Abstract
We present a genome assembly from an individual male Pyrgus malvae (the grizzled skipper; Arthropoda; Insecta; Lepidoptera; Hesperiidae). The genome sequence is 725 megabases in span. The majority (99.97%) of the assembly is scaffolded into 31 chromosomal pseudomolecules, with the Z sex chromosome assembled.
Keywords: Pyrgus malvae, grizzled skipper, genome sequence, chromosomal, Lepidoptera
Species taxonomy
Eukaryota; Metazoa; Ecdysozoa; Arthropoda; Hexapoda; Insecta; Pterygota; Neoptera; Endopterygota; Lepidoptera; Glossata; Ditrysia; Papilionoidea; Hesperiidae; Pyrginae; Pyrgus; Pyrgus malvae (Linnaeus, 1758) (NCBI:txid218760).
Background
The grizzled skipper, Pyrgus malvae, is a small butterfly, characteristic of chalk downland and woodland clearings, and other grassland habitats. Not to be confused with the term ‘grisly’ (i.e. extremely unpleasant or gruesome), P. malvae gets its common name from the tufts of long grey hair that cover its body and inner wings. Its wings bear a striking black and white checkerboard pattern, with alternating black and white stripes on the wing fringes and antennae. Notoriously difficult to follow, P. malvae has a fast and darting, low flight pattern. Pyrgus malvae is found throughout Europe, except for northern Scandinavia, several Mediterranean Islands and Iberia, southern France and Italy (where it is replaced by its sister species P. malvoides), with a range that extends eastwards across temperate Asia to Northern China and Korea ( Tolman & Lewington, 2008). In the UK the species is found mainly in central and southern England, with a patchy distribution in Wales and the southwest. P. malvae typically exists in small populations (<100 adults) that are thought to form metapopulations across its range ( Asher et al., 2001).
In the UK, P. malvae typically emerges in April and flies until June, although the date of first emergence is advancing, and in warm years may occur as early as March . It is univoltine in northern Europe and at higher altitudes, but is bivoltine elsewhere, and in the north it may be bivoltine when weather conditions are particularly favourable ( Asher et al., 2001).
Pyrgus malvae larvae feed on a variety of host plants in the Rosaceae family, particularly agrimony ( Agrimonia eupatoria), creeping cinquefoil ( Potentilla reptans) and wild strawberry ( Fragaria vesca) ( Asher et al., 2001). When fully-grown, the larva constructs a cocoon at the base of low vegetation, where it overwinters as a pupa. Adults feed on a wide variety of nectar sources, including Bird’s foot trefoil ( Lotus corniculatus), bugle ( Ajuga reptans), buttercup ( Ranunculus species), daisy ( Bellis perennis), and dandelion ( Taraxacum officinale). Males are territorial, and exhibit either perching or patrolling behaviour according to habitat type ( Brereton et al., 1998), and have two scent organs: the forewing costal fold and tibial tufts composed of specialised setae on the hind leg, which appear to be used to waft pheromones towards the female during courtship ( Hernández-Roldán et al., 2014). Eggs are laid singly on the leaf underside of larval host plants, with the majority deposited on short vegetation in locations with a favourably warm microclimate and/or elevated nutritional content ( Brereton et al., 1998).
Populations of P. malvae in the UK have declined markedly in the twentieth century ( Brereton et al., 1998) and the species is a conservation priority in the UK ( Brig, 2007). Encouragingly, P. malvae appears to be positively associated with grazed vegetation, and implementing grazing in habitat restoration regimes may offer a means to help reverse population declines ( Wallis De Vries & Raemakers, 2001). At European level this species is listed as Least Concern in the IUCN Red List ( Van Swaay et al., 2010). Pyrgus malvae has been reported as having 33 ( Bigger, 1960; England) and 31 ( Federley, 1938; Finland) chromosome pairs. The assembly described herein contains 31 chromosome pairs.
