalence as indicated in Fig. 2 are included. CAZy families were identified in the genomes using Hidden Markov Models. Asterisks indicate families with statistically different copy counts (FDR < 0.001 Kruskal Wallis test with false discovery rate adjustment) and with at least one copy in any one of the three groups (copri, stercorea or other). See Table S7 for copy numbers. CE carbohydrate esterase, GH glycoside hydrolase, GT glycosyl transferase, PL polysaccharide lyase.
To examine whether diet may influence the distribution of gut Prevotella species, we assessed dietary intake information collected as part of the Yunnan gut microbiome study25 together with abundance values obtained from metagenome read mapping (see section above). The Yunnan survey was comprised of 881 individuals representing six ethnic groups in Yunnan province; 427 of the 881 were recruited from rural counties and another 454 from urban Kunming (capital of Yunnan province). A total of 676 individuals provided their past month dietary intake through a binary response questionnaire. We first established that diet primarily differed among the ethnic groups followed by rural/urban status (p < 0.05, PERMANOVA) (Table S8). While the Zang (Tibetan) diet was most distinct compared with other ethnic groups (Fig. S3A), total Prevotella relative abundance in the Zang was comparable to several ethnicities (11.7% vs. 10.9–15.5%) whereas the Dai (8.0%) and Han (3.9%) had significantly lower relative abundances (Fig. S3B). Across all ethnicities, only coffee consumption was associated with an increased Prevotella relative abundance (p < 0.05, generalised linear model with FDR adjustment) (Table S9) although coffee consumption was reported in only 19 individuals. Other foods in the questionnaire were not correlated with either total Prevotella relative abundance or with abundances of the two stercorea and copri lineages identified above. Similarly, the low relative abundances of Prevotella in the Dai and Han could not be attributed to any specific foods included in this questionnaire. These observations indicate that non-dietary influences (e.g. genetics, lifestyle) could play a role in gut Prevotella associations in the Yunnan cohort, although more in-depth and quantitative dietary information in addition to existing data provided in that Yunnan study may be required to resolve exact correlations between these factors and diet.
Discussion
The Prevotella genus is a lineage of interest as its member species are a common feature of the human gut microbiome and have been repeatedly implicated in health and disease. P. copri is often the primary focus of the Prevotella genus, but there is increasing incentive to catalogue and characterise other gut Prevotella species. For example, a gut microbiome survey of competitive cyclists revealed enrichment of P. copri and several unidentified Prevotella species correlated with exercise duration21. Another study of gut microbiome development in malnourished children reported that children who received a food formula designed to stimulate development of a healthy gut microbiota54 showed greater enrichment of P. copri and other unnamed Prevotella species compared with existing nutrition regimes22. To capture gut Prevotella diversity without relying on known marker genes (e.g. MetaPhlAn) or genomes deposited in RefSeq (Kraken2), our workflow in this study classifies metagenome reads against a large collection of human gut-derived Prevotella MAGs to better resolve and estimate abundances of Prevotella species in the human gut15,17,18.
We show that the most numerically abundant Prevotella species in the human gut are largely comprised of two distinct lineages, one closely related to and including P. copri while the other to P. stercorea. Both the copri and stercorea groups were more relatively abundant in rural non-Westernised human populations, suggesting that the presence of stercorea-related lineages is also driven by diet and lifestyle factors akin to P. copri2–4. Using the P. copri phylogenetic framework inferred by Tett et al.1, the copri group of species we identified here largely corresponded with the four clades circumscribed in that study and the potential for carbohydrate metabolism detected in this group also broadly mirrors that reported by Tett et al.1. While P. copri are known hemicellulose degraders55,56, compared with P. copri the stercorea group occupies a separate functional niche categorised by a lack of hemicellulolytic gene families. In addition, the stercorea group possesses carbohydrate esterase families that were comparatively much less common in the copri group. From these features, we posit that a possible role of the stercorea group is to remove ester-based modifications from dietary carbohydrates to facilitate hydrolysis by gut species with the necessary hydrolytic capacity52. Additionally, the stercorea group lineages possess sialidases. Sialic acids have been demonstrated to play roles in processes such as mediating interaction between immune cells, pathogen binding to human cells57 and inflammation in the gut58. The presence of sialidases in the stercorea group therefore suggests that stercorea-related bacteria have important functions that could impact human health.
