Abstract
Recombinant plasmids were constructed to secrete mouse tumor necrosis factor alpha (mTNF-α) from Clostridium acetobutylicum. The shuttle plasmids contained the clostridial endo-β1,4-glucanase (eglA) promoter and signal sequence that was fused in frame to the mTNF-α cDNA. The construction was first tested in Escherichia coli and then introduced in C. acetobutylicum DSM792 by electroporation. Controls confirmed the presence and stability of the recombinant plasmids in this organism. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and an in vitro cytotoxic assay were used to monitor expression and secretion of mTNF-α during growth. Significant levels of biologically active mTNF-α were measured in both lysates and supernatants. The present report deals with investigations on the elaboration of a gene transfer system for cancer treatment using anaerobic bacteria.
The genus Clostridium comprises a heterogeneous group of rod-shaped, anaerobic, gram-positive, endospore-forming bacteria. Understanding the molecular biology of clostridia brings with it improved prospects for biotechnological exploitation. The development of suitable molecular tools, especially transformation procedures and specialized shuttle vectors, made it possible to introduce and express both homologous and heterologous genes in these microorganisms (20). Recently, attention has been paid to the use of nonpathogenic (24) or avirulent bacteria (18, 27) as delivery systems of therapeutic genes in anticancer therapy. These strategies have been shown to be safe in use. Colonization occurs selectively in tumors, not in normal tissues (12), and treatment can be stopped with suitable antibiotics (11).
Taken together, these technologies provide a new approach to use genetically engineered bacteria in the treatment of cancer. So far, constructs based on the pMTL500F shuttle vector with the Escherichia coli genes cytosine deaminase and nitroreductase have been developed (4, 17). The expression of the cloned genes has been evaluated in Clostridium beijerinckii, but the therapeutic value of these achievements has yet to be realized. In this context, the present investigations were aimed at the establishment of a recombinant Clostridium acetobutylicum that secretes mouse tumor necrosis factor alpha (mTNF-α). This therapeutic agent was selected because it is a cytokine with multiple antitumor effects (28). These include selective action on the neovasculature of tumors, stimulation of T-cell-mediated immunity, and direct cytotoxicity to tumor cells, mainly through induction of apoptosis (3, 15, 16, 40). Moreover, enhancement of the antiproliferative effect of tumor cells was demonstrated in vivo when TNF-α was combined with irradiation (6, 31). However, systemic toxicity hampers its straight use. Therefore, innovative strategies are required to increase the therapeutic efficacy of TNF-α treatment. One approach is to increase the effective concentration to which the tumor is exposed by using local treatment as proposed with the bacterial gene transfer system. Since we are specifically interested in studying the effects of introducing mTNF-α in the tumor microenvironment, it was necessary to construct a vector which after introduction in Clostridium provoked the secretion of the therapeutic agent.
This report describes the cloning of mTNF-α cDNA in a stable E. coli-Clostridium shuttle vector. As a proof of principle that mTNF-α can be produced by C. acetobutylicum DSM792, mTNF-α cDNA was placed under transcriptional control of the endo-β1,4-glucanase (eglA) promoter of C. acetobutylicum P262 and fused to the eglA signal sequence. Data on mTNF-α production by recombinant C. acetobutylicum DSM792 are presented.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
C. acetobutylicum DSM792 was obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (Braunschweig, Germany). The strain was grown in 2× YT medium (26) or reinforced clostridial medium (Difco Laboratories, Detroit, Mich.) at 37°C in an anaerobic system (model 1024; Forma Scientific, Marietta, Ohio) with 90% N2 and 10% H2 with palladium as the catalyst. For analysis of mTNF-α production, Clostridium was cultivated in 5-ml aliquots of 2× YT medium after 1/10 inoculation with an overnight culture. The medium was buffered at pH 7.2 with filter-sterilized morpholinepropanesulfonic acid (MOPS) (Sigma) added at a final concentration of 50 mM.
For primary vector constructions, E. coli TG1 (29) was used. In vitro mutagenesis was carried out with E. coli JM109 (29) and E. coli BMH 71-18mut S (37). These strains were regularly grown in Luria-Bertani broth at 37°C (300 rpm). Prior to electroporation, plasmids were methylated in vivo in E. coli ER2275(pAN1) (22). After isolation, plasmid DNA preparations were desalted by two spins in Microcon 100 microconcentrators (Amicon, Inc., Beverly, Mass.) as recommended by the manufacturer.
Media were supplemented, when applicable, with erythromycin (25 μg/ml), ampicillin (50 μg/ml), chloramphenicol (35 μg/ml), isopropyl-β-thiogalactopyranoside (IPTG; 50 μg/ml), or 5-bromo-4-chloro-3-indolyl-β-galactoside (X-Gal; 40 μg/ml).