Genome sequence report
The genome was sequenced from a single male P. malvae ( Figure 1) collected from Suatu, Cluj County, Romania (latitude 46.7648, longitude 23.9845). A total of 69-fold coverage in Pacific Biosciences single-molecule circular consensus (HiFi) long reads and 49-fold coverage in 10X Genomics read clouds were generated. Primary assembly contigs were scaffolded with chromosome conformation Hi-C data. Manual assembly curation corrected 2 missing/misjoins and removed 1 haplotypic duplication, reducing the assembly length by 0.01% and the scaffold number by 7.69%.
Figure 1. Fore and hind wings of the Pyrgus malvae specimen from which the genome was sequenced.
Top: Dorsal (left) and ventral (right) surface view of wings from specimen RO_PM_973 (ilPyrMalv3) from Suatu, Romania, used to generate Pacific Biosciences and 10X genomics data. Bottom: Dorsal (left) and ventral (right) surface view of wings from specimen RO_PM_838 (ilPyrMalv2) from Cluj-Napoca, Romania, used to generate Hi-C data.
The final assembly has a total length of 725 Mb in 36 sequence scaffolds with a scaffold N50 of 27.0 Mb ( Table 1). The majority, 99.97%, of assembly sequence was assigned to 31 chromosomal-level scaffolds, representing 30 autosomes (numbered by sequence length), and the Z sex chromosome ( Figure 2– Figure 5; Table 2). The assembly has a BUSCO v5.1.2 ( Manni et al., 2021) completeness of 98.8% (single 98.3%, duplicated 0.4%) using the lepidoptera_odb10 reference set (n=5286). While not fully phased, the assembly deposited is of one haplotype. Contigs corresponding to the second haplotype have also been deposited.
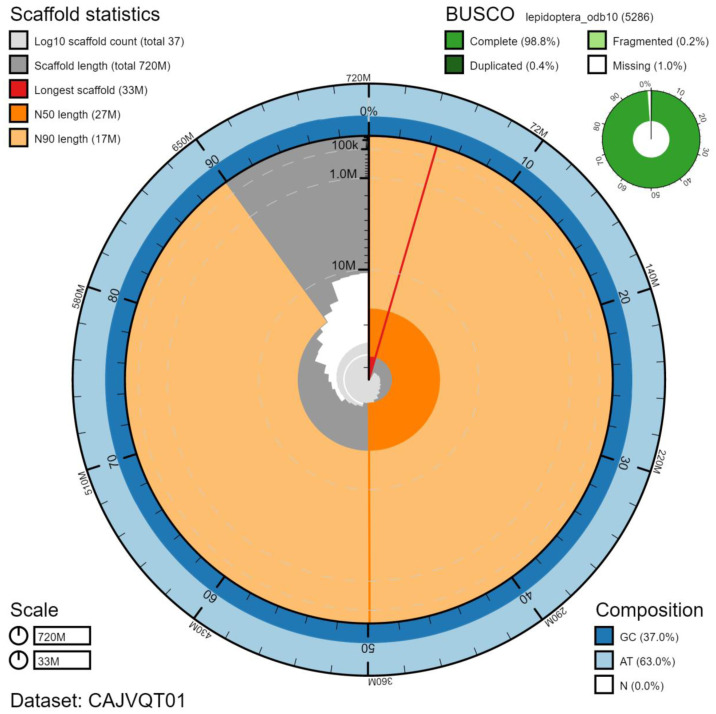
Figure 2. Genome assembly of Pyrgus malvae, ilPyrMalv3.1: metrics.