The main limitation of this work is the lack of detailed lifestyle and dietary information needed to investigate whether distribution of the several gut Prevotella identified here is influenced by lifestyle and/or diet. Existing evidence indicate that P. copri is associated with dietary fibre intake59, but we did not observe such associations with the limited dietary data (yes or no responses) provided in the Yunnan study25. In addition, our metabolic inferences of the various Prevotella species are based on genome sequences and thus need to be validated by isolation and laboratory culture experiments. Nevertheless, since low fibre diets can result in poor gastrointestinal health60 and Prevotella are one of the major fibrolytic bacteria in human guts61, we foresee that there will be increased interest in exploiting Prevotella to potentially improve human health and nutrition. Our findings of distinct Prevotella species in the human population underscores the need to classify the human gut microbiome in greater resolution to understand and exploit the metabolic potential of gut microorganisms.
Supplementary Information
Acknowledgements
This work was supported by InnoHK (The Government of Hong Kong, Special Administrative Region of the People’s Republic of China), the National Natural Science Foundation of China (U1802282, 82060107), Applied Basic Research Projects of Yunnan Province, China (202001AY070001-202), and a seed fund from the Centre for Gut Microbiota Research (Faculty of Medicine, The Chinese University of Hong Kong).
Author contributions
Y.K.Y. conceived the project. Data analyses was performed by Y.K.Y. and L.Y.T.I. Manuscript was written by Y.K.Y. with input from Y.S., L.Y.T.I., L.W., F.K.L.C., Y.M., and S.C.N.
Data availability
Metagenome-assembled genomes used in this study can be downloaded from https://drive.google.com/file/d/1hLaPiR1qHKBeDsK0A3WLzk3ADgCFkrNH/view?usp=sharing. Codes used to generate genomes are provided in supplementary file 1.
Competing interests
S.C.N. and F.K.L.C. are the scientific co-founders and sit on the board of Directors of GenieBiome Ltd. S.C.N. has served as an advisory board member for Pfizer, Ferring, Janssen, and Abbvie and a speaker for Ferring, Tillotts, Menarini, Janssen, Abbvie, and Takeda. S.C.N. has received research grants from Olympus, Ferring, Janssen and Abbvie. F.K.L.C. has served as an advisor and lecture speaker for Eisai Co. Ltd., AstraZeneca, Pfizer Inc., Takeda Pharmaceutical Co., and Takeda (China) Holdings Co. Ltd.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yinglei Miao, Email: miaoyinglei@yeah.net.
Siew C. Ng, Email: siewchienng@cuhk.edu.hk
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-12721-4.
References
- 1.Tett A, et al. The Prevotella copri complex comprises four distinct clades underrepresented in westernized populations. Cell Host Microbe. 2019;26:666–679. doi: 10.1016/j.chom.2019.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu GD, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.David LA, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Filippis F, et al. High-level adherence to a Mediterranean diet beneficially impacts the gut microbiota and associated metabolome. Gut. 2016;65:1812–1821. doi: 10.1136/gutjnl-2015-309957. [DOI] [PubMed] [Google Scholar]
- 5.Kovatcheva-Datchary P, et al. Dietary fiber-induced improvement in glucose metabolism is associated with increased abundance of prevotella. Cell Metab. 2015;22:971–982. doi: 10.1016/j.cmet.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 6.Asnicar F, et al. Microbiome connections with host metabolism and habitual diet from 1098 deeply phenotyped individuals. Nat. Med. 2021;27:321–332. doi: 10.1038/s41591-020-01183-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pedersen HK, et al. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature. 2016;535:376–381. doi: 10.1038/nature18646. [DOI] [PubMed] [Google Scholar]
- 8.Li J, et al. Gut microbiota dysbiosis contributes to the development of hypertension. Microbiome. 2017;5:14. doi: 10.1186/s40168-016-0222-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dillon SM, et al. Gut dendritic cell activation links an altered colonic microbiome to mucosal and systemic T-cell activation in untreated HIV-1 infection. Mucosal. Immunol. 2016;9:24–37. doi: 10.1038/mi.2015.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fehlner-Peach H, et al. Distinct polysaccharide utilization profiles of human intestinal Prevotella copri isolates. Cell Host Microbe. 2019;26:680–690. doi: 10.1016/j.chom.2019.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Filippis F, et al. Distinct genetic and functional traits of human intestinal Prevotella copri strains are associated with different habitual diets. Cell Host Microbe. 2019;25:444–453. doi: 10.1016/j.chom.2019.01.004. [DOI] [PubMed] [Google Scholar]
