Abstract
Induction and suppression of antiviral RNA interference (RNAi) has been observed in mammals during infection with at least seven distinct RNA viruses, including some that are pathogenic in humans. However, while the cell‐autonomous immune response mediated by antiviral RNAi is gradually being recognized, little is known about systemic antiviral RNAi in mammals. Furthermore, extracellular vesicles (EVs) also function in viral signal spreading and host immunity. Here, we show that upon antiviral RNAi activation, virus‐derived small‐interfering RNAs (vsiRNAs) from Nodamura virus (NoV), Sindbis virus (SINV), and Zika virus (ZIKV) enter the murine bloodstream via EVs for systemic circulation. vsiRNAs in the EVs are biologically active, since they confer RNA–RNA homology‐dependent antiviral activity in both cultured cells and infant mice. Moreover, we demonstrate that vaccination with a live‐attenuated virus, rendered deficient in RNAi suppression, induces production of stably maintained vsiRNAs and confers protective immunity against virus infection in mice. This suggests that vaccination with live‐attenuated VSR (viral suppressor of RNAi)‐deficient mutant viruses could be a new strategy to induce immunity.
Keywords: antiviral RNAi, extracellular vesicles, live‐attenuated virus vaccine, systemic antiviral RNAi, Zika virus
Subject Categories: Immunology, RNA Biology
EV‐associated vsiRNAs confer homology‐dependent antiviral activity in both cell culture and infant mice.
Introduction
Extracellular vesicles (EVs) are small membrane structures containing a complex cargo of proteins, lipids, and nucleic acids (Théry et al, 2018). EVs are produced by different types of cells and can be detected in most body fluids including blood, saliva, urine, and cerebrospinal fluid (Srinivasan et al, 2019). EVs are classified as apoptotic bodies, microvesicles, or exosomes. Cells undergoing apoptosis release apoptotic bodies 1–5 µm in diameter. Microvesicles are generated via shedding/budding from the plasma membrane and are between 150 nm and 1 µm in diameter. Exosomes originate from multivesicular bodies (MVBs) with the cellular plasma membrane and small dimensions (30–150 nm) (Théry et al, 2018). EVs have essential roles in intercellular communication, both locally and systemically. Recent studies have demonstrated that EVs with viral components play dichotomous roles in viral infections and pathology (Nolte et al, 2016; Raab‐Traub & Dittmer, 2017; Caobi et al, 2020; McNamara & Dittmer, 2020). Viruses often exploit the EV biogenesis pathway to promote viral infection and propagation. On the other hand, EVs can help the host cell to suppress virus infection by triggering antiviral responses and cytokine secretion. Sometimes, this enigmatic dual roles of EVs are observed in infections by the same virus (Caobi et al, 2020; Martins & Alves, 2020). Therefore, the role of EVs during viral infections needs to be further investigated.
Recent studies including ours have demonstrated production of abundant virus‐derived small‐interfering RNAs (vsiRNAs) to target several distinct RNA viruses, such as influenza A virus (IAV), Zika virus (ZIKV), and Dengue virus (DENV), in mammalian cells (Li et al, 2013, 2017; Maillard et al, 2013; Qiu et al, 2017, 2020; Xu et al, 2019; Zhang et al, 2020). Infection of mouse embryonic stem cells (mESCs) and human neural progenitor cells (hNPCs) respectively by wild‐type encephalomyocarditis virus (EMCV) and ZIKV triggers Dicer‐mediated biogenesis of abundant vsiRNAs only before the cells are induced to differentiate (Maillard et al, 2013; Xu et al, 2019). Reis e Sousa and colleagues have recently demonstrated a critical role for a Dicer isoform in vsiRNA production to initiate antiviral RNAi in mouse and human stem cells (Poirier et al, 2021). In contrast, vsiRNAs accumulate to high levels in both mESCs and mature baby hamster cells as well as infant mice in response to the infection with Nodamura virus (NoV) only after its VSR protein B2 is rendered either non‐expressing or non‐functional (Li et al, 2013; Maillard et al, 2013). Similarly, abundant vsiRNAs are produced by Dicer from IAV mutants not expressing its VSR non‐structural protein 1 (NS1) in mature human and monkey cells after infection (Li et al, 2017). Zhou and colleagues also showed that the 3A protein of human enterovirus 71 (HEV71) is a dsRNA‐binding VSR that suppresses Dicer‐mediated production of vsiRNAs like B2 and NS1 despite no sequence similarity among the three mammalian VSRs (Qiu et al, 2017; Fang et al, 2021).
The role of antiviral RNAi in the innate immunity and its relationship to the interferon (IFN) response remain poorly understood (Berkhout, 2018; Ding et al, 2018; Guo et al, 2019; Maillard et al, 2019). It is well known that the IFN response directs a potent innate antiviral pathway. Independent studies have shown that the RNAi response is antiviral in undifferentiated cells, in which the canonical IFN system is not active (Maillard et al, 2013, 2019; Wu et al, 2019; Xu et al, 2019). In differentiated mammalian cells, antiviral RNAi is active against only mutant viruses defective in RNAi suppression (Li et al, 2013, 2017; Qiu et al, 2017, 2020; Han et al, 2020). Moreover, Dicer‐mediated siRNA production and sequence‐specific RNAi can be triggered by artificial long dsRNAs only when the IFN pathway is defective (Kennedy et al, 2015; Maillard et al, 2016). Notably, activation of siRNA production in cells from artificially introduced long dsRNA confers specific antiviral resistance to subsequent infections, indicating that mammalian cells can be “vaccinated” by RNA in a virus‐specific manner (Maillard et al, 2016). Subsequently, Reis e Sousa and colleagues further showed that binding of LGP2 to Dicer inhibited cleavage of dsRNA into siRNAs in vitro and in cells but did not inhibit Dicer‐dependent pre‐miRNA processing (Van der Veen et al, 2018). Interestingly, a recent study found abundant accumulation of vsiRNAs from Dicer processing of viral dsRNA replicative intermediates in adult mice either wild‐type or defective in the signaling of IFN, suggesting that vsiRNA biogenesis is distinct from Dicer processing of artificial long dsRNA (Han et al, 2020). Our recent work also revealed that viral dsRNA replicative intermediates made during authentic infection of mature somatic cells are efficiently processed by Dicer into vsiRNAs to direct antiviral RNAi (Zhang et al, 2021).
The control of viral infections in mammals requires signaling molecules to elicit an effective response and to establish systemic immunity at the organismal level. These signals must be amplified and disseminated throughout the organism to inhibit pathogen propagation and establishment of the infection (Hu & Shu, 2018; Hur, 2019). In mammals, cells contain pattern‐recognition receptors (PRRs) to detect molecules associated with infection. These PRRs, once stimulated by their appropriate ligands, activate intracellular signaling cascades leading to transcription of IFN genes. Once secreted, IFN binds to the IFN receptor on neighboring uninfected cells and activates an intracellular signaling cascade leading to upregulation of several hundred IFN‐stimulated genes (ISGs), many of which have direct or indirect antiviral activities (Schneider et al, 2014). Cell‐autonomous antiviral RNAi has been shown to be functional in mammals, but whether there is a systemic immune response mediated by RNAi needs to be investigated (Berkhout, 2018; Ding et al, 2018; Maillard et al, 2019).
We have shown that infection with the VSR‐deficient mutant of NoV, NoVΔB2, induces production of abundant vsiRNAs and is rapidly cleared in suckling mice in contrast to lethal infection by wild‐type NoV (Li et al, 2013). Intriguingly, we subsequently found that infant mice became fully protected against lethal NoV challenge after immunization with NoVΔB2 for only two days, suggesting rapid induction of protective immunity at whole organism level. A recent study has shown that infection of fruit flies with Sindbis virus (SINV) induces secretion of vsiRNAs‐containing exosome‐like vesicles circulating in the hemolymph sufficient to confer passive protection against SINV challenge in naive flies (Tassetto et al, 2017). This prompted us to investigate a potential role of EVs in NoVΔB2‐triggered protection in mice. We demonstrate that immunization of suckling mice with NoVΔB2 induced accumulation of EV‐associated vsiRNAs in the blood stream capable of conferring resistance against NoV challenge in naïve mice. Notably, our findings indicate that murine EV‐associated vsiRNAs mediate Ago2‐dependent RNAi in cultured cells. We propose that the cell‐autonomous and systemic antiviral RNAi are both active in mammals and that mutant viruses rendered deficient to suppress vsiRNA biogenesis can be explored as a new class of live‐attenuated vaccines against animal and human RNA viruses.
Results
Vaccination with a live‐attenuated, VSR‐deficient NoV (NoV∆B2) confers complete protective immunity in mice against lethal infection
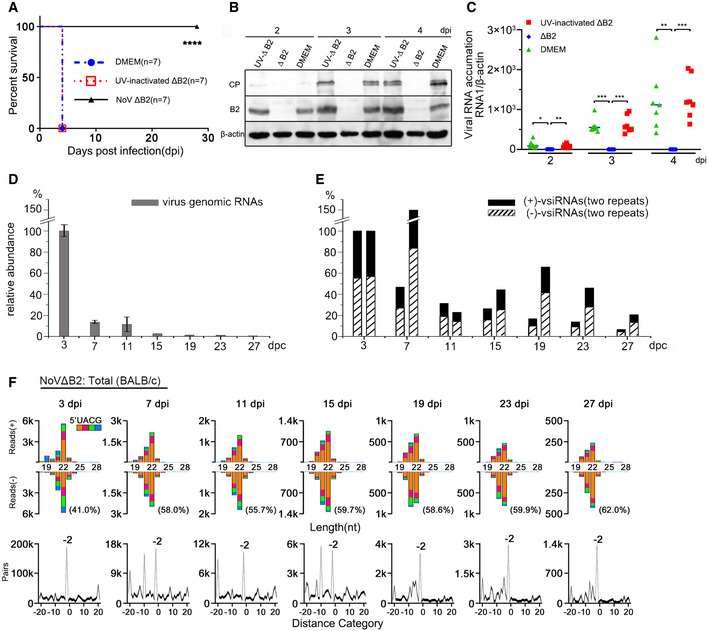
Our previous study have shown that unlike the 100% mortality observed 5 days post‐NoV infection, suckling mice challenged by NoVΔB2 remained healthy for the duration of the experiment, up to 4 weeks post‐inoculation (Li et al, 2013). If the viral siRNAs produced de novo in NoV∆B2‐infected mice directed the clearance of NoV∆B2 by RNAi, they might confer specific protective immunity against secondary infection by NoV since NoV∆B2 and NoV differ only by three nucleotides. To test this hypothesis, we inoculated 6‐day‐old mice with buffer (mock), NoV∆B2, or NoV∆B2 inactivated by ultraviolet light (UV) irradiation, and two days later, challenged the mice with a lethal dose of NoV. The timing of NoV challenge was chosen according to the accumulation profile of viral siRNAs in the NoV∆B2‐immunized neonatal mice. We found that all of the suckling mice pre‐inoculated with either buffer or UV‐inactivated NoV∆B2 developed hind limb paralysis 4 days after NoV challenge before death by 5 dpi. By contrast, none of the suckling mice immunized with NoV∆B2 exhibited any signs of disease up to 4 weeks after the secondary infection, indicating complete protection of the NoV∆B2‐immunized mice (Fig 1A). Western blotting detected abundant expression of both the B2 and coat proteins, encoded by the NoV genomic RNAs 1 and 2, respectively, after NoV infection in the control mice pre‐inoculated with either buffer or UV‐inactivated NoV∆B2 (Fig 1B). However, neither viral protein was detectable in the NoV∆B2‐immunized mice after NoV challenge (Fig 1B). RT–qPCR further verified successful infection by NoV in the two control groups of mice, but not in the mice immunized with NoV∆B2 (Fig 1C).
Figure 1. Vaccination with a VSR‐deficient mutant virus induces protection against secondary infection in neonatal mice.
-
AAt 2 days post vaccination with NoVΔB2, UV‐inactivated NoVΔB2 (UV‐ ΔB2) or culture medium DMEM, BALB/c mice were challenged with WT NoV. The survival data were analyzed by the log rank test, **** indicates P < 0.0001, n = 7 per group.
-
B, CNoV levels in the vaccinated mice hindlimb muscle tissue at 2, 3, or 4 days post infection (dpi) with WT NoV were determined by Western blotting to detect the viral coat protein (CP) and B2 (B) or quantitative RT–qPCR to detect the viral genomic RNA1, n = 7 per group (C). Staining of β‐actin was used as a loading control (B). The viral RNA1 level was normalized by β‐actin mRNA levels (C). n = 7 per group. Horizontal bars represent for means. * indicates P < 0.05, ** indicates P < 0.01, and *** indicates P < 0.001 by Student’s t‐test.
-
D, ETime course analysis of the accumulation of NoVΔB2 RNA1 determined by RT–qPCR (D) and 21‐ to 23‐nt vsiRNAs determined by deep sequencing (E, two independent sets) from BALB/c suckling mice infected with NoVΔB2. Mice, n = 3, in each independent analysis. Data are shown as mean ± SD. Normalization was done by β‐actin mRNA levels for NoVΔB2 RNA1 (D) or by mature miRNAs for vsiRNAs (E). Accumulation levels of NoVΔB2 RNA1 or vsiRNAs at 3 dpi are set as 100%, respectively.
-
FProperties of viral siRNAs (per million mature miRNAs) cloned and sequenced from the time course series of NoVΔB2‐infected BALB/c suckling mice. Size distribution, 5’ terminal nucleotide, and duplexes by 22‐nt vsiRNAs are indicated. The percentage of 1 U vsiRNAs (21‐ to 23‐nt) in each library is shown in parentheses.
We showed recently that NoV∆B2‐immunized suckling mice support significantly reduced replication of Sindbis virus (SINV) when engineered to contain a NoV fragment targeted by high densities of vsiRNAs in the immunized mice (Zhang et al, 2021), suggesting a role of vsiRNAs in the induced protection. Thus, we further investigated the in vivo stability of the vsiRNAs in suckling mice following the clearance of NoV∆B2. As described previously (Li et al, 2013), NoV∆B2 was rapidly cleared from the infected mice following peak replication at 3 dpi (Fig 1D). Cloning and sequencing of the small RNAs from NoV∆B2‐infected suckling mice at 3, 7, 11, 15, 19, 23, and 27 dpi in two independent repeats demonstrated accumulation of vsiRNAs throughout the time course (Figs 1E and F, and EV1). The vsiRNAs cloned after virus clearance were similar to those cloned at earlier time points in the size distribution and the strand ratios. We found that the vsiRNAs sequenced from mice at 7 dpi and afterward were as highly enriched for 1 U vsiRNAs (Figs 1F and EV1) as found for those co‐immunoprecipitated with AGOs (Li et al, 2017; Zhang et al, 2020). Consistently, we observed de‐enrichment of the canonical vsiRNA duplexes among the total vsiRNAs cloned from the mice from later time points (Figs 1F and EV1). These findings indicate that the vsiRNAs remain stable in vivo up to two weeks following the clearance of the virus, which is likely a result of their association with Argonaute complexes.
Figure EV1. Long maintenance of vsiRNAs in neonatal mice.
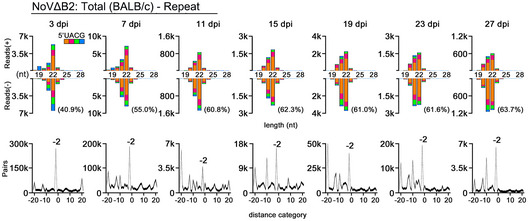
Properties of viral siRNAs (per million mature miRNAs) cloned and sequenced from the time course series of NoVΔB2‐infected BALB/c suckling mice (a repeat of Fig 1F). Size distribution, 5’ terminal nucleotide, and duplexes by 22‐nt vsiRNAs are indicated. The 1 U% of 21‐ to 23‐nt vsiRNAs in each library is shown in parentheses.
The vsiRNA profiles of serum EVs upon NoV∆B2 infection
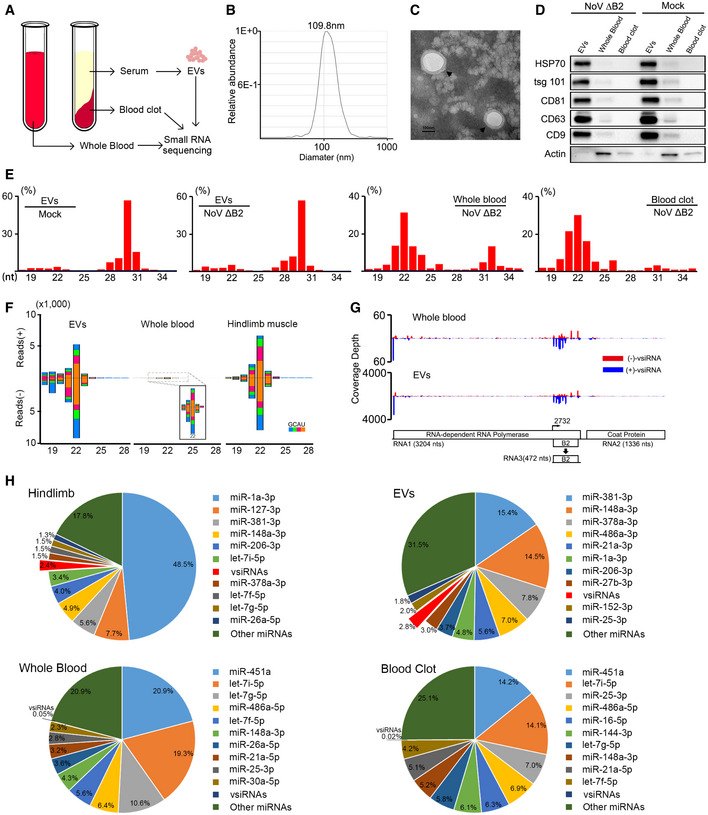
We next investigated whether murine vsiRNAs enter the blood stream for systemic circulation by sequencing the total small RNAs extracted from the whole blood upon NoV∆B2 infection. The results showed that vsiRNAs were detectable by small RNA sequencing (Table EV1). To clarify the origin of the vsiRNAs, exosome‐enriched EVs were precipitated from mouse serum through exosome isolation kit (Fig 2A). These EVs were physically homogenous, with a size distribution peaking at 110 nm in diameter as determined by nanoparticle tracking analysis (NTA) and electron microscopy (Fig 2B and C). To further confirm that the isolated particles were exosome‐enriched EVs, we examined the accumulation of five proteins known as exosomal biomarkers, CD9, CD63, CD81, HSP70, and tsg101, by Western blotting. The results showed that these proteins are readily detectable in the isolated preparations, indicating that EVs isolated from the serum were enriched for exosomes (Fig 2D).
Figure 2. Characterization of viral siRNAs in murine EVs.
-
AModel of sample preparation for small RNA deep sequencing.
-
BSize distribution of the purified EVs analyzed by NTA.
-
CTEM images of purified EVs. Scale bar: 100 nm.
-
DWestern blotting of HSP70, tsg 101, CD81, CD63, and CD9 in EVs, whole blood, and blood clot derived from suckling mice infected with DMEM or NoVΔB2. Staining of β‐actin was used as a loading control.
-
ESize distribution of total sequenced small RNAs in the libraries constructed from EVs of DMEM challenged BALB/c suckling mice, and EVs, whole blood, blood clot of BALB/c suckling mice infected with NoVΔB2 at 3 dpi.
-
F, GSize distribution of 18‐ to 28‐nt vsiRNAs (F) and genomic coverage depth of each nucleotide position by 21‐ to 23‐nt vsiRNAs (G) were analyzed from BALB/c suckling mice libraries infected by NoVΔB2 at 3 dpi.
-
HPie charts of the top ten most abundant miRNAs and vsiRNAs in hindlimb, EVs, whole blood, blood clot from BALB/c suckling mice infected by NoVΔB2 at 3 dpi. Red charts indicate 18‐ to 28‐nt vsiRNAs abundance. The percentage of miRNA reads was shown above. The abundance of total miRNAs and 18‐ to 28‐nt vsiRNAs reads was set as 1.
The small RNA sequencing was performed to characterize the RNA species in the EVs from mock and NoV∆B2 infection. The results indicated that the length of these small RNAs formed two peaks at approximately 21–23 nt and 29–31 nt (Figs 2E and EV2A). We found that miRNAs and transfer RNA (tRNA) contents (tRNA‐derived fragments, tRFs and tRNA‐halves, tiRNAs) were also the two main types of small RNA species in the EVs as previously reported (El‐Mogy et al, 2018; Srinivasan et al, 2019; Zhao et al, 2020). However, tRNA contents were highly enriched in the EVs to levels higher than those of miRNAs (Fig 2E). In contrast, miRNAs were the most abundant small RNAs in the whole blood and blood clot samples. Notably, we detected the accumulation of the vsiRNAs not only in the whole blood, but also in the purified EVs from NoV∆B2‐infected mice (Figs 2F and G, and EV2A). The EV‐associated vsiRNAs were similar in the size distribution, strand ratio, 1 U preference, and abundance to those sequenced from mouse hindlimb muscle tissues (Figs 2F and EV3). By comparison with the respective total miRNAs, the vsiRNAs were approximately 60‐fold more abundant in the EVs (2.8% of the total) than those in the whole blood (0.05% of the total) (Fig 2F and H). Moreover, the distribution patterns of vsiRNA hot spots on the two positive‐strand viral genomic RNAs were similar between the whole blood and EV libraries, indicating enrichment of the vsiRNAs in the EVs (Fig 2G).
Figure EV2. Properties of total small RNA reads and vsiRNAs.
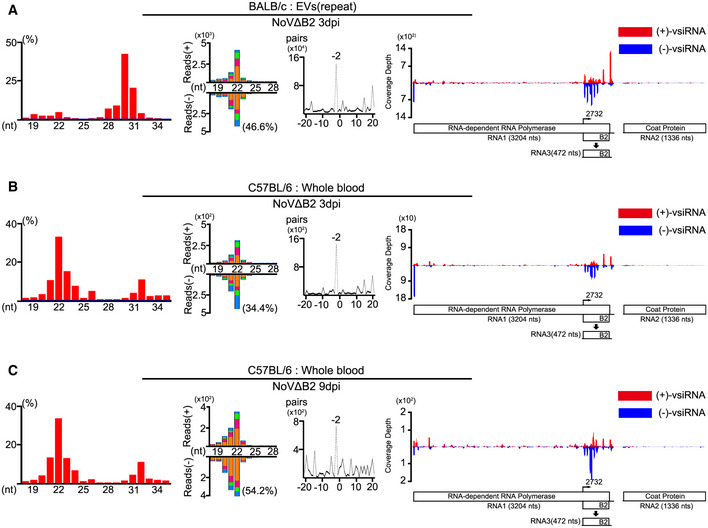
-
A–CProperties of total small RNA reads and vsiRNAs sequenced from the EVs in BALB/c suckling mice (A, a repeat of Fig 3), whole blood in C57BL/6 suckling mice infected with NoVΔB2 at 3 dpi (B) and 9 dpi (C). Length distribution of total 18‐ to 35‐nt reads in each library, size distribution of virus‐derived siRNAs, duplexes by 22‐nt vsiRNAs, and viral genomic coverage depth of each nucleotide position by 21‐ to 23‐nt vsiRNAs are presented as described in previous figures. Reads are shown as per million total mature miRNAs. 5’ terminal nucleotide and 1 U % of 21‐ to 23‐nt vsiRNAs are indicated.
Figure EV3. Duplex pattern of 22‐nt vsiRNAs in EVs, whole blood, and hindlimb muscle tissue from NoV∆B2 infected suckling mice.
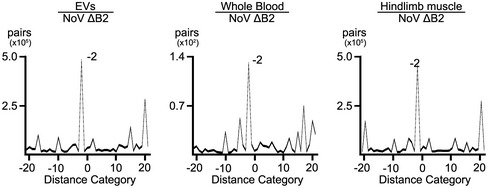
Peaks at −2 nt and 20 nt indicated a typical duplex model by cleavage of endoribonuclease Dicer.
To track the source of the EVs, we further compared the frequency of known miRNAs among the hindlimb, EVs, whole blood, and blood clot samples. In hindlimb muscle tissue, miR‐1a‐3p, which has been reported as one of the most highly expressed miRNAs in differentiated muscle tissues, accounted for 48.5% of the total miRNAs (Fig 2H). In contrast, the accumulation of miR‐1a‐3p in whole blood and blood clot was relatively low. Instead, the miR‐451a was the most abundant miRNA in these two blood samples. The most abundant EVs miRNA was miR‐381‐3p (15.4% of the total), whose expression level was low in the two blood samples, but ranked third (5.6% of the total) in the muscle tissue (Fig 2H). The accumulation patterns of specific cellular miRNAs and vsiRNAs indicate that although additional studies are necessary to conclude an origin from the virus‐infected muscle tissues, EVs‐associated vsiRNAs are unlikely from blood cells.
The induction of EV‐vsiRNAs in distinct mouse strains by NoV∆B2, SINV, and ZIKV
To determine whether the accumulation of EV‐associated vsiRNAs is mouse strain‐specific, we profiled the vsiRNAs in the blood and the EVs in C57BL/6 infant mice infected with NoV∆B2. Similar to BALB/c mice, a typical population of vsiRNAs was detected in both the whole blood and the purified EVs in C57BL/6 mice after infection with NoV∆B2 at 3 and 9 dpi (Figs 3A and B, and EV2B and C, Table EV1). Interestingly, we noted that the abundance of EV‐associated vsiRNAs from NoV∆B2‐infected C57BL/6 mice (1.1% of the total miRNAs at 3 dpi and 1.0% of the total miRNAs at 9 dpi) was similar to that from NoV∆B2‐infected BALB/c mice.
Figure 3. Characterization of EV‐vsiRNAs with distinct virus infections.
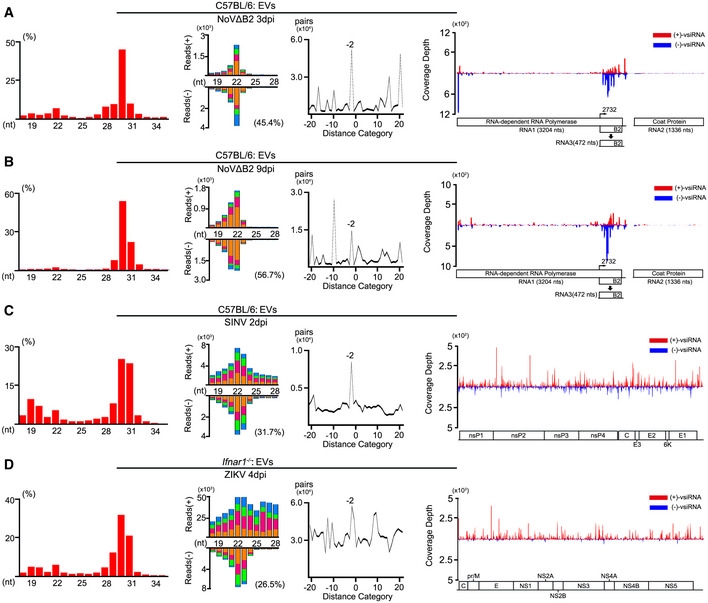
-
A–DProperties of total small RNA reads and vsiRNAs sequenced from EVs in C57BL/6 suckling mice infected with NoVΔB2 at 3 dpi (A) and 9 dpi (B), SINV at 3 dpi (C) and Ifnar −/− mice infected with ZIKV at 4 dpi (D). Length distribution of total 18‐ to 35‐nt reads in each library, size distribution of 18‐ to 28‐nt virus‐derived siRNAs, and viral genomic coverage depth of each nucleotide position by 21‐ to 23‐nt vsiRNAs are presented as described in previous figures. Reads are shown as per million total mature miRNAs. 5’ terminal nucleotide and the percentage of 1 U vsiRNAs and duplex pattern of the 22‐nt vsiRNAs are indicated.
We have shown that the vsiRNAs are induced with SINV or ZIKV infections in vivo (Zhang et al, 2020, 2021). Thus, we next purified the EVs from the mouse serum after infection with SINV or ZIKV using the exosome isolation kit. We detected the accumulation of abundant SINV vsiRNAs predominantly within 21 to 23‐nt size range in the EVs of C57BL/6 mice infected with SINV at 2 dpi (Fig 3C and Table EV1). The 22‐nt vsiRNAs of SINV found in the EVs were highly enriched for canonical siRNA duplexes with 2‐nt 3’ overhangs, and its abundance equivalent to 4.9% of the total EV‐associated miRNAs (Fig 3C and Table EV1). We also isolated the EVs from the serum of A6 (type I IFN receptor knockout C57BL/6) mice infected with ZIKV at 4 dpi. We also detected the accumulation of abundant vsiRNAs predominantly in 21 to 23‐nt size range in EVs from ZIKV‐infected A6 mice (Fig 3D and Table EV1). The vsiRNAs of ZIKV (21–23‐nt) found in the EVs were highly enriched for 22‐nt canonical siRNA, and its abundance equivalent to 14.9% of the total EVs miRNAs (Fig 3D and Table EV1). These results indicate the circulation of abundant vsiRNAs via the EVs in mice after infection with NoV∆B2, SINV, or ZIKV.
The EV‐associated vsiRNAs confer homology‐dependent antiviral activity in vitro and in vivo
We next explored whether the systemic spread of the vsiRNAs in NoV∆B2‐infected mice provides protection against secondary virus infection. To this end, we investigated whether the vsiRNAs in the circulating EVs of NoV∆B2‐immunized mice play a role in the activation of specific virus resistance at the organismal level. In the first set of in vitro experiments, we examined NoV infection of baby hamster kidney fibroblasts (BHK‐21 cells) after prior incubation with the EVs purified from NoV∆B2‐infected infant mice (Fig 4A and B). As controls, we purified EVs from infant mice either mock‐inoculated or infected with SINV that also induced systemic spread of SINV vsiRNAs via EVs (Fig 3C). We found that NoV replicated to significantly reduced levels in BHK‐21 cells treated by two different concentrations of the EVs purified from NoV∆B2‐infected infant mice compared to the EVs from mock‐inoculated mice (Fig 4B). In contrast, we detected no significant differences in the accumulation of NoV RNAs in BHK‐21 cells treated with EVs from either mock‐inoculated or SINV‐infected mice (Fig 4C). These results demonstrate that pre‐incubation with the EVs circulating in mice immunized with NoV∆B2 but not SINV inhibited NoV infection of BHK‐21 cells, suggesting a specific antiviral activity of the EV‐associated vsiRNAs.
Figure 4. Passive protection in vitro and in vivo conferred by isolated EV.
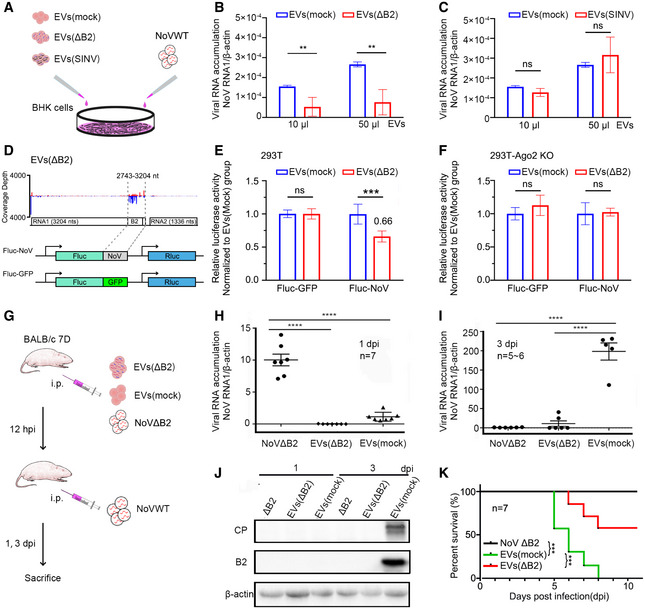
-
AScheme for protocol to determine the in vitro antiviral function of EVs from NoVΔB2‐infected BALB/c suckling mice.
-
B, C10 μl or 50 μl EVs stock from naïve [EVs (mock)] or NoVΔB2‐infected [EVs (ΔB2), B] or SINV‐infected [EVs (SINV), C] mice were added to BHK cell culture medium along with NoV‐WT, and then BHK cells were harvested at 24 hpi. Accumulation of NoV RNA1 in BHK cells were measured by RT–qPCR. Normalization was done by β‐actin mRNA levels. Data from at least three independent experiments were combined. Data are shown as mean ± SD. ** indicates P < 0.01, ns indicates no significant difference by Student’s t‐test.
-
DDiagram showing the 462‐nt sequence in length from NoV∆B2 RNA1 was inserted into the 3’ untranslated region (UTR) of firefly luciferase reporter mRNA, to be targeted by antisense vsiRNAs with EVs from NoV∆B2‐immunized BALB/c suckling mice. A translation initiation codon‐deleted GFP sequence of the same length was inserted into the same site as control.
-
E, FRelative luciferase activity of the two reporter constructs after transfection of 293T cells (E) and 293T‐Ago2 KO cells (F) and treatment by different pools of EVs, EVs (∆B2), or EVs (Mock). Error bars indicate standard deviation of three independent replicates. *** indicates P < 0.001, ns indicates no significant difference by Student’s t‐test.
-
GExperimental design for the in vivo functional analysis of EVs. In brief, mice were first immunized with live NoVΔB2, EVs (mock), or EVs (ΔB2) and challenged with WT NoV at 12 h post immunization. The NoV‐challenged mice were sacrificed at 1 and 3 days after challenge infection for determining the viral load or monitored daily for survival.
-
H, IAccumulation of NoV RNA1 was determined by RT–qPCR in NoVWT‐infected BALB/c suckling mice at 1 (H) or 3 dpi (I). n = 5–7 per group. Data are shown as mean ± SD. **** indicates P < 0.0001 by Student’s t‐test.
-
JTotal proteins were extracted from the hindlimb muscle tissue of NoVWT‐infected BALB/c suckling mice to measure the viral coat protein (CP) and B2 protein by Western blotting. Staining of β‐actin was used as a loading control.
-
KSurvival curve of WT NoV‐challenged suckling mice immunized with live NoVΔB2, EVs (ΔB2), or EVs (mock). n = 7 per group. *** indicates P < 0.001. All data were analyzed by log rank test.
We performed another set of experiments to examine the RNAi activity of EV‐associated vsiRNAs in human 293T cells using a dual luciferase reporter system. A fragment of NoV RNA1 in sense orientation known to be targeted by high densities of EV‐associated vsiRNAs from NoV∆B2‐infected infant mice was inserted into the 3′ UTR of a dual luciferase reporter plasmid and the same length of GFP mRNA sequence was used in a control construct (Fig 4D). Four hours after transfection with one of the reporter constructs, the human 293T cells were treated by the EVs purified from NoV∆B2‐infected infant mice or mock‐inoculated mice. We observed significant suppression of the luciferase reporter containing the NoV fragment treated by the EVs purified from NoV∆B2‐infected infant mice but not from mock‐inoculated mice (Fig 4E). In contrast, treatment with the two different pools of EVs did not cause significant difference in the expression of the luciferase reporter containing the GFP fragment (Fig 4E). We further generated an Argonaute‐2 knockout (Ago2 KO) line of 293T cells (Fig EV4) and performed the same luciferase reporter experiment using these cells. We found that the specific suppression of the luciferase reporter containing the NoV fragment by the EVs purified from NoV∆B2‐infected infant mice was abolished in 293T‐Ago2 KO cells (Fig 4F). These findings suggest that the specific antiviral effect of EVs from NoV∆B2‐immunized infant mice is mediated by RNAi.
Figure EV4. Functional assay of 293T Ago2‐KO cells.
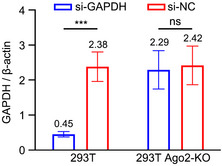
293T and 293T Ago2‐KO cells were transfected with siRNAs targeting GAPDH mRNA (si‐GAPDH) or control siRNAs (si‐NC). The accumulation levels of GAPDH mRNA were quantified by RT–qPCR. Normalization was done by β‐actin mRNA. Three biological replicates were performed, and data are shown as mean ± SD. *** indicates P < 0.001, ns indicates no significant difference by Student’s t‐test.
The next set of experiments examined the antiviral activity of the EV against NoV infection in vivo (Fig 4G). We challenged the infant mice with a lower dose of NoV (1/10 dose of vaccination experiment as described above) one day after i.p. injection with the EVs purified from NoV∆B2‐ or mock‐inoculated mice, measured NoV titers at 1 and 3 days post challenge (dpc) by quantitative RT–PCR (RT–qPCR) detection of the NoV RNA1, and monitored survival of the challenged mice. As a control, infant mice immunized with NoV∆B2 for 1 day were also challenged by the same dose of NoV. At 1 dpc with NoV, accumulation of the viral RNA1 in both groups of mice injected with EVs was significantly lower than that in NoV∆B2‐immunized mice (Fig 4H), possibly resulting from the on‐going replication of NoV∆B2 rather than NoV. By 3 dpc, however, whereas NoV replicated to high levels in mice pre‐injected with the EVs from mock‐inoculated mice, NoV titers in mice pre‐injected with the EVs from NoV∆B2‐infected mice were as low as those in NoV∆B2‐immunized mice (Fig 4I). Among the three groups of mice challenged with NoV, viral B2 and coat proteins were detectable by Western blotting only in mice pre‐injected with EVs from mock‐infected mice (Fig 4J), indicating suppression of NoV infection by prior injection with either NoV∆B2 or the EVs from NoV∆B2‐immunized mice. Notably, after NoV challenge, none of the seven infant mice immunized with NoV∆B2 for 1 day exhibited any signs of disease, and the mice pre‐injected with the EVs purified from NoV∆B2‐infected mice also survived significantly longer compared to those pre‐injected with EVs from mock‐infected mice (Fig 4K). We found that mice at 21 days post immunization with either live NoV∆B2 or EVs from NoV∆B2‐immunized mice remained highly resistant to NoV challenge compared to those immunized with DMEM, UV‐inactivated ∆B2, or EVs from mock‐immunized mice (Fig EV5). These results show that the EVs circulating in mice during the course of antiviral RNAi activation and NoV∆B2 clearance conferred passive protection against NoV infection in naïve mice, suggesting an in vivo antiviral role of the EV in NoV∆B2 immunization‐induced antiviral protection.
Figure EV5. Long‐term protection by NoVΔB2 and EVs (∆B2).
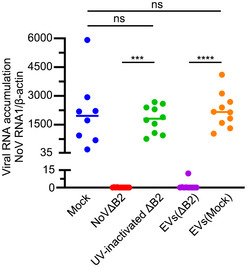
BABL/c suckling mice were immunized with DMEM, live NoV∆B2, UV‐inactivated NoV∆B2, or EVs from mock or NoV∆B2‐immunized mice. At 21 days post immunization, the mice were challenged with WT NoV by intraperitoneal injection. These challenged mice were sacrificed at 3 dpi, and NoV RNA1 levels in hindlimb muscle tissue were determined by RT–qPCR. Normalization was done by β‐actin mRNA. *** indicates P < 0.001, **** indicates P < 0.0001, ns indicates no significant difference by Student’s t‐test, n = 8–10 per group. Data are shown as mean ± SD.
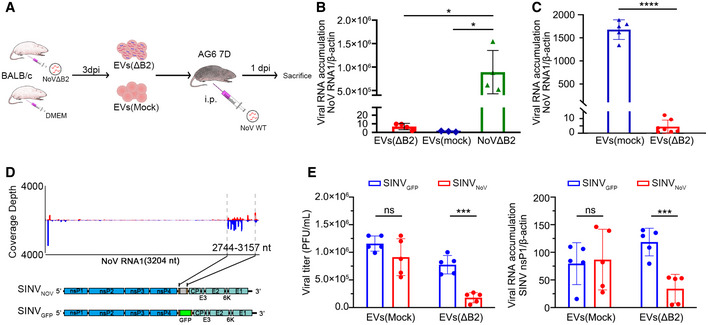
Extracellular vesicles can mediate communication between immune cells and other cell types upon virus infection (McNamara & Dittmer, 2020). EVs carry cytokines and cytokine‐related RNAs that can affect recipient cells in the antiviral response (Nolte et al, 2016; Raab‐Traub & Dittmer, 2017; Caobi et al, 2020). It is therefore possible that EVs from NoV∆B2‐infected mouse serum may carry ISGs or dsRNAs and induce an IFN response to suppress reinfection. To investigate this possibility, we performed the EV immunization experiments in infant AG6 mice, which are deficient of both the type I and II IFN receptors in C57BL/6 background (Fig 5A). In control experiments, we detected only extremely low levels of NoVΔB2 RNA1 in mice injected with the EVs from either naïve or NoVΔB2‐infected BALB/c suckling mice compared to those injected directly with NoVΔB2 (Fig 5B), suggesting presence of very few NoVΔB2 virions in the EVs purified from NoVΔB2‐infected BALB/c suckling mice. We immunized infant AG6 mice for 12 h with the EVs purified from NoV∆B2‐ or mock‐inoculated BALB/c mice before challenge with WT NoV, and compared NoV accumulation by RT–qPCR detection of the NoV RNA1 between the two groups of mice at 1 dpc (Fig 5A). We found that the accumulation of the viral RNA1 in mice immunized with the EVs from NoV∆B2‐infected mice was significantly lower than those immunized with EVs from mock‐inoculated mice (Fig 5C).
Figure 5. The EV vsiRNAs of NoV induce homology‐dependent virus resistance in AG6 mice.
-
ASchematic overview of antiviral function of EVs. Briefly, EVs from naïve or NoVΔB2‐infected BALB/c suckling mice were extracted 3 days post infection, and then injected into AG6 suckling mice. After 12 h post immunization, AG6 suckling mice were challenged by WT NoV and then sacrificed at 1 dpi.
-
BNoV replication was determined by RT–qPCR from hindlimb muscle of AG6 suckling mice challenged with NoVΔB2 as control or EVs from naïve or NoVΔB2‐infected BALB/c suckling mice at 3 dpi. Normalization was done by β‐actin mRNA levels. * indicates P < 0.05 by Student’s t‐test. n = 3–4 per group. Data are shown as mean ± SD.
-
CRelative viral RNA1 accumulation levels at 1 dpi in NoV WT‐infected AG6 mice 12 h after injection with EVs from NoVΔB2 or mock‐inoculated BALB/c mice were measured by RT–qPCR. Normalization was done by β‐actin mRNA levels. **** indicates P < 0.0001 by Student’s t‐test. n = 5–6 per group. Data are shown as mean ± SD.
-
DDiagram of recombinant SINVNoV (top) and SINVGFP (bottom). The gene of B2 protein was inserted at downstream of the duplicated subgenomic promotor sequence as shown.
-
EAG6 suckling mice were challenged with EVs from naïve or NoVΔB2‐infected BALB/c suckling mice and then inoculated by SINVNoV and SINVGFP respectively. Viral titer (PFU/ml) in the hindlimb muscle of infected mice was measured by a standard plaque assay and normalized by tissue mass, and the viral genomic RNA accumulation was determined by RT–qPCR amplification of the viral nsP1 coding region. Normalization was done by β‐actin mRNA levels. *** indicates P < 0.001, ns indicates no significant difference by Student’s t‐test, n = 5 per group. Data are shown as mean ± SD.
Furthermore, we determined whether vsiRNAs produced from NoV∆B2‐infected mice trigger the homology‐dependent viral RNA degradation guided by the vsiRNAs using Sindbis virus recombinants described recently (Zhang et al, 2021). SINVNoV contains an insert corresponding to a region of NoV genomic RNA1 targeted by high densities of vsiRNAs in NoV∆B2‐infected mice whereas SINVGFP contains the sequence of GFP (Fig 5D). SINVNoV replicated to significantly lower levels than SINVGFP in the AG6 suckling mice immunized with the EVs from NoV∆B2‐infected infant mice, but not in mice pre‐inoculated with the control EVs, suggesting the EV‐containing vsiRNAs were able to guide the homology‐dependent antiviral response in the absence of IFN signaling (Fig 5E).
Discussion
It is known that the EV composition changes drastically during virus infection (Nolte et al, 2016; Raab‐Traub & Dittmer, 2017). EVs carry cytokines and cytokine‐related RNAs that can affect viral pathogenicity, restrict viral propagation in recipient cells, and play an important role in the antiviral response. Different types of nucleic acids have been detected in EVs, such as miRNAs, lncRNA, mRNA, and various viral RNAs (Nolte et al, 2016; Raab‐Traub & Dittmer, 2017). EV‐associated host miRNAs produced by virus‐resistant cells can confer resistance against various viruses (Delorme‐Axford et al, 2013). Notably, a previous study demonstrated EV‐mediated delivery of herpesvirus‐encoded miRNAs from infected cells into non‐infected recipient cells (Kalamvoki et al, 2014). In this work, we show for the first time that abundant vsiRNAs accumulate in serum EVs upon infections by three distinct RNA viruses. We further show that both BHK cells and mice appear to be partially protected from reinfection with WT NoV when immunized by the EVs originated from the NoVΔB2‐infected mice. The induced protection comes at least in part from nucleotide sequence homology‐dependent antiviral immunity, suggesting a role for the systemic accumulation of vsiRNAs induced by NoVΔB2. However, other proteins or nucleic acids from the EVs with NoVΔB2 infection may also simultaneously vaccinate the naïve mice, allowing the mice to establish an immune protective effect.
In our current work, EVs were isolated from the serum by an exosome‐enriched protocol. Several established methods used for EV isolation, such as differential ultracentrifugation, immunomagnetic‐bead separation, density gradient centrifugation, chromatography, precipitation‐based separation, and ultrafiltration, can also be used to isolate virions (Théry et al, 2018; Caobi et al, 2020). Thus, it is often difficult to fully separate high‐purity EVs from virions since they are of similar size, density, and biochemical content (Caobi et al, 2020). However, our isolated EVs from the serum of NoVΔB2‐immunized suckling mice contained little infection‐competent virions as verified in AG6 infant mice (Fig 5B). The small non‐enveloped particles (28 nm in diameter) of NoV (Newman et al, 1978) and the severe in vivo defects of NoVΔB2 in replication (Li et al, 2013) may contribute to our successful separation of the EVs from the virions. In contrast, our use of the same isolation protocol in this study did not allow the purification of the EVs from the larger enveloped virions (65 nm in diameter) of SINV (Griffin, 2013), suggesting that optimization is necessary to obtain virion‐free EVs from SINV‐infected cells.
In addition to the ZIKV‐induced antiviral RNAi response characterized in human NPCs (Xu et al, 2019), our recent study demonstrates that abundant vsiRNAs targeting ZIKV were also produced in the central neuron system (CNS) and muscle tissues of Ifnar1 −/− mice upon virus infection (Zhang et al, 2020). A growing body of evidence indicates that EVs mediate the restriction of ZIKV infection (Caobi et al, 2020). For instance, Avraham et al showed that exosomes carrying the chromosome 19 microRNA cluster (C19MC) of miRNAs attenuate ZIKV infection of U2OS cells in an IFN‐independent manner (Bayer et al, 2018). Janis et al showed that semen prevents ZIKV attachment to the target cells by the anti‐ZIKV activity of EVs; however, which components of semen EVs confer this inhibition remains unknown (Müller et al, 2018). Here, we detected abundant 21‐ to 23‐nt vsiRNAs (equivalent to 14.9% of the total EVs miRNAs) in the serum EVs from ZIKV‐infected mice, indicating that these EVs‐associated vsiRNAs may have potential antiviral functions. Recently, Wu and colleagues provide evidence that EVs engineered to contain a host restriction factor, IFN‐induced transmembrane protein 3 (IFITM3), can effectively move across the placental barrier to suppress ZIKV infection of fetuses (Zou et al, 2021). Thus, knowledge of the phenotypes and functions of EVs generated in response to natural viral infection may be applied to improve anti‐virus treatment in the future.
Fang et al (2021) have recently achieved antiviral protection in mice using synthetic peptides designed to specifically inactivate the VSR of HEV71, demonstrating the functional importance of antiviral RNAi in mammals and a new therapeutic strategy of antiviral treatment. Maillard et al (2016) have shown that transfection with artificial long dsRNA specifically vaccinates cultured IFN‐deficient cells against infection with viruses bearing a homologous sequence by RNAi. However, whether viral dsRNA replicative intermediates made during the authentic virus infections can trigger similar immune protection in an IFN‐competent animal is not known. Results from this study show that infant mice were fully protected against lethal NoV challenge only one or two days after immunization with NoVΔB2, a live‐attenuated mutant of NoV rendered defective to suppress vsiRNA biogenesis. Importantly, the vsiRNAs accumulated in the hindlimb muscle tissues and the serum EVs of NoVΔB2‐immunized mice were similar in the size distribution, strand ratio, 1 U preference, and abundance and these vsiRNAs were stably maintained in the immunized mice up to 27 days post vaccination and at least more than one week after the mutant virus is cleared. These findings suggest a role for the EVs in the spread of the protective immunity induced by NoVΔB2 immunization to uninfected cells and a new strategy for the development of animal and human RNA virus vaccines using mutant viruses attenuated by inactivating their VSRs.
In summary, we show for the first time that mammalian vsiRNAs of three positive‐strand RNA viruses entered the blood stream for systemic circulation via EVs. The population of EV‐associated vsiRNAs is indistinguishable from those found in the limb muscle tissues of the same mice immunized with NoVΔB2. Notably, immunization of infant mice with either NoVΔB2 or vsiRNA‐containing EVs induced specific protective immunity against challenge infection, suggesting a critical role for the systemic spread of vsiRNAs. Similar to antiviral RNAi in fruit flies (Li et al, 2002; Galiana‐Arnoux et al, 2006; Van Rij et al, 2006; Wang et al, 2006; Tassetto et al, 2017), antiviral RNAi is not only active in the infected cell but also sends immune signals to prevent viral infection in distant non‐infected cells in mammals (Fig 6). In addition to the biogenesis of vsiRNAs from Dicer processing of the viral dsRNA replicative intermediates, insect vsiRNAs are further amplified using viral DNA templates reverse‐transcribed from the invading RNA viruses (Goic et al, 2013; Tassetto et al, 2017; Poirier et al, 2018). There are reports of viral DNA synthesis from non‐retroviral RNA viruses by endogenous reverse transcriptases in mammalian cells (Klenerman et al, 1997; Geuking et al, 2009; Shimizu et al, 2014). Thus, it will be interesting to investigate a possible contribution of an insect‐like vsiRNA amplification mechanism to the prolonged accumulation of vsiRNAs and the spread of the induced protection in mice immunized by VSR‐deficient mutant of NoV.
Figure 6. The EV‐vsiRNAs confer antiviral activity upon antiviral RNAi activation.
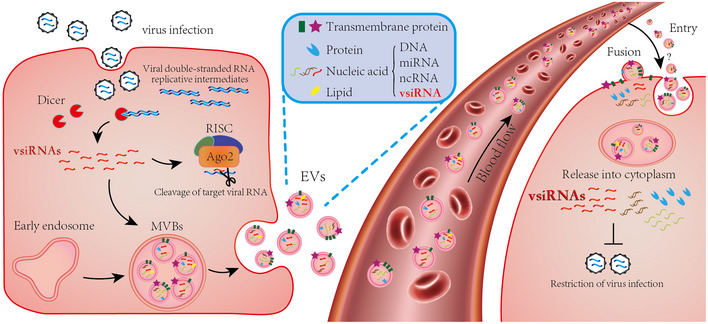
VsiRNAs are induced in virus‐infected cells by dicing the viral double‐stranded RNA replicative intermediates and loaded into RISC for subsequent cleavage of viral RNA genomes. Part of the vsiRNAs enter the EVs together with miRNAs, ncRNAs, proteins, and lipids. EVs in MVBs are secreted to the extracellular system. When attached to recipient cells, vsiRNAs in EVs are released into the cytoplasm to restrict the virus replication.
Materials and Methods
Viruses and cell culture
Wild‐type NoV and mutant NoV∆B2 strains used in this study were as described previously (Li et al, 2013). SINV was rescued from the plasmid of pSVN1, a gift from Dr. C.M. Rice. ZIKV strain (SZ01) was provided from Shanghai Public Health Clinical Center, Fudan University. ZIKV stocks were propagated in Vero cells after inoculating at a multiplicity of infection (MOI) of 0.01 and harvesting supernatants after 5 days post infection. Viruses were harvested and tittered as previously described (Zhang et al, 2020). Human embryonic kidney (293T) cell was gift from Dr. B. Cullen. Baby hamster kidney cells (BHK) and African green monkey kidney epithelial cells (Vero) were purchased from the American Type Culture Collection (ATCC). All cells were cultured in Dulbecco's modified Eagle's medium (DMEM, Gibco) containing 10% fetal bovine serum (Gibco).
Animals
BALB/c and C57BL/6 mice were purchased from Shanghai SLAC Laboratory Animal Co., Ltd. and Jackson labs (Bar Harbor, ME). Ifnar1 −/− mice (A6) were bought from Cyagen Biosciences (Suzhou, China). Ifnar1/2 −/− mice (AG6) were a gift from Dr. Y. Xu. All the animal experiments in China were carried out under the guidelines of the Institutional Animal Care and Use Committee, Fudan University of China. Animals in USA were housed in the Animal Resources Facility according to the guidelines described under the federal Animal Welfare Regulations Act and the procedures were approved by the Institutional Animal Care and Use Committee at the University of California, Riverside.
NoV infection
WT NoV or NoV∆B2 preparations shown to contain 7 × 106 copies of genomic RNA1 from the titrated set of stocks was inoculated to each of suckling mice of 6 to 8 days old after birth by intraperitoneal injection (i.p.) as described previously (Li et al, 2013). Total RNAs were extracted from the hindlimb muscle tissues of mice after infection with NoV∆B2, which were used to determine the accumulation of NoV∆B2 RNA1 by RT–qPCR and the construction of small RNA libraries as described. Whole blood samples were obtained from NoV∆B2 infected suckling mice at 3 dpi and 9 dpi, and then used to isolate EVs.
Vaccination
For the suckling mouse immunization and challenge study, 6‐day‐old BALB/c mice were inoculated by i.p. with NoV∆B2 (shown to contain 7 × 106 copies of genomic RNA1) either with or without UV‐inactivation, or with the same volume of DMEM (mock). To inactivate NoV∆B2 by UV, virus stocks were placed on ice in a XL‐1000 UV crosslinker (Spectronics corporation, Rolling Meadows, IL), and irradiated twice at 1 J/cm2 for 30 min. Virus inactivation was verified by in vivo infection. Two days after immunization, mice were challenged by i.p. inoculation with WT NoV (shown to contain 7 × 106 copies of genomic RNA1 for BALB/c suckling mice). One suckling mouse immunized with DMEM, UV‐inactivated NoV∆B2, or NoV∆B2 was euthanized 2, 3, or 4 days after WT NoV challenge to determine virus titers in the hind limb tissues by RT–qPCR and Western blotting. All suckling mice immunized with DMEM or UV‐inactivated NoV∆B2 were euthanized 4 days after WT NoV challenge since they began to show hind limb paralysis. The vaccination experiments were repeated two additional times.
Construction of SINVNoV
The pSVN1 and pTE/5’2J/GFP (SINV expression EGFP) and pTE/5’2J were gifts from Dr C.M. Rice (Rockefeller University, New York, NY, USA). In pTE/5’2J, the gene of interest can be inserted at a multiple cloning site (MCS) downstream of the duplicated subgenomic promotor sequence (Pierro et al, 2003). pTE/5’2J/NoV were constructed by ligating PCR products of NoV RNA1 flanked by XbaI sites at 5’ and ApaI sites at 3’ into the MCS of pTE/5’2J.
SINV Viruses production
SINV, SINVGFP, and SINVNoV viruses were rescued separately from the plasmids of pSVN1, pTE/5’2J/GFP, and pTE/5’2J/NoV. Briefly, plasmids containing SINV genomic RNAs were linearized with XhoI and transcribed in vitro using an SP6 mMESSAGE mMACHINE kit (Ambion). Purified SINV genomic RNAs were transfected into BHK cells by TransIT®‐mRNA Transfection Kit (Mirus Bio, WI). Viruses were harvested and tittered as previously described (Zhang et al, 2021).
SINV and ZIKV infection
50 PFU of SINV was inoculated to C57BL/6 suckling mice and 104 PFU of ZIKV was inoculated to Ifnar1 −/− suckling mice by i.p. Whole blood samples were obtained from SINV‐infected suckling mice at 2 dpi and ZIKV‐infected suckling mice at 4 dpi, and then used to isolate EVs.
EVs isolation
Whole blood samples were obtained from infected or mock‐infected suckling mice at respective dpi for each virus. The whole blood samples were placed in 37°C, thermostated for 15 min, then spun at 3,000 g, 30 min to collect serum. Centrifugation was performed twice to remove cells and cell debris, and serum were used to isolate EVs according to manufacturer’s protocol of Exosome isolation kits: ExoQuick Exosome Isolation Reagent (System Biosciences, Palo Alto, CA, USA). Serum was placed into 1.5 ml tube with ExoQuick Exosome Precipitation Solution at a ratio of 4:1. Mixture were thermostated at 4°C for an hour then centrifuged at 1,500 g for 30 min and supernatant were aspirated. EVs were resuspended in 1× PBS. The isolated EVs were prepared for Transmission Electron Microscope (TEM) (FEI, Holland) and Nanoparticle Tracking Analysis (NTA) (Paricle Metrix‐PMX, Germany). The presence of five exosome marker proteins, HSP70, tsg101, CD81, CD63, and CD9, were confirmed by Western blotting. EVs obtained by precipitation were quantified for particle number by NTA. The concentration of EVs stock was suspended in PBS to 8 × 1011 particles/ml. EVs, whole blood, and blood clot derived from different virus‐infected suckling mice were used for RNA extraction by TRIzol (Invitrogen, Carlsbad, CA). Total RNAs obtained were used for the construction of small RNA libraries as described, and proteins samples were used for Western blot analysis (Li et al, 2017).
Function assay of EV in vitro and vivo
For in vitro study, BHK cells were seeded in a 12‐well plate at a density of 5 × 105/well one day before infection. Cell culture medium was removed to opti‐MEM and then the cells was inoculated with 10 μl or 50 μl EVs stock (stock concentration, 8 × 1011 particles/ml) and then infected with NoV WT (shown to contain 7 × 105 copies of genomic RNA1). The cells were cultured at 30°C for next 24 h, and were harvested for the extraction of total RNAs using TRIzol according to the manufacturer's protocol. For in vivo study, 7‐day‐old BALB/c and AG6 suckling mice were inoculated with NoVΔB2 (shown to contain 7 × 106 copies of genomic RNA1), 2.4 × 1010 EVs (from NoVΔB2‐infected mice), or 2.4 × 1010 EVs (from mock‐infected mice). 12 h after inoculation, the mice were infected with WT NoV (shown to contain 7 × 105 copies of genomic RNA1; note, it is the 1/10 dose of vaccination experiment as described previously) by i.p. The hindlimb muscle tissues were isolated to obtain total RNAs and proteins for detecting RNA1 accumulation, Capsid, and B2 protein expression at 1 and 3 dpi. Survival was monitored daily. To exclude the effect of NoV particle existing in EVs, NoV replication was determined by RT–qPCR from hindlimb muscle of AG6 suckling mice challenged with NoVΔB2 as control or EVs from naïve or NoVΔB2‐infected BALB/c suckling mice at 3 dpi.
293T Ago2‐KO cell generation
293T cells were seeded in a 60 mm dish the day before transfection with 6 µg eIF2c (Ago2)‐targeting CRISPR plasmids (Santa Cruz sc‐400813) by lipofectamine 2000 according to the manufacturer’s instructions. Two days after transfection, culture medium was replaced with DMEM containing puromycin at a dose of 2 µg/ml. Single clones were separated at 2 weeks post treatment. Positive Ago2‐KO clones were identified and confirmed by siRNA knockdown analysis. In brief, siRNAs targeting GAPDH mRNA or control siRNAs were transfected into the Ago2‐KO cells and the loss of knockdown ability was determined by comparing the expression levels of GAPDH mRNA in 293T and Ago2‐KO cells after transfection with the specific and control siRNAs.
Luciferase reporter assay
The pmirGLO‐NoV/GFP reporters were constructed by cloning a fragment (nt 2743–3204) of NoV∆B2 genomic RNA1 in sense orientation (to be targeted by the antisense vsiRNAs) or GFP (nt 4‐465 of the coding sequence) into pmirGLO plasmid at XbaI site, by ClonExpress II One Step Cloning Kit (Vazyme Biotech Co., Ltd). Luciferase reporter assay was performed as described by the manufacturer’s instructions. Briefly, 293T cells in 12‐well plates were transfected with 1 μg of pmirGLO‐NoV/GFP plasmid DNA by lipofectamine 2000. At 4 h post‐transfection, culture medium was replaced with fresh medium containing pre‐diluted EVs at a density of 5 × 109 particles/well. Luciferase activity was analyzed using the Dual Luciferase Reporter Assay Kit (Vazyme Biotech Co., Ltd) and measured on Fluoreskan Ascent FL (Thermo Scientific) after 24 h post‐transfection. The normalized relative luciferase activity was calculated by dividing the firefly luciferase value with Renilla luciferase value.
Western and Northern blot analyses
Western blotting analysis was performed as described previously (Li et al, 2017). Antibodies to NoV B2 and coat protein were described previously (Li et al, 2013). Antibodies to exosomal marker proteins, CD9, CD63, CD81, HSP 70, and tsg 101 (sc‐13118, sc‐5275, sc‐166029, sc‐24, sc‐7964), were purchased from Santa Cruz Biotechnology, Santa Cruz, CA.
In vivo recombinant SINV reporter experiments
For AG6 suckling mice, 7‐day‐old mice were inoculated by i.p. with 2.4 × 1010 EVs (from NoVΔB2‐infected mice) or 2.4 × 1010 EVs (from mock‐infected mice). 12 h after inoculation, the mice were infected by i.p. with SINVGFP or SINVNoV viruses of 500 PFU. Each group of four suckling mice were euthanized one day after SINV infection to determine virus titers in the hind limb tissue by viral plaque assays as described (Zhang et al, 2021). In brief, hindlimb muscles were isolated from infected or mock‐infected suckling mice. After homogenization, 1 ml DMEM were added to per 50 mg tissue. The tissue homogenates were used for plague assay. BHK cells were plated at a density of 5.0 × 105 cells/ well in 3 ml DMEM, 10% FBS, on 6‐well plates and incubated at 37°C. On the next day, 1 ml of 10‐fold gradient dilutions of supernatants was added to each well. After incubation of 1 h, supernatants were discarded and the cells were overlaid with 5 ml 1×MEM containing 0.5% agarose in each well. One day after incubation, the agarose gels were removed, and cell layer was stained with Coomassie Brilliant Blue staining solution. Virus content of the supernatants was calculated as plaque forming units (PFU)/ml.
Long‐term protection
Six‐day‐old BALB/c suckling mice were vaccinated with NoV∆B2, UV‐inactivated ∆B2, EVs (∆B2), or EVs (Mock) as the same dose of NoV vaccination or in vivo function assay of EVs. At 21 days post treatment, the mice were infected with WT NoV by intraperitoneal injection (shown to contain 2.8 × 106 copies of NoV genomic RNA1, grown mice were challenged four times dose of virus as much as suckling mice based on body weight). Infected mice were sacrificed at 3 dpi, and relative accumulations of NoV RNA1 were determined by RT–qPCR.
RNA extraction, RT–qPCR, and data analysis
For virus infection and exosomal function assay in vitro and in vivo, total RNAs were extracted using Trizol reagent (Thermo) from hindlimb muscle of infected mice according to the manufacturer’s instructions as described (Zhang et al, 2020) and reverse‐transcription were done by 1st strand synthesis kit (Vazyme Biotech Co., Ltd). Virus accumulation of mice tissues was determined by RT–qPCR using ChamQ Universal SYBR qPCR Master Mix (Vazyme Biotech Co., Ltd). Primers were shown in Table EV2. GraphPad Prism was used for all statistical analyses, which was carried out by unpaired t‐test or the log rank test for the survival data. All experiments were repeated at least three times.
Construction of small RNA libraries
RNA preparations in this study were used for the construction of small RNA libraries by the method that depends on the 5’ monophosphate of small RNAs as described previously with the TruSeq Small RNA Sample Preparation Kit of Illumina (San Diego, CA).
Deep sequencing and bioinformatic analysis of small RNAs
Libraries of small RNAs were cloned from the RNA samples (mice n = 3, in each independent analysis) and sequenced by Illumina HiSeq 2000/2500. 26 libraries in total were sequenced from this work (Table EV1). Adapters were removed and reads containing any N base were removed. Trimmer reads were then mapped to virus genome references or compared to mature miRNAs of mouse (mmu21‐). Alignment was done by Bowtie 1.2.2 with perfect match. Reads mapped to positive and negative strands were used to define positive and negative vsRNAs separately. For scheme of vsRNAs size distribution and genome mapping results, reads were normalized to per million miRNA. All the references used were downloaded from web sources. Subsequent bioinformatic analysis of virus‐derived small RNAs was carried out using in‐house Perl scripts as described previously. 22‐nt vsiRNAs in each library were used to calculate overhang length of complemented duplexes by a previously described algorithm (Li et al, 2017). Detailed information of vsRNAs in each library are shown in Table EV1. The reference sequences used in this study are either identical with those described previously or as listed below:
NoV RNAs 1 and 2: AF174533.1 and AF174534.1
NoV∆B2 RNAs 1 and 2: the same as NoV except for 3 substitutions in RNA1: U2745C, U2754C, C2757G.
SINV: J02363.1
ZIKV: KX253996.1
Mature miRNAs and miRNA precursors: miRBase 21 (http://www.mirbase.org/).
Author contributions
Yang Li: Conceptualization; Resources; Data curation; Formal analysis; Supervision; Funding acquisition; Validation; Investigation; Methodology; Writing—original draft; Project administration; Writing—review & editing. Yuqiang Zhang: Data curation; Formal analysis; Validation; Methodology; Writing—original draft. Yunpeng Dai: Data curation; Software; Formal analysis; Validation; Visualization; Methodology. Jiaxin Wang: Formal analysis; Validation; Methodology. Yan Xu: Software; Formal analysis. Zhe Li: Formal analysis; Methodology. Jinfeng Lu: Resources; Software. Yongfen Xu: Resources. Jin Zhong: Resources. Shou‐Wei Ding: Conceptualization; Supervision; Funding acquisition; Writing—original draft; Writing—review & editing.
In addition to the CRediT author contributions listed above, the contributions in detail are:
YZ, YD, JW, ZL, and YL performed all infection experiments. YD and YX performed all bioinformatics analyses of small RNA libraries. YFX and JZ assisted with infection experiments in mice. JL assisted with writing the manuscript. YL designed experiments and interpreted results. YL and S‐WD conceived the study and wrote the final manuscript.
Disclosure and competing interests statement
SWD and YL declare to be named as inventors on an issued patent (US 10,034,929B2), claiming subject matter related to the use of an attenuated virus defective in RNAi suppression as a vaccine to induce antiviral protection described in this paper. The other authors declare that they have no conflict of interest.
Supporting information
Expanded View Figures PDF
Table EV1
Table EV2
Acknowledgements
The authors thank Dr. C. Rice, M. McDonald, and B. Cullen for providing materials. This work was supported by the Innovation Program of Shanghai Municipal Education Commission (2017‐01‐07‐00‐07‐E00015) and National Natural Science Foundation of China (NSFC) (32192423, 91640111, and 31770179) to YL, NIH grants AI52447, AI110579, and AI141887 (to S‐WD).
The EMBO Journal (2022) 41: e109902.
See also: KL Schierhorn et al (June 2022)
Contributor Information
Shou‐Wei Ding, Email: shou-wei.ding@ucr.edu.
Yang Li, Email: yangli15@fudan.edu.cn.
Data availability
The RNA sequencing data have been deposited to the database under the accession number PRJNA716839. All SRR data can be achieved through SRA‐toolkit. Go to: https://www.ncbi.nlm.nih.gov/Traces/study/?acc=PRJNA716839
References
- Bayer A, Lennemann NJ, Ouyang Y, Sadovsky E, Sheridan MA, Roberts RM, Coyne CB, Sadovsky Y (2018) Chromosome 19 microRNAs exert antiviral activity independent from type III interferon signaling. Placenta 61: 33–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkhout B (2018) RNAi‐mediated antiviral immunity in mammals. Curr Opin Virol 32: 9–14 [DOI] [PubMed] [Google Scholar]
- Caobi A, Nair M, Raymond AD (2020) Extracellular vesicles in the pathogenesis of viral infections in humans. Viruses 12: 1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delorme‐Axford E, Donker RB, Mouillet J‐F, Chu T, Bayer A, Ouyang Y, Wang T, Stolz DB, Sarkar SN, Morelli AE et al (2013) Human placental trophoblasts confer viral resistance to recipient cells. Proc Natl Acad Sci USA 110: 12048–12053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding SW, Han Q, Wang J, Li WX (2018) Antiviral RNA interference in mammals. Curr Opin Immunol 54: 109–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El‐Mogy M, Lam B, Haj‐Ahmad TA, McGowan S, Yu D, Nosal L, Rghei N, Roberts P, Haj‐Ahmad Y (2018) Diversity and signature of small RNA in different bodily fluids using next generation sequencing. BMC Genom 19: 1–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, Liu Z, Qiu Y, Kong J, Fu Y, Liu Y, Wang C, Quan J, Wang Q, Xu W (2021) Inhibition of viral suppressor of RNAi proteins by designer peptides protects from enteroviral infection in vivo . Immunity 54: 2231–2244 [DOI] [PubMed] [Google Scholar]
- Galiana‐Arnoux D, Dostert C, Schneemann A, Hoffmann JA, Imler JL (2006) Essential function in vivo for Dicer‐2 in host defense against RNA viruses in drosophila. Nat Immunol 7: 590–597 [DOI] [PubMed] [Google Scholar]
- Geuking MB, Weber J, Dewannieux M, Gorelik E, Heidmann T, Hengartner H, Zinkernagel RM, Hangartner L (2009) Recombination of retrotransposon and exogenous RNA virus results in nonretroviral cDNA integration. Science 323: 393–396 [DOI] [PubMed] [Google Scholar]
- Goic B, Vodovar N, Mondotte JA, Monot C, Frangeul L, Blanc H, Gausson V, Vera‐Otarola J, Cristofari G, Saleh MC (2013) RNA‐mediated interference and reverse transcription control the persistence of RNA viruses in the insect model Drosophila . Nat Immunol 14: 396–403 [DOI] [PubMed] [Google Scholar]
- Griffin D (2013) Alphaviruses. In Fields virology, Knipe DM, Howley PM (eds), 6 th edn, pp 651–686. Philadelphia, PA: Wolters Kluwer Health Adis (ESP). [Google Scholar]
- Guo Z, Li Y, Ding SW (2019) Small RNA‐based antimicrobial immunity. Nat Rev Immunol 19: 31–44 [DOI] [PubMed] [Google Scholar]
- Han Q, Chen G, Wang J, Jee D, Li W‐X, Lai EC, Ding S‐W (2020) Mechanism and function of antiviral RNA interference in mice. MBio 11: e03278‐19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu M‐M, Shu H‐B (2018) Cytoplasmic mechanisms of recognition and defense of microbial nucleic acids. Annu Rev Cell Dev Biol 34: 357–379 [DOI] [PubMed] [Google Scholar]
- Hur S (2019) Double‐stranded RNA sensors and modulators in innate immunity. Annu Rev Immunol 37: 349–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalamvoki M, Du T, Roizman B (2014) Cells infected with herpes simplex virus 1 export to uninfected cells exosomes containing STING, viral mRNAs, and microRNAs. Proc Natl Acad Sci USA 111: E4991–E4996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy EM, Whisnant AW, Kornepati AV, Marshall JB, Bogerd HP, Cullen BR (2015) Production of functional small interfering RNAs by an amino‐terminal deletion mutant of human Dicer. Proc Natl Acad Sci USA 112: E6945–E6954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenerman P, Hengartner H, Zinkernagel RM (1997) A non‐retroviral RNA virus persists in DNA form. Nature 390: 298–301 [DOI] [PubMed] [Google Scholar]
- Li H, Li WX, Ding SW (2002) Induction and suppression of RNA silencing by an animal virus. Science 296: 1319–1321 [DOI] [PubMed] [Google Scholar]
- Li Y, Lu JF, Han YH, Fan XX, Ding SW (2013) RNA interference functions as an antiviral immunity mechanism in mammals. Science 342: 231–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Basavappa M, Lu J, Dong S, Cronkite DA, Prior JT, Reinecker H‐C, Hertzog P, Han Y, Li W‐X et al (2017) Induction and suppression of antiviral RNA interference by influenza A virus in mammalian cells. Nat Microbiol 2: 16250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillard PV, Ciaudo C, Marchais A, Li Y, Jay F, Ding SW, Voinnet O (2013) Antiviral RNA interference in mammalian cells. Science 342: 235–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillard PV, Van der Veen AG, Deddouche‐Grass S, Rogers NC, Merits A, Reis e Sousa C (2016) Inactivation of the type I interferon pathway reveals long double‐stranded RNA‐mediated RNA interference in mammalian cells. EMBO J 35: 2505–2518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillard PV, van der Veen AG, Poirier EZ, Reis e Sousa C (2019) Slicing and dicing viruses: antiviral RNA interference in mammals. EMBO J 38: e100941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins SdT, Alves LR (2020) Extracellular vesicles in viral infections: two sides of the same coin? Front Cell Infect Microbiol 10: 737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara RP, Dittmer DP (2020) Extracellular vesicles in virus infection and pathogenesis. Curr Opin Virol 44: 129–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller JA, Harms M, Krüger F, Groß R, Joas S, Hayn M, Dietz AN, Lippold S, von Einem J, Schubert A et al (2018) Semen inhibits Zika virus infection of cells and tissues from the anogenital region. Nat Commun 9: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman J, Matthews T, Omilianowski D, Salerno T, Kaesberg P, Rueckert R (1978) In vitro translation of the two RNAs of Nodamura virus, a novel mammalian virus with a divided genome. J Virol 25: 78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolte E, Cremer T, Gallo RC, Margolis LB (2016) Extracellular vesicles and viruses: are they close relatives? Proc Natl Acad Sci USA 113: 9155–9161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierro D, Myles K, Foy B, Beaty B, Olson K (2003) Development of an orally infectious Sindbis virus transducing system that efficiently disseminates and expresses green fluorescent protein in Aedes aegypti . Insect Mol Biol 12: 107–116 [DOI] [PubMed] [Google Scholar]
- Poirier EZ, Goic B, Tomé‐Poderti L, Frangeul L, Boussier J, Gausson V, Blanc H, Vallet T, Loyd H, Levi LI et al (2018) Dicer‐2‐dependent generation of viral DNA from defective genomes of RNA viruses modulates antiviral immunity in insects. Cell Host Microbe 23: 353–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier EZ, Buck MD, Chakravarty P, Carvalho J, Frederico B, Cardoso A, Healy L, Ulferts R, Beale R, Reis e Sousa C (2021) An isoform of Dicer protects mammalian stem cells against multiple RNA viruses. Science 373: 231–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y, Xu Y, Zhang Y, Zhou H, Deng Y‐Q, Li X‐F, Miao M, Zhang Q, Zhong BO, Hu Y et al (2017) Human virus‐derived small RNAs can confer antiviral immunity in mammals. Immunity 46: 992–1004 [DOI] [PubMed] [Google Scholar]
- Qiu Y, Xu Y‐P, Wang M, Miao M, Zhou H, Xu J, Kong J, Zheng DA, Li R‐T, Zhang R‐R et al (2020) Flavivirus induces and antagonizes antiviral RNA interference in both mammals and mosquitoes. Sci Adv 6: eaax7989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab‐Traub N, Dittmer DP (2017) Viral effects on the content and function of extracellular vesicles. Nat Rev Microbiol 15: 559–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider WM, Chevillotte MD, Rice CM (2014) Interferon‐stimulated genes: a complex web of host defenses. Annu Rev Immunol 32: 513–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu A, Nakatani Y, Nakamura T, Jinno‐Oue A, Ishikawa O, Boeke JD, Takeuchi Y, Hoshino H (2014) Characterisation of cytoplasmic DNA complementary to non‐retroviral RNA viruses in human cells. Sci Rep 4: 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan S, Yeri A, Cheah PS, Chung A, Danielson K, De Hoff P, Filant J, Laurent CD, Laurent LD, Magee R et al (2019) Small RNA sequencing across diverse biofluids identifies optimal methods for exRNA isolation. Cell 177: 446–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassetto M, Kunitomi M, Andino R (2017) Circulating immune cells mediate a systemic RNAi‐based adaptive antiviral response in Drosophila . Cell 169: 314–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, Antoniou A, Arab T, Archer F, Atkin‐Smith GK et al (2018) Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles 7: 1535750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Veen AG, Maillard PV, Schmidt JM, Lee SA, Deddouche‐Grass S, Borg A, Kjær S, Snijders AP, Reis e Sousa C (2018) The RIG‐I‐like receptor LGP2 inhibits Dicer‐dependent processing of long double‐stranded RNA and blocks RNA interference in mammalian cells. EMBO J 37: e97479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Rij RP, Saleh M‐C, Berry B, Foo C, Houk A, Antoniewski C, Andino R (2006) The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster . Genes Dev 20: 2985–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X‐H, Aliyari R, Li W‐X, Li H‐W, Kim K, Carthew R, Atkinson P, Ding S‐W (2006) RNA interference directs innate immunity against viruses in adult Drosophila . Science 312: 452–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Kwong AC, Rice CM (2019) Antiviral resistance of stem cells. Curr Opin Immunol 56: 50–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y‐P, Qiu Y, Zhang B, Chen G, Chen QI, Wang M, Mo F, Xu J, Wu J, Zhang R‐R et al (2019) Zika virus infection induces RNAi‐mediated antiviral immunity in human neural progenitors and brain organoids. Cell Res 29: 265–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Li Z, Ye Z, Xu Y, Wang B, Wang C, Dai Y, Lu J, Lu B, Zhang W et al (2020) The activation of antiviral RNA interference not only exists in neural progenitor cells but also in somatic cells in mammals. Emerg Microbes Infect 9: 1580–1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Xu Y, Dai Y, Li Z, Wang J, Ye Z, Ren Y, Wang H, Li W‐X, Lu J et al (2021) Efficient Dicer processing of virus‐derived double‐stranded RNAs and its modulation by RIG‐I‐like receptor LGP2. PLoS Pathog 17: e1009790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao F, Cheng LI, Shao Q, Chen Z, Lv X, Li J, He LI, Sun Y, Ji Q, Lu P et al (2020) Characterization of serum small extracellular vesicles and their small RNA contents across humans, rats, and mice. Sci Rep 10: 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou X, Yuan M, Zhang T, Zheng N, Wu Z (2021) EVs containing host restriction factor IFITM3 inhibited ZIKV infection of fetuses in pregnant mice through trans‐placenta delivery. Mol Ther 29: 176–190 [DOI] [PMC free article] [PubMed] [Google Scholar]


