Abstract
Expression of chlorophyll (Chl) catabolic genes during leaf senescence is tightly controlled at the transcriptional level. Here, we identified a NAC family transcription factor, LpNAL, involved in regulating Chl catabolic genes via the yeast one-hybrid system based on truncated promoter analysis of STAYGREEN (LpSGR) in perennial ryegrass (Lolium perenne L.). LpNAL was found to be a transcriptional repressor, directly repressing LpSGR as well as the Chl b reductase gene, NONYELLOWING COLORING1. Perennial ryegrass plants over-expressing LpNAL exhibited delayed leaf senescence or stay-green phenotypes, whereas knocking down LpNAL using RNA interference accelerated leaf senescence. Comparative transcriptome analysis of leaves at 30 d after emergence in wild-type, LpNAL-overexpression, and knock-down transgenic plants revealed that LpNAL-regulated stay-green phenotypes possess altered light reactions of photosynthesis, antioxidant metabolism, ABA and ethylene synthesis and signaling, and Chl catabolism. Collectively, the transcriptional repressor LpNAL targets both Chl a and Chl b catabolic genes and acts as a brake to fine-tune the rate of Chl degradation during leaf senescence in perennial ryegrass.
A senescence-inducible NAC transcription factor gene, LpNAL, encodes a transcriptional repressor targeting chlorophyll catabolic genes and suppresses leaf senescence in perennial ryegrass.
Introduction
Leaf senescence is a plant developmental process that is accelerated by external stresses. Natural or stress-induced leaf senescence typically features loss of chlorophyll (Chl), which is regulated by Chl catabolic genes (CCGs), including STAYGREEN (SGR, also known as NYE1), NONYELLOWING COLORING 1 (NYC1), NYC1-LIKE (NOL), and PHEOPHYTIN PHEOPHORBIDE HYDROLYASE (PPH) (Zhang et al., 2015; Kim et al., 2018a; Sade et al., 2018; Xu et al., 2018, 2019; Woo et al., 2019; Guo et al., 2021; Yu et al., 2021). Tight control of these CCGs’ expression is an important regulatory mechanism of leaf senescence. For example, SGR is one of the first well-characterized CCGs, dechelating Mg atom out of Chl a, and knockdown (KD) or knockout of this gene results in cosmetic SGR in model plants and crop species (Shimoda et al., 2016; Wu et al., 2016; Xu et al., 2019). Identifying the key upstream regulatory factors of SGR will provide further insights into molecular signaling networks regulating leaf senescence and facilitate molecular breeding of stay-green plants.
To date, transcriptional factors (TFs) identified upstream of SGR belong to the TF families of No Apical Meristem (NAM), ATAF1/2, and Cup-shaped cotyledon 2 (NAC); basic region/leucine zipper motif; phytochrome interacting factor; and WRKY (Woo et al., 2019; Guo et al., 2021). Among them, the NAC family represents one of the largest plant-specific central regulators of leaf senescence (Jensen et al., 2010; Podzimska-Sroka et al., 2015; Kim et al., 2018a, 2018b). In Arabidopsis (Arabidopsis thaliana), NAP (NAC-LIKE, ACTIVATED BY AP3/PI) (Guo and Gan, 2006; Liang et al., 2014; Yang et al., 2014), ORE1 (ANAC092) (Kim et al., 2009, 2018b; Qiu et al., 2015), ANAC016 (Kim et al., 2013; Sakuraba et al., 2016), and ANAC072 (Li et al., 2016) have been identified as transcriptional activators upstream of CCGs regulating leaf senescence. In wheat (Triticum aestivum), NAM-B1, a finely mapped worthy allele, functions in accelerating leaf yellowing across all life stages (Uauy et al., 2006). Other members, such as ORS1/AtNAC059 (Balazadeh et al., 2011), ZmNAC126 (Yang et al., 2020), and ANAC046 (Oda-Yamamizo et al., 2016) were also reported to play positive roles in accelerating leaf senescence, although they do not directly target CCGs. Identification and characterization of these TFs provided insights into mechanisms of developmental and external cues-induced expression of CCGs, hence loss of Chl and leaf senescence. However, studies on upstream regulation of SGR mainly focused on transactivation of these CCGs, but less is known about transcriptional suppression of Chl catabolism that may suppress leaf senescence and develop the stay-green phenotype (Woo et al., 2019).
Several NAC negative regulators of leaf senescence were identified, such as VND-INTERACTING2 (VNI2) (Yang et al., 2011), JUNGBRUNNEN1 (JUB1) (Wu et al., 2012; Mayta et al., 2019), ONAC106 (Sakuraba et al., 2015), and NAC075 (Kan et al., 2021). JUB1 was responsive to both H2O2 and ABA, yet constitutive over-expression of JUB1 led to delayed leaf senescence in Arabidopsis and tomato (Lycopersicon esculentum) (Wu et al., 2012; Thirumalaikumar et al., 2018; Mayta et al., 2019). VNI2 functioned in repressing ABA-mediated leaf senescence by directly regulating a subset of COLD-REGULATED (COR) and RESPONSIVE TO DEHYDRATION genes (Yang et al., 2011). Arabidopsis NAC075 was characterized as a transcriptional activator that directly bound the promoter of catalase 2, suppressed the over-production of reactive oxygen species (ROS), and thereby delayed the progression of leaf senescence (Kan et al., 2021). In rice (Oryza sativa), ONAC106 was transcriptionally induced by senescence signal and repressed the expression of CCGs (Sakuraba et al., 2015). Additional TFs were yet to be identified that could orchestrate with the transcription activators upstream of CCGs to fine-tune their expression during leaf senescence.
Perennial ryegrass (Lolium perenne) is one of the most important forage and turfgrass species grown in temperate zones worldwide. One feature of perennial ryegrass is its fast leaf senescence rate that the grass remains only 3.5 green leaves in each tiller throughout its life stages (Xu et al., 2019; Yu et al., 2021). Reducing leaf senescence rate is important for the genetic improvement of forage quality as well as turf amenity in perennial ryegrass and other grass species. In our previous studies, a series of CCGs such as LpPPH, NONYELLOWING COLORING1 (LpNYC1), LpSGR, and LpNOL have been identified in regulating Chl degradation associated with cosmetic or functional SGR in its KD transgenic plants (Zhang et al., 2015; Xu et al., 2018, 2019; Yu et al., 2021). The objective of this study was to identify negative regulators in leaf senescence by screening upstream transcription factors regulating the expression of CCGs. Here, we identified a novel NAC, LpNAL, that acts as a transcriptional repressor on LpSGR and LpNYC1 and negatively regulates leaf senescence in perennial ryegrass.
Results
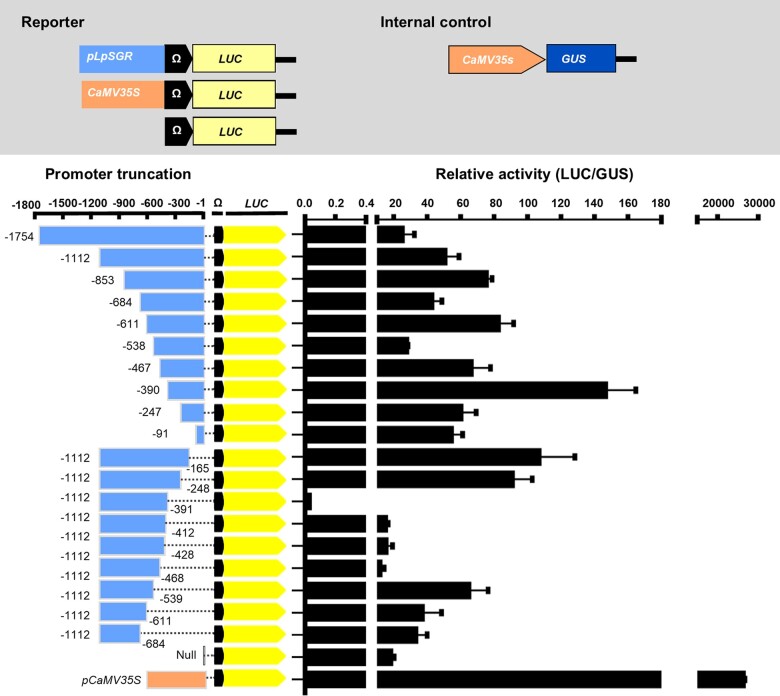
Presence of transcriptional suppression regions in LpSGR promoter
Approximately 1.7 kb long promoter of LpSGR (abbreviated as pLpSGR hereafter) was cloned using anchored-PCR, based on specific primers located in a known sequence in 5′-untranslated region (UTR) and the cloned genomic sequence of LpSGR. In silico promoter analysis of orthologous SGRs revealed the presence of common cis-elements including NAC binding site (NACBS, CGT[G/A]) (Puranik et al., 2012; Qiu et al., 2015; Supplemental Figure S1).
A bi-directional truncational analysis of pLpSGR was carried out for their transcriptional activity in Nicotiana benthamiana, and the result pinpointed three transcriptional suppression regions of pLpSGR, namely −411 to −390, −538 to −467, and −684 to −611, and three transcriptional activation regions (TARs), namely −390 to −248, −611 to −538, and −853 to −684. As for the three repression regions of −684 to −611, −538 to −467, and −411 to −390, their transcriptional activities were 2, 4, and 500 times lower than the negative control with the empty vector (Figure 1). The NACBS was found in both TAR and transcriptional suppression region of pLpSGR (Supplemental Figure S1).
Figure 1.
Promoter activity analysis of LpSGR in in N. benthamiana. The reporter and internal control were co-injected in mature N. benthamiana leaves with the same concentration at OD600 = 0.6. Relative transcriptional activities of different truncated promoters were displayed as LUC/GUS. Data are means ± standard deviation (sd) (n = 10).
