Abstract
The barley MLA nucleotide-binding leucine-rich-repeat (NLR) receptor and its orthologs confer recognition specificity to many fungal diseases, including powdery mildew, stem-, and stripe rust. We used interolog inference to construct a barley protein interactome (Hordeum vulgare predicted interactome, HvInt) comprising 66,133 edges and 7,181 nodes, as a foundation to explore signaling networks associated with MLA. HvInt was compared with the experimentally validated Arabidopsis interactome of 11,253 proteins and 73,960 interactions, verifying that the 2 networks share scale-free properties, including a power-law distribution and small-world network. Then, by successive layering of defense-specific “omics” datasets, HvInt was customized to model cellular response to powdery mildew infection. Integration of HvInt with expression quantitative trait loci (eQTL) enabled us to infer disease modules and responses associated with fungal penetration and haustorial development. Next, using HvInt and infection–time–course RNA sequencing of immune signaling mutants, we assembled resistant and susceptible subnetworks. The resulting differentially coexpressed (resistant – susceptible) interactome is essential to barley immunity, facilitates the flow of signaling pathways and is linked to mildew resistance locus a (Mla) through trans eQTL associations. Lastly, we anchored HvInt with new and previously identified interactors of the MLA coiled coli + nucleotide-binding domains and extended these to additional MLA alleles, orthologs, and NLR outgroups to predict receptor localization and conservation of signaling response. These results link genomic, transcriptomic, and physical interactions during MLA-specified immunity.
Keywords: interologs, interactome inference, Hordeum vulgare, immunity, disease modules, yeast-two-hybrid, systems biology, powdery mildew
Introduction
Plants attain a focused immune response via interconnected signaling networks (Yuan et al. 2020; Ngou et al. 2022). These networks are activated upon recognition of conserved pathogen molecules termed pathogen-associated molecular patterns (Lo-Presti et al. 2015; Toruño et al. 2016; Bentham et al. 2020). Pathogens have adapted to suppress this response by secreting effectors into the host cell. As a counter mechanism, families of resistance (R) proteins have evolved, encoding a series of intracellular receptors that are associated with immune activation (Lo-Presti et al. 2015; Toruño et al. 2016). Experimental and computational evidence have shown that most R-proteins converge to a group of intracellular nucleotide-binding leucine-rich-repeat (NLR) receptors, one of the largest plant protein families (Baggs et al. 2017; Monteiro and Nishimura 2018; Sun et al. 2020; Tamborski and Krasileva 2020; Van-Wersch et al. 2020; Barragan and Weigel 2021). The protein structure of this family consists of an N-terminal domain [coiled-coil (CC) or Toll-like Interleukin-1 receptor], a conserved nucleotide-binding (NB) domain shared with apoptotic protease activating factor 1, various R-proteins and cell death protein-4 (ARC), and a C-terminal leucine-rich repeat domain (LRR) (Baggs et al. 2017; Monteiro and Nishimura 2018; Van-Wersch et al. 2020). Since R-proteins have a prevalent role in determining the outcome of plant–pathogen interactions, they are ideal probes to dissect host immunity as well as basic plant cell function (Bray Speth et al. 2007; Krattinger and Keller 2016; Sun et al. 2020; Sánchez-Martín and Keller 2021).
Obligate biotrophic fungi, which include mildews and rusts, cause some of the most detrimental impacts to crop production (Dean et al. 2012). In the interaction between Blumeria graminis f. sp. hordei (Bgh), the causal agent of powdery mildew, and its cereal host plant, barley [Hordeum vulgare L. (Hv)], disease is blocked by the action of specific R-proteins, designated ML (for mildew resistance locus), that respond to corresponding Bgh avirulence effectors, designated AVR (Ridout et al. 2006; Lu et al. 2016; Saur et al. 2019; Bauer et al. 2021). Diversification of the MLA NLR (Wei et al. 2002; Seeholzer et al. 2010) has generated up to 30 variants that confer specific recognition to different AVR-associated Bgh isolates (Ridout et al. 2006; Lu et al. 2016; Saur et al. 2019; Bauer et al. 2021). In addition, NLRs encoded by mildew resistance locus a (Mla) alleles or orthologs appear to recognize effector targets from evolutionary diverged fungi, and once triggered, confer resistance to powdery mildew in wheat and transgenic Arabidopsis (Jordan et al. 2011; Maekawa et al. 2012), wheat and rye Ug99 stem rust (Periyannan et al. 2013; Mago et al. 2015; Cesari et al. 2016) and wheat stripe rust (Bettgenhaeuser et al. 2021).
MLA-mediated immunity involves the stabilization of the protein through interactions with the heat shock protein 90 (HSP90), required for MLA12 resistance 1 (RAR1), and suppressor of G-two allele of Skp1 (SGT1) complex, with differential requirements for the various components, depending on the Mla allele (Shen et al. 2003; Bieri et al. 2004; Halterman and Wise 2004; Shirasu 2009; Chapman et al. 2021, 2022). A RING-type E3 ligase (MIR1) has also been reported as an interactor of MLA, attenuating defense signaling by degradation of the receptor via the ubiquitin proteasome system (Wang et al. 2016). Upon recognition of Bgh AVR effectors, MLA accumulates in the nucleus, followed by its association with multiple transcription factors (TFs), including WRKY1, WRKY2, and MYB6 (Shen et al. 2007; Chang et al. 2013).
In order to establish a comprehensive view of the regulatory programs that render a plant resistant to pathogens, deeper insight into R-gene activation and the resulting signaling cascades is needed (Martin et al. 2003; Deng et al. 2020). In this regard, genome-wide protein interactomes are advantageous to interpret the molecular mechanisms in immune cell signaling (Mukhtar et al. 2011; Weßling et al. 2014). Yet, building interactomes from experimental data is a cost and labor intensive task, and as a result, they are incomplete and only available for few model organisms (Matthews et al. 2001; McWhite et al. 2020). To circumvent this challenge, predictive approaches have been developed. One of these is interologs inference, which consists of mining protein interactions using ortholog information between the species of interest and model organisms that possess experimentally validated interactions (Matthews et al. 2001). This approach has been successfully used to generate predicted interactomes in Arabidopsis and several crop species including rice, maize, and tulsi (Geisler-Lee et al. 2007; Ho et al. 2012; Musungu et al. 2015; Singh et al. 2020). Then, integration of interactomes with context-specific data types can be performed to highlight nodes, edges, structures, and modules associated with a phenotype of interest (Randhawa and Pathania 2020). For example, infection time–course transcriptomes enable the use of gene coexpression as evidence to increase confidence of predicted protein–protein interactions (PPIs) and build phenotype-specific subnetworks (Braun et al. 2013; Petrey and Honig 2014; Jiang et al. 2016). In addition, expression quantitative trait locus (eQTL) data can be used to obtain disease modules (DMs) in the interactome and provide genetic and physical connections between nodes, avoiding biases given by the amount of information of well-studied disease genes (Dwivedi et al. 2020; Wang et al. 2020).
To discover new connections in NLR-based immunity, we used interolog inference to develop a predicted interactome for the Triticeae grain crop, barley, using experimentally validated interactions in several model organisms and crop species. Then, using time–course gene expression and eQTL data, we assembled several subnetworks associated with resistance, susceptibility, and the dynamics of defense during key stages in powdery mildew infection. Lastly, we used yeast-two-hybrid (Y2H) next-generation-interaction screens (NGIS) to confirm 15 PPIs between the MLA6 NLR-receptor and barley proteins, including a new polyamine factor 1-binding protein. We then extended these interactions to additional synthesized MLA alleles, wheat and rye orthologs, and NLR outgroups. Fourteen (9 validated by Y2H-NGIS and 5 from previous literature) of the 21 MLA interactors could be incorporated into the barley interactome and by extension, cellular compartments. These analyses enabled us to propose a predictive model of MLA cellular localization and to link the receptor with gene coexpression during resistance.
Methods
Collection of infection time course RNA-Seq and differential expression analysis
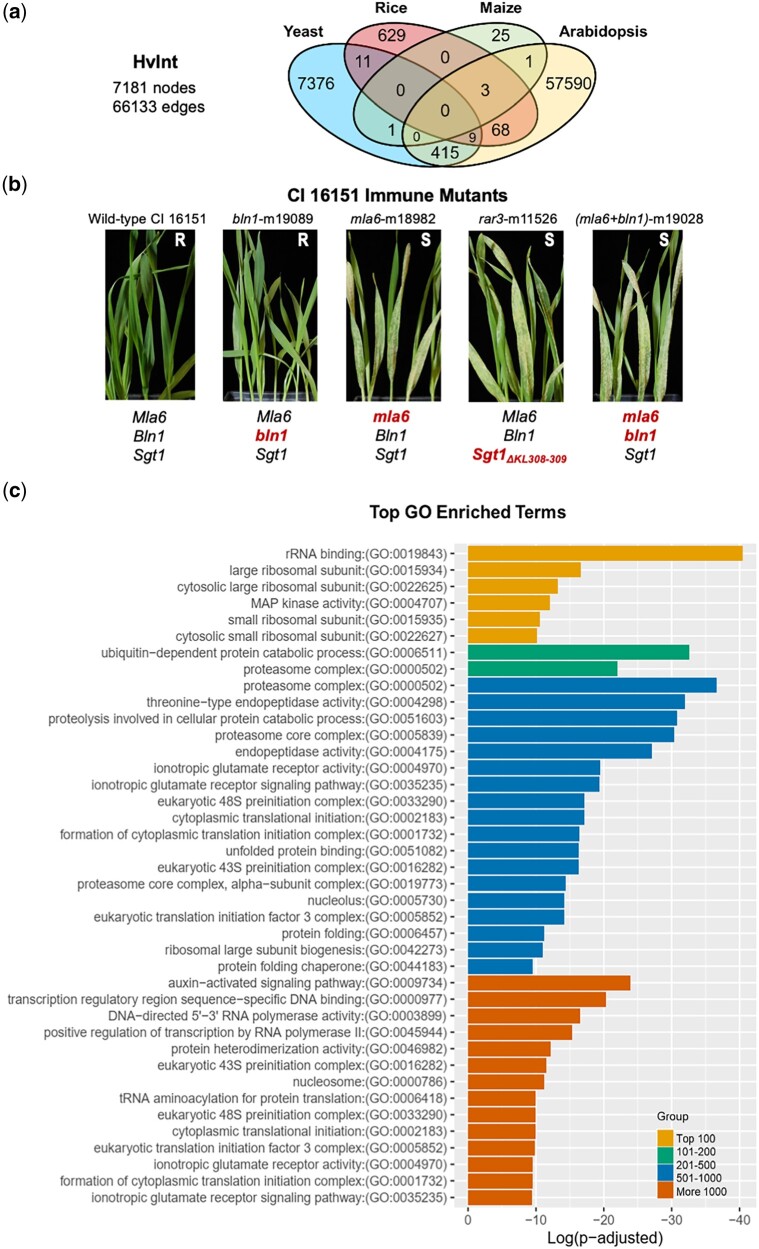
An infection time course of CI 16151 (Mla6, Bln1, Sgt1) and derived fast-neutron mutants bln1-m19089 (Mla6, bln1, Sgt1), mla6-m18982 (mla6, Bln1, Sgt1), rar3-m11526 (Mla6, Bln1, Sgt1ΔKL308-309), and (mla6+bln1)-m19028 (mla6, bln1, Sgt1) was used for RNA-Seq analysis. CI 16151 contains the functional Mla6 R gene and is resistant to Bgh isolates that contain the effector, AVRa6. Bln1 (Blufensin1) is a negative regulator of pattern-triggered immunity (PTI), whose silencing results in down-regulation of genes associated with basal defense (Meng et al. 2009; Xu et al. 2015). The resistant bln1 mutant, m19089, exhibits enhanced basal defense. Mla6 is deleted in the m18982 mutant, which is thus, susceptible. Rar3 (required for Mla6 resistance3) is required for Mla6-mediated generation of H2O2 and the hypersensitive response (HR). This susceptible mutant contains a Lys–Leu deletion in the SGT1-specific domain of SGT1, which interacts with NLR proteins (Chapman et al. 2021, 2022). The (mla6 + bln1) double mutant is susceptible as it contains the same Mla6 deletion as in m18982.