The BlobToolKit Snailplot shows N50 metrics and BUSCO gene completeness. The main plot is divided into 1,000 size-ordered bins around the circumference with each bin representing 0.1% of the 724,649,524 bp assembly. The distribution of chromosome lengths is shown in dark grey with the plot radius scaled to the longest chromosome present in the assembly (33,217,309 bp, shown in red). Orange and pale-orange arcs show the N50 and N90 chromosome lengths (26,976,370 and 16,663,010 bp), respectively. The pale grey spiral shows the cumulative chromosome count on a log scale with white scale lines showing successive orders of magnitude. The blue and pale-blue area around the outside of the plot shows the distribution of GC, AT and N percentages in the same bins as the inner plot. A summary of complete, fragmented, duplicated and missing BUSCO genes in the lepidoptera_odb10 set is shown in the top right. An interactive version of this figure is available at https://blobtoolkit.genomehubs.org/view/ilPyrMalv3.1/dataset/CAJVQT01/snail.
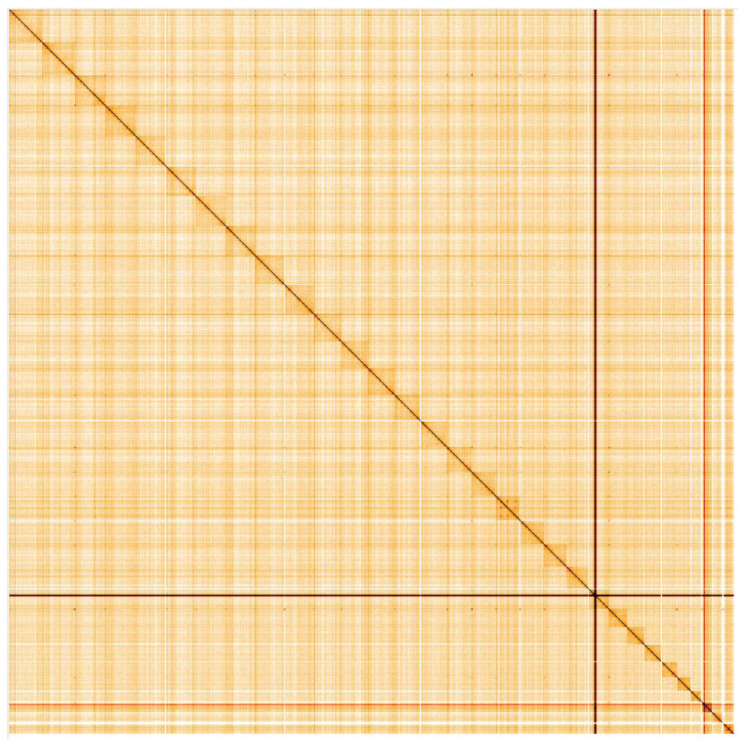
Figure 5. Genome assembly of Pyrgus malvae, ilPyrMalv3.1: Hi-C contact map.
Hi-C contact map of the ilPyrMalv3.1 assembly, visualised in HiGlass. Chromosomes are shown in size order from left to right and top to bottom. The interactive Hi-C map can be viewed here.
Table 1. Genome data for Pyrgus malvae, ilPyrMalv3.1.