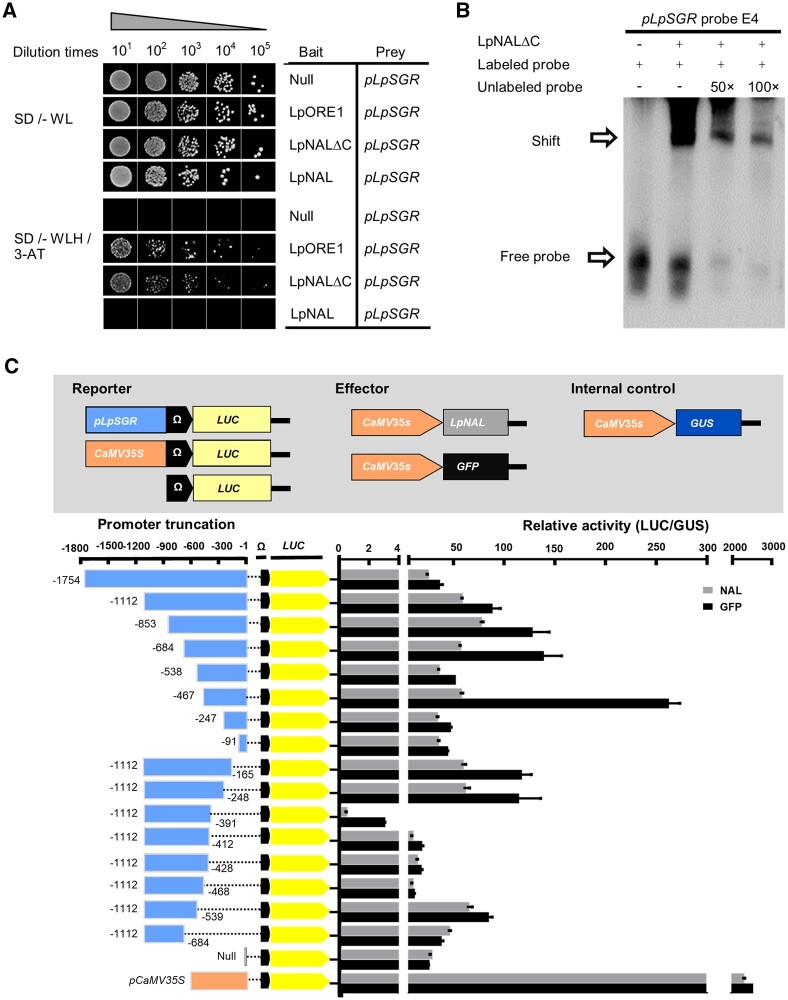
A NAC family transcription factor, LpNAL, directly repressed transcription of LpSGR
Based on the truncational promoter analysis, we hypothesized that there were transcription factors specifically repressing the expression of LpSGR. In our previous study (Yu et al., 2021), differentially expressed TFs in mature and old leaves of perennial ryegrass were identified (Supplemental Table S1). Taking these differentially expressed TFs as the potential targets, we performed yeast one-hybrid (Y1H) analysis using pLpSGR as the bait and identified a putative NAC TF, LpNAL, in the Y1H screen. Phylogenetic analysis showed that LpNAL belonged to the ATAF subfamily (Supplemental Figure S2).
LpNAL had the classical NAM and DNA-binding domain (DBD) at its N-terminus and TAR/transcriptional suppression region domain at the C-terminus (Supplemental Figure S3). The C-terminus truncated protein, LpNALΔC, trans-activated pLpSGR in Y1H, while the whole protein of LpNAL did not (Figure 2A). In vitro electrophoretic mobility shift assay (EMSA) showed that both the truncated LpNALΔC and the whole LpNAL proteins bound and shifted two probes of pLpSGR (E3 and E4 corresponding to −519 to −465 and −635 to −567 of pLpSGR, respectively) where NACBS were identified (Figure 2B;Supplemental Figure S4). The in vivo assay by co-expressing 35S::LpNAL and pLpSGR::LUC in N. benthamiana showed that the transcriptional suppression effect was most remarkable on −467 to −247 and −684 to −538 promoter regions of pLpSGR, which covered the E3 and E4 probes (Figure 2C). Collectively, these results proved that LpNAL directly bound the promoter of and repressed the transcription of LpSGR.
Figure 2.
LpNAL directly repressed transcription of LpSGR. A, Binding activity of both LpNAL and LpNALΔC on pLpSGR by Y1H assay. SD, selection medium with dropout amino acid; W, L, and H are abbreviations of Trp, Leu, and His amino acid, respectively. B, LpNALΔC directly binds the E4 fragment of pLpSGR. C, LpNAL could repress the transcription of LpSGR in vivo by transient co-expression in mature leaves of N. benthamiana. Relative transcriptional activities of different truncated promoters were displayed as LUC/GUS. Data in (C) are means ± sd (n = 8).
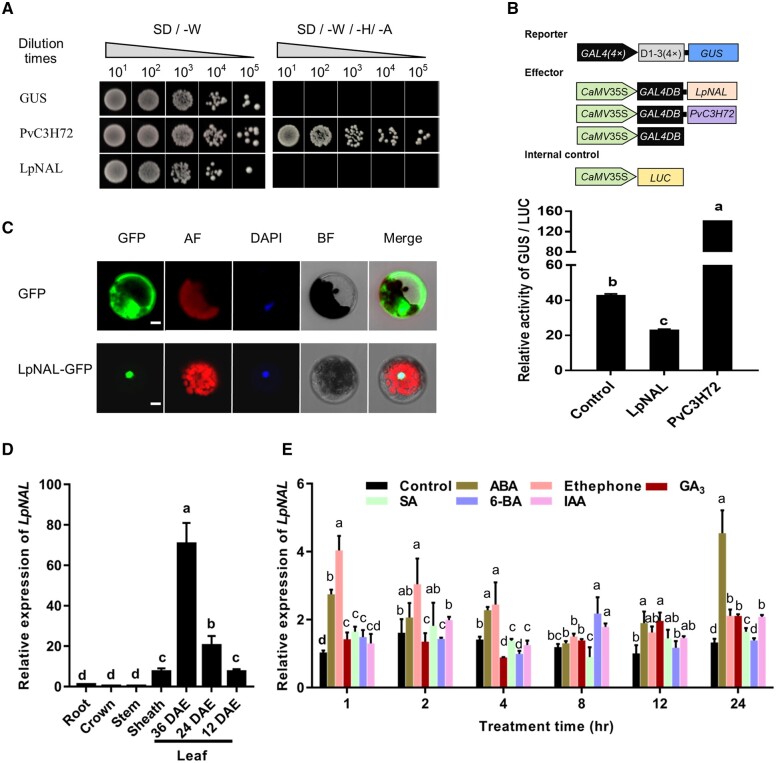
LpNAL acts as a transcriptional repressor of leaf senescence
To confirm the transcriptional repression activity of LpNAL, we firstly used the yeast-based transcriptional assay. As shown in Figure 3A, LpNAL and the negative control did not show self-activation when fused to the GAL4 DBD (GAL4DB), while a known transcription activator PvC3H72 (the positive control) did (Xie et al., 2019), indicating that LpNAL had no trans-activation activity. Furthermore, in planta transcriptional activity assay in protoplasts of perennial ryegrass showed that, compared with the empty vector control, LpNAL infusion with GAL4DB significantly repressed the expression of the β-glucuronidase (GUS) reporter gene, while the contrary was true for the control TF, PvC3H72 (Figure 3B). Green fluorescence signal of the LpNAL-green fluorescent protein (GFP) fusion protein merged with the DAPI stained nucleus signal, while fluorescence signal of the GFP-only control dispersed in protoplasts (Figure 3C). Collectively, these results demonstrated that LpNAL was a nuclear-localized TF with transcriptional repression activity.
Figure 3.
LpNAL acts as a transcriptional repressor associated with leaf senescence. A, LpNAL showed no self-transactivation activity according to the auto-transactivation assay in yeast. A, L, and H are the abbreviations of Ade, Leu, and His amino acid, respectively. B, LpNAL is a transcriptional repressor according to the in planta trans-activity assay in perennial ryegrass protoplast. C, Subcellular localization of LpNAL. The green color represents the localization of GFP or LpNAL-GFP, DAPI-stained nucleus is shown in blue color, and Chl auto-fluorescence is in red. GFP, AF, DAPI, and BF represent location of green fluorescent protein, auto-fluorescence, 4,6-diamino-2-phenyl indole, and bright field, respectively. The white bars represent 10 µm. D, LpNAL mainly expressed in senescing and senescent leaves in perennial ryegrass by RT-qPCR analysis. E, LpNAL is responsive to two senescence-associated phytohormones (ABA and Ethephone, the Ethylene releaser). Columns marked with different letters in (B, D, and E) indicate significant differences among treatments based on the LSD values (P ≤ 0.05). Data are means ± sd (n = 4).
RT-qPCR was performed to further understand the expression profile of LpNAL associated with leaf senescence. The expression of LpNAL was relatively lower in nongreen organs (e.g. roots, crown, and stems) than in leaves, especially in early (24 d after leaf emergence [DAE]) and late senescent leaves (36 DAE) (Xu et al., 2019; Figure 3D). We also characterized the transcriptional response of LpNAL to senescence-related hormones’ treatments, including ethylene releaser (ethephon), salicylic acid (SA), abscisic acid (ABA), gibberellic acid (GA3), 6-benzylaminopurine (6-BA), and indole-3-acetic acid (IAA). As shown in Figure 3E, the transcription of LpNAL was quickly upregulated by senescence-promoting hormones, ABA and ethylene, within 1 h, and was continually upregulated after 24 h in response to ABA treatment.