Seven-day-old first leaves were challenged with Bgh isolate 5874 (AVRa1, AVRa6, AVRa12) according to Caldo et al. (2004), positioned randomly in a controlled growth chamber (18°C, 8 h darkness, 16 h light), and harvested into liquid nitrogen from a split-plot design at 0, 16, 20, 24, 32, and 48 h after inoculation (HAI) (5 genotypes × 6 time points × 3 biological replications). Total RNA was extracted using a hot (60°C) phenol/guanidine thiocyanate method as previously described (Caldo et al. 2004, 2006). Single-end mRNA libraries were prepared with the Illumina TruSeq stranded RNA sample preparation kit (Illumina, Inc., San Diego, CA, USA) and sequenced at the Iowa State University DNA facility (Ames, IA, USA) using 100-bp reads and the Illumina HiSeq2500 system. Raw reads (NCBI-GEO accession GSE101304) were processed using Trimmomatic V0.40 (Bolger et al. 2014) and Salmon V1.8.0 (Patro et al. 2017), taking as references the barley TRITEX (Monat et al. 2019) and Bgh DH14 (Spanu et al. 2010; Frantzeskakis et al. 2018) annotations. Taxon-specific normalization to the raw counts was applied: Salmon raw count matrices were separated for barley and Bgh and size factors were calculated using median-of-ratios normalization (Anders and Huber 2010), which were then combined to calculate the final normalized counts matrix (Klingenberg and Meinicke 2017). Differentially expressed (DE) genes were identified using a DESeq2 V1.34.0 (Love et al. 2014) model with read counts as response, and timepoint and genotype terms as explanatory variables. We controlled for multiple testing using Benjamin and Hochberg methodology (Benjamini and Hochberg 1995), calling DE genes with an adjusted P-value of <0.001.
Interactome reconstruction
We constructed a Hordeum vulgare predicted interactome (HvInt), by adapting the interolog method outlined by Matthews et al. (2001), Geisler-Lee et al. (2007), and Nakajima et al. (2018). Homologs of H. vulgare with Arabidopsis thaliana, Oryza sativa, and Zea mays were obtained from the Plant Compara tables V96 from Ensembl Plants, filtering those that met high confidence similarity scores from the gene tree (>25%) (Howe et al. 2020). Homologs between barley and Saccharomyces cerevisiae were determined by InParanoid8 (Sonnhammer and Östlund 2015) by applying the maximum score cutoff of 100%. Next, experimentally validated PPIs for the reference organisms were mined from BioGRID V4.1.190 (Stark et al. 2006), the PPI database for Maize (PPIM) (Zhu et al. 2016), and the Predicted Rice Interactome Network (PRIN) database (Gu et al. 2011). Further PPIs were obtained from the pan–plant protein interactome (McWhite et al. 2020) and research literature (Bieri et al. 2004; Shen et al. 2007; Mukhtar et al. 2011; Chang et al. 2013; Weßling et al. 2014; Wang et al. 2016; Trigg et al. 2017; Smakowska-Luzan et al. 2018; Wierbowski et al. 2020). For each species, interactions were updated to the most recent Ensembl genome annotation and then, the compiled list was converted to barley gene IDs using the high confidence homologs. The final nonredundant interactome was obtained by removing duplicated interactions and tracking the species where they were originally reported. Code used to generate HvInt is posted at the GitHub page https://github.com/Wiselab2/Barley_Interactome [accessed 15 April 2022].
The infection–time–course expression dataset was integrated with HvInt by adding information to the edges and the nodes. We first assigned the weight of the edges as the pairwise distance correlation calculated from the complete expression dataset, or subsets of it based on the infection phenotype using a permutation test as described below (see Construction of resistant and susceptible interactomes). Second, we adapted the DiffSLC (Mistry et al. 2017) pipeline to use expression data from RNA-Seq and compute the node essentiality of the proteins in the predicted barley interactome (deletion of an essential protein is associated with lethality). We characterized the predicted network, using walktrap clustering (Székely et al. 2007) and Gene Ontology (GO) term enrichment using the reported TRITEX annotation and clusterProfiler V4.2.2 (Yu et al. 2012; Monat et al. 2019). Scale-free and small-world network properties were tested for the final predicted interactome using the igraph R package V1.2.7 (Watts and Strogatz 1998; Albert et al. 1999; Csardi and Nepusz 2006). Network topological properties such as protein essentiality with DiffSLC (Mistry et al. 2017), degree, betweenness (Csardi and Nepusz 2006) and distance correlation (Székely et al. 2007) were used to characterize the subnetworks obtained from HvInt.
DM prediction using eQTL and interactome data
We used Node2vec-Hierarchical Clustering (N2V-HC; method of disease module identification for multi-layer biological networks) (Wang et al. 2020) to integrate HvInt with eQTL data collected during Bgh appressorial penetration (16 HAI) and haustorial development (32 HAI) (Surana et al. 2017). All genetic markers with significant eQTL associations were used as input, removing associations with adjusted P-values larger than 0.001. The SNP and eQTL matrices used as input to the software were built to match the chromosome markers with linked genes (input as SNP table) and eQTL associations (input as eQTL table). The resulting modules for each timepoint were analyzed using GO enrichment. Results were compared and classified as core response (common between the 2 timepoints) and unique to each infection stage. Core and unique GO terms for the DMs were analyzed using DE enrichment between the resistant CI 16151 (Mla6, Bln1, Sgt1) and the susceptible m18982 (mla6, Bln1, Sgt1) genotypes, using an adjusted P-value threshold <0.001. If a gene associated with the GO term was DE, then it was called DM DE. Results from these analyses were summarized with a schema using Biorender.com. Expression profiles of the DM DE genes associated with each group were plotted using R V4.1.3 (RCoreTeam 2013).
Construction of resistant and susceptible interactomes
Resistant and susceptible barley interactomes were obtained using expression distance correlation (Székely et al. 2007). The infection–time–course RNA-Seq was separated by disease phenotype and used to generate conditional subnetworks consisting of significantly coexpressed interactions (Jiang et al. 2016). The coexpression significance threshold used to generate each subnetwork was obtained by building a null distribution of this parameter. The RNA-Seq count data, grouped by phenotype, were permuted for each timepoint leaving the replicates as a block. The resulting distribution built from 10,000 permutations was used to calculate empirical P-values and correlation thresholds for significance of the pairwise correlation values. Using a P-value of 0.05, we calculated the significance correlation threshold for the resistant and susceptible subnetworks. Edges with values below the correlation thresholds were removed from the network to obtain the resistant [HvInt(R)] and susceptible [HvInt(S)] interactomes.
The differentially coexpressed resistant [HvInt(R–S)] and susceptible [HvInt(S–R)] subnetworks were obtained by removing the common interactions between HvInt(R) and HvInt(S), [HvInt(R = S)], from these subnetworks, respectively. The differentially coexpressed interactomes were further characterized using topological properties. Significance of the differences in these properties were calculated using Wilcoxon rank sum tests. Hypergeometric tests were applied to look for enrichment of eQTL associations in the HvInt(R), HvInt(S), HvInt(R–S), HvInt(S–R), and HvInt(R = S) subnetworks. The resistant subnetworks HvInt(R) and HvInt(R–S) were clustered using a walktrap algorithm (Csardi and Nepusz 2006), and clusters were analyzed using GO enrichment with clusterProfiler V4.2.2 (Yu et al. 2012). Visualization was done using Cytoscape V3.7 (Shannon et al. 2003) and the R package RCy3 V2.14.2 (Gustavsen et al. 2019).
Building an MLA protein–protein interactome subnetwork anchored with validated Y2H-NGIS data
MLA6 aa 1–225 (Halterman et al. 2001), representing the CC and NB domains, was screened as bait using Y2H-NGIS, with a 3-frame cDNA prey library of 1.1× 107 primary clones generated from the infection time course (Surana 2017; Velásquez-Zapata et al. 2021). A top list of candidate interactors was selected from the Borda ensemble provided by Y2H-SCORES (Velásquez-Zapata et al. 2021). Interacting prey fragments were determined using Integrative Genomics Viewer (IGV) alignments obtained from NGPINT (Banerjee et al. 2021), reported in-frame prey transcripts with the highest in-frame score from Y2H-SCORES, and clones inserted into the prey vector. After transforming the candidates into yeast, binary Y2H tests (Dreze et al. 2010) were performed under 3 levels of selective media: Diploid selection (SC–LW), interaction selection (SC–LWH), and selection (SC–LWH) using 3 dilutions (10°, 10−1, and 10−2) and 0.5 mM 3-AT. Validated interactions were extended to the CC + NB domain sequences encoded by Mla alleles Mla1, Mla3, Mla7, Mla7-2, Mla8, Mla9, Mla10, Mla12, Mla13 (Zhou et al. 2001; Halterman et al. 2003; Shen et al. 2003; Halterman and Wise 2004; Seeholzer et al. 2010; Bettgenhaeuser et al. 2021), and orthologs including TmMla, Sr33, and Sr50 (Jordan et al. 2011; Periyannan et al. 2013; Mago et al. 2015). Sr22, Pbr1, Pm3a, and Zar1 were used for the outgroup (Salanoubat et al. 2000; Srichumpa et al. 2005; Steuernagel et al. 2016; Carter et al. 2019). Protein sequences of the CC + NB domains from these proteins were aligned MUSCLE V5 (Edgar 2004), and a phylogenetic tree constructed using maximum likelihood with a Jones–Taylor–Thornton substitution model and a gamma-distributed-rate among sites model using MEGA-X (Tamura et al. 2011).
The MLA protein–protein interactome (MLAInt) subnetwork was built by retaining the nodes in the shortest paths computed for any pair of MLA interactors. First and second neighbors to the nodes in this subnetwork were added to complete the MLAInt. This subnetwork was clustered using a fast-greedy algorithm, and clusters were analyzed using clusterProfiler V4.2.2 for GO (Yu et al. 2012) and a hypergeometric test for eQTL (Surana et al. 2017) enrichment. Topological properties were used to characterize MLAInt. Visualization was done using igraph V1.2.7 (Csardi and Nepusz 2006).
Phylogenetic trees of predicted barley TF families
Basic helix–loop–helix (bHLH) and homeobox (HB) TFs were predicted, using the TRITEX Morex r2 barley high confidence protein reference and PlantTFDB V4.0 (Jin et al. 2017; Monat et al. 2019). The sequences were aligned using MUSCLE V5 (Edgar 2004), and then the trees were calculated using maximum likelihood with a Jones–Taylor–Thornton substitution model and a gamma-distributed-rate among sites model. The resulting trees were annotated using the ggtree R package V3.2.1 (Yu et al. 2017). The annotation consisted in plotting: (1) genes with position weight matrix (PWM) DNA-binding domain according to our database search (Matys et al. 2006; Weirauch et al. 2014; Jin et al. 2017); (2) log2 of the foldchange of the RNA-Seq expression data for the comparison between CI 16151 (Mla6, Bln1, Sgt1) wild-type genotype and the m18982 (mla6, Bln1, Sgt1) mutant in a time course experiment of infection (0, 16, 20, 24, 32, and 48 HAI); and (3) previously characterized TFs in plant immunity.