| Project accession data | |
|---|---|
| Assembly identifier | ilPyrMalv3.1 |
| Species | Pyrgus malvae |
| Specimen | ilPyrMalv3 (genome assembly); ilPyrMalv2 (Hi-C);
ilPyrMalv1 (RNA-Seq) |
| NCBI taxonomy ID | NCBI:txid111923 |
| BioProject | PRJEB46857 |
| BioSample ID | SAMEA7523296 |
| Isolate information | Male, whole organism (ilPyrMalv3); unknown sex,
whole organisms (ilPyrMalv1, ilPyrMalv2) |
| Raw data accessions | |
| PacificBiosciences SEQUEL II | ERR6606794-ERR6606796 |
| 10X Genomics Illumina | ERR6363273-ERR6363276 |
| Hi-C Illumina | ERR6363278 |
| Illumina polyA RNA-Seq | ERR6363277 |
| Genome assembly | |
| Assembly accession | GCA_911387765.1 |
| Accession of alternate haplotype | GCA_911387725.2 |
| Span (Mb) | 725 |
| Number of contigs | 41 |
| Contig N50 length (Mb) | 26.0 |
| Number of scaffolds | 36 |
| Scaffold N50 length (Mb) | 27.0 |
| Longest scaffold (Mb) | 33.2 |
| BUSCO * genome score | C:98.8%[S:98.3%,D:0.4%],F:0.2%,M:1.0%,n:5286 |
| Genome annotation | |
| Number of protein-coding genes | 12,096 |
| Average length of coding sequence (bp) | 1,534.63 |
| Average number of exons per transcript | 7.83 |
| Average exon size (bp) | 207.85 |
| Average intron size (bp) | 2,914.74 |
*BUSCO scores based on the lepidoptera_odb10 BUSCO set using v5.1.2. C= complete [S= single copy, D=duplicated], F=fragmented, M=missing, n=number of orthologues in comparison. A full set of BUSCO scores is available at https://blobtoolkit.genomehubs.org/view/ilPyrMalv3.1/dataset/CAJVQT01/busco.
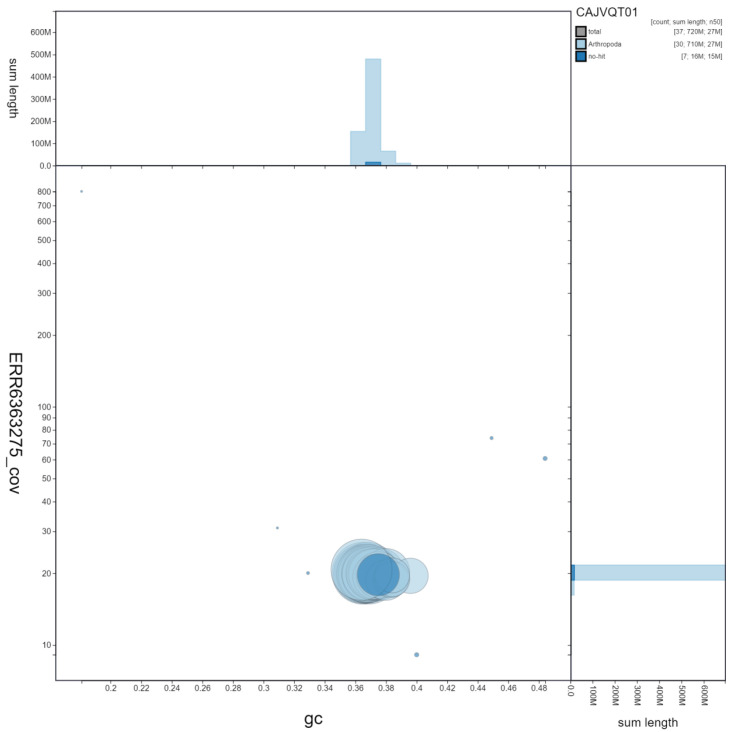
Figure 3. Genome assembly of Pyrgus malvae, ilPyrMalv3.1: GC coverage.
BlobToolKit GC-coverage plot. Scaffolds are coloured by phylum. Circles are sized in proportion to scaffold length. Histograms show the distribution of scaffold length sum along each axis. An interactive version of this figure is available at https://blobtoolkit.genomehubs.org/view/ilPyrMalv3.1/dataset/CAJVQT01/blob.