Shuttle vectors used in this study are listed in Table 1. Plasmid pHZ117, containing the eglA gene of C. acetobutylicum P262, was a gift from H. Zappe (39). The mTNF-α cDNA was available on plasmid pIG2mTNF (Innogenetics, Ghent, Belgium).
TABLE 1.
Shuttle vectors used in this study
| Plasmid | Size (kb) | Parent plasmid
|
Selectable markera
|
Reference | ||
|---|---|---|---|---|---|---|
| Gram + | Gram − | Gram + | Gram − | |||
| pKNT19 | 4.9 | pIM13 | pUC19 | EM (MLSr) | AP | 2 |
| pIMP1 | 4.7 | pIM13 | pUC9 | EM (MLSr) | AP | 21 |
| pMTL500E | 6.4 | pAMβ1 | pMTL20 | EM | AP | 23 |
EM, erythromycin; AP, ampicillin, MLSr, macrolide-lincosamide-streptogramin B resistance.
Transformation procedures and DNA manipulations.
E. coli was transformed by using chemically competent cells obtained with the RbCl method. Transformation of C. acetobutylicum was carried out by electroporation as recently published (25). In brief, cells were grown in 50 ml of reinforced clostridial medium until mid-log phase, washed with ice-cold electrotransformation buffer (270 mM sucrose, 0.6 mM Na2HPO4, 4.4 mM NaH2PO4, 10 mM MgCl2 [pH 6]), and finally resuspended in 2 ml of electrotransformation buffer without MgCl2. A 0.6-ml sample of the cell suspension was transferred to a 0.4-cm electroporation cuvette containing 1 to 5 μl of methylated plasmid DNA (0.1 to 1.5 μg). After the pulse (1.8 kV, 600 Ω, 50 μF), cell suspensions were diluted with 1.4 ml of 2× YT medium and incubated for 4 h at 37°C, before the cells were spread on selective plates and incubated for 3 to 5 days at 37°C.
All general DNA manipulations in E. coli were carried out essentially as described by Sambrook et al. (29). Restriction endonucleases and DNA-modifying enzymes were purchased from Roche Diagnostics (Brussels, Belgium), GIBCO BRL (Gaithersburg, Md.), and Eurogentec (Seraing, Belgium) and used as indicated by the suppliers.
DNA plasmid isolation from E. coli was performed with the Wizard Plus SV miniprep kit (Promega Inc., Madison, Wis.). Plasmid DNA was isolated from C. acetobutylicum by the alkaline lysis procedure described by Nakotte et al. (25).
Mutations were carried out with the Altered Sites in vitro mutagenesis kit from Promega. Oligonucleotides for mutagenesis were purchased from Eurogentec or Amersham Pharmacia Biotech. Oligonucleotides used were 5′-GCTGAAGCTTCAACAACATC-3′ (for introduction of the HindIII site in eglA) and 5′-GACTACTTGATCTTACGTAGATTTAAACCTCCTG-3′ (for introduction of the SnaBI site in mTNF-α cDNA). The DNA fragments containing the introduced mutations were subcloned in pUC19 to verify the mutations by DNA sequencing with an automated laser fluorescent ALF DNA sequencer (Amersham Pharmacia Biotech). Primers used for sequencing were the fluorolabeled M13 universal and reverse primer.
DNA probes for Southern blotting experiments were labeled with digoxigenin. Labeling efficiency was checked by using chemiluminescent detection as recommended by the manufacturer (Roche Diagnostics). Southern blotting was performed with an optimized hybridization protocol (9).
Vector construction.