Results
The barley predicted interactome shows protein essentiality in defense
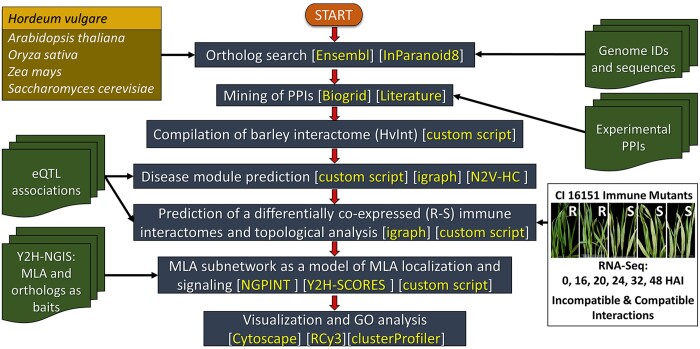
As outlined in Fig. 1, we generated a high-quality, barley interactome, HvInt, based on the interologs method of Matthews et al. (2001). Only experimentally validated interactions were used for the prediction, including PPIs across several plant species reported in the pan–plant protein interactome (Bieri et al. 2004; Stark et al. 2006; Shen et al. 2007; Gu et al. 2011; Mukhtar et al. 2011; Chang et al. 2013; Weßling et al. 2014; Wang et al. 2016; Zhu et al. 2016; Trigg et al. 2017; Smakowska-Luzan et al. 2018; McWhite et al. 2020; Wierbowski et al. 2020). HvInt (Supplementary Data 1) contains 66,133 edges and 7,181 nodes and can be used as baseline network to investigate different signaling events in the barley cell. Here, we show how HvInt can be customized to study immune response, via integration of defense-specific datasets (Fig. 1). Figure 2a illustrates the number of interactions mined from the different species used for interologs prediction. About 87.8% of the predicted interactions are supported by experimental validations in Arabidopsis thaliana, followed by S. cerevisiae (11.8%), O. sativa (0.012%), and Z. mays (0.0005%). The relative percentages of interologs are directly correlated to the number of validated PPI available for each species at the time of compiling HvInt, with model organisms (Arabidopsis and yeast) foremost over cereal crops. Therefore, as a quality control, properties of HvInt were compared with the collected interactions in A. thaliana [Arabidopsis thaliana interactome (AtInt)], which comprise 73,960 interactions and 11,253 proteins (Supplementary Data 1). This analysis demonstrated that HvInt is robust, as it maintains the power-law and small-world properties of the AtInt network (P-value >0.05 for the Kolmogorov–Smirnov tests, global clustering coefficient ratio >1 and an average shortest path length ratio ∼1) (Watts and Strogatz 1998; Albert et al. 1999).
Fig. 1.
HvInt bioinformatic workflow. HvInt was generated from experimentally validated orthologs from A. thaliana, O. sativa, Z. mays, and S. cerevisiae and followed by integration of context-specific “omics” datasets associated with response to the powdery mildew pathogen. An eQTL dataset was used for DM prediction, time–course RNA-Seq of wildtype and fast-neutron derived immune mutants to build resistant and susceptible subnetworks, and yeast-two-hybrid next-generation-interaction screens (Y2H-NGIS) to build an MLA associated subnetwork. R, resistant; S, susceptible; HAI, hours after inoculation.
Fig. 2.
HvInt, the predicted barley interactome. a) Venn diagram with the number of interactions in HvInt across the species of origin. b) Wild-type CI 16151 (Mla6, Bln1, Sgt1) and derived immune mutants bln1-m19089 (Mla6, bln1, Sgt1), mla6-m18982 (mla6, Bln1, Sgt1), rar3-m11526 (Mla6, Bln1, Sgt1ΔKL308-309, and (mla6+bln1)-m19028 (mla6, bln1, Sgt1) 7 days after inoculation with Bgh isolate 5874 (AVRa6). R, resistant; S, susceptible; mutant alleles are designated below the images in red. c) GO term enrichment of the top essential proteins in HvInt.
We applied a walktrap clustering algorithm on HvInt using distance correlation from expression data as edge weights (Székely et al. 2007). For this purpose, we used RNA-Seq data from our infection time course of the resistant barley line CI 16151 (Mla6, Bln1, Sgt1) and 4 fast-neutron derived mutants challenged with Bgh isolate 5,874 (AVRa6), as shown in Fig. 2b (Hunt et al. 2019; Chapman et al. 2021). We acquired 54 clusters comprising 1–2,313 proteins, with 97.6% classified in the top 5 clusters (2, 8, 11, 13, and 15). We then calculated node essentiality using DiffSLC (Supplementary Data 2), which correlates with functionally important nodes, whose deletion is associated with lethality (Mistry et al. 2017). The top 1,000 essential proteins, which are involved in 80.7% of the interactions in HvInt, were separated into 4 groups (top 100, 101–200, 201–500, and 501–1,000), distributed in 8 clusters and significantly enriched in clusters 13 and 15 (Supplementary Data 2). Lastly, essential proteins were analyzed using GO terms (Fig. 2e). The top 100 were enriched with terms associated with ribosome and MAP kinase activity; 101–200 were associated with channel activity, proteasome complex and ubiquitin-dependent catabolism; and 201–500 were associated with chaperone-mediated protein folding, auxin-activated signaling pathway, and ionotropic glutamate receptor (GLR) activity. Finally, 501–1,000 were associated with proton-exporting ATPase activity and translation. Some of the top essential proteins are linked with fundamental cellular processes while others have functions associated with plant immunity (Zeng et al. 2006; Forde and Roberts 2014; Lee et al. 2015). Key examples are described in subsequent sections.
Disease modules (DM) reveal barley cellular responses at Bgh penetration and haustorial development
Proteins associated with disease phenotypes tend to be tightly connected at the physical level, generating interaction modules and pathways (Barabási et al. 2011; Sharma et al. 2015). The concept of DM emerges then to describe this phenomenon, i.e. when interactome clusters are functionally linked to a disease phenotype through genetic and physical associations (Sharma et al. 2015). We adapted the disease-module concept, originally developed in the context of network medicine (Barabási et al. 2011), to add temporal-defense information to HvInt at 2 key infection stages of barley by Bgh. We used N2V-HC (Wang et al. 2020) to integrate HvInt with eQTL data derived from transcriptome analysis of the barley Q21861 × SM89010 doubled-haploid population during appressorial penetration (16 HAI) and haustorial development (32 HAI) (Surana et al. 2017). The dataset contained 317 markers mapped to 1,009 genes associated with 16,943 eQTLs. From those, we filtered out associations with adjusted P-values larger than 0.001, ending up with 4,357 eQTLs at 16 HAI and 6,375 at 32 HAI. This resulted in DMs with proteins enriched with eQTLs by timepoint (Supplementary Data 3).
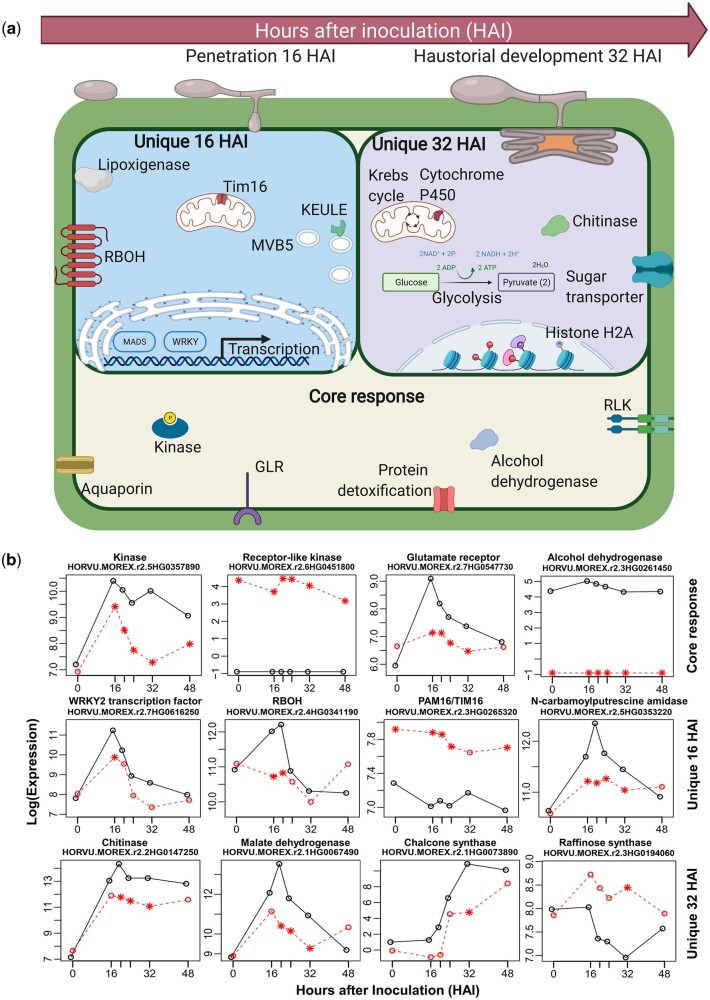
We performed GO analyses of the DMs to further understand the temporal response to powdery mildew. Modules were associated with individual timepoints, and a core response common to both timepoints. At penetration, we found 85 unique GO terms (adjusted P-values <0.05) while haustorial development had 274, and 499 were associated with the core response. The derived GO terms were explored further by surveying for enrichment of DE genes (adjusted P-values <0.001) extracted from our RNA-Seq data described above. By comparing the resistant CI 16151 (Mla6, Bln1, Sgt1) and the derived susceptible mutant, mla6-m18982 (mla6, Bln1, Sgt1), we could focus on gene sets that were functionally specific and displayed expression differences between compatible and incompatible interactions connected to the Mla6 NLR. The core response consisted of 110 GO terms that had DE genes associated at both 16 HAI and 32 HAI (shown in Supplementary Fig. 1). The most significant were associated with vesicle trafficking, transporter activity, serine/threonine kinase signaling, and response to biotic stimulus, which is consistent for a host–pathogen interaction scenario. Unique GO terms at appressorial penetration included protein import into mitochondrial matrix and the polyamine biosynthetic process. In contrast, haustorial development was characterized by the unique GO terms cell wall macromolecule catabolic process, chitin catabolic process, and pyruvate metabolic process.
An overview of key DE genes associated with core and unique GO terms by time point is illustrated in Fig. 3a. We designated these genes as DM DE genes (Supplementary Fig. 1 and Data 3). Seven DM DE genes are included in the combined 16- and 32-HAI core response: alcohol dehydrogenase, 3 kinases, receptor-like protein kinase, GLR, and aquaporin. Thirty unique DM DE genes were identified during fungal penetration (16 HAI), including 2 WRKY, 1 agamous MADS-box TF, 2 DETOXIFICATION proteins and several kinases. Lastly, 67 additional DM DE genes were identified during haustorial development (32 HAI), including chitinase, mildew resistance locus o (MLO)-like protein, LRR receptor-like protein, malate dehydrogenase, and several transporters. Then, to interrogate activity over time, DM DE genes were visualized by expression profile plots, contrasting the resistant progenitor, CI 16151 (Mla6, Bln1, Sgt1), to the susceptible mutant, m18982 (mla6, Bln1, Sgt1) (selected examples are shown in Fig. 3b; the complete set is presented in Supplementary Fig. 2). Because DM DE genes were initially selected based on eQTL associations, and not clustered by gene expression, the patterns of transcript accumulation were connected to phenotype, and thus characterized by significant differences between the 2 genotypes across all 6 timepoints, or in contrast, an abrupt expression change at penetration, or haustorial development, in one of the genotypes.