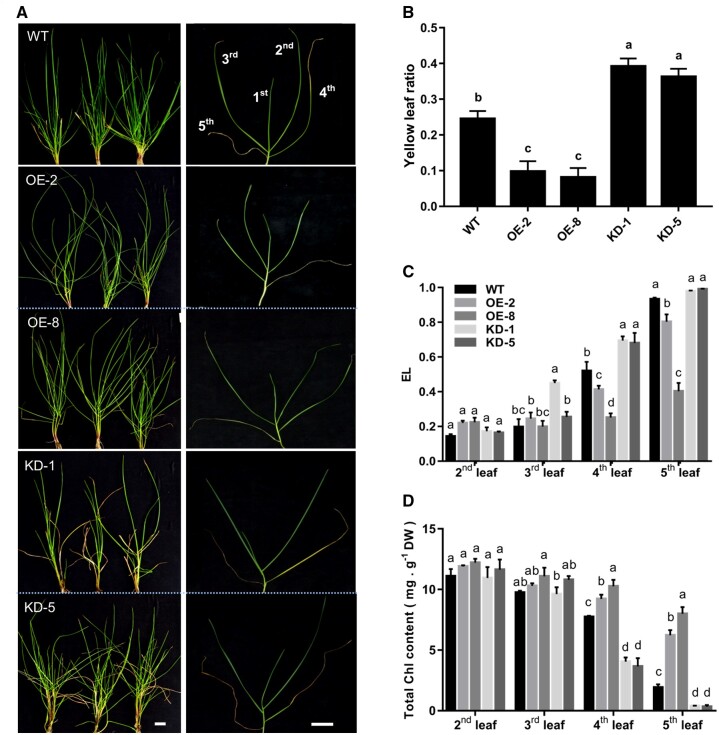
LpNAL suppressed leaf senescence in perennial ryegrass
Then we generated LpNAL overexpression (OE) and RNAi transgenic ryegrass (abbreviated as OE and KD lines, respectively) using the Agrobacterium-mediated transformation. Two OE lines (OE-2 and OE-8) and two KD lines (KD-1 and KD-5), confirmed by GUS staining, regular PCR, and RT-qPCR (Supplemental Figure S5), were used in phenotypic characterization. Overall, the leaves of OE lines showed much greener appearances than wild-type (WT) and the contrary was true for the KD lines that showed accelerated senescence (Figure 4A). OE lines had the lowest ratio of yellow leaves to the total number of leaves per plant, which were ∼30% of WT and ∼16% of the KD lines (Figure 4, B–D). In particular, in the fourth to fifth leaves from the top of each tiller, OE lines had significantly lower electrolyte leakage (EL) rates and higher Chl contents than those of WT. On the contrary, the fourth leaf of KD lines showed premature senescence with significantly higher EL values and lower Chl contents than WT (Figure 4, C and D).
Figure 4.
LpNAL suppressed developmental leaf senescence. A and B, Phenotype and green leaves ratio in each tiller in WT, LpNAL OE and KD lines. White bars equal to 3.2 cm. (C) EL rate and (D) Chl content of the second to fifth leaves from the top. Columns marked with different letters in (B–D) indicate significant differences among treatments based on the LSD values (P ≤ 0.05). Data are means ± sd (n = 4).
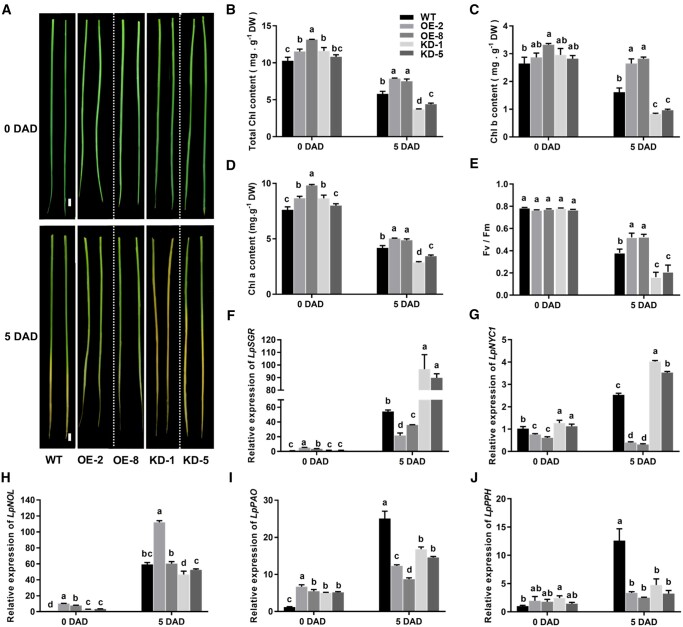
Under dark-induced leaf senescence, detached leaves of the OE lines exhibited delayed leaf senescence, while KD lines had accelerated leaf senescence compared with the WT (Figure 5A). Contents of total Chl, Chl a & b, and Fv/Fm of OE lines were significantly higher than those of WT after 5 d of dark (DAD) treatment, while the contrary was true for the KD lines (Figure 5, B–E). Moreover, we detected expression levels of five Chl catabolic genes and found that LpSGR and LpNYC1 were expressed at lower levels in OE lines but higher levels in KD lines than in WT (Figure 5, F–J). Together, these results showed that LpNAL was a suppressor of leaf senescence in perennial ryegrass.
Figure 5.
LpNAL repressed dark-induced leaf senescence. A, Dark induced leaf senescence of WT, LpNAL OE, and KD transgenic ryegrass. B–D, Total Chl content, Chl a and Chl b contents, and (E) photochemical efficiency (Fv/Fm) of WT, OE, and KD plants before and 6 DAD incubation. Fv, variable fluorescence; Fm, maximal fluorescence. F–J, Relative expression levels of CCGs in leaves including LpSGR, LpNYC1, LpNOL, LpPAO, and LpPPH using RT-qPCR. Columns marked with different letters in (B–J) indicate significant differences among treatments based on the LSD values (P ≤ 0.05). Data are means ± sd (n = 4).
LpNAL directly bound to pLpSGR and pLpNYC1
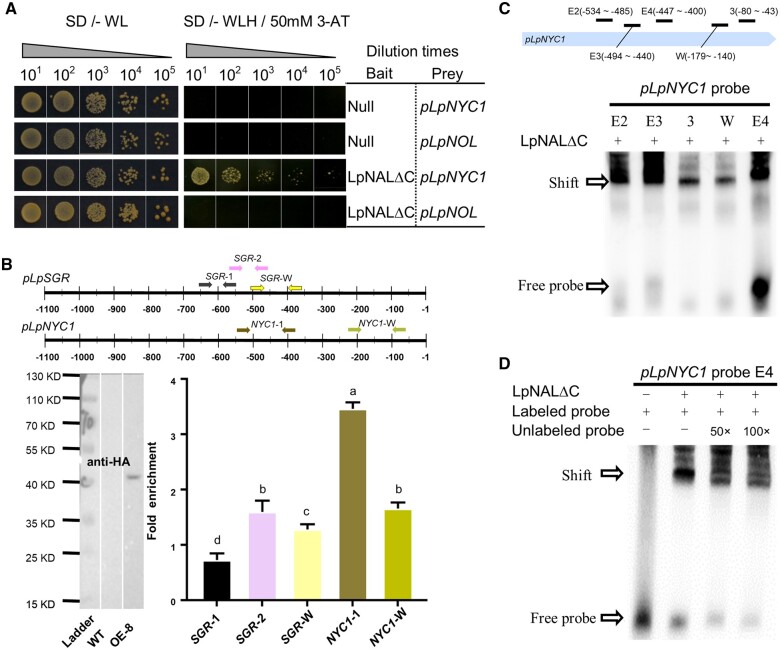
Considering LpSGR and LpNYC1 showed opposite expression patterns to LpNAL in transgenic lines, we postulated that LpNAL might directly bind to the promoter of LpNYC1 (pLpNYC1) as well. To confirm this hypothesis, we performed the Y1H assay using LpNALΔC as bait, and pLpNYC1, pLpNOL as the prey. As shown in Figure 6A, only the pair of LpNALΔC and pLpNYC1 showed a positive result, indicating that LpNALΔC could bind pLpNYC1 in the yeast system.
Figure 6.
LpNAL directly bound pLpSGR and pLpNYC1. A, LpNAL’s binding activity on pLpNOL and pLpNYC1 using Y1H assay. The bait represents the recombinant protein that is infusion with the AD domain in pGADT7. B, Western blot identification of OE-8 plant using hemagglutinin antibody (left) and ChIP-qPCR (top and right) for pLpSGR and pLpNYC1. C and D, EMSA confirmation for the binding of LpNAL to pLpNYC1. Shifted probes and free probes are noted with arrows. E2, E3, E4, 3, and W represent different candidate probes. Columns marked with different letters in (B) indicate significant differences among amplicons based on the LSD values (P ≤ 0.05). Data are means ± sd (n = 3).
Taking advantage of the OE plants where LpNAL contained a HA tag at the C terminus, we carried out chromatin immunoprecipitation quantitative PCR (ChIP-qPCR) to analyze the enriched binding DNA fragments immunoprecipitated together with LpNAL-HA proteins (Figure 6B). As shown in Figure 6B, two promoter regions of pLpSGR (SGR-1 and SGR-W) and two of pLpNYC1 (NYC1-1 and NYC1-W) were significantly enriched. In detail, at least two repressing regions, −611 to −538 and −390 to −248, and half of the region −684 to −611 were covered in SGR-1 and SGR-W amplicons. Collectively, both in vitro and in vivo experimental results demonstrated that LpNAL directly bound pLpNYC1 and pLpSGR.