- 12.Hayashi H, Shibata K, Sakamoto M, Tomita S, Benno Y. Prevotella copri sp. nov. and Prevotella stercorea sp. nov., isolated from human faeces. Int. J. Syst. Evol. Microbiol. 2007;57:941–946. doi: 10.1099/ijs.0.64778-0. [DOI] [PubMed] [Google Scholar]
- 13.Belkacemi S, et al. Description of Prevotella rectalis sp. nov., a new bacterium isolated from human rectum. New Microbes New Infect. 2020;36:100703. doi: 10.1016/j.nmni.2020.100703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shahi SK, et al. Prevotella histicola, a human gut commensal, is as potent as COPAXONE® in an animal model of multiple sclerosis. Front. Immunol. 2019;10:462. doi: 10.3389/fimmu.2019.00462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Almeida A, et al. A unified catalog of 204,938 reference genomes from the human gut microbiome. Nat. Biotechnol. 2021;39:105–114. doi: 10.1038/s41587-020-0603-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tett A, Pasolli E, Masetti G, Ercolini D, Segata N. Prevotella diversity, niches and interactions with the human host. Nat. Rev. Microbiol. 2021;19:585–599. doi: 10.1038/s41579-021-00559-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nayfach S, Shi ZJ, Seshadri R, Pollard KS, Kyrpides NC. New insights from uncultivated genomes of the global human gut microbiome. Nature. 2019;568:505–510. doi: 10.1038/s41586-019-1058-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pasolli E, et al. Extensive unexplored human microbiome diversity revealed by over 150,000 genomes from metagenomes spanning age, geography, and lifestyle. Cell. 2019;176:649–662. doi: 10.1016/j.cell.2019.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pasolli E, et al. Accessible, curated metagenomic data through ExperimentHub. Nat. Methods. 2017;14:1023–1024. doi: 10.1038/nmeth.4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anani H, et al. Prevotella ihumii sp. nov., a new bacterium isolated from a stool specimen of a healthy woman. New Microbes New Infect. 2019;32:100607. doi: 10.1016/j.nmni.2019.100607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petersen LM, et al. Community characteristics of the gut microbiomes of competitive cyclists. Microbiome. 2017;5:98. doi: 10.1186/s40168-017-0320-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen RY, et al. A microbiota-directed food intervention for undernourished children. N. Engl. J. Med. 2021;384:1517–1528. doi: 10.1056/NEJMoa2023294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parks DH, et al. A complete domain-to-species taxonomy for Bacteria and Archaea. Nat. Biotechnol. 2020;38:1079–1086. doi: 10.1038/s41587-020-0501-8. [DOI] [PubMed] [Google Scholar]
- 24.Yeoh YK, et al. Southern Chinese populations harbour non-nucleatum Fusobacteria possessing homologues of the colorectal cancer-associated FadA virulence factor. Gut. 2020;69:1998–2007. doi: 10.1136/gutjnl-2019-319635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun Y, et al. Population-level configurations of gut mycobiome across 6 ethnicities in urban and rural China. Gastroenterology. 2021;160:272–286. doi: 10.1053/j.gastro.2020.09.014. [DOI] [PubMed] [Google Scholar]
- 26.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li D, Liu CM, Luo R, Sadakane K, Lam TW. MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 2015;31:1674–1676. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- 28.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kang DD, et al. MetaBAT 2: An adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ. 2019;7:e7359. doi: 10.7717/peerj.7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu YW, Simmons BA, Singer SW. MaxBin 2.0: An automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics. 2016;32:605–607. doi: 10.1093/bioinformatics/btv638. [DOI] [PubMed] [Google Scholar]
- 31.Sieber CMK, et al. Recovery of genomes from metagenomes via a dereplication, aggregation and scoring strategy. Nat. Microbiol. 2018;3:836–843. doi: 10.1038/s41564-018-0171-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chaumeil PA, Mussig AJ, Hugenholtz P, Parks DH. GTDB-Tk: A toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics. 2020;36:1925–1927. doi: 10.1093/bioinformatics/btz848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wood DE, Lu J, Langmead B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019;20:257. doi: 10.1186/s13059-019-1891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Minh BQ, et al. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020;37:1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feng Q, et al. Gut microbiome development along the colorectal adenoma–carcinoma sequence. Nat. Commun. 2015;6:6528. doi: 10.1038/ncomms7528. [DOI] [PubMed] [Google Scholar]
- 37.Li J, et al. An integrated catalog of reference genes in the human gut microbiome. Nat. Biotechnol. 2014;32:834–841. doi: 10.1038/nbt.2942. [DOI] [PubMed] [Google Scholar]