Fig. 3.
Identification of DMs at Bgh appressorial penetration and haustorial development. a) Schematic summary of the DM DEs at penetration (16 HAI) and haustorial development (32 HAI), separated by core and unique responses. Created with BioRender.com. b) Examples of time–course expression patterns of DM DE genes by type of response; CI 16151 (Mla6, Bln1, Sgt1) in black and m18982 (mla6, Bln1, Sgt1) in red, significant differences (adjusted P-values <0.001) are designated by an asterisk. The complete set of DM DE transcript patterns is presented in Supplementary Fig. 2.
Coexpressed interactomes during MLA-specified resistance and susceptibility
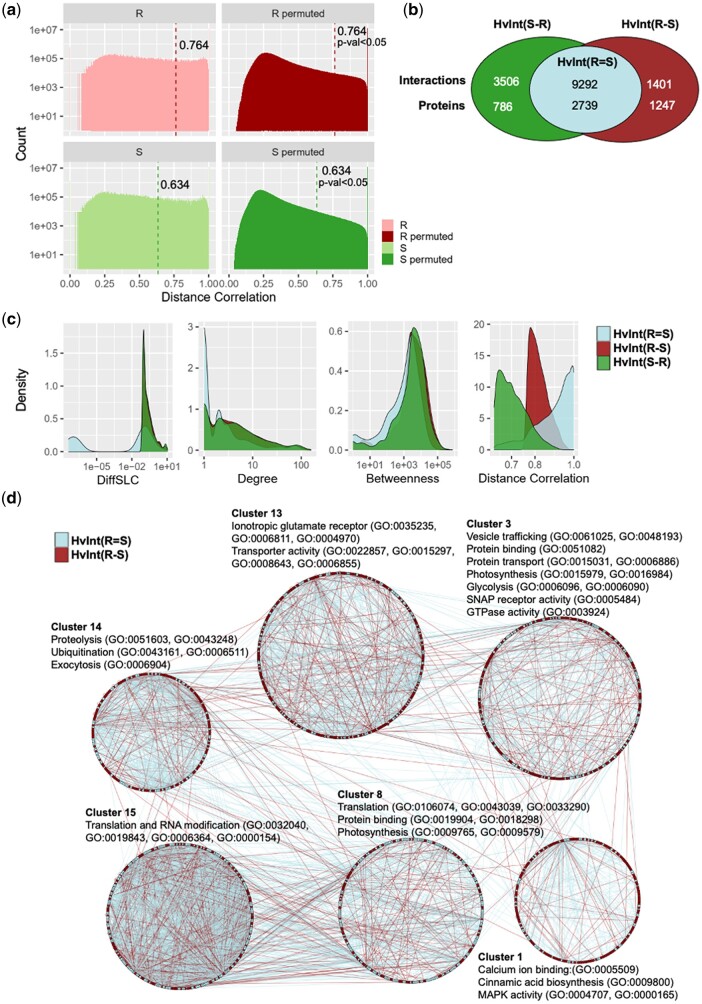
Time–course RNA-Seq data empowers rapid visualization of differential transcript accumulation in wild-type vs mutant backgrounds and thus demonstrates the impact of pathogens on immune signaling pathways. To complement the DM analysis above, we identified immune-active subnetworks under resistant and susceptible disease outcomes by integrating HvInt with our infection–time–course RNA-Seq data. The RNA-Seq read counts for each group of infection phenotypes in Fig. 2b were used to calculate coexpression values defined as pairwise distance correlations across genes. HvInt was then subset into the resistant [HvInt(R)] and susceptible [HvInt(S)] interactomes by retaining significantly coexpressed pairs under each phenotype. We defined significant correlation values for each subnetwork using a permutation test (Fig. 4a; see Methods).
Fig. 4.
Construction of resistant and susceptible interactomes. a) Experimental and permuted distributions of the expression distance correlations in HvInt by disease phenotype, Resistant (R) or Susceptible (S); significance thresholds (P-value <0.05) are marked. b) Distribution of interactions and proteins in the resistant [HvInt(R)] and susceptible [HvInt(S)] interactomes, separating by common [HvInt(R = S)] and difference [HvInt(R–S), HvInt(S–R)] subnetworks. c) Network properties of the resistant interactome, separated by common interactions with the susceptible network [HvInt(R = S)] and the resistant unique interactions [HvInt(R–S)]. d) Clustering and GO annotation of the resistant interactome HvInt(R), separating the differentially coexpressed subnetwork HvInt(R–S) and the common HvInt(R = S). Each cluster is grouped in a circular layout.
The resistant HvInt(R) subnetwork includes all edges with high expression distance correlation for the resistant genotypes. This subnetwork contains 3,120 nodes and 10,693 interactions (Supplementary Data 4) and was clustered into 33 groups with GO enrichment annotations as shown in Supplementary Data 4. The filtered susceptible subnetwork HvInt(S) has 12,798 interactions and 3,525 proteins, clustered in 45 modules with GO annotations reported in Supplementary Data 4. There is an overlap of 9,292 interactions and 2,739 proteins between the HvInt(R) and HvInt(S) subnetworks [Fig. 4b; HvInt(R = S)]. We then removed the common interactions HvInt(R = S) from HvInt(R) to obtain the differentially coexpressed resistant interactome HvInt(R–S) with 1,401 interactions and 1,247 proteins (Supplementary Data 4). We postulate that HvInt(R–S) shows unique, coexpressed interactions associated with the barley immune response to Bgh, therefore, represents the focus of our following analyses.
We tested for differences among the common HvInt(R = S), the differentially coexpressed resistant HvInt(R–S), and susceptible HvInt(S–R) interactomes by computing different topological properties that can be associated with the information content and flow in the networks. DiffSLC, degree, betweenness, and expression distance correlation between the 3 subnetworks were compared as shown in Fig. 4c. In addition, Wilcoxon rank sum tests were performed to quantify the significance of the differences. These analyses showed that HvInt(R–S) and HvInt(S–R) had significantly higher DiffSLC, degree and betweenness and lower expression distance correlation than HvInt(R = S) (P-values <1×10−100). This indicates that the proteins in the unique HvInt(R–S) and HvInt(S–R) have higher essentiality than the common resistant and susceptible HvInt(R = S) subnetwork. Importantly, these differences are associated with higher degree centrality, and not higher distance correlation, as the latter was lower in the differentially coexpressed subnetworks. We focused the following analyses on the resistant interactomes, as we were interested in exploring their functional significance.
The differentially coexpressed, resistant subnetwork HvInt(R–S) was analyzed using GO term enrichment. Figure 4d shows the HvInt(R), differentiating nodes and edges from HvInt(R–S) in red, and HvInt(R = S) in light blue. Community analysis revealed different distributions of these subnetworks in each cluster, retaining GO enriched terms that have been associated with plant immunity including: calcium ion binding, protein ubiquitination, and MAP kinase activity for the HvInt(R) cluster 1; vesicle trafficking, protein binding and transport, SNAP receptor activity, and GTPase activity for HvInt(R) cluster 3; translation, protein binding, and photosynthesis for HvInt(R) cluster 8; ionotropic GLR signaling pathway and transporter activity for HvInt(R) cluster 13. All terms are reported in Supplementary Data 4. Finally, using hypergeometric tests, enrichment of eQTL associations was calculated for each subnetwork, taking HvInt as reference. eQTLs were split by timepoint, using genome-wide or only Mla associations, a previously reported trans eQTL hotspot (Surana et al. 2017). From all the listed tests, significant enrichment (adjusted P-values <0.001) of Mla eQTL associations was only identified in the resistant networks HvInt(R) and HvInt(R–S) at 32 HAI during haustorial development (test adjusted P-values 7.76× 10−5 and 5.56×10−13, respectively). From a total of 1,247 proteins in HvInt(R–S), 299 are associated with the Mla trans eQTL, accounting for 23.9% of the total number of nodes (1.7 times more than HvInt which contains only 14% of associations).
Interactors of MLA are conserved across orthologs and other plant NLRs
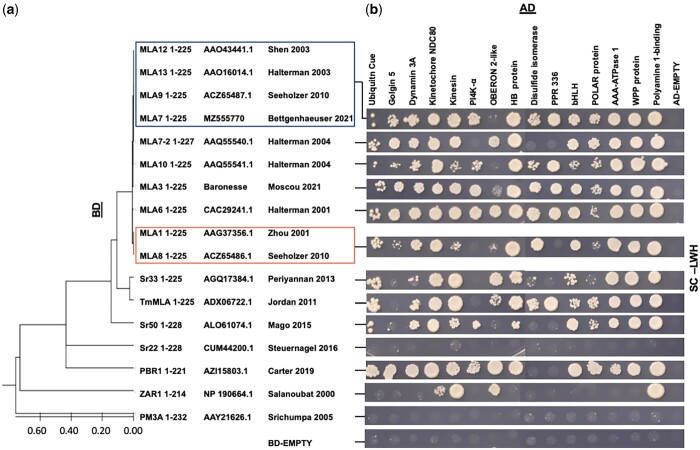
Previously, we developed the Y2H-SCORES statistical software, and as proof of concept, used a Y2H-NGIS to identify 14 new interacting proteins of the MLA6 CC and NB domains (Velásquez-Zapata et al. 2021). To continue to build HvInt resources in this study, we also confirmed a new polyamine factor 1-binding protein, predicted to mediate translocation of NLRs between the nucleus and the cytoplasm, as an additional interactor for the MLA6CC+NB domain. Then, because isoforms of the MLA immune receptor have been shown to function in a number of plant–fungal interactions (Jordan et al. 2011; Periyannan et al. 2013; Mago et al. 2015; Cesari et al. 2016; Krattinger and Keller 2016; Bettgenhaeuser et al. 2021; Sánchez-Martín and Keller 2021), we further tested these 15 MLA6CC+NB interactors against 9 additional MLA alleles, 3 MLA orthologs, as well as 4 diverse NLRs to explore the extent of signaling domain binding. Binary Y2H tests were performed with synthesized CC + NB domains that correspond to specific subclades of MLA alleles as shown in the phylogenetic tree in Fig. 5a, i.e. MLA12, MLA13, MLA9, and MLA7 (designated MLA13-type); MLA7-2, MLA10, MLA3, and MLA6; as well as MLA1 and MLA8 (MLA1-type). The MLA13-type CC + NB domain present in MLA7, MLA9, MLA12, and MLA13 differs from MLA6 by Q91K, P150T, and D177G amino-acid substitutions. MLA7-2 differs from the MLA7 variant by an EL110 insertion. MLA3 has a hybrid structure—the same as MLA13 at Q91K and P150T, but switches to MLA6 at D177. MLA10 differs from MLA13-type by a single E41D change in the CC domain, and MLA1 and MLA8 are further diverged from MLA6, with S92F, L102F, and P150T (Halterman et al. 2001, 2003; Zhou et al. 2001; Shen et al. 2003; Halterman and Wise 2004; Seeholzer et al. 2010; Bettgenhaeuser et al. 2021). Positive interactions were found for all the validated targets with MLA3, MLA6, and the MLA13-type CC + NB baits as shown in Fig. 5b and Supplementary Fig. 3, with some variation in the strength of the interactions. MLA10 had 1 negative interaction with the OBERON 2-like protein while the MLA7-2 CC + NB lost 3 interactions with PI4K-α, disulfide isomerase and PPR 336, and the MLA1-type had 2 negative confirmations with PI4K-α and PPR 336.