Then we screened five candidate fragments of pLpNYC1 (E2, E3, 3, W, and E4) by EMSA to narrow down the possible binding sequence. We found that LpNAL bound all five probes (Figure 6C). Furthermore, we showed that the binding between LpNAL and the E4-probe was repressed by the unlabeled competitors (Figure 6D), confirming that LpNAL directly bound the E4 region of pLpNYC1. Moreover, fragments sequence alignment of pLpNYC1 and pLpSGR showed that the sequence (CGT{6,8,9}ACG or CTG{8,9}ACG or CTG{8,9}CAAG) could be the putative binding cis-element of LpNAL. Exceptions were found with two fragments of pLpSGR, suggesting that flanking sequences of the core cis-element were another factor required for the recognition (Supplemental Figure S4).
Transcriptomic comparison between WT and transgenic lines
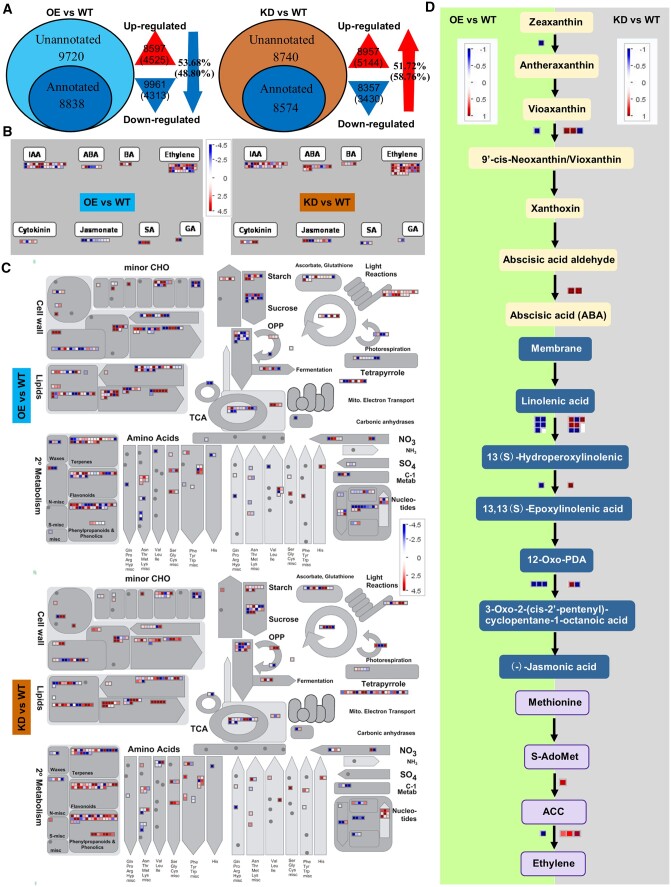
Transcriptomic comparisons among OE, KD, and WT from 30-d-old leaves were conducted by using Illumina- HiSeq 4000. The contigs were assembled into 85,630 unigenes with an N50 length of 1,730 bp and a mean unigene size of 986 bp (Supplemental Figure S6). By Venn analysis, a total of 18,558, 17,314, and 9,686 differentially expressed genes (DEGs) were found in the pair-wise comparison of transcriptomes of OE versus WT, KD versus WT, and KD versus OE, respectively, with the cut-off value set at |log2ratio| ≥ 1 and FDR < 0.001 (Supplemental Figure S7). A total of 9,961 out of 18,558 (53.68%) DEGs were downregulated in OE versus WT, while 7,078 out of 9,686 (73.07%) and 8,957 out of 17,314 (51.72%) ones were upregulated in KD versus OE and KD versus WT (Figure 7;Supplemental Figure S7).
Figure 7.
Transcriptomic analysis of OE versus WT and KD versus WT in 30 DAE leaves. A, Percentage of upregulated or downregulated DEGs in OE versus WT and KD versus WT. Numbers in parenthesis are the numbers or the percentages of the annotated DEGs. B and C, Cell-function overview and Metabolism overview of DEGs in OE versus WT and KD versus WT. G, Comparison of DEGs in ABA, JA, and ethylene biosynthesis in OE versus WT and KD versus WT. The red color means upregulation while the blue color means downregulation. BA, brassinosteroid; TCA, tricarboxylic acid; CHO, carbohydrate.
To identify DEGs responsive to the biological function of LpNAL involved in regulating leaf senescence, we conducted gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses on three sets of comparative transcriptome analysis (KD versus WT, OE versus WT, and KD versus OE). The GO analysis results showed that most upregulated DEGs due to LpNAL expression were mainly enriched in “intrinsic component of membrane,” “plastid thylakoid,” “cytoskeleton,” and “ribosomal subunit” while most downregulated DEGs were mainly included in the ribosomal subunit of OE versus WT (Supplemental Table S2). Similar GO terms were also identified in KD versus WT and KD versus OE. According to KEGG analysis, common pathways were mainly enriched in phenylpropanoid biosynthesis and DNA replication for up and downregulated DEGs in OE versus WT, respectively, compared with KD versus WT and KD versus OE (Supplemental Table S2).
Further MapMan analysis showed that 94.7% DEGs regulated by LpNAL in photosystem showed higher expression levels in OE plants including CABP, PsbP domain-OEC23 like protein, Psb28, oxygen-evolving enhancer 3 (PsbQ), PsbS, photosystem II 11 kDa, one helix protein (OHP), PsaP, cytochrome c6 (ATC6), ferredoxin-related protein, ferredoxin-NADP(+)-oxidoreductase 2 (FNR2), chlororespiratory reduction 23 (CRR23), and NAD(P)H dehydrogenase subunit H protein (NAHH). And up to 71.4% DEGs in “ascorbate and glutathione metabolism” upregulated in OE plants, such as APX, dehydroascorbate reductase 2 (DHAR2), ascorbate peroxidase 6 (APX6), vitamin C defective 5, and gamma-glutamyl transpeptidase 4 (GGT4), when compared with WT and KD plants. While “hormone regulation overview” showed that DEGs related to ABA (like ABA1, CCD1, NCED1, NCED9, and AAO2), JA (LOX2, LOX5, AOS, and OPPR1), and ethylene (ACO1 and ACS) synthesis pathways were downregulated in OE but upregulated in KD compared with WT (Figure 7B; Supplemental Figure S8). In addition, senescence-associated genes, such as SAG12, FRS5, PAP10, SGR, and NYC1, were downregulated in OE plants but upregulated in KD plants (Supplemental Figure S8). These results indicate that the regulation of LpNAL in leaf senescence mainly influences light reactions of photosynthesis, senescence-related hormones (ABA and ethylene), and the ascorbate and glutathione reduction pathways of antioxidant metabolism.
DEGs participating in cell wall breakdown, lipid metabolism, and nucleotide degradation processes showed different transcriptional levels in KD, OE, and WT plants (Figure 7C). Three DEGs encoding (1,4)-beta-mannan endohydrolases (β-MEH), beta-xylosidase 1 (BXL1), and two DEGs responsive to pectate lyase (PEL) in cell wall breakdown showed lower expression in OE plants than those in WT, among which β-MEH and two PEL genes showed much higher expression levels in KD plants than in WT (Figure 7C;Supplemental Table S3). Ten DEGs responsive to lipid biosynthesis showed lower expression levels in OE versus WT, including 3-ketoacyl-acyl-carrier protein synthase I, beta-hydroacyl-ACP dehydratase, mosaic death 1, acyl activating enzyme1 (AAE1), AAE7, 3-ketoacyl-COA synthase 2 (KCA2), KCA4, and KCA6 compared with those in KD versus WT (Figure 7C).
Additionally, transcriptional levels of 16 DEGs were confirmed by RT-qPCR to verify the transcriptome data, including three ABA-related pathway genes (ABA1, AAO2, and SnRK2.2,), two ethylene genes (ACO1 and EIN3-LIKE 1 [EIL1]), two JA-related genes (LOX5 and JAZ2), five senescence marker genes (SGR, NYC1, CABP, SAG12, and FRS5), and five candidate target genes (SAG12, C3H39, PAP10, GST39, and NAC087). The RT-qPCR result was consistent with the Illumina-seq result, supporting the reliability of the transcriptome data (Supplemental Figure S8).
Discussion
LpNAL directly represses expression of two Chl catabolic genes, LpSGR and LpNYC1
The CCGs, including SGR and NYC1, are known as positive regulators of leaf senescence as KD or knockout of those genes results in stay-green phenotypes in different plant species (Thorogood, 1996; Armstead et al., 2006; Jiang et al., 2007; Ren et al., 2007; Fang et al., 2014; Xu et al., 2019). Several activators upstream of such CCGs directly bind pSGR and pNYC1, while few repressors have been reported and verified as targeting such CCGs (Sakuraba et al., 2015; Woo et al., 2019). This study confirmed the presence of a transcriptional repressor that targets SGR and NYC1 promoters, repressing their expression during leaf senescence. We found that three suppression regions (−411 to −390, −538 to −467, and −684 to −611) of pLpSGR in ryegrass were recognized by LpNAL, all of which harbored the NACBS (Puranik et al., 2012). Furthermore, we also found that LpNAL could directly bind pLpNYC1 besides pLpSGR. Our study demonstrated that LpNAL acted as a transcriptional repressor upstream of the LpSGR and LpNYC1.
NAC family represents one of the largest plant-specific regulators of leaf senescence (Jensen et al., 2010; Podzimska-Sroka et al., 2015; Kim et al., 2018a, 2018b). Based on the knowledge of NAC TFs involved in age or abiotic stress-induced leaf senescence, most NAC TFs were found as positive regulators, except for JUB 1 (Wu et al., 2012; Mayta et al., 2019), VIN2 (Yang et al., 2011), ONAC106 (Sakuraba et al., 2015), and NAC075 (Kan et al., 2021). And none of them were proven to be transcriptional repressors directly binding promoters of these CCGs. The rice ONAC106 was another TF that could bind pOsSGR and pOsNYC1 according to the Y1H result, yet no further in vitro or in vivo experimental result was available to characterize whether it could repress these CCGs’ expression (Sakuraba et al., 2015). Comparatively, LpNAL was a novel NAC TF acting as a repressor for leaf senescence, at least partially, by directly suppressing two CCGs (LpSGR and LpNYC1).