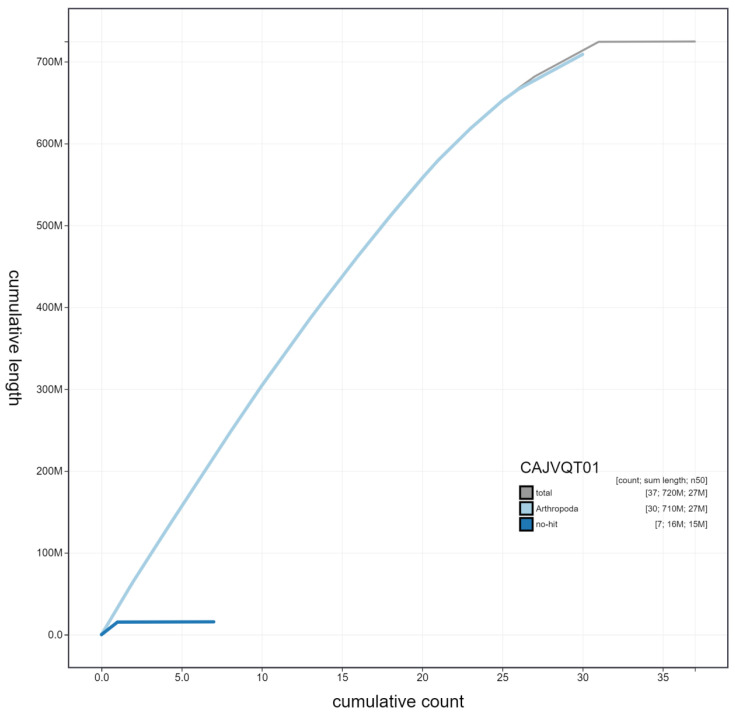
Figure 4. Genome assembly of Pyrgus malvae, ilPyrMalv3.1: cumulative sequence.
BlobToolKit cumulative sequence plot. The grey line shows cumulative length for all scaffolds. Coloured lines show cumulative lengths of scaffolds assigned to each phylum using the buscogenes taxrule. An interactive version of this figure is available at https://blobtoolkit.genomehubs.org/view/ilPyrMalv3.1/dataset/CAJVQT01/cumulative.
Table 2. Chromosomal pseudomolecules in the genome assembly of Pyrgus malvae, ilPyrMalv3.1.
| INSDC accession | Chromosome | Size (Mb) | GC% |
|---|---|---|---|
| OU426946.1 | 1 | 33.22 | 36.5 |
| OU426948.1 | 2 | 30.61 | 36.9 |
| OU426949.1 | 3 | 30.48 | 36.8 |
| OU426950.1 | 4 | 30.18 | 36.6 |
| OU426951.1 | 5 | 30.06 | 36.5 |
| OU426952.1 | 6 | 29.97 | 36.8 |
| OU426953.1 | 7 | 29.61 | 36.7 |
| OU426954.1 | 8 | 29.23 | 36.7 |
| OU426955.1 | 9 | 28.53 | 36.8 |
| OU426956.1 | 10 | 27.64 | 36.6 |
| OU426957.1 | 11 | 27.02 | 36.7 |
| OU426958.1 | 12 | 26.98 | 36.9 |
| OU426959.1 | 13 | 25.95 | 36.9 |
| OU426960.1 | 14 | 25.59 | 36.9 |
| OU426961.1 | 15 | 25.29 | 36.9 |
| OU426962.1 | 16 | 24.25 | 37.1 |
| OU426963.1 | 17 | 24.17 | 37.5 |
| OU426964.1 | 18 | 23.61 | 37.2 |
| OU426965.1 | 19 | 22.98 | 37.1 |
| OU426966.1 | 20 | 21.93 | 37.2 |
| OU426967.1 | 21 | 19.36 | 38.0 |
| OU426968.1 | 22 | 19.17 | 37.5 |
| OU426969.1 | 23 | 17.50 | 37.3 |
| OU426970.1 | 24 | 16.66 | 37.5 |
| OU426971.1 | 25 | 15.31 | 37.5 |
| OU426972.1 | 26 | 14.10 | 38.0 |
| OU426974.1 | 27 | 10.67 | 39.6 |
| OU426973.1 | 28 | 10.82 | 38.3 |
| OU426975.1 | 29 | 10.54 | 38.4 |
| OU426976.1 | 30 | 10.42 | 38.4 |
| OU426947.1 | Z | 32.47 | 36.4 |
| OU426977.1 | MT | 0.02 | 18.4 |
| - | Unplaced | 0.34 | 41.9 |
Genome annotation report
The ilPyrMalv3.1 genome has been annotated using the Ensembl rapid annotation pipeline ( Table 1; https://rapid.ensembl.org/Pyrgus_malvae_GCA_911387765.1/). The resulting annotation includes 23,484 transcribed mRNAs from 12,096 protein-coding and 2,976 non-coding genes. There are 1.66 coding transcripts per gene and 7.83 exons per transcript.