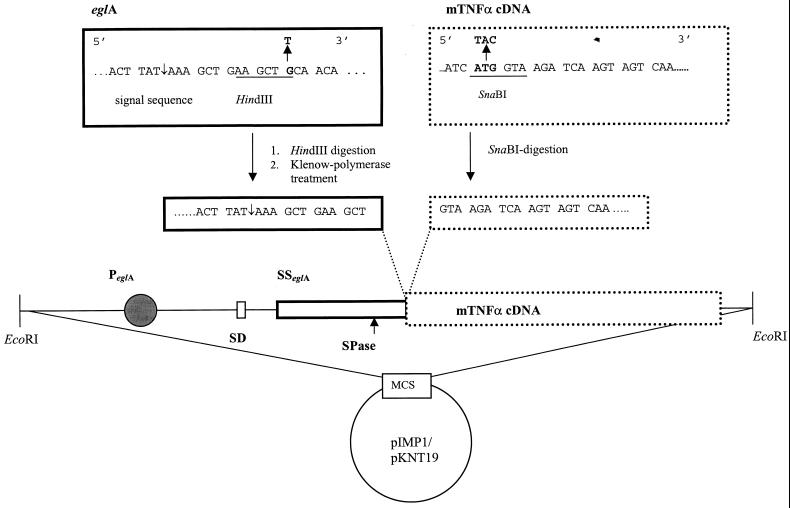
pHZ117 was digested with HindIII/SacI. The resulting 1.2-kb eglA fragment was isolated and subcloned in pSelect. A HindIII site was introduced four codons upstream of the signal sequence. A SnaBI site was created at the first codon of the mTNF-α cDNA, available on the pIG2mTNF plasmid, resulting in pIG2mTNFSnaBI (13). A 0.5-kb EcoRI/HindIII fragment containing the eglA promoter, ribosome binding site, and signal sequence was next cloned in pBR322. Restriction digestion with HindII and HindIII, followed by Klenow polymerase treatment, resulted in a 0.9-kb blunt-ended fragment. This fragment was isolated and ligated in the pIG2mTNFSnaBI vector that was previously digested with SnaBI. The obtained construct was designated pIG2eglAmTNF. Subsequently, the eglAmTNF fragment was digested from the pIG2eglAmTNF plasmid by EcoRI and inserted into the EcoRI site of the different shuttle vectors (Fig. 1).
FIG. 1.
Schematic representation of the construction of the pIMP1eglATNF and the pKNT19eglATNF shuttle vector and of the mutations introduced in the original eglA sequence and the mTNF-α cDNA. PeglA, promoter region of eglA; SSeglA, signal sequence of eglA; SD, Shine-Dalgarno sequence; SPase, signal peptidase cleavage site (indicated by ↑).
Determination of segregational plasmid stability.
Five milliliters of fresh 2× YT medium containing erythromycin was inoculated with 0.5 ml of late-exponential-growth-phase cultures of recombinant C. acetobutylicum DSM792 strains. Cultures were subsequently subcultivated every day into 5 ml of fresh 2× YT medium without erythromycin pressure over a 6-day period. These cultures were diluted and subsequently plated onto both selective and nonselective 2× YT plates. The numbers of colonies on selective and nonselective growth media were compared to determine plasmid stability [expressed as a percentage; (number of Emr colonies on selective plates)/(total number of colonies on nonselective plates) × 100]. Randomly isolated colonies from selective plates were examined for the presence of plasmid DNA.
Detection and quantification of mTNF-α.
For mTNF-α quantification, cells were harvested at various time periods during growth. At each interval, 0.5 ml of cell suspension was pelleted by centrifugation (Sorvall MC; 12 V, 12,000 rpm, 2 min). The supernatant was removed and immediately stored at −80°C. The cell pellet was resuspended in 0.5 ml of 100 mM Tris-HCl (pH 7.4), and the cells were lysed by sonication with two pulses of 20 s. Cellular debris was thereafter pelleted by centrifugation (12,000 rpm, 2 min), and the cleared lysate was stored at −80°C. Subsequently, the amount of biologically active mTNF-α in lysates and supernatants was determined as described by Heremans et al. (8) by using recombinant mTNF-α (specific activity, 2.5 × 108 U mg−1) produced in E. coli as the standard. Briefly, cytotoxicity of mTNF-α towards WEH164 clone 13 cells was spectrophotometrically evaluated via the in situ reduction of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), measuring the percentage of dead cells.
Immunoblot analysis with polyclonal rabbit anti-mTNF-α antibodies was carried out by the method of Van Mellaert et al. (35).
RESULTS
Construction of shuttle vectors containing the mTNF-α cDNA and Clostridium transformation.
The vectors used to construct recombinant plasmids containing the mTNF-α cDNA are listed in Table 1. To investigate the possibility of expressing and secreting mTNF-α in Clostridium, the promoter and signal sequence of the endo-β1,4-glucanase (eglA) gene of C. acetobutylicum P262 were chosen as a model. As outlined in Fig. 1, to obtain in-frame fusions between the eglA signal sequence and the mTNF-α coding sequence, restriction sites were created in the 3′ end of eglA as well as in the 5′ end of the mTNF-α cDNA. The introduction of the mutated restriction sites to fuse the regulatory sequence with the region at the start of the coding sequence ensured that the authenticity of the nucleotide sequence remained. Correct in-frame fusion was controlled by restriction digestion and by DNA sequence analysis. The eglA promoter is active in E. coli. As a consequence, it was possible to test lysates for the production of mTNF-α. Immunoblotting of the proteins of the cell lysates clearly demonstrated the presence of mTNF-α (data not shown).