LpNAL represses senescence-associated pathways and candidate target genes
Several NAC members directly or indirectly mediate CCGs in the regulation of leaf senescence, altering cell division, ROS accumulation, and biotic or abiotic stress responses in addition to the regulation of Chl degradation (Fujita et al., 2004; Zhao et al., 2008; Wu et al., 2012; Kim et al., 2016; Oda-Yamamizo et al., 2016; Duan et al., 2017; Woo et al., 2019; Yang et al., 2020, Kan et al., 2021). To further understand the regulatory mechanisms of LpNAL-induced repression of leaf senescence in addition to its direct control of Chl catabolism, we compared the transcriptomic profiles of senescent leaves in OE, KD, and WT plants as LpNAL was only highly expressed in senescent leaves. The comparative transcriptomic analysis revealed that LpNAL-regulated DEGs were mainly involved in light reactions of photosynthesis, antioxidant metabolism, ABA, and ethylene synthesis and signaling, although a small number (27) of LpNAL-responsive DEGs were involved in amino acid transportation and degradation, cell organization, glutathione metabolism, energy metabolism, nucleotide degradation, protein degradation, posttranslational modification, RNA transcriptional regulation, and secondary metabolism and signaling.
At the initiation of leaf senescence, photosynthetic activity declines rapidly due to the breakdown of reaction center complexes with Chl degradation (Yoshida 2003). Therefore, the regulation of genes involved in photosynthesis during the early stage of leaf senescence is critical. Notably, ∼95% of DEGs involved in light reactions of photosynthesis exhibited higher expression levels in OE plants than those in WT and KD plants, including those encoding light-harvesting proteins (e.g. Lhcb1&7), electron transport proteins (e.g. Chl a/b binding protein, FNR2, and CRR23), reduction-oxidation reaction proteins (e.g. PsbP domain-OEC23 like protein, Psb28, PsbQ, PsbS, photosystem II 11 kDa, OHP, PsaP, ATC6), and ATP production proteins (e.g. NAHH). Those genes play key roles in light harvesting, electron transport, reduction-oxidation reactions, and ATP production (Hihara and Sonoike, 2001; Taiz and Zeiger, 2002; Knoop et al., 2013). The upregulation of those genes by overexpressing LpNAL provides strong evidence supporting the positive functions of LpNAL in delaying leaf senescence and maintaining the photosynthetic activity of leaves, mainly by acting on light reactions of photosynthesis.
During the decline in photosynthetic activity during leaf senescence, ROS is produced, which has also been attributed to accelerated leaf senescence with natural leaf aging or induced by environmental stresses (Asthir 2015; Rogers and Munné-Bosch, 2016). In this study, over 70% of DEGs involved in ascorbate and glutathione metabolism were upregulated by overexpressing LpNAL, including thylakoid APX, DHAR2, APX6 (also called APX-R), and GGT4. APX and DHAR are key enzymes in the ascorbate-glutathione pathways for scavenging H2O2 (Hasanuzzaman et al., 2019), and APX6 was also reported as a SAG alleviating oxidative damage and delaying the onset of age-dependent leaf senescence (Chen et al., 2014, 2021). In plants, the GGT enzyme is involved in glutathione catabolism to produce glutamate, cysteine, and glycine which are involved in a number of important cellular functions, such as the protection of cells against oxidative stress (Schmitt et al., 2015; Bachhawat and Yadav 2018). The upregulation of these antioxidant enzymes indicated that LpNAL could regulate ROS scavenging and reduction of oxidation-caused leaf senescence during natural or stress-induced leaf senescence.
ABA, ethylene, and jasmonic acid (JA) induce leaf senescence and their hormonal levels increase during natural or stress-induced leaf senescence (Kim et al., 2016; Woo et al., 2019). Many TF families (e.g. WRKY, TCP, MYC, CCCH, and NAC) are involved in these hormonal signaling pathways to regulate leaf senescence (Ullah et al., 2019; Woo et al., 2019). KD or knockout TF genes, such as ABI5, EIN3, EIL1, OsDOS, MYC2, or ORE1 that are downstream of ABA, ethylene, or JA signaling pathways, produced stay-green phenotypes in model plants (Breeze et al., 2011; Woo et al., 2019). In this study, all the DEGs encoding key enzymes in ABA, ethylene, and JA biosynthesis were repressed at least two times more in OE plants than in WT, while most of the DEGs involved in catalyzing these three hormones showed >1.5 times higher expression in KD plants than in WT. Consistently, several TFs downstream of these hormones, including the candidate targets (e.g. JAZ2, NAC087, and C3H39), were all downregulated in OE plants but upregulated in KD plants compared with WT. These results demonstrated that suppression of leaf senescence by LpNAL could be related to its repression of ABA, ethylene, or JA biosynthesis and signaling pathways in perennial ryegrass.
Several DEGs that participate in cell wall breakdown processes, such as β-MEH and PEL, were downregulated by overexpressing LpNAL but upregulated by suppressing LpNAL. β-MEHs are hydrolytic enzymes that cleave the mannan backbone from cell wall polysaccharides and are activated during seed germination and plant senescence (Schröder et al., 2009). Recent discoveries showed that β-MEH was associated with flowering and fruit softening in barley and tomato (Hrmova et al., 2006; Vicente et al., 2007). PEL is an enzyme catalyzing pectate cleavage and is involved in the maceration and soft rotting of plant tissue (Sun et al., 2018). Dwarf and early-senescence leaf 1, a PEL-like homolog, maintained normal cell division and the induction of leaf senescence (Leng et al., 2017). These results suggest that the effect of LpNAL on leaf senescence suppression might also involve the maintenance of cell wall integrity.
Conclusion
In conclusion, LpNAL acts as a repressor for LpSGR and LpNYC1 by directly binding to their promoters to suppress leaf senescence. Over-expressing LpNAL in perennial ryegrass led to the stay-green phenotype with inhibited dark- and age-induced leaf senescence. LpNAL-regulated stay-green phenotypes might also be altered in their regulation of metabolic processes other than that of Chl degradation, including light reactions of photosynthesis, antioxidant metabolism, ABA, and ethylene synthesis and signaling.
The progression of leaf senescence is a well-programmed process where both “accelerators” and “brakes” are necessary to drive it to the destination. In contrast to the senescence-promoting “accelerators,” and “brakes” (e.g. CLE14) were induced by senescence signals but functioned to repress age-dependent and/or stress-induced leaf senescence (Zhang et al., 2022). Here, we propose that LpNAL, a senescence-responsive TF, is another “brake” during leaf senescence that works with other TFs to coordinate the degradation of Chl and progression of leaf senescence. This finding provided additional knowledge on the leaf senescence mechanism for the genetic improvement of perennial ryegrass and other forage- and turfgrass species.
Materials and methods
Gene cloning and plasmid construction
About 1.5 kb LpSGR promoter sequence was cloned using a Tail-PCR method (GenBank assembly accession: GCA_001735685.1). pLpNOL and pLpNYC1 were cloned by referring to the publicly available genome dataset of perennial ryegrass (GenBank assembly accession: GCA_001735685.1). The coding sequence (CDS) of LpNAL was amplified from senescent leaves cDNA of perennial ryegrass. The primers used for cloning are listed in Supplemental Table S4.
CDS of LpNAL was firstly cloned into Gateway entry vector pENTR/D (Invitrogen, Carlsbad, CA, USA) at the EcoRI and HindIII sitesand then subcloned into pEarleyGate103 (Earley et al., 2006) for transient OE in N. benthamiana in studying promoter repression by LpNAL, into pVT1629 (Xu et al., 2011) for the generation of LpNAL OE transgenic lines, and into P2GWF7.0 (Yu et al., 2017) for the study of subcellular functional localization, respectively. A LpNAL-specific fragment (Supplemental Figure S2) was chosen for the RNAi vector construction using the pEnD-Kannibal vector and then recombined to pVT1629 (Xu et al., 2011) to produce RNA interference transgenic plants. LpNAL and its C-terminal truncated gene (LpNALΔC) were subcloned into the Y1H prey vector pGADT7 (Clonetech, Palo Alto, CA, USA) at NdeI and EcoRI sites, into the prokaryotic protein expression vector pCold-TF vector (TaKaRa Bio, Inc., Shiga, Japan), and into the pZB369 vector (Xie et al., 2019) for the transcriptional activity assay in plant cells. pLpNYC1, pLpPPH, and pLpSGR were subcloned into Y1H bait vector pHIS2.1 (Clonetech, Mountain View, CA, USA) at SpeI and EcoRI sites, respectively. pLpSGR was also subcloned into pCambia1381Z at BamHI and HindIII sites to drive the Luciferase (LUC) reporter gene.
Cis-elements in pLpSGR and the repressing activity of LpNAL in N. benthamiana
By PLACE (http://www.dna.affrc.go.jp/PLACE/), a promoter cis-element analyzing online software, a series of senescence-associated cis-elements were targeted and truncated according to the analysis. Bidirectional truncations of pLpSGR were cloned and recombined into the pCambia1381Z reporter vector in front of the 35S Ω sequence to drive the LUC reporter gene. The pEarleyGate103-LpNAL was used as the effector vector with pEarleyGate103-eGFP as the negative control. The vector pCambia1302-GUS-polyA was used as the internal control in which the GUS gene (UidA) was driven under the 35S promoter. The resulted vectors were transformed into Agrobacterium tumefaciens strain “AGL1.” Before inoculation into leaves of N. benthamiana, Agrobacterium carrying corresponding vectors were normalized to the same concentration of OD600 at 0.8 with infection medium. Three kinds of Agrobacterium harboring the effector, the reporter, and the internal control vectors were mixed with equal volume, centrifuged, and resuspended with the final OD600 adjusted to 1.8. The fourth leaf from the top of 2-month-old N. benthamiana was injected with the Agrobacterium solution using a one ml Micro syringe. One day later, the treated leaves were cut and separated in liquid nitrogen and stored at −80°C for LUC and GUS protein activity analysis. The protocol for protein extraction and enzyme activity analysis was according to Yoo et al. (2007). The ratio of LUC to GUS in the sample injected with the effector gene indicated the relative activation/repression effect of the effector (e.g. LpNAL or GFP as the control).