Methods
Sample acquisition and nucleic acid extraction
A male P. malvae specimen (ilPyrMalv3, male, genome assembly) was collected from Suatu, Cluj County, Romania (latitude 46.7648, longitude 23.9845) using a net by Konrad Lohse, Alex Hayward Dominik Laetsch and Roger Vila, who also identified the sample. A further two specimens (ilPyrMalv2, unknown sex, Hi-C; ilPyrMalv1, RNA-Seq) were collected from Baciu, Cluj County, Romania (latitude 46.8, longitude 23.5) using a net and were identified by the same team. All samples were snap-frozen at -80°C.
DNA was extracted from the whole organism of ilPyrMalv3 at the Wellcome Sanger Institute (WSI) Scientific Operations core from the whole organism using the Qiagen MagAttract HMW DNA kit, according to the manufacturer’s instructions. RNA (from the whole organism of ilPyrMalv1) was extracted in the Tree of Life Laboratory at the WSI using TRIzol, according to the manufacturer’s instructions. RNA was then eluted in 50 μl RNAse-free water and its concentration RNA assessed using a Nanodrop spectrophotometer and Qubit Fluorometer using the Qubit RNA Broad-Range (BR) Assay kit. Analysis of the integrity of the RNA was done using Agilent RNA 6000 Pico Kit and Eukaryotic Total RNA assay.
Sequencing
Pacific Biosciences HiFi circular consensus and 10X Genomics read cloud DNA sequencing libraries were constructed according to the manufacturers’ instructions. Poly(A) RNA-Seq libraries were constructed using the NEB Ultra II RNA Library Prep kit. DNA and RNA sequencing was performed by the Scientific Operations core at the WSI on Pacific Biosciences SEQUEL II (HiFi), Illumina HiSeq X (10X) and Illumina HiSeq 4000 (RNA-Seq) instruments. Hi-C data were also generated from whole organism tissue of ilPyrMalv2 using the Arima v1 Hi-C kit and sequenced on an Illumina NovaSeq 6000 instrument.
Genome assembly
Assembly was carried out with Hifiasm ( Cheng et al., 2021); haplotypic duplication was identified and removed with purge_dups ( Guan et al., 2020). One round of polishing was performed by aligning 10X Genomics read data to the assembly with longranger align, calling variants with freebayes ( Garrison & Marth, 2012). The assembly was then scaffolded with Hi-C data ( Rao et al., 2014) using SALSA2 ( Ghurye et al., 2019). The assembly was checked for contamination and corrected using the gEVAL system ( Chow et al., 2016) as described previously ( Howe et al., 2021). Manual curation ( Howe et al., 2021) was performed using gEVAL, HiGlass ( Kerpedjiev et al., 2018) and Pretext. The mitochondrial genome was assembled using MitoHiFi ( Uliano-Silva et al., 2021), which performed annotation using MitoFinder ( Allio et al., 2020). The genome was analysed and BUSCO scores generated within the BlobToolKit environment ( Challis et al., 2020). Table 3 contains a list of all software tool versions used, where appropriate.