Fig. 5.
Interactors of MLA CC+NB alleles, orthologs and NLR outgroups. a) Phylogenetic analysis of CC+NB domains of MLA-associated alleles and NLRs using a protein alignment with MUSCLE (Edgar 2004) followed by a maximum likelihood tree estimation; identical sequences are enclosed in boxes. Evolutionary distance is shown at the bottom of the tree. b) Y2H binary confirmation of the interactors (across the top) aligned with the phylogenetic tree in (a) (top to bottom). AD designates activation domain (left); BD designates binding domain (top). Empty designates vector without insert. SC-LW media was used as control for diploid growth, and the interaction was tested using selection with SC-LWH media using 3 dilutions (100, 10−1, 10−2). The image shown was taken from the 100 dilution at 0.5 OD and photographed 7 days after plating. The full experiment with all 3 dilutions and the 3-AT specificity control is shown in Supplementary Fig. 3.
As illustrated in Fig. 5a, phylogenetic analysis of the CC + NB domains of MLA orthologs and other diverse NLRs grouped Sr33 and TmMLA in a cluster, followed by Sr50, Sr22, PBR1, ZAR1, and PM3A. Sr33 (Periyannan et al. 2013), Sr50 (Mago et al. 2015), and TmMla (Jordan et al. 2011) had several positive interactions, excepting golgin 5, and differential strength with OBERON 2-like, PI4K-α, disulfide isomerase, PPR 336, and the POLAR protein. Unexpectedly, we also identified interactions with some outgroup NLRs, including 13 with PBR1 (Carter et al. 2019) and 4 with ZAR1 (Salanoubat et al. 2000). Interestingly, the CC + NB of the wheat powdery mildew resistance protein, PM3A (Srichumpa et al. 2005), and the wheat stem rust NLR, Sr22 (Steuernagel et al. 2016) displayed no PPI with these MLA targets. Nevertheless, these results suggest that many of the identified interactors are MLA-associated and that they can be conserved across this NLR family. All bait and prey sequences are reported in Supplementary Text 1. Differences among MLA and the other NLR baits are reported in the protein alignment in Supplementary Text 2 and the full Y2H experiment is shown in Supplementary Fig. 3.
Phylogenetic and network analysis of MLA interactors predict NLR localization and signaling
To further explore signaling events triggered by MLA, we gathered all reported MLA protein interactors for further analysis. These included HSP90, SGT1, MIR1, WRKY1, WRKY2, and MYB6 (Bieri et al. 2004; Shen et al. 2007; Chang et al. 2013; Wang et al. 2016). We started by positioning the interactors in HvInt, for a total of 14 targets, including 9 validated by us using Y2H (Dreze et al. 2010), and 5 previously reported in the literature. Table 1 summarizes all previous and novel interactions for MLA and other NLR proteins, their descriptions, conservation of the interaction, as well as A. thaliana orthologs and the predicted cellular location in the TAIR database (Berardini et al. 2015). Predicted cellular localizations included nuclear, cytoplasmic, and organelle-associated proteins. Predicted functions included transcription, vesicle transport, protein folding and degradation, among others. These analyses suggest novel localizations of the MLA receptor and molecular mechanisms that regulate its function over the course of fungal infection and disease resistance.
Table 1.
MLA and NLR conserved interactors.
| Gene ID and description | Conservation of PPIs with MLA alleles and NLRs as tested in Fig. 5 or inferred from the literature | A. thaliana ortholog and cellular locationa | MLAInt clusterb |
|---|---|---|---|
| HORVU.MOREX.r2.3HG0259240 | MLACC+NB: 1-type, 3, 6, 7-2, 10, 13-type | AT4G33650 | C2 |
| Dynamin 3A | NLRCC+NB: Sr33, TmMLA, Sr50, PBR1 | Cytoplasm, plasma membrane | |
| HORVU.MOREX.r2.7HG0598540 | MLACC+NB: 1-type, 3, 6, 7-2, 10, 13-type | AT2G17990 | C2 |
| Polyamine 1-binding protein | NLRCC+NB: Sr33, TmMLA, Sr50, PBR1, ZAR1 | Cytosol, plasma membrane | |
| HORVU.MOREX.r2.3HG0192030c | MLACC+NB: 1-type (Wang et al. 2016) | AT3G54360 | C3 |
| MIR1 | Cytosol, nucleus | ||
| HORVU.MOREX.r2.3HG0226630c | MLA1LRR, MLA6Full length D502V autoactive (Bieri et al. 2004; Chapman et al. 2022) | AT4G23570 | C3 |
| SGT1 | Cytosol, nucleus | ||
| HORVU.MOREX.r2.5HG0406220c | MLA1LRR, MLA6LRR (Bieri et al. 2004) | AT5G56030 | C3 |
| HSP90 protein | Golgi, cytoplasm | ||
| HORVU.MOREX.r2.5HG0439020 | MLACC+NB: 1-type, 3, 6, 7-2, 10, 13-type | AT3G54630 | C3 |
| Kinetochore NDC80 | NLRCC+NB: Sr33, TmMLA, Sr50, PBR1, ZAR1 | Nucleus | |
| HORVU.MOREX.r2.6HG0471210c | MLACC: 1-type, 3, 6, 7-2, 10, 13-type (Shen et al. 2007) | AT1G80840 | C3 |
| WRKY1 | Nucleus | ||
| HORVU.MOREX.r2.7HG0616250c | MLACC: 1-type, 3, 6, 7-2, 10, 13-type (Shen et al. 2007) | AT4G31800 | C3 |
| WRKY2 | Nucleus | ||
| HORVU.MOREX.r2.7HG0616280 | MLACC+NB: 1-type, 3, 6, 7-2, 10, 13-type | AT4G31805 | C3 |
| POLAR protein | NLRCC+NB: Sr50, TmMLA, PBR1 | Cell periphery, cytoplasm, nucleus | |
| HORVU.MOREX.r2.7HG0618510 | MLACC+NB: 1-type, 3, 6, 7-2, 10, 13-type | AT5G32440 | C3 |
| Ubiquitin Cue protein | NLRCC+NB: Sr33, TmMLA, Sr50, PBR1 | Nucleus | |
| HORVU.MOREX.r2.5HG0370990 | MLACC+NB: 6, 7-2 | AT5G48160 | C5 |
| OBERON 2-like protein | NLRCC+NB: TmMLA, Sr50, PBR1, ZAR1 | Nucleus | |
| HORVU.MOREX.r2.1HG0038710 | MLACC+NB: 1-type, 3, 6, 7-2, 10, 13-type | AT4G39050 | C5 |
| Kinesin | NLRCC+NB: Sr33, TmMLA, Sr50, PBR1, ZAR1 | Microtubule, nucleus | |
| HORVU.MOREX.r2.3HG0255360d | MLACC+NB: 3, 6, 10, 13-type | AT3G02650 | C7 |
| Pentatricopeptide repeat 336 protein (PPR 336) | NLRCC+NB: TmMLA | Mitochondrion | |
| HORVU.MOREX.r2.1HG0071390 | MLACC+NB: 3, 6, 7-2, 10, 13-type | AT1G79830 | C8 |
| Golgin 5 | NLRCC+NB: PBR1 | Golgi, cytoplasm, nucleus | |
| HORVU.MOREX.r2.1HG0058670 | MLACC+NB: 1-type, 3, 6, 7-2, 10, 13-type | AT3G24140 | NA |
| Basic helix–loop–helix (bHLH) | NLRCC+NB: Sr33, TmMLA, Sr50, PBR1 | Nucleus | |
| HORVU.MOREX.r2.3HG0208220 | MLACC+NB: 1-type, 3, 6, 7-2, 10, 13-type | AT2G34730 | NA |
| WPP protein | NLRCC+NB: Sr33, TmMLA, Sr50, PBR1 | Mitochondrion, nucleus | |
| HORVU.MOREX.r2.4HG0338560 | MLACC+NB: 1-type, 3, 6, 7-2, 10, 13-type | Nucleus | NA |
| Homeobox (HB) protein | NLRCC+NB: Sr33, TmMLA, Sr50, PBR1 | ||
| HORVU.MOREX.r2.5HG0402280 | MLACC+NB: 1-type, 3, 6, 10, 13-type | AT1G04980 | NA |
| Disulfide isomerase | NLRCC+NB: TmMLA | Chloroplast, ER, cytosol, nucleus | |
| HORVU.MOREX.r2.5HG0404360 | MLACC+NB: 3, 6,10, 13-type | Cytosol, ER | NA |
| PI 4-kinase alpha (PI4K-α) | NLRCC+NB: Sr50, PBR1 | ||
| HORVU.MOREX.r2.7HG0598410 | MLACC+NB: 1-type, 3, 6, 7-2, 10, 13-type | AT4G27680, AT5G53540 | NA |
| AAA-ATPase 1 | NLRCC+NB: Sr33, TmMLA, Sr50, PBR1 | Plasma membrane | |
| HORVU.MOREX.r2.6HG0484160c | MLACC+NB: 1-type (Chang et al. 2013) | Nucleus | NA |
| MYB6 |
Information including gene ID, description, conservation of PPIs across tested NLRs, A. thaliana ortholog, TAIR predicted cellular localization, and MLAInt cluster number.
Predicted localization based on experimental evidence of Arabidopsis orthologs.
NA designates not assembled into the original HvInt, and thus, not present in MLAInt.
Interaction confirmed in independent investigation.
The PPR 336 prey (HORVU6Hr1G017930) is not reported in TRITEX (Monat et al. 2019). HORVU.MOREX.r2.3HG0255360 is the closest gene ID in this assembly.