Phylogenetic analysis
The NAC family proteins in Arabidopsis and a series of well-characterized NAC proteins in rice were downloaded from the Plant Transcription Factor Database (http://planttfdb.gao-lab.org). LpNAL and NAC family proteins were firstly aligned by the Align module “Align by ClustalW” with a nonconserved C-terminal domain excluded for further analysis. Then, the distance MEGA file was further analyzed for constructing a neighbor-joining phylogenetic tree with 1,000 bootstrap replicates. Branches corresponding to partitions reproduced in ˂50% bootstrap replicates are collapsed. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method and are in the units of the number of amino acid substitutions per site. The analysis involved 104 amino acid sequences. All positions containing gaps and missing data were eliminated. There was a total of 32 positions in the final dataset.
Plant growth conditions
In this study, both N. benthamiana and perennial ryegrass (cv. “Buena vista”) were planted in mixed soil (peat: vermiculite = 3:1; v/v) in each plastic pot (20 cm in diameter and 25 cm in height) and maintained in a well-controlled growth chamber (25°C/20°C (day/night), 70% relative humidity, and 12-h photoperiod with photosynthetic active radiation of 700 µmol photons m−2 s−1).
Genetic transformation of perennial ryegrass
LpNAL OE and KD transgenic perennial ryegrass were generated by Agrobacterium-mediated genetic transformation following the protocol described by Zhang et al. (2013). The Hygromycin-resistant positive plant lines were further confirmed by GUS staining and regular PCR for the presence of the HPTII gene.
Transactivation assay
LpNAL and the positive control, PvC3H72, were subcloned and fused with the GAL4 DBD. The recombined vectors were then transformed into the yeast strain “Y2HGold” (Clonetech) and screened on SD/-Trp medium. Then, positive clones were diluted and grown on plates containing SD/-Trp-Leu-His and SD/-Trp-Leu-His-Ade for auto-transactivation assay.
For the transcriptional activity assay of LpNAL and LpNALΔC in plant cells, the pZB369-LpNAL and -LpNALΔC vectors were used as the effectors, and the pZB369-PvC3H72 vector was used as the positive control following the same procedure reported before (Xie et al., 2019).
Subcellular localization of LpNAL in protoplasts of perennial ryegrass
LpNAL was cloned into the p2GWF version 7.0 vector to generate the LpNAL-GFP fusion gene. The resultant vector was transformed into ryegrass protoplasts for subcellular localization observation following a protocol reported previously (Yu et al., 2017). GFP signal (excitation at 488 nm, scanning at 505–530 nm) and DAPI staining (excitation at 405 nm, and scanning at 430 nm) were visualized under a confocal laser scanning microscope (Zeiss LSM780 Exciter).
Y1H assay
For the Y1H screen, firstly, we identified the putative senescence-responsive TFs by analyzing transcriptomes of early senescent and mature leaves (Yu et al., 2021), cloned most of these senescence-responsive TFs (or partial sequences covering the putative DNA-binding motif), and recombined them into the pGADT7 vector to construct a “mini-library.” Then, each of these mini-library vectors and the prey vector (pHIS2.1-pLpSGR) were co-transformed into the yeast strain Y187. The transformed yeast cells were recovered in YPDA plus medium (Clonetech) at 30°C for 1 h and screened on SD/-Leu-Trp. Positive clones were further tested according to the method depicted before (Xu et al., 2018). Full length CDS of the identified “prey” were further cloned according to our newly constructed transcriptome (Yu et al., 2021). For verification, the recombined pGADT7 vectors (pGADT7-LpNAL, -LpNALΔC, and -LpORE1) and pHIS2.1 vectors (pHIS2.1-pLpNYC1, -pLpNOL, and -pLpSGR) were co-transformed into the yeast strain Y187 and grown on the selection plates with SD/-Trp-Leu-His + 50 mM 3-AT.
ChIP-PCR
The fourth detached leaves from the top of 2-month-old LpNAL OE transgenic plants in which a C′-HA tag was infusion with LpNAL were sampled for the analysis. The ChIP DNA fractions were normalized to the input DNA (ΔCt) to avoid preparation errors of the chromatin sample. Input samples and immunoprecipitated samples were analyzed by qPCR. Input normalized LpNAL ChIP fractions were then adjusted for the normalized negative control (IgG) giving the ΔΔCt value. qPCR reactions were performed three times for each sample, and the expression levels were normalized to the input sample for the enrichment detection. The fold enrichment was calculated against the IgG reference by raising 2 to the ΔΔCt power (Walley et al., 2008). Two or three primers across the putative binding sites or nonbinding sites in the promoters of three CCGs were used for quantification. All primers’ information is listed in Supplemental Table S4.
EMSA
pCold TF-LpNAL and -LpNALΔC vectors were constructed to generate LpNAL-HIS and LpNALΔC-HIS fusion genes. These recombinant proteins were expressed in Escherichia coli strain “Rosetta BL21 (DE3)” under induced by 0.5 mM isopropyl β-D-thiogalactoside at 16°C for 10 h and then purified with Ni-IDA resin (Transgene, China) according to the manufacturer’s instructions. The EMSA was carried out using a Light Shift Chemiluminescent EMSA Kit (Thermo Scientific, Waltham, MA, USA), and bind shift results were detected using a chemiluminescence imaging system (Vilber Lourmat, Marne la Vallée, France). Unlabeled competitor probes were added in a 50- or 100-fold molar excess.
Dark-induced leaf senescence
The first fully expanded leaf was detached and sandwiched in two-layer filter paper prewetted in distilled water. Then, the leaves were incubated in dark and kept in humidity and sampled before and after dark incubation.
Transcriptional response of LpNAL to senescence-related hormones and developmental senescence signal
Two-week-old seedlings cultured in liquid half strength-Hogland nutrient solution were treated with water, 50 µM ABA, 200 µM ethephone (ethylene releaser), 100 µM GA3, 100 µM SA, 25 µM 6-BA, and 20 µM IAA. The first fully expanded leaves were taken at six-time points (1, 2, 4, 8, 12, and 24 h) with three replicates for each treatment. The samples were immediately frozen in liquid nitrogen and stored in the −80°C refrigerator before the RNA extraction.
The emerged leaves on top were labeled as 0 DAE and three stages of leaves (12, 24, and 36 DAE) representing mature, early senescent, and late-senescent leaves (Xu et al., 2019). Leaves, roots, crowns, stems, and sheaths were sampled for the RT-qPCR analysis with three biological replicates.
RT-qPCR analysis
Total RNA was extracted using the Plant RNA Kit (Omega Biotech, Norcross, GA, USA) according to the manufacturer’s protocol. The first-strand cDNA was synthesized with 1 μg RNA using the PrimeScript RT reagent Kit with gDNA Eraser (Perfect Real Time) (Takara, Otsu, Japan). For RT-qPCR, the reaction was performed with the SYBR Green PCR Master Mix (Applied Biosystems, Waltham, MA, USA) using a Roche Light Cycler 480 II Real-Time PCR System. Primers used for RT-qPCR are listed in Supplemental Table S4. eIHF4A was used as the reference gene (Huang et al., 2014). Relative expression levels were determined by the 2−ΔΔCT method. The RT-qPCR reactions were performed with three biological replicates and two technical repeats for each biological replicate. Detailed information of primers used for RT-qPCR is listed in Supplemental Table S4.
Physiological analysis of leaf senescence in perennial ryegrass
Chl content measurement was performed by DMSO extraction from about 0.1 g fresh leaf samples under 4 d darkness and measuring at the absorbance of 663 and 645 nm, then the blades were dried at 80°C for 3 d for dry weight (DW) measurement. Chl content calculation followed the description in Arnon (1949). For leaf photochemical efficiency (Fv/Fm), the leaves after 30 min dark acclimation were measured using a fluorescence induction monitor (Bioscientific Ltd., Herts, UK). EL was measured by following the procedure described in Martin et al. (1987). For the leaf yellowing rate, leaves containing one-fourth or more yellowing part of the whole leaf were labeled as the yellow leaf. Then, numbers of yellow leaves and green leaves in a single tiller were recorded. Ten tillers were counted in each pot for the calculation of the yellow leaf ratio.
Transcriptomic analysis
The RNA-Seq was carried out by Gene Denovo Co. (Guangzhou, China) using the paired-end technology of Illumina HiSeq 2000. Leaves of WT and each transgenic line were used for RNA-seq with three biological replicates. Two independent OE (OE2 and OE8) and KD (KD1 and KD8) lines were used in this study. Detailed information about library construction, sequencing, and transcriptomic analysis (e.g. GO and KEGG analyses) were the same as described before (Yu et al., 2021). The original transcriptome data were deposited at NCBI (BioProject: PRJNA780368; SRA: SUB10670514). Among the set of genes expressed above the minimum expression cutoff, we selected those with at least two-fold expression change and P-value < 0.001 for further analysis. The DEGs among OE, WT, and KD were classified functionally using the biological process category referenced to Brachypodium distachyon (http://plants.ensembl.org/index.html). Differentially expressed unigenes were analyzed with the Veen module using the TBtools platform (Chen et al., 2018). Mapman overview was generated with MAPMAN3.5.1R2 software (https://mapman.gabipd.org/).
Statistical analysis
Data in this study were statistically analyzed using the JMP software (version 10, SAS Institute Inc., Cary, NC, USA) for ANOVA at a significance level of 0.05. The data are expressed as means ± standard deviation (sd).