Table 3. Software tools used.
| Software tool | Version | Source |
|---|---|---|
| Hifiasm | 0.15.1 | Cheng et al., 2021 |
| purge_dups | 1.2.3 | Guan et al., 2020 |
| SALSA2 | 2.2 | Ghurye et al., 2019 |
| longranger align | 2.2.2 | https://support.10xgenomics.com/genome-exome/software/pipelines/latest/advanced/other-pipelines |
| freebayes | 1.3.1-17-gaa2ace8 | Garrison & Marth, 2012 |
| MitoHiFi | 2 | Uliano-Silva et al., 2021 |
| gEVAL | N/A | Chow et al., 2016 |
| HiGlass | 1.11.6 | Kerpedjiev et al., 2018 |
| PretextView | 0.2.x | https://github.com/wtsi-hpag/PretextView |
| BlobToolKit | 2.6.4 | Challis et al., 2020 |
Genome annotation
The Ensembl gene annotation system ( Aken et al., 2016) was used to generate annotation for the Pyrgus malvae assembly ( GCA_911387765.1). Annotation was created primarily through alignment of transcriptomic data to the genome, with gap filling via protein-to-genome alignments of a select set of proteins from UniProt ( UniProt Consortium, 2019).
Data availability
European Nucleotide Archive: Pyrgus malvae (grizzled skipper). Accession number PRJEB45665; https://identifiers.org/ena.embl/PRJEB45665.
The genome sequence is released openly for reuse. The P. malvae genome sequencing initiative is part of the Darwin Tree of Life (DToL) project. All raw sequence data and the assembly have been deposited in INSDC databases. Raw data and assembly accession identifiers are reported in Table 1.
Funding Statement
This work was supported by Wellcome through core funding to the Wellcome Sanger Institute (206194) and the Darwin Tree of Life Discretionary Award (218328). Fieldwork was supported by an ERC grant (ModelGenom Land 757648) to KL. AH is supported by a Biotechnology and Biological Sciences Research Council (BBSRC) David Phillips Fellowship (BB/N020146/1). RV was supported by the Spanish government through grant PID2019-107078GB-I00/ MCIN/AEI/ 10.13039/501100011033.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 1; peer review: 2 approved, 1 approved with reservations]
Author information
Members of the Darwin Tree of Life Barcoding collective are listed here: https://doi.org/10.5281/zenodo.5744972.
Members of the Wellcome Sanger Institute Tree of Life programme are listed here: https://doi.org/10.5281/zenodo.6125027.
Members of Wellcome Sanger Institute Scientific Operations: DNA Pipelines collective are listed here: https://doi.org/10.5281/zenodo.5746904.
Members of the Tree of Life Core Informatics collective are listed here: https://doi.org/10.5281/zenodo.6125046.
Members of the Darwin Tree of Life Consortium are listed here: https://doi.org/10.5281/zenodo.5638618.
References
- Aken BL, Ayling S, Barrell D, et al. : The Ensembl Gene Annotation System. Database (Oxford). 2016;2016:baw093. 10.1093/database/baw093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allio R, Schomaker-Bastos A, Romiguier J, et al. : MitoFinder: Efficient Automated Large-Scale Extraction of Mitogenomic Data in Target Enrichment Phylogenomics. Mol Ecol Resour. 2020;20(4):892–905. 10.1111/1755-0998.13160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asher J, Warren M, Fox R, et al. : The Millennium Atlas of Butterflies in Britain and Ireland. Oxford University Press.2001. Reference Source [Google Scholar]
- Bigger TRL: Chromosome Numbers of Lepidoptera. Part I. Entomologist’s Gazette. 1960;11:149–52. [Google Scholar]
- Brereton TM, Bourn NAD, Warren MS: Grizzled Skipper Action Plan. Butterfly Conserv. 1998. 10.13140/RG.2.1.3636.8407 [DOI] [Google Scholar]