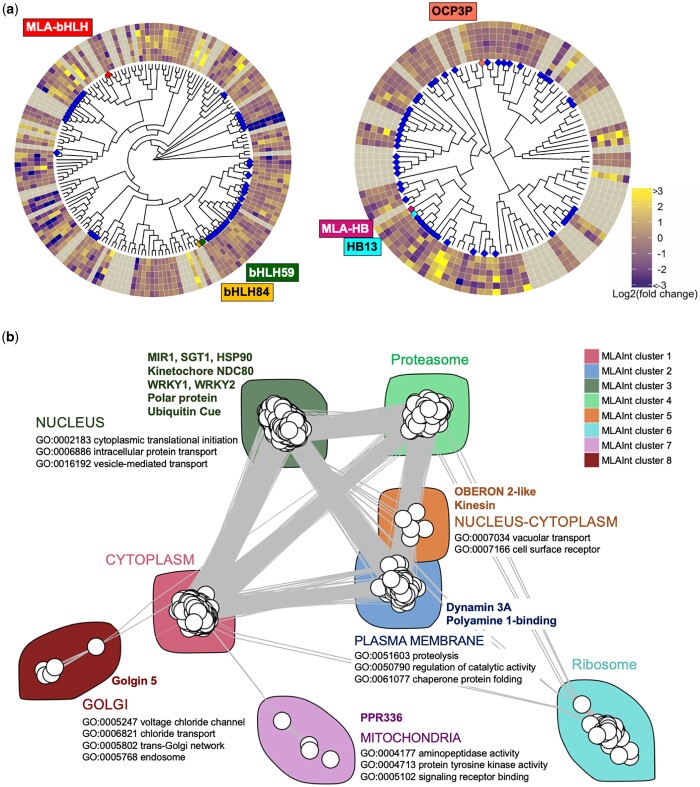
Two MLA interacting TFs, bHLH and HB, were not present in the current HvInt. Therefore, these were characterized using phylogenetics and annotation of the TF gene families (Fig. 6a). Annotation included available knowledge of a role in immunity (Coego et al. 2005; Cabello et al. 2012; Gao et al. 2014; Xu et al. 2014; Bruessow et al. 2021), the presence of a PWM (Matys et al. 2006; Weirauch et al. 2014; Jin et al. 2017), and the expression fold-change values between the wild-type CI 16151 (Mla6, Bln1, Sgt1) and susceptible m18982 (mla6, Bln1, Sgt1) mutant, covering 0–48 HAI. We found 173 members of the bHLH family in barley, as shown in Fig. 6a. The MLA-interacting bHLH interactor is divergent from other TFs characterized in defense response or possessing a PWM. The MLA-interacting bHLH appears to have a similar expression pattern in CI 16151 (Mla6, Bln1, Sgt1) and m18982 (mla6, Bln1, Sgt1) across the time course. We performed a similar analysis for the HB protein finding 109 family members. The MLA-interacting HB belongs to a previously studied clade, with a PWM reported and one related TF involved in plant resistance (Gao et al. 2014). The expression heatmap shows overexpression of the MLA-interacting HB in CI 16151 (Mla6, Bln1, Sgt1) when compared with m18982 (mla6, Bln1, Sgt1).
Fig. 6.
Phylogenetic and network analysis of MLA interactors to predict receptor localization and signaling response. a) A total of 173 and 109 TFs from the bHLH and HB families were compared, annotating the resulting trees with the log2 of the fold change expression of CI 16151 (Mla6, Bln1, Sgt1) when compared with the m18982 (mla6, Bln1, Sgt1) genotype at each timepoint [0, 16, 20, 24, 32, 48 HAI (with 48 HAI on the outside)]. Information about the availability of PWMs id marked in blue. Gray color in the heatmap indicates that the gene was not present in the expression dataset. TFs involved in plant defense are highlighted with different colors in the phylogenetic tree and with the name on the outer edge of the trees. b) Clustering of the MLA-associated interactome (MLAInt). Proteins in MLAInt were classified in 8 clusters (showed in colors) that were associated with different marked cellular localizations and functions based on GO analysis. MLA interactors are shown by the cluster where they are positioned.
We then computed an MLA-associated interactome (MLAInt), which consisted of proteins in the shortest paths between each pair of MLA interactors that do not pass-through MLA, adding the first and second neighbors to the resulting network (Supplementary Data 5). MLAInt has 1,566 nodes and 13,203 edges clustered into 8 groups, with the 14 MLA targets distributed in 5 of the MLAInt clusters (Fig. 6b and Table 1). As suggested by Table 1, MLA interactors point to different possible cellular localizations through experimental evidence of the Arabidopsis orthologs. These were further supported by the predicted function and localization of the MLAInt clusters. MLA interactors and other proteins in MLAInt were clustered in different cellular localizations, giving them functional independence within the network. GO term analysis of the MLAInt clusters highlights cellular processes associated with MLA-target signaling including vesicle-mediated transport (cluster 2), MAP kinase cascades (cluster 3), vacuolar transport, endosome, transmembrane receptor protein serine/threonine kinase activity (cluster 5), signaling and innate immune response (cluster 7), and trans-Golgi network and endosome (cluster 8).
MLAInt has significantly higher essentiality than HvInt in immunity (DiffSLC), degree centrality, betweenness and expression distance correlation, as supported by Wilcoxon rank sum tests (P-values 8.68× 10−93, 0, 2.58×10−29, 1.04×10−158, respectively). The expression distance correlation was also compared by separating the datasets by defense phenotype for each subnetwork, finding higher values for MLAInt in all cases (Wilcoxon rank sum tests P-values 2.71×10−53 for resistant genotypes and 4.7×10−33 for susceptible genotypes). Then, we measured the shortest paths between MLA and the proteins in the HvInt, HvInt(R = S), and HvInt(R–S) networks. Distances between MLA and HvInt(R–S) are significantly lower than between MLA and HvInt or HvInt(R = S) (t-test P-values 2.4×10−10 and 1.2×10−10, respectively). MLA and 4 of its interactors were found as part of HvInt(R–S) including SGT1, HSP90, WKRY2, and the ubiquitin Cue protein. Among them, SGT1 and HSP90 had the highest values of protein essentiality, degree and betweenness (within the top 20% of the values for the nodes in the network) and had significant eQTL associations with Mla. These observations link MLAInt and MLA interactors with resistant subnetworks.
Discussion
In this report, we aimed to create a comprehensive overview of the different mechanisms that govern NLR-specified immunity in the large-genome cereal, barley (Deng et al. 2020) and extend these to species and fungal interactions within the Triticeae. We used interolog inference to develop a predicted interactome (HvInt), which was integrated with context-specific datasets to model the immune response of barley to the powdery mildew pathogen. There are a few caveats associated with this analysis, however. First, there were much greater numbers of experimentally validated PPIs in the model organisms, Arabidopsis and yeast; this resulted in a bias toward these species for the number of interactions in HvInt. Secondly, there was a possibility of ancient genome duplications present in the lineages used (Paterson et al. 2004); this could lead to recovery of both orthologs and their diversified copies, although we selected the top candidates based on protein similarity and gene trees (Sonnhammer and Östlund 2015; Howe et al. 2020). Nevertheless, conservation of protein interactions across plant species has been shown by McWhite et al. (2020), a principle that we could leverage to move forward with the subsequent analyses of HvInt. To start, we measured general network properties of HvInt and compared it with the Arabidopsis experimental interactome (AtInt), which we obtained by collecting experimentally validated interactions in this organism (Supplementary Data 1). We found they have a power-law distribution (scale-free) and are small-world networks. These properties are important as they account for robustness of the interactome, buffering perturbation effects and optimizing information transfer (Watts and Strogatz 1998; Albert et al. 1999). We extended these principles for the subsequent analyses that we performed using the HvInt and subnetworks, integrating them with barley defense-associated datasets, including results from eQTL, immune signaling mutants, infection–time–course transcriptome, and Y2H PPI interactions.
Integration of eQTL and interactome data predicts DMs during Bgh penetration and haustorial development
Disease-modules (Barabási et al. 2011) were used to characterize immune signaling at 2 key infection stages of barley by Bgh. Using GO terms and DE enrichment, we separated predicted module functions into core and unique responses. Among unique DM DE genes at Bgh penetration (16 HAI) we found WRKY2, a previously reported interactor of MLA. WRKY2 has been shown to regulate the expression of genes involved in PAMP-triggered defense (Shen et al. 2007) and to be associated with race-nonspecific resistance mediated by MLO (Spies et al. 2012). The identification of WRKY2 validated our computational approach, which we then went forward to identify additional disease interactions. Other responses were associated with the regulation of reactive oxygen species (ROS), including mitochondrial import inner membrane translocase subunit Tim16/Pam16, respiratory burst oxidase homolog (RBOH), and the N-carbamoyl-putrescine amidase. Transitioning to Bgh haustorial development (32 HAI), we found chalcone synthase, a gene involved in phytoalexin metabolism, to be DM DE. Chitinases, which we also found overexpressed at this timepoint, are secreted by the host cell to degrade fungal walls and elicit PAMP-triggered immunity (PTI) through chitin recognition (Pusztahelyi 2018). Accumulation of chitinases has also been reported in the extra-haustorial complex during Bgh infection (Lambertucci et al. 2019). Similarly, other protein families that were also reported in the extra-haustorial complex were also found as DM DE at 32 HAI including genes associated with the TCA cycle and pyruvate metabolism. Remarkably, among this group there is also a malate dehydrogenase (HORVU.MOREX.r2.1HG0067490) that was characterized as target of the Bgh effector BEC1054/CSEP0064 (Pennington et al. 2016).
We postulate that common GO terms at both stages of fungal development represent core responses that the host cell maintains during immunity, and these may be driven by the same or different genes. Often these core responses had annotations directly related to plant defense, including vacuolar transport, lignification, and GLR signaling. DM DE genes associated with vacuolar transport included KEULE and charged multivesicular body protein 5 (MVB5). Previous studies have found that the KEULE interacts with the SYP121 SNARE protein (ROR2), which confers penetration resistance of barley to Bgh and control the formation of the SNARE complex (Heese et al. 2001; Collins et al. 2003). MVBs have also been found to be transported to Bgh penetration sites and contribute to callose deposition (Böhlenius et al. 2010). Cinnamyl alcohol dehydrogenase was also found as DM DE in both timepoints, an enzyme that is involved in the lignification of the cell wall and in the activation of the salicylic acid (SA) defense pathway (Stadnik and Buchenauer 2000). Lastly, we identified a GLR involved in potassium transport to be DM DE at both stages. This family of proteins has been found to act as sensors in plant resistance (Forde and Roberts 2014). The glr3.3 knock out mutant in Arabidopsis has shown increased susceptibility to obligate biotrophs such as Hyaloperonospora arabidopsidis and modulation of ROS and nitric oxide production (Manzoor et al. 2013), while the triple mutant glr2.7 2.8 2.9 is more susceptible to bacterial infection, dampening PTI (Bjornson et al. 2021). The above examples illustrate the power of the DM concept for targeted mining of interactomes, yet many more DM DE remain to be characterized during disease development (Supplementary Data 3).
Resistant coexpressed interactomes are significantly associated with Mla eQTL
We used coexpression data to identify PPI subnetworks associated with disease phenotypes (Dong et al. 2015; Jiang et al. 2016; Mishra et al. 2017). Using distance correlation, we measured the nonmonotonic behavior of our expression dataset (Székely et al. 2007) to obtain the resistant HvInt(R), susceptible HvInt(S) and the differentially coexpressed interactomes HvInt(R–S) and HvInt(S–R). The biological significance of these subnetworks lays on the assumption that coexpressed proteins are part of the same or overlapping pathways and coordinated by a common set of transcriptional regulators. At the physical level, the interactions in these networks may be part of the same protein complexes and then, high coexpression values increase the confidence of the predicted interactions. Our analyses indicate that the differentially coexpressed subnetworks, HvInt(R–S) and HvInt(S–R), have properties that predict their importance and role in defining the barley response to Bgh. For example, higher essentiality (DiffSLC), degree centrality and betweenness, and lower distance correlation, provide more robustness in signaling and carry more information (Koschützki and Schreiber 2008; Mistry et al. 2017). Lower distance correlation values indicate a higher variability in the gene expression of the nodes, which is consistent with a differential response during defense between resistant and susceptible genotypes. Yet, despite the topological similarities between HvInt(R–S) and HvInt(S–R), only the resistant networks showed significant enrichment of eQTL associations with Mla.
GO enrichment of HvInt(R–S) indicates that this network is associated with multiple biological processes in plant defense, such as MAP kinase activity (Cui et al. 2019). Other GO terms including protein binding, folding, and ubiquitination were also enriched. The chaperone DnaK (Hsp70) and calreticulin were associated with these terms. Chaperones stabilize protein complexes and interactions. One Hsp70 isoform has already been shown to participate in the response to salt, drought and heavy metal stress in barley, while another isoform is associated with response to fungal attack (Landi et al. 2019). Calreticulin is involved in calcium signaling and folding of glycoproteins, proteins from this family have been involved in the defense response to biotrophic pathogens, and to the stabilization of the EFR NLR in Arabidopsis (Qiu et al. 2012). Lastly, protein degradation via ubiquitination/proteasome 26S controls the dynamics of cellular functions such as hormone signaling, transcription, and NLR-triggered immune response, including accumulation of MLA (Wang et al. 2016).