Verification of the recombinant constructs in C. acetobutylicum DSM792.
The recombinant constructs were methylated in vivo before they were introduced into Clostridium by electroporation. The obtained electroporation efficiency was ∼102 transformants μg of DNA−1. The recombinant plasmids were isolated from C. acetobutylicum by using a modified plasmid isolation protocol and visualized after electrophoresis on a 1% agarose gel and staining with ethidium bromide (Fig. 2). The pIMP1eglATNF and pKNT19eglATNF constructs were stably maintained in the clostridial host. This was confirmed by restriction digestion analysis and by transformation of E. coli with the isolated plasmid DNA. Southern blotting of isolated plasmid DNA either from Clostridium or from the transformed E. coli cultures with a digoxigenin-labeled specific mTNF-α probe resulted in the expected signal for the construct with pIMP1 and pKNT19 but not for the construct with pMTL500E (data not shown).
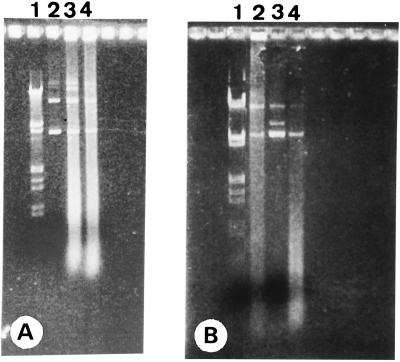
FIG. 2.
Photograph of isolated plasmids pIMP1eglATNF and pKNT19eglATNF after gel electrophoresis on a 1% agarose gel stained with ethidium bromide. (A) Lanes: 1, λ DNA digested with EcoRI and HindIII; 2, pIMP1eglATNF isolated from recombinant E. coli TG1; 3 and 4, pIMP1eglATNF isolated from recombinant C. acetobutylicum DSM792. (B) Lanes: 1, λ DNA digested with EcoRI and HindIII; 3, pKNT19eglATNF isolated from recombinant E. coli TG1; 2 and 4, pKNT19eglATNF isolated from recombinant C. acetobutylicum DSM792.
The segregational stability of both the pIMP1eglATNF and the pKNT19eglATNF construct was examined. Both plasmids were stably maintained following repeated culture transfer over a 6-day period in the absence of antibiotic pressure. The presence of plasmid DNA isolated from randomly selected colonies was confirmed by agarose gel electrophoresis (data not shown).
Immunoblot detection of mTNF-α in C. acetobutylicum(pIMP1eglATNF) and C. acetobutylicum(pKNT19eglATNF) cultures was carried out as described previously (35). Cultures of C. acetobutylicum(pIMP1), C. acetobutylicum(pKNT19), and plasmid-free C. acetobutylicum were taken as a control. Lysates and supernatants from overnight cultures were assessed for mTNF-α production. mTNF-α was detected in lysates of the recombinant cultures both as a preprotein (21 kDa) and as the mature, processed form (17 kDa). In the supernatant, however, only the mature form was present (data not shown). No mTNF-α was detected in lysates or supernatants of the Clostridium control cultures. These results clearly showed the functionality of the promoter and signal sequence preceding the mTNF-α cDNA.
Biological activity of produced mTNF.
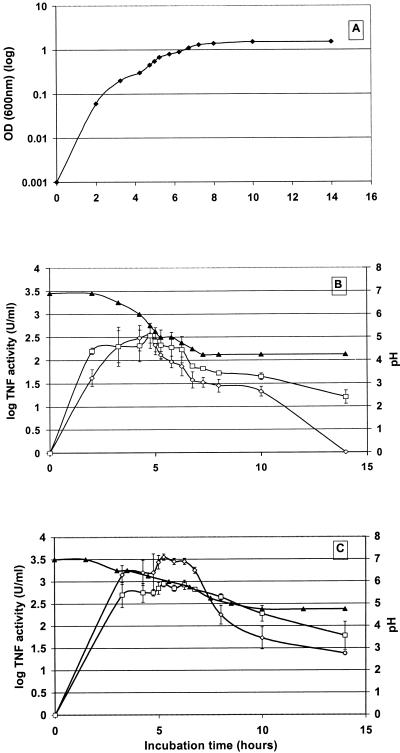
Samples of growing recombinant bacteria containing the pIMP1eglATNF or pKNT19eglATNF construct were taken at various stages of growth (Fig. 3A), and the biological activity of mTNF-α in lysates and supernatants was measured. Bacteria without a plasmid and bacteria with the pIMP1 or the pKNT19 vector alone were selected for controls. The activity was quantified by titration with the mTNF-α standard of known concentration (0.9 μg ml−1). Several independent experiments were performed, and the results were normalized towards the titer of the first experiment so that the data could be pooled. The normalization also allowed the interexperiment comparison, showing confirmation for quantitative reproducibility.