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL under accession numbers KX791203, KX686491, KX686493, KX686495, and KT345726.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Cis-element analysis of SGR promoters in perennial ryegrass and other plant species.
Supplemental Figure S2. Phylogenetic tree of LpNAL with NAC family proteins in A. thaliana and rice (O. sativa).
Supplemental Figure S3. Domain analysis of LpNAL according to Ooka et al. (2003).
Supplemental Figure S4. Both LpNAL and LpNALΔC specifically bound the probes of E3 and E4 of pLpSGR in vitro in the EMSA. E3, E4, 4, 2/W, and W represent different candidate probes, respectively.
Supplemental Figure S5. Verification of OE and KD transgenic perennial ryegrasses.
Supplemental Figure S6. Distribution of unigene size, the number of reads, and the assembled information of unigenes.
Supplemental Figure S7. Venn analysis of OE versus WT, KD versus WT, and KD versus OE.
Supplemental Figure S8. Association analysis between RPKM values and relative expression levels of the RT-qPCR.
Supplemental Table S1. Assembled information of DEGs in the transcriptomes of 30 DAE leaves in WT, OE, and KD plants.
Supplemental Table S2. Differentially expressed transcription factors in mature (WTM) and old (WTO) leaves.
Supplemental Table S3. The annotations of DEGs encoding β-MEH, BXL1, and PEL.
Supplemental Table S4. Primers and probes used in the study.
Supplementary Material
Acknowledgments
We thank Dr. Wang Hui for technical guidance.
Funding
This study was supported by National Natural Science Foundation of China (NSFC) (31772659 and 31971757) and by Natural Science Foundation of Jiangsu Province (BK20190536). Funding to conduct part of the collaborative work at Rutgers University was supported through postdoctoral fellowship for G.Y. and student stipend for Z.X. from Chinese Scholarship Council.
Conflict of interest statement. The authors declared that they have no conflicts of interest to this work.
B.X. developed the research and experimental designs. Z.X., G.Y., S.L., and H.L. performed the experiments. G.Y., B.X., and B.H. analyzed all data and wrote the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Bin Xu (binxu@njau.edu.cn).
References
- Armstead I, Donnison I, Aubry S, Harper J, Hörtensteiner S, James C, Mani J, Moffet M, Ougham H, Roberts L, et al. (2006) From crop to model to crop: identifying the genetic basis of the staygreen mutation in the Lolium/Festuca forage and amenity grasses. New Phytol 172: 592–597 [DOI] [PubMed] [Google Scholar]
- Arnon DI (1949) Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol 24: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asthir B (2015) Protective mechanisms of heat tolerance in crop plants. J Plant Interact 10: 202–210 [Google Scholar]
- Bachhawat AK, Yadav S (2018) The glutathione cycle: Glutathione metabolism beyond the γ‐glutamyl cycle. IUBMB Life 7: 585–592 [DOI] [PubMed] [Google Scholar]
- Balazadeh S, Kwasniewski M, Caldana C, Mehrnia M, Zanor MI, Xue GP, Mueller-Roeber B (2011) ORS1, an H2O2-responsive NAC transcription factor, controls senescence in Arabidopsis thaliana. Mol Plant 2: 346–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breeze E, Harrison E, McHattie S, Hughes L, Hickman R, Hill C, Kiddle S, Kim Y, Penfold CA, Jenkins D, et al. (2011) High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 23: 873–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R (2020) TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol Plant 13: 1194–1202 [DOI] [PubMed] [Google Scholar]
- Chen C, Galon Y, Rahmati Ishka M, Malihi S, Shimanovsky V, Twito S, Rath A, Vatamaniuk OK, Miller G (2021) ASCORBATE PEROXIDASE6 delays the onset of age-dependent leaf senescence. Plant Physiol 185: 441–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Letnik I, Hacham Y, Dobrev P, Ben-Daniel BH, Vanková R, Amir R, Miller G (2014) ASCORBATE PEROXIDASE6 protects Arabidopsis desiccating and germinating seeds from stress and mediates cross talk between reactive oxygen species, abscisic acid, and auxin. Plant Physiol 166: 370–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan M, Zhang R, Zhu F, Zhang Z, Gou L, Wen J, Dong J, Wang T (2017) A lipid-anchored NAC transcription factor is translocated into the nucleus and activates glyoxalase I expression during drought stress. Plant Cell 29: 1748–1772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Fang C, Li C, Li W, Wang Z, Zhou Z, Shen Y, Wu M, Wu Y, Li G, Kong LA, et al. (2014) Concerted evolution of D1 and D2 to regulate chlorophyll degradation in soybean. Plant J 77: 700–712 [DOI] [PubMed] [Google Scholar]
- Fujita M, Fujita Y, Maruyama K, Seki M, Hiratsu K, Ohme-Takagi M, Phan Tran LS, Yamaguchi-Shinozaki K, Shinozaki K (2004) A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J 39: 863–876 [DOI] [PubMed] [Google Scholar]
- Guo Y, Gan S (2006) AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J 46: 601–612 [DOI] [PubMed] [Google Scholar]
- Guo Y, Ren G, Zhang K, Li Z, Miao Y, Guo H (2021) Leaf senescence: Progression, regulation, and application. Mol Hortic 1: 1–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasanuzzaman M, Bhuyan MHM, Anee TI, Parvin K, Nahar K, Mahmud JA, Fujita M (2019) Regulation of ascorbate–glutathione pathway in mitigating oxidative damage in plants under abiotic stress. Antioxidants 8: 384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hihara Y, Sonoike K (2001) Regulation, inhibition and protection of photosystem I. In EM Aro, B Andersson, eds, Regulation of Photosynthesis, Springer, Dordrecht, Netherlands, pp 507–531 [Google Scholar]
- Huang L, Yan H, Jiang X, Yin G, Zhang X, Qi X, Zhang Y, Yan Y, Ma X, Peng Y (2014) Identification of candidate reference genes in perennial ryegrass for quantitative RT-PCR under various abiotic stress conditions. PLoS ONE 9: e93724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrmova M, Burton RA, Biely P, Lahnstein J, Fincher GB (2006) Hydrolysis of (1, 4)-β-d-mannans in barley (Hordeum vulgare L.) is mediated by the concerted action of (1, 4)-β-d-mannan endohydrolase and β-d-mannosidase. Biochem J 399: 77–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Li M, Liang N, Yan H, Wei Y, Xu X, Liu J, Xu Z, Chen F, Wu G (2007) Molecular cloning and function analysis of the stay green gene in rice. Plant J 52: 197–209 [DOI] [PubMed] [Google Scholar]
- Jensen MK, Kjaersgaard T, Nielsen MM, Galberg P, Petersen K, O’Shea C, Skriver K (2010) The Arabidopsis thaliana NAC transcription factor family: structure–function relationships and determinants of ANAC019 stress signalling. Biochem J 426: 183–196 [DOI] [PubMed] [Google Scholar]
- Kan C, Zhang Y, Wang HL, Shen Y, Xia X, Guo H, Li Z (2021) Transcription factor NAC075 delays leaf senescence by deterring reactive oxygen species accumulation in Arabidopsis. Front Plant Sci 12: 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Park JH, Kim J, Kim JJ, Hong S, Kim J, Hong S, Kim J, Kim JH, Hye Ryun Woo HR, et al. (2018a) Time-evolving genetic networks reveal a NAC troika that negatively regulates leaf senescence in Arabidopsis. Proc Natl Acad Sci USA 115: E4930–E4939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Kim HJ, Vu QT, Jung S, McClung CR, Hong S, Nam HG (2018b) Circadian control of ORE1 by PRR9 positively regulates leaf senescence in Arabidopsis. Proc Natl Acad Sci USA 115: 8448–8453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Nam HG, Lim PO (2016) Regulatory network of NAC transcription factors in leaf senescence. Curr Opin Plant Biol 33: 48–56 [DOI] [PubMed] [Google Scholar]
- Kim JH, Woo HR, Kim J, Lim PO, Lee IC, Choi SH, Hwang D, Nam HG (2009) Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis. Science 323: 1053–1057 [DOI] [PubMed] [Google Scholar]
- Kim YS, Sakuraba Y, Han SH, Yoo SC, Paek NC (2013) Mutation of the Arabidopsis NAC016 transcription factor delays leaf senescence. Plant Cell Physiol 54: 1660–1672 [DOI] [PubMed] [Google Scholar]
- Knoop H, Gründel M, Zilliges Y, Lehmann R, Hoffmann S, Lockau W, Steuer R (2013) Flux balance analysis of cyanobacterial metabolism: The metabolic network of Synechocystis sp. PCC 6803. PLoS Comput Biol 9: e1003081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng Y, Yang Y, Ren D, Huang L, Dai L, Wang Y, Chen L, Tu Z, Gao Y, Li X, et al. (2017) A rice PECTATE LYASE-LIKE gene is required for plant growth and leaf senescence. Plant Physiol 2: 1151–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, Wang Y, Zhu Y, Tang J, Hu B, Liu L, Ou S, Wu H, Sun X, Chu J, et al. (2014) OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. Proc Natl Acad Sci USA 111: 10013–10018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Gao J, Yao L, Ren G, Zhu X, Gao S, Qiu K, Zhou X, Kuai B (2016) The role of ANAC072 in the regulation of chlorophyll degradation during age-and dark-induced leaf senescence. Plant Cell Rep 35: 1729–1741 [DOI] [PubMed] [Google Scholar]
- Martin U, Pallardy SG, Bahari ZA (1987) Dehydration tolerance of leaf tissues of six woody angiosperm species. Physiol Plant 69: 182–186 [Google Scholar]