- Brig: Report on the Species and Habitat Review (Report by the Biodiversity Reporting and Information Group (BRIG) to the UK Standing Committee). JNCC Peterborough. 2007. Reference Source [Google Scholar]
- Challis R, Richards E, Rajan J, et al. : BlobToolKit - Interactive Quality Assessment of Genome Assemblies. G3 (Bethesda). 2020;10(4):1361–74. 10.1534/g3.119.400908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Concepcion GT, Feng X, et al. : Haplotype-Resolved de Novo Assembly Using Phased Assembly Graphs with Hifiasm. Nat Methods. 2021;18(2):170–75. 10.1038/s41592-020-01056-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow W, Brugger K, Caccamo M, et al. : gEVAL — a Web-Based Browser for Evaluating Genome Assemblies. Bioinformatics. 2016;32(16):2508–10. 10.1093/bioinformatics/btw159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federley H: Chromosomenzahlen Finnlän-Discher Lepidopteren. Hereditas. 1938;24(4):397–464. 10.1111/j.1601-5223.1938.tb03219.x [DOI] [Google Scholar]
- Garrison E, Marth G: Haplotype-Based Variant Detection from Short-Read Sequencing. arXiv: 1207.3907.2012. 10.48550/arXiv.1207.3907 [DOI] [Google Scholar]
- Ghurye J, Rhie A, Walenz BP, et al. : Integrating Hi-C Links with Assembly Graphs for Chromosome-Scale Assembly. PLoS Comput Biol. 2019;15(8):e1007273. 10.1371/journal.pcbi.1007273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan D, McCarthy SA, Wood J, et al. : Identifying and Removing Haplotypic Duplication in Primary Genome Assemblies. Bioinformatics. 2020;36(9):2896–98. 10.1093/bioinformatics/btaa025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Roldán JL, Bofill R, Dapporto L, et al. : Morphological and Chemical Analysis of Male Scent Organs in the Butterfly Genus Pyrgus (Lepidoptera: Hesperiidae). Org Divers Evol. 2014;14(3):269–78. 10.1007/s13127-014-0170-x [DOI] [Google Scholar]
- Howe K, Chow W, Collins J, et al. : Significantly Improving the Quality of Genome Assemblies through Curation. GigaScience. 2021;10(1):giaa153. 10.1093/gigascience/giaa153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerpedjiev P, Abdennur N, Lekschas F, et al. : HiGlass: Web-Based Visual Exploration and Analysis of Genome Interaction Maps. Genome Biol. 2018;19(1):125. 10.1186/s13059-018-1486-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manni M, Berkeley MR, Seppey M, et al. : BUSCO Update: Novel and Streamlined Workflows along with Broader and Deeper Phylogenetic Coverage for Scoring of Eukaryotic, Prokaryotic, and Viral Genomes. Mol Biol Evol. 2021;38(10):4647–54. 10.1093/molbev/msab199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SSP, Huntley MH, Durand NC, et al. : A 3D Map of the Human Genome at Kilobase Resolution Reveals Principles of Chromatin Looping. Cell. 2014;159(7):1665–80. 10.1016/j.cell.2014.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolman T, Lewington R: Collins Butterfly Guide.HarperCollins UK.2008. Reference Source [Google Scholar]
- Uliano-Silva M, Nunes JGF, Krasheninnikova K, et al. : marcelauliano/MitoHiFi: mitohifi_v2.0.2021. 10.5281/zenodo.5205678 [DOI] [Google Scholar]
- UniProt Consortium: UniProt: A Worldwide Hub of Protein Knowledge. Nucleic Acids Res. 2019;47(D1):D506–15. 10.1093/nar/gky1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Swaay C, Wynhoff I, Verovnik R, et al. : IUCN Red List of Threatened Species: Pyrgus Malvae. IUCN Red List of Threatened Species. 2010. Reference Source [Google Scholar]
- Wallis De Vries MF, Raemakers I: Does Extensive Grazing Benefit Butterflies in Coastal Dunes? Restor Ecol. 2001;9(2):179–88. 10.1046/j.1526-100x.2001.009002179.x [DOI] [Google Scholar]