The enrichment of Mla eQTL associations in HvInt(R–S) indicates that the expression of these genes is controlled by signaling and/or transcriptional cascades in which Mla participates. Support for this hypothesis can be found when we consider that HvInt(R–S) was built from RNA-Seq data with active Mla6 transcript accumulation [CI 16151 (Mla6, Bln1, Sgt1) and m19089 (Mla6, bln1, Sgt1)]. HvInt(S–R), in contrast, was generated from mutants with inactive or depleted Mla6-associated expression [m18982 (mla6, Bln1, Sgt1), m19028 (mla6, bln1, Sgt1), m11526 (Mla6, Bln1, Sgt1ΔKL308-309)] and had no enrichment of Mla eQTL associations. Further characterization of the predicted barley interactome can help to explain the association between MLA and proteins that regulate the expression of these genes. Incompleteness of the HvInt network limits this approach, as no interologs were found for most of the TFs that interact physically with MLA. However, as genome resources increase (Sánchez-Martín and Keller 2021), these gaps will continue to fill in. Inference of gene regulatory networks can then be used to model the regulation mechanisms associated with MLA and to complement the analyses that we performed using PPI networks.
Conservation of MLA interactors with other NLRs provides clues as to their signaling specificity
Based on the functional evidence that links MLACC+NB self-association and activation of cell death, the reported interactors represent an important part of the MLA-immune signaling response. Previous studies have shown that MLA10CC+NB self-interacts in yeast while shorter CC domains failed to reconstruct the associations, suggesting that MLA10CC+NB has a higher stability (Maekawa et al. 2011). In addition, self-association of the CC domain is essential for immune signaling of the Mla orthologs Sr33 and Sr50 in wheat and rye, respectively (Casey et al. 2016) while triggering cell death in wheat (Cesari et al. 2016). At the structural level, available data shows different polymerization states across MLA and its orthologs. MLA10CC appeared as a dimer in its crystal structure while Sr33CC appeared as a tetramer (Maekawa et al. 2011; Casey et al. 2016). Higher order polymerization states have been reported for other NLRs, such as the pentamer obtained from the full length ZAR1 protein (Wang et al. 2019). It also has been shown that a variety of CC-NLRs use their N-terminal domains to transactivate other receptors (Wróblewski et al. 2018). Our interaction data suggest that the functionality of the CC domain may be linked to the NB domain, therefore it is possible that the polymerization of MLA reaches higher levels if a longer sequence of the protein is considered. Many studies have shown that the N-terminal region of NLRs is involved in triggering cell death (Casey et al. 2016; Cesari et al. 2016; Wróblewski et al. 2018). The MLA10CC domain activates cell death in N. benthamiana, and mutations of this domain impair the response (Maekawa et al. 2011).
In this study, we used phylogenetics of the MLA6CC+NB domain, in concert with the 15 interactors, as a preface to determining MLA signaling networks. Among MLA alleles, 4 of the 15 preys had differential negative/weak interactions—these help to determine specificity of the MLA isoforms based on their polymorphisms. Taking MLA6 as reference, we found that interactions with PI4K-α and PPR 336 appear to be dependent on mutations in the CC domain, for example, the EL110 insertion of MLA7-2 or the S92F and L102F mutations in the MLA1-type group. The MLA1-type group also contains MLA8, an allele that possesses dual specificity with Rps7 for recognition to wheat stripe rust (Bettgenhaeuser et al. 2021). We observed that the interaction between MLA and disulfide isomerase is disrupted by the EL110 insertion; also, interactions between MLA and the OBERON 2-like protein were absent or very weak in alleles that contained the P150T mutation positioned between the CC and the NB domains of MLA.
The next step was to investigate the more distantly related CC + NB in wheat orthologs TmMLA (Jordan et al. 2011) (powdery mildew resistance); wheat Sr33 (Periyannan et al. 2013) and rye Sr50 (Mago et al. 2015) (Ug99 stem rust resistance). Here, as expected, we identified additional negative/weak interactions, notably the golgin 5 and POLAR proteins. Because the number of polymorphisms between MLA and these proteins are higher, we did not have resolution at the amino acid level to determine what mutations triggered these differential interactions. Despite this, superposition of the structures of Sr33 and MLA (Casey et al. 2016) led us to conclude that the interaction with golgin 5 may be dependent of the amino acids that comprise the alpha 1 or alpha 4 domains of these NLRs. Lastly, we encountered interactions with 2 NLRs from the outgroup; barley PBR1 (AvrPphB Response 1) and Arabidopsis ZAR1. Protease activity mediated by AvrPphB from Pseudomonas syringae pv. phaseolicola cleaves barley PBS1, thereby activating HvPBR1, similar to Arabidopsis RPS5 (Carter et al. 2019). ZAR1 regulates resistance to P. syringae in Arabidopsis through the recognition of the HopZ1a effector (Lewis et al. 2010). The high overlap of interactions with HvPBR1 points to similarity of its structure with MLA, despite their differences at the amino acid level. Positive interactions with ZAR1, which were also conserved across all the NLRs that had positive interactions, may provide clues on conservation of NLR signaling across different species.
MLA interactors confer information about NLR functionalities and cellular localizations
Using ortholog-aided association to its protein targets, we obtained a predictive model of MLA cellular localization during Bgh infection. Studies on MLA10 showed that cell death is triggered while the protein is localized to the cytosol, while disease resistance requires nuclear localization (Shen et al. 2007; Bai et al. 2012; Cesari 2018). Our model indicates that one likely localization of MLA during the infection time course is the nucleus. This result is supported by previous evidence that points to an early nuclear localization of MLA during Bgh infection (Shen et al. 2007). Our model suggests that MLA could localize to the cytosol and possibly to other compartments as well, including the plasma membrane, the Golgi apparatus, the endoplasmic reticulum (ER), and mitochondria. Positive interactions for Sr33 and Sr50 suggest that these receptors may localize to the nucleus, the cytosol, and the plasma membranes while TmMLA and PBR1 may also localize to the mitochondria and Golgi, respectively (TmMLA interacts strongly with PPR 336, and PBR1 with golgin 5).
Predicted localizations were also supported by the MLAInt clusters; groups of proteins within the clusters were associated with different organelles or functional cellular substructures. For example, as illustrated in Fig. 6b, MLAInt cluster 3 has the highest number of MLA interactors with 8 proteins predicted to be localized to the nucleus, including previously reported TFs WRKY1/2 and MYB6 (Shen et al. 2007; Chang et al. 2013). MLAInt cluster 5, the second group that contained nuclear MLA interactors, contained the OBERON and kinesin proteins. Functional annotations of this cluster indicate these proteins may be involved in pathways different than the other nuclear MLA interactors, with enrichment in vesicle transport and cell surface receptor signaling. According to our predictions, these 2 MLA targets share a series of receptor-like kinase interactors, important drivers of defense response. Here, we linked these proteins with NLR and effector signaling in Arabidopsis by showing their interactions with ZAR1. More experimental evidence points to this conclusion as OBERON and kinesin are hubs targeted by pathogen effectors from P. syringae and H. arabidopsidis (Mukhtar et al. 2011).
Two MLA TF interactors with nuclear localization were characterized using phylogenetic analysis, as they did not have interactors reported in HvInt. The MLA-interacting bHLH occupies an unexplored clade, separated from the orthologs of bHLH84, a TF that enhances autoimmunity of the NLR mutant snc1 (Xu et al. 2014), and from bHLH059, a temperature–responsive SA immunity regulator in in Arabidopsis. The MLA-interacting HB protein was found to be closely related to the barley ortholog of AtHB13, a TF involved in tolerance to cold stress (Cabello et al. 2012) and further showed to confer resistance to powdery mildew, downy mildew, and green peach aphid in Arabidopsis (Gao et al. 2014). The clade that contains the MLA-interacting HB and the barley ortholog of AtHB13 is separated from OCP3, another HB protein that confers resistance to necrotrophic pathogens in Arabidopsis, suggesting that the MLA-interacting HB may have specificity in the response to biotrophs.
Lastly, we report a conserved MLA interactor, a polyamine-modulated factor 1-binding protein, that may mediate the translocation of the MLA and other NLRs between the nucleus and the cytoplasm. The Arabidopsis ortholog of this protein, AT2G17990, is a calcium-dependent kinase adaptor protein involved in vacuolar biogenesis and trafficking (Kwon et al. 2018). This protein has 1 interactor reported in HvInt, the nuclear pore complex protein MOS7/NUP88 (Park et al. 2014), which is a constituent of a nuclear pore involved in the translocation and nuclear concentration of the R-protein SNC1, and the immune regulators EDS1 and NPR1 (Wiermer et al. 2010). This link between MLA, TmMLA, Sr33, Sr50, PBR1, ZAR1, and MOS7/NUP88 may indicate this nuclear pore mediates the translocation of these receptors to the nucleus.
Presently, only the nucleus and the cytosol are reported cellular localizations of the MLA immune receptor (Shen et al. 2007). Our localization model points to other structures as well, although these are based on evidence from other species, and thus, need further study. First, we found a group of targets that shared nuclear and other cellular localizations such as the cell periphery and the ER. The plasma membrane, as a novel NLR cellular localization, helps to explain cell death by triggering changes of electrochemical potential (Dangl and Jones 2019; Van-Wersch et al. 2020). The POLAR protein in Arabidopsis relocates the BIN2 protein and other GSK3-like kinases from the nucleus to the plasma membrane regulating their activity and attenuating MAPK signaling (Houbaert et al. 2018). This mechanism may also mediate the translocation of MLA to the plasma membrane, considering that the NLR ZAR1 is positioned to this location to activate the immune response (Wang et al. 2019). The MLA interactor dynamin 3A, also predicted to localize to the plasma membrane, belongs to a protein family that in Arabidopsis controls cell death after powdery mildew infection (Tang et al. 2006). Another MLA target, the AAA-ATPase (not present in HvInt), was also predicted to be localized to the plasma membrane. This protein belongs to a group recently found to interact with NLRs and associated with triggering the HR by mediating electrochemical gradients (Lee et al. 2022).
The ER was also found to be associated with MLA-signaling since the interactor disulfide isomerase (PDI) is predicted to be localized to this compartment sometime during the infection cycle. PDI is an enzyme that catalyzes protein disulfide bonds, helping to the correct folding and aggregation of proteins at this compartment (Ray et al. 2003). PDIs also have a role in the response to abiotic and biotic stresses (Kayum et al. 2017). Finally, we found 2 MLA interactors with unique predicted cellular localizations, different than those already discussed: the pentatricopeptide repeat protein, assigned to MLAInt cluster 7, localized to the mitochondrion, and golgin 5 (MLAInt cluster 8), localized to the Golgi apparatus. Fluorescent expression of MLA10 indicates a protein distribution similar to the morphology of the Golgi apparatus (Shen et al. 2007); golgin 5, localized to this compartment, is involved in vesicle tethering and regulation of intraorganelle transport by interacting with the small GTPase Rab6 (Latijnhouwers et al. 2007; Muschalik and Munro 2018).