FIG. 3.
Amount of biologically active mTNF-α in supernatant (◊) and lysates (□) of C. acetobutylicum DSM792 transformed with pIMP1eglATNF and evolution of pH (▴) in nonbuffered (B) and buffered (C) medium as a function of growth time (⧫) (A). Error bars represent standard deviations.
The mTNF-α concentration in lysates and supernatants of recombinant clostridia containing the pIMP1eglATNF plasmid increased until the growing cells reached mid-log phase (optical density at 600 nm, ≅0.6). Thereafter, the amount of the mTNF-α present in the supernatant decreased below the detection limit (3.1 U ml−1) after 12 h, whereas there was still biologically active mTNF-α in lysates for up to 20 h. Both in lysate and supernatant, a maximum of ∼103 U of mTNF-α activity ml−1 was found (Fig. 3B). Very similar time-related mTNF-α activity measurements were obtained with the pKNT19eglATNF construct. In supernatants and lysates of cultures without the plasmid or with the plasmid not containing the mTNF-α gene, no mTNF-α activity was detected.
To test whether the decrease in activity was due to the action of proteases or due to the formation of acidic fermentation products from the recombinant clostridia, causing a pH decrease in the medium, MOPS (pH 7.2) was added to buffer the culture medium. This resulted in an elevated level of biologically active mTNF-α (Fig. 3C) in both lysates and supernatants (measured within a follow-up period of 24 h). From these data, we can conclude that the higher amount of active mTNF-α clearly corresponds with the slower decrease in pH in the buffered medium. This is completely in accordance with the literature describing the stability of TNF-α within a pH region of 10 to 5.5 (36).
DISCUSSION
The present experiments were carried out in the context of a tumor-specific gene transfer system using apathogenic clostridia (11, 12). We assessed whether the eukaryotic mTNF-α cDNA could be expressed in Clostridium and whether mTNF-α could subsequently be secreted. The mTNF-α production from the colonizing recombinant bacteria may lead to an improved in vivo antitumor response. This would likely occur in the absence of systemic toxicity because of the selective intratumoral deposition of the cytokine.
Therefore, C. acetobutylicum DSM792 was genetically engineered to produce mTNF-α. With these experiments, the presence of biologically active mTNF-α in culture supernatants and cell lysates of C. acetobutylicum DSM792(pIMP1eglATNF) and C. acetobutylicum DSM792(pKNT19eglATNF), containing the mTNF-α cDNA fused to the signal sequence of eglA and preceded by the eglA promoter, was clearly demonstrated both by Western blotting with mTNF-α antibodies and in a bioassay using WEH164 clone 13 cells. Notwithstanding that the mTNF-α gene was preceded by a signal sequence originating from Clostridium, not all of the mTNF-α expressed was also secreted. Secretion efficiency might possibly be improved by using a different signal peptide or by modifying, e.g., the number of positive charges in the signal peptide, as shown for other organisms (13).
Our experiments showed that the pIMP1 and pKNT19 derivatives were segregationally stable in C. acetobutylicum DSM792. Both constructs were structurally more stable than the construct with pMTL500E. It has been suggested that vectors based on the pAMβ1 replicon such as pMTL500E would be structurally more stable and thus more suitable for general use in Clostridium (38). This hypothesis relies on the absence of highly recombinogenic single-stranded DNA intermediates that would interfere with the structural stability of the plasmid. Our experiments, however, do not confirm this general hypothesis. Since other derivatives of pMTL500E have been shown in vitro to be stably maintained in C. beijerinckii (4, 17), these findings and our data considered together possibly reflect a strain- and/or sequence-specific character of vector stability.
Clostridium is characterized by a low G+C content with a strongly biased codon usage towards codons in which A and U predominate. However, since biologically active mTNF-α was detected in our experiments, the low G+C content of the host organism (28 to 29 mol%) seemingly has no implications for the expression of the cloned mTNF-α gene, at least in the strain used. Moreover, functional mTNF-α was secreted. Secretion was possible because of the presence of a clostridial signal sequence (39). To our knowledge, this is the first report describing the secretion of a eukaryotic protein from Clostridium.
Anaerobic bacteria selectively colonize the hypoxic-necrotic areas of solid tumors. This has been demonstrated with rodent tumor models (11, 19) and is documented for some cancer patients (10, 32). Since these hypoxic-necrotic regions are not present in normal tissues, this transfer system is very tumor specific (12). As a consequence, it should be possible to increase the local concentration of therapeutic agents by using genetically modified anaerobic bacteria expressing and secreting these compounds. The antitumor effectiveness will obviously depend on the stability of the recombinant constructs and on the expression of the therapeutic genes in an in vivo tumor system. In vivo results described by Minton et al., using the EMT6 mouse tumor model, indicated the difficulty in obtaining sufficient amounts of therapeutic proteins (24). This might relate to the stability of the constructs and/or to the insufficient Clostridium colonization of the tumor. Using another tumor model and different therapeutic gene products, we aim to improve the therapeutic efficiency in vivo. Separately, the ameliorated tumor colonization of rat rhabdomyosarcomas using a vascular targeting compound has recently been demonstrated in our laboratories (14).
The clinical usefulness of systemically administered mTNF-α is limited due to hepatotoxicity and hypotension as major dose-limiting side effects (28, 30). The strategy introduced in the present investigations aims to restrict the antitumor potential of this cytokine solely in the tumor site, thereby bypassing the systemic toxicity.
TNF-α has been shown to be cytotoxic towards many transformed cell lines in vitro (5, 33). Antitumor activity against a variety of murine as well as human tumors has also been documented with in vivo investigations (1, 7, 34). Experiments are in progress to analyze in vivo the qualitative and quantitative effects of mTNF-α expression in rodent tumor models.
ACKNOWLEDGMENTS
We appreciate the fruitful discussions with Elke Lammertyn on the present research.
We acknowledge financial support from Het Fonds voor Wetenschappelijk Onderzoek-Vlaanderen, De Vlaamse Kankerliga, Verkennende Internationale Samenwerking, Het K.U.Leuven Onderzoeksfonds, and Sportvereniging tegen Kanker of Belgium. Jan Theys and Sandra Nuyts are research fellows of I.W.T. (Vlaams Instituut voor de bevordering van het Wetenschappelijk-Technologisch onderzoek in de industrie).
REFERENCES
- 1.Asher A, Mulé J J, Reichert C M, Shiloni E, Rosenberg S A. Studies on the anti-tumor efficacy of systemically administered recombinant tumor necrosis factor against several murine tumors in vivo. J Immunol. 1987;138:963–974. [PubMed] [Google Scholar]
- 2.Azzedoug H, Hubert J, Reysset G. Stable inheritance of shuttle vectors based on plasmid pIM13 in a mutant strain of Clostridium acetobutylicum. J Gen Microbiol. 1992;138:1371–1378. doi: 10.1099/00221287-138-7-1371. [DOI] [PubMed] [Google Scholar]
- 3.Fiers W. Tumor necrosis factor. Characterization at the molecular, cellular and in vivo level. FEBS Lett. 1991;285:199–212. doi: 10.1016/0014-5793(91)80803-b. [DOI] [PubMed] [Google Scholar]
- 4.Fox M E, Lemmon M J, Mauchline M L, Davis T O, Giaccia A J, Minton N P, Brown J M. Anaerobic bacteria as a delivery system for cancer gene therapy: in vitro activation of 5-fluorocytosine by genetically engineered clostridia. Gene Ther. 1996;3:173–178. [PubMed] [Google Scholar]
- 5.Fransen L, Ruysschaert M R, Van Der Heyden J, Fiers W. Recombinant tumor necrosis factor: species specificity for a variety of human and murine transformed cell lines. Cell Immunol. 1986;100:260–267. doi: 10.1016/0008-8749(86)90025-0. [DOI] [PubMed] [Google Scholar]
- 6.Hallahan D E, Mauceri H J, Seung L P, Dunphy E J, Wayne J D, Hanna N D, Toledano A, Hellman S, Kufe D W, Weichselbaum R R. Spatial and temporal control of gene therapy using ionizing radiation. Nature Med. 1995;1:786–791. doi: 10.1038/nm0895-786. [DOI] [PubMed] [Google Scholar]
- 7.Haranaka K, Satomi N, Sakurai A. Antitumor activity of murine tumor necrosis factor (TNF) against transplanted murine tumors and heterotransplanted human tumors in nude mice. Int J Cancer. 1984;34:263–267. doi: 10.1002/ijc.2910340219. [DOI] [PubMed] [Google Scholar]
- 8.Heremans H, Van Damme J, Dillen C, Dijkmans R, Billiau A. Interferon-γ, a mediator of lethal lipopolysaccharide-induced Shwartzman-like shock reactions in mice. J Exp Med. 1990;171:1853–1869. doi: 10.1084/jem.171.6.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoeltke H-J, Schneider S, Ettl I, Binsack R, Obermaier I, Seller M, Sagner G. Rapid, highly sensitive detection of digoxigenin-labeled nucleic acids by improved chemiluminescent alkaline phosphatase substrates. Biochemica (Boehringer) 1995;1:17–20. [Google Scholar]
- 10.Kornbluth A A, Danzig J B, Bernstein L H. Clostridium septicum infection and associated malignancy. Medicine. 1989;68:30–37. doi: 10.1097/00005792-198901000-00002. [DOI] [PubMed] [Google Scholar]
- 11.Lambin, P., W. Landuyt, J. Theys, S. Nuyts, L. Van Mellaert, and J. Anné. Specific tumor colonization by Clostridium saccharoperbutylacetonicum. Submitted for publication.
- 12.Lambin P, Theys J, Landuyt W, Rijken P, van der Kogel A, van der Schueren E, Hodgkiss R, Fowler J, Nuyts S, de Bruijn E, Van Mellaert L, Anné J. Colonization of Clostridium in the body is restricted to hypoxic and necrotic areas of tumors. Anaerobe. 1998;4:183–188. doi: 10.1006/anae.1998.0161. [DOI] [PubMed] [Google Scholar]
- 13.Lammertyn E, Van Mellaert L, Schacht S, Dillen C, Sablon E, Van Broekhoven A, Anné J. Evaluation of a novel subtilisin inhibitor gene and mutant derivatives for the expression and secretion of mouse tumor necrosis factor alpha by Streptomyces lividans. Appl Environ Microbiol. 1997;63:1808–1813. doi: 10.1128/aem.63.5.1808-1813.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Landuyt, W., J. Theys, S. Nuyts, A. Reijnders, J. W. Fowler, W. Van Den Bogaert, L. Van Mellaert, J. Anné, and P. Lambin. Tumor colonisation with anaerobic bacteria following vascular targeting treatment: improvement for bacterial gene therapy developments.
- 15.Larrick J W, Wright S C. Cytotoxic mechanism of tumor necrosis factor-α. FASEB J. 1990;4:3215–3223. doi: 10.1096/fasebj.4.14.2172061. [DOI] [PubMed] [Google Scholar]
- 16.Laster S M, Wood J G, Goodding L R. Tumor necrosis factor can induce both apoptic and necrotic forms of cell lysis. J Immunol. 1988;141:2629–2634. [PubMed] [Google Scholar]
- 17.Lemmon M J, van Zijl P, Fox M E, Mauchline M L, Giaccia A J, Minton N P, Brown J M. Anaerobic bacteria as a gene delivery system that is controlled by the tumor microenvironment. Gene Ther. 1997;4:791–796. doi: 10.1038/sj.gt.3300468. [DOI] [PubMed] [Google Scholar]
- 18.Low K B, Ittensohn M, Le T, Platt J, Sodi S, Amoss M, Ash O, Carmichael E, Chakraborty A, Fisher J, Lin S L, Luo X, Miller S I, Zheng L M, King I, Pawelek J M, Bermudes D. Lipid A mutant Salmonella with suppressed virulence and TNFα induction retain tumor-targeting in vivo. Nature Biotechnol. 1999;17:37–41. doi: 10.1038/5205. [DOI] [PubMed] [Google Scholar]
- 19.Malmgren R A, Flanigan C C. Localization of the vegetative form of Clostridium tetani in mouse tumors following intravenous spore administration. Cancer Res. 1955;15:473–478. [PubMed] [Google Scholar]
- 20.Mauchline M L, Davis T, Minton N P. Clostridia. In: Demain A L, Davies J E, editors. Manual of industrial microbiology and biotechnology. Oxford United Kingdom: Blackwell Science; 1999. pp. 475–490. [Google Scholar]
- 21.Mermelstein L D, Welker N E, Bennett G N, Papoutsakis E T. Expression of cloned homologous fermentative genes in Clostridium acetobutylicum ATCC 824. Bio/Technology. 1992;10:190–195. doi: 10.1038/nbt0292-190. [DOI] [PubMed] [Google Scholar]
- 22.Mermelstein L D, Papoutsakis E T. In vivo methylation in Escherichia coli by the Bacillus subtilis phage φ3T I methyltransferase to protect plasmids from restriction upon transformation of Clostridium acetobutylicum ATCC824. Appl Environ Microbiol. 1993;59:1077–1081. doi: 10.1128/aem.59.4.1077-1081.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Minton N P, Oultram J D. Host:vector systems for gene cloning in Clostridium. Microbiol Sci. 1988;5:310–315. [PubMed] [Google Scholar]
- 24.Minton N P, Mauchline M L, Lemmon M J, Brehm J K, Fox M, Michael N P, Giaccia A, Brown J M. Chemotherapeutic tumor targeting using clostridial spores. FEMS Microbiol Rev. 1995;17:357–364. doi: 10.1111/j.1574-6976.1995.tb00219.x. [DOI] [PubMed] [Google Scholar]
- 25.Nakotte S, Schaffer S, Böhringer M, Dürre P. Electroporation of plasmid isolation from and plasmid conservation in Clostridium acetobutylicum DSM792. Appl Microbiol Biotechnol. 1998;50:564–567. doi: 10.1007/s002530051335. [DOI] [PubMed] [Google Scholar]
- 26.Oultram J D, Loughlin M, Swinfield T J, Brehm J K, Thompson D E, Minton N P. Introduction of plasmids into whole cells of Clostridium acetobutylicum by electroporation. FEMS Microbiol Lett. 1988;56:83–88. [Google Scholar]
- 27.Pawelek J M, Low K B, Bermudes D. Tumor-targeted Salmonella as a novel anticancer vector. Cancer Res. 1997;57:4537–4544. [PubMed] [Google Scholar]
- 28.Raijinder S S, Bollon A P. Tumor necrosis factor activities and cancer therapy—a perspective. Pharmacol Ther. 1993;57:79–128. doi: 10.1016/0163-7258(93)90037-e. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 30.Schiller J H, Storer B E, Witt P L, Alberti D, Tombes M B, Arzoomanian R, Proctor R A, McCarthy D, Brown R R, Voss S D, Remick S C, Grem J L, Borden E C, Trump D L. Biological and clinical effects of intravenous tumor necrosis factor α administered three times weekly. Cancer Res. 1991;51:1651–1658. [PubMed] [Google Scholar]
- 31.Sersa G, Willingham V, Milas L. Anti-tumor effects of tumor necrosis factor alone or combined with radiotherapy. Int J Cancer. 1988;42:129–134. doi: 10.1002/ijc.2910420124. [DOI] [PubMed] [Google Scholar]
- 32.Sjöstedt J, Kager L, Heimdahl A, Nord C E. Microbial colonization of tumors in relation to the upper gastrointestinal tract in patients with gastric carcinoma. Ann Surg. 1988;207:341–346. doi: 10.1097/00000658-198803000-00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sugarman B J, Aggarwal B B, Hass P E, Figari I S, Palladino M A, Shepard H M. Recombinant human tumor necrosis factor α: effects on proliferation of normal and transformed cells in vitro. Science. 1985;230:943–948. doi: 10.1126/science.3933111. [DOI] [PubMed] [Google Scholar]
- 34.Teng M N, Park B H, Koeppen H K W, Tracey K J, Fendly B M, Schreiber H. Long-term inhibition of tumor growth by tumor necrosis factor in the absence of cachexia or T-cell immunity. Proc Natl Acad Sci USA. 1991;88:3535–3539. doi: 10.1073/pnas.88.9.3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Mellaert L, Dillen C, Proost P, Sablon E, Deleys R, Van Broekhoven A, Heremans H, Van Damme J, Eyssen H, Anné J. Efficient secretion of biologically active mouse tumor necrosis factor α by Streptomyces lividans. Gene. 1994;150:153–158. doi: 10.1016/0378-1119(94)90876-1. [DOI] [PubMed] [Google Scholar]
- 36.Van Ostade X, Tavernier J, Fiers W. Structure-activity studies of human tumor necrosis factors. Protein Eng. 1994;7:5–22. doi: 10.1093/protein/7.1.5. [DOI] [PubMed] [Google Scholar]
- 37.Wallace R B, Johnson M J, Suggs S V, Miyoshi K, Bhatt R, Itakura K. A set of synthetic oligodeoxyribonucleotide primers for DNA sequencing in the plasmid vector pBR322. Gene. 1981;16:21–26. doi: 10.1016/0378-1119(81)90057-3. [DOI] [PubMed] [Google Scholar]
- 38.Young M, Minton N P, Staudenbauer W L. Recent advances in the genetics of the clostridia. FEMS Microbiol Rev. 1989;63:301–326. doi: 10.1111/j.1574-6968.1989.tb03402.x. [DOI] [PubMed] [Google Scholar]
- 39.Zappe H, Jones W A, Jones D T, Woods D R. Structure of an endo-β-1,4-glucanase gene from Clostridium acetobutylicum P262 showing homology with endoglucanase genes from Bacillus spp. Appl Environ Microbiol. 1988;54:1289–1292. doi: 10.1128/aem.54.5.1289-1292.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zheng L X, Fisher G, Miller R E, Peschon J, Lynch D H, Lenardo M J. Induction of apoptosis in mature T cells by tumour necrosis factor. Nature. 1995;377:348–351. doi: 10.1038/377348a0. [DOI] [PubMed] [Google Scholar]