- 38.Zeller G, et al. Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol. Syst. Biol. 2014;10:766. doi: 10.15252/msb.20145645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bäckhed F, et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe. 2015;17:690–703. doi: 10.1016/j.chom.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 40.Qin N, et al. Alterations of the human gut microbiome in liver cirrhosis. Nature. 2014;513:59–64. doi: 10.1038/nature13568. [DOI] [PubMed] [Google Scholar]
- 41.Yachida S, et al. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat. Med. 2019;25:968–976. doi: 10.1038/s41591-019-0458-7. [DOI] [PubMed] [Google Scholar]
- 42.Pehrsson EC, et al. Interconnected microbiomes and resistomes in low-income human habitats. Nature. 2016;533:212–216. doi: 10.1038/nature17672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brito IL, et al. Mobile genes in the human microbiome are structured from global to individual scales. Nature. 2016;535:435–439. doi: 10.1038/nature18927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zeevi D, et al. Personalized nutrition by prediction of glycemic responses. Cell. 2015;163:1079–1094. doi: 10.1016/j.cell.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 45.Liu W, et al. Unique features of ethnic mongolian gut microbiome revealed by metagenomic analysis. Sci. Rep. 2016;6:34826. doi: 10.1038/srep34826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rampelli S, et al. Metagenome sequencing of the Hadza hunter-gatherer gut microbiota. Curr. Biol. 2015;25:1682–1693. doi: 10.1016/j.cub.2015.04.055. [DOI] [PubMed] [Google Scholar]
- 47.Smits SA, et al. Seasonal cycling in the gut microbiome of the Hadza hunter-gatherers of Tanzania. Science. 2017;357:802–806. doi: 10.1126/science.aan4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mehta RS, et al. Stability of the human faecal microbiome in a cohort of adult men. Nat. Microbiol. 2018;3:347–355. doi: 10.1038/s41564-017-0096-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.The Human Microbiome Project Consortium Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tonkin-Hill G, et al. Producing polished prokaryotic pangenomes with the Panaroo pipeline. Genome Biol. 2020;21:180. doi: 10.1186/s13059-020-02090-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oksanen, J. et al. Package ‘vegan’. Community ecology package, version 2.5-7 (2020).
- 52.Nakamura AM, Nascimento AS, Polikarpov I. Structural diversity of carbohydrate esterases. Biotechnol. Res. Innov. 2017;1:35–51. doi: 10.1016/j.biori.2017.02.001. [DOI] [Google Scholar]
- 53.Zaramela LS, et al. Gut bacteria responding to dietary change encode sialidases that exhibit preference for red meat-associated carbohydrates. Nat. Microbiol. 2019;4:2082–2089. doi: 10.1038/s41564-019-0564-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gehrig JL, et al. Effects of microbiota-directed foods in gnotobiotic animals and undernourished children. Science. 2019;365:139. doi: 10.1126/science.aau4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Accetto T, Avguštin G. The diverse and extensive plant polysaccharide degradative apparatuses of the rumen and hindgut Prevotella species: A factor in their ubiquity? Syst. Appl. Microbiol. 2019;42:107–116. doi: 10.1016/j.syapm.2018.10.001. [DOI] [PubMed] [Google Scholar]
- 56.Emerson EL, Weimer PJ. Fermentation of model hemicelluloses by Prevotella strains and Butyrivibrio fibrisolvens in pure culture and in ruminal enrichment cultures. Appl. Microbiol. Biotechnol. 2017;101:4269–4278. doi: 10.1007/s00253-017-8150-7. [DOI] [PubMed] [Google Scholar]
- 57.Varki A. Sialic acids in human health and disease. Trends Mol. Med. 2008;14:351–360. doi: 10.1016/j.molmed.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang YL, Chassard C, Hausmann M, von Itzstein M, Hennet T. Sialic acid catabolism drives intestinal inflammation and microbial dysbiosis in mice. Nat. Commun. 2015;6:8141. doi: 10.1038/ncomms9141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vangay P, et al. US immigration westernizes the human gut microbiome. Cell. 2018;175:962–972. doi: 10.1016/j.cell.2018.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Desai MS, et al. A dietary fiber-deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell. 2016;167:1339–1353. doi: 10.1016/j.cell.2016.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Flint HJ, Scott KP, Duncan SH, Louis P, Forano E. Microbial degradation of complex carbohydrates in the gut. Gut Microbes. 2012;3:289–306. doi: 10.4161/gmic.19897. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Metagenome-assembled genomes used in this study can be downloaded from https://drive.google.com/file/d/1hLaPiR1qHKBeDsK0A3WLzk3ADgCFkrNH/view?usp=sharing. Codes used to generate genomes are provided in supplementary file 1.