Our predictive model for MLA signaling suggests that there is a transcriptional network mediated by MLA, which is likely triggered at early timepoints of infection (Moscou et al. 2011). This response leads to the accumulation of MLA itself at about 16 HAI (at Bgh penetration). This model also suggests that the immune receptor has functions in the cytoplasm and other cellular compartments, where it contributes to cell death. Further questions remain in this model, including where and when the recognition of effectors occurs and its influence in the translocation of MLA to different cellular compartments. Topological properties of MLAInt indicate a cohesive response of the subnetwork in defense signaling. In addition, distances between MLA and the proteins in HvInt(R–S) were shorter than for any other subnetwork, and enrichment of Mla eQTL associations was only found with resistant coexpressed subnetworks. This could indicate a potential involvement of the resistance protein in the signaling and transcriptional regulation of the proteins in HvInt(R–S). These results support a link among Mla trans eQTLs, MLA signaling and gene coexpression during resistance to Bgh.
Data availability
Strains and plasmids are available upon request. Supplemental files available at figshare: https://doi.org/10.25386/genetics.19089992. Supplementary Data 1 contain the predicted barley interactome (HvInt) and experimentally validated Arabidopsis interactome (AtInt). Supplementary Data 2 contain node properties of HvInt and enrichment of essential proteins across clusters. Supplementary Data 3 contain DMs and DM DE genes at Bgh penetration and haustorial development. Supplementary Data 4 contain resistant and susceptible interactomes. Supplementary Data 5 contain the MLA interactome (MLAInt). Supplementary Figure 1 contains core and unique DM GO terms associated with DE genes by timepoint. Supplementary Figure 2 contains complete time–course expression patterns DM DE genes by type of response. Supplementary Figure 3 contains complete Y2H validation of interactions for the CC + NB domains of Mla alleles, orthologs and outgroup NLRs. Supplementary Text 1 contains bait and prey sequences used for Y2H binary validation among Mla alleles, orthologs, and outgroup NLRs. Supplementary Text 2 contains the protein alignment of the Mla alleles, orthologs, and outgroup used for Y2H tests. Code used to support the main findings of this manuscript are at the GitHub page https://github.com/Wiselab2/Barley_Interactome [accessed 15 April 2022]. MIAME-compliant Barley1 GeneChip profiling data (Affymetrix part number 900515) from the Q21861 × SM89010 eQTL analysis are available as accession GSE68963 at NCBI’s Gene Expression Omnibus (GEO). Conversion of the GeneChip Probe sets to gene IDs was done using Ensemble and Biomart. Infection–time–course RNA-Seq datasets are available in NCBI-GEO under accession number GSE101304 (https://www.ncbi.nlm.nih.gov/gds/?term=GSE101304). R code and the ReadMe file for the NGPINT and Y2H-SCORES software used to identify MLA interactors are provided at the GitHub page https://github.com/Wiselab2/ [accessed 15 April 2022]. Raw Y2H-Seq reads are at NCBI-GEO under the accession number GSE164815 (MLA6CC+NB).
Su pplemental material is available at figshare DOI: https://doi.org/10.25386/genetics.19089992
Acknowledgments
The authors thank Sagnik Banerjee for his invaluable assistance mapping of the RNA-Seq raw reads to the TRITEX genome, and Ana Mía Corujo Ramirez, Morgan Bixby, Jessica Faust, and Stephanie Schuler for technical assistance with the Y2H-NGIS screens. They thank Matt Moscou for sharing sequences corresponding to the CC+NB of MLA3 and MLA7, and Peter Dodds and Sam Periyannan for the CC+NB of Sr33 and Sr50.
VVZ, JME, and RPW designed the research; VVZ, JME, and GF performed the research; VVZ contributed in new analytic/computational tools, analyzed data, and wrote the manuscript; VVZ, GF, and RPW edited the manuscript.
Funding
Research supported in part by Fulbright—Minciencias 2015 & Schlumberger Faculty for the Future fellowships to VVZ, USDA-Agricultural Research Service Postdoctoral Research Associateship and USDA-National Institute of Food and Agriculture-Education and Literacy Initiative Postdoctoral Fellowship 2017-67012-26086 to JME, and National Science Foundation—Plant Genome Research Program grant 13-39348, USDA-National Institute of Food and Agriculture grant 2020-67013-31184 and USDA-Agricultural Research Service project 3625-21000-067-00D to RPW. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the USDA, National Institute of Food and Agriculture, Agricultural Research Service, or the National Science Foundation. USDA is an equal opportunity provider and employer.
Conflicts of interest
None declared.
Contributor Information
Valeria Velásquez-Zapata, Program in Bioinformatics & Computational Biology, Iowa State University, Ames, IA 50011, USA; Department of Plant Pathology & Microbiology, Iowa State University, Ames, IA 50011, USA.
James Mitch Elmore, Department of Plant Pathology & Microbiology, Iowa State University, Ames, IA 50011, USA; Corn Insects and Crop Genetics Research, USDA-Agricultural Research Service, Ames, IA 50011, USA.
Gregory Fuerst, Department of Plant Pathology & Microbiology, Iowa State University, Ames, IA 50011, USA; Corn Insects and Crop Genetics Research, USDA-Agricultural Research Service, Ames, IA 50011, USA.
Roger P Wise, Program in Bioinformatics & Computational Biology, Iowa State University, Ames, IA 50011, USA; Department of Plant Pathology & Microbiology, Iowa State University, Ames, IA 50011, USA; Corn Insects and Crop Genetics Research, USDA-Agricultural Research Service, Ames, IA 50011, USA.
Literature cited
- Albert R, Jeong H, Barabási AL. Diameter of the world-wide web. Nature. 1999;401(6749):130–131. doi: 10.1038/4360. [DOI] [Google Scholar]
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106.doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baggs E, Dagdas G, Krasileva KV. NLR diversity, helpers and integrated domains: making sense of the NLR identity. Curr Opin Plant Biol. 2017;38:59–67. doi: 10.1016/j.pbi.2017.04.012. [DOI] [PubMed] [Google Scholar]
- Bai S, Liu J, Chang C, Zhang L, Maekawa T, Wang Q, Xiao W, Liu Y, Chai J, Takken FLW, et al. Structure-function analysis of barley NLR immune receptor MLA10 reveals its cell compartment specific activity in cell death and disease resistance. PLoS Pathog. 2012;8(6):e1002752. doi: 10.1371/journal.ppat.1002752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S, Velásquez-Zapata V, Elmore JM, Fuerst G, Wise RP. NGPINT: a next-generation protein–protein interaction software. Brief Bioinform. 2021;22(4):bbaa351. doi: 10.1093/bib/bbaa351. [DOI] [PubMed] [Google Scholar]
- Barabási AL, Gulbahce N, Loscalzo J. Network medicine: a network-based approach to human disease. Nat Rev Genet. 2011;12(1):56–68. doi: 10.1038/nrg2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barragan AC, Weigel D. Plant NLR diversity: the known unknowns of pan-NLRomes. Plant Cell. 2021;33(4):814–831. doi: 10.1093/plcell/koaa002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer S, Yu D, Lawson AW, Saur IML, Frantzeskakis L, Kracher B, Logemann E, Chai J, Maekawa T, Schulze-Lefert P, et al. The leucine-rich repeats in allelic barley MLA immune receptors define specificity towards sequence-unrelated powdery mildew avirulence effectors with a predicted common RNase-like fold. PLoS Pathog. 2021;17(2):e1009223. doi: 10.1371/journal.ppat.1009223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995;57(1):289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- Bentham AR, De la Concepcion JC, Mukhi N, Zdrzałek R, Draeger M, Gorenkin D, Hughes RK, Banfield MJ. A molecular roadmap to the plant immune system. J Biol Chem. 2020;295(44):14916–14935. doi: 10.1074/jbc.REV120.010852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berardini TZ, Reiser L, Li D, Mezheritsky Y, Muller R, Strait E, Huala E. The Arabidopsis information resource: making and mining the “gold standard” annotated reference plant genome. Genesis. 2015;53(8):474–485. doi: 10.1002/dvg.22877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettgenhaeuser J, Hernández-Pinzón I, Dawson AM, Gardiner M, Green P, Taylor J, Smoker M, Ferguson JN, Emmrich P, Hubbard A, et al. The barley immune receptor Mla recognizes multiple pathogens and contributes to host range. Nat Commun. 2021;12(1):6915. doi: 10.1038/s41467-021-27288-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieri S, Mauch S, Shen Q-H, Peart J, Devoto A, Casais C, Ceron F, Schulze S, Steinbiss H-H, Shirasu K, et al. RAR1 positively controls steady state levels of barley MLA resistance proteins and enables sufficient MLA6 accumulation for effective resistance. Plant Cell. 2004;16(12):3480–3495. doi: 10.1105/tpc.104.026682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjornson M, Pimprikar P, Nürnberger T, Zipfel C. The transcriptional landscape of Arabidopsis thaliana pattern-triggered immunity. Nat Plants. 2021;7(5):579–586. doi: 10.1101/2020.11.30.404566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhlenius H, Mørch SM, Godfrey D, Nielsen ME, Thordal-Christensen H. The multivesicular body-localized GTPase ARFA1b/1c is important for callose deposition and ROR2 syntaxin-dependent preinvasive basal defense in barley. Plant Cell. 2010;22(11):3831–3844. doi: 10.1105/tpc.110.078063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun P, Aubourg S, Van Leene J, De Jaeger G, Lurin C. Plant protein interactomes. Annu Rev Plant Biol. 2013;64(1):161–187. doi: 10.1146/annurev-arplant-050312-120140. [DOI] [PubMed] [Google Scholar]
- Bray Speth E, Lee YN, He SY. Pathogen virulence factors as molecular probes of basic plant cellular functions. Curr Opin Plant Biol. 2007;10(6):580–586. doi: 10.1016/j.pbi.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruessow F, Bautor J, Hoffmann G, Yildiz I, Zeier J, Parker JE. Natural variation in temperature-modulated immunity uncovers transcription factor bHLH059 as a thermoresponsive regulator in Arabidopsis thaliana. PLoS Genet. 2021;17(1):e1009290.doi: 10.1371/journal.pgen.1009290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabello JV, Arce AL, Chan RL. The homologous HD-Zip I transcription factors HaHB1 and AtHB13 confer cold tolerance via the induction of pathogenesis-related and glucanase proteins. Plant J. 2012;69(1):141–153. doi: 10.1111/j.1365-313X.2011.04778.x. [DOI] [PubMed] [Google Scholar]
- Caldo RA, Nettleton D, Peng J, Wise RP. Stage-specific suppression of basal defense discriminates barley plants containing fast- and delayed-acting Mla powdery mildew resistance alleles. Mol Plant Microbe Interact. 2006;19(9):939–947. doi: 10.1094/MPMI-19-0939. [DOI] [PubMed] [Google Scholar]
- Caldo RA, Nettleton D, Wise RP. Interaction-dependent gene expression in Mla-specified response to barley powdery mildew. Plant Cell. 2004;16(9):2514–2528. doi: 10.1105/tpc.104.023382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter ME, Helm M, Chapman AVE, Wan E, Restrepo Sierra AM, Innes RW, Bogdanove AJ, Wise RP. Convergent evolution of effector protease recognition by Arabidopsis and barley. Mol Plant Microbe Interact. 2019;32(5):550–565. doi: 10.1094/MPMI-07-18-0202-FI. [DOI] [PubMed] [Google Scholar]
- Casey LW, Lavrencic P, Bentham AR, Cesari S, Ericsson DJ, Croll T, Turk D, Anderson PA, Mark AE, Dodds