- Mayta ML, Hajirezaei MR, Carrillo N, Lodeyro AF (2019) Leaf senescence: The chloroplast connection comes of age. Plants 11: 495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda-Yamamizo C, Mitsuda N, Sakamoto S, Ogawa D, Ohme-Takagi M, Ohmiya A (2016) The NAC transcription factor ANAC046 is a positive regulator of chlorophyll degradation and senescence in Arabidopsis leaves. Sci Rep 6: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooka H, Satoh K, Doi K, Nagata T, Otomo Y, Murakami K, Matsubara K, Osato N, Kawai J, Carninci P, et al. (2003) Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res 10: 239–247 [DOI] [PubMed] [Google Scholar]
- Podzimska-Sroka D, O’Shea C, Gregersen PL, Skriver K (2015) NAC transcription factors in senescence: from molecular structure to function in crops. Plants 4: 412–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puranik S, Sahu PP, Srivastava PS, Prasad M (2012) NAC proteins: Regulation and role in stress tolerance. Trends Plant Sci 17: 369–381 [DOI] [PubMed] [Google Scholar]
- Qiu K, Li Z, Yang Z, Chen J, Wu S, Zhu X, Gao S, Gao J, Ren G, Kuai B, et al. (2015) EIN3 and ORE1 accelerate degreening during ethylene-mediated leaf senescence by directly activating chlorophyll catabolic genes in Arabidopsis. PLoS Genet 11: e1005399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren G, An K, Liao Y, Zhou X, Cao Y, Zhao H, Cao Y, Zhao H, Ge X, Kuai B (2007) Identification of a novel chloroplast protein AtNYE1 regulating chlorophyll degradation during leaf senescence in Arabidopsis. Plant Physiol 144: 1429–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers H, Munné-Bosch S (2016) Production and scavenging of reactive oxygen species and redox signaling during leaf and flower senescence: Similar but different. Plant Physiol 171: 1560–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sade N, del Mar Rubio-Wilhelmi M, Umnajkitikorn K, Blumwald E (2018) Stress-induced senescence and plant tolerance to abiotic stress. J Exp Bot 69: 845–853 [DOI] [PubMed] [Google Scholar]
- Schröder R, Atkinson RG, Redgwell RJ (2009) Re-interpreting the role of endo-β-mannanases as mannan endotransglycosylase/hydrolases in the plant cell wall. Ann Bot 2: 197–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt B, Vicenzi M, Garrel C, Denis FM (2015) Effects of N-acetylcysteine, oral glutathione (GSH) and a novel sublingual form of GSH on oxidative stress markers: A comparative crossover study. Redox Biol 6: 198–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoda Y, Ito H, Tanaka A (2016) Arabidopsis STAY-GREEN, Mendel’s green cotyledon gene, encodes magnesium-dechelatase. Plant Cell 28: 2147–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuraba Y, Han SH, Lee SH, Hörtensteiner S, Paek NC (2016) Arabidopsis NAC016 promotes chlorophyll breakdown by directly upregulating STAYGREEN1 transcription. Plant Cell Rep 35: 155–166 [DOI] [PubMed] [Google Scholar]
- Sakuraba Y, Piao W, Lim JH, Han SH, Kim YS, An G, Paek NC (2015) Rice ONAC106 inhibits leaf senescence and increases salt tolerance and tiller angle. Plant Cell Physiol 56: 2325–2339 [DOI] [PubMed] [Google Scholar]
- Sun H, Hao P, Ma Q, Zhang M, Qin Y, Wei H, Su J, Wang H, Gu L, Wang N, et al. (2018) Genome-wide identification and expression analyses of the pectate lyase (PEL) gene family in cotton (Gossypium hirsutum L.). BMC Genome 19: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taiz L, Zeiger E (2002) Photosynthesis: physiological and ecological considerations. Plant Physiol 9: 172–174 [Google Scholar]
- Thirumalaikumar VP, Devkar V, Mehterov N, Ali S, Ozgur R, Turkan I, Mueller-Roeber B, Balazadeh S (2018) NAC transcription factor JUNGBRUNNEN 1 enhances drought tolerance in tomato. Plant Biotechnol J 2: 354–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorogood D (1996) Varietal colour of Lolium perenne L. turfgrass and its interaction with environmental conditions. Plant Var Seeds 9: 15–20 [Google Scholar]
- Uauy C, Distelfeld A, Fahima T, Blechl A, Dubcovsky J (2006) A NAC gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science 314: 1298–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah A, Akbar A, Yang X (2019) Jasmonic acid (JA)-mediated signaling in leaf senescence. In M Sarwat, N Tuteja, eds, Senescence Signalling and Control in Plants, Academic Press, Cambridge, MA, pp 111–123 [Google Scholar]
- Vicente AR, Saladie M, Rose JK, Labavitch JM (2007) The linkage between cell wall metabolism and fruit softening: looking to the future. J Sci Food Agric 87: 1435–1448 [Google Scholar]
- Walley JW, Rowe HC, Xiao Y, Chehab EW, Kliebenstein DJ, Wagner D, Dehesh K (2008) The chromatin remodeler SPLAYED regulates specific stress signaling pathways. PLoS Pathog 4: e1000237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo HR, Kim HJ, Lim PO, Nam HG (2019) Leaf senescence: systems and dynamics aspects. Annu Rev Plant Biol 70: 347–376 [DOI] [PubMed] [Google Scholar]
- Wu A, Allu AD, Garapati P, Siddiqui H, Dortay H, Zanor MI, Asensi-Fabado AM, Munné-Bosch S, Antonio C, Tohge T, et al. (2012) JUNGBRUNNEN1, a reactive oxygen species-responsive NAC transcription factor, regulates longevity in Arabidopsis. Plant Cell 24: 482–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Li Z, Yang L, Xie Z, Chen J, Zhang W, Liu T, Gao S, Gao J, Zhu Y, et al. (2016) NON-YELLOWING2 (NYE2), a close paralog of NYE1, plays a positive role in chlorophyll degradation in Arabidopsis. Mol Plant 9: 624–627 [DOI] [PubMed] [Google Scholar]
- Xie Z, Lin W, Yu G, Cheng Q, Xu B, Huang B (2019) Improved cold tolerance in switchgrass by a novel CCCH-type zinc finger transcription factor gene, PvC3H72, associated with ICE1–CBF–COR regulon and ABA-responsive genes. Biotechnol Biofuels 12: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Escamilla-Treviño LL, Sathitsuksanoh N, Shen Z, Shen H, Percival Zhang YH, Dixon RA, Zhao B (2011) Silencing of 4-coumarate: coenzyme A ligase in switchgrass leads to reduced lignin content and improved fermentable sugar yields for biofuel production. New Phytol 192: 611–625 [DOI] [PubMed] [Google Scholar]
- Xu B, Yu G, Li H, Xie Z, Wen W, Zhang J, Huang B (. 2019) Knockdown of STAYGREEN in perennial ryegrass (Lolium perenne L.) leads to transcriptomic alterations related to suppressed leaf senescence and improved forage quality. Plant Cell Physiol 60: 202–212 [DOI] [PubMed] [Google Scholar]
- Xu B, Li H, Li Y, Yu G, Zhang J, Huang B (2018) Characterization and transcriptional regulation of chlorophyll b reductase gene NON-YELLOW COLORING 1 associated with leaf senescence in perennial ryegrass (Lolium perenne L.). Environ Exp Bot 149: 43–50 [Google Scholar]
- Yang J, Worley E, Udvardi M (2014) A NAP-AAO3 regulatory module promotes chlorophyll degradation via ABA biosynthesis in Arabidopsis leaves. Plant Cell 26: 4862–4874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SD, Seo PJ, Yoon HK, Park CM (2011) The Arabidopsis NAC transcription factor VNI2 integrates abscisic acid signals into leaf senescence via the COR/RD genes. Plant Cell 23: 2155–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Wang C, Qiu K, Chen H, Li Z, Li X, Song J, Wang X, Gao J, Kuai B, et al. (2020) The transcription factor ZmNAC126 accelerates leaf senescence downstream of the ethylene signaling pathway in maize. Plant Cell Environ 43: 2287–2300 [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565. [DOI] [PubMed] [Google Scholar]
- Yoshida S (2003) Molecular regulation of leaf senescence. Curr Opin Plant Biol 1: 79–84 [DOI] [PubMed] [Google Scholar]
- Yu G, Cheng Q, Xie Z, Xu B, Huang B, Zhao B (2017) An efficient protocol for perennial ryegrass mesophyll protoplast isolation and transformation, and its application on interaction study between LpNOL and LpNYC1. Plant Methods 13: 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Xie Z, Zhang J, Lei S, Lin W, Xu B, Huang B (2021) NOL-mediated functional stay-green traits in perennial ryegrass (Lolium perenne L.) involving multifaceted molecular factors and metabolic pathways regulating leaf senescence. Plant J 106: 1219–1232 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Liu C, Li K, Li X, Xu M, Guo Y (2022) CLE14 functions as a “brake signal” to suppress age-dependent and stress-induced leaf senescence by promoting JUB1-mediated ROS scavenging in Arabidopsis. Mol Plant 15: 179–188 [DOI] [PubMed] [Google Scholar]
- Zhang J, Yu G, Wen W, Ma X, Xu B, Huang B (2015) Functional characterization and hormonal regulation of the PHEOPHYTINASE gene LpPPH controlling leaf senescence in perennial ryegrass. J Exp Bot 67: 935–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang WJ, Dewey RE, Boss W, Phillippy BQ, Qu R (2013) Enhanced Agrobacterium-mediated transformation efficiencies in monocot cells is associated with attenuated defense responses. Plant Mol Biol 81: 273–286 [DOI] [PubMed] [Google Scholar]
- Zhao C, Avci U, Grant EH, Haigler CH, Beers EP (2008) XND1, a member of the NAC domain family in Arabidopsis thaliana, negatively regulates lignocellulose synthesis and programmed cell death in xylem. Plant J 53: 425–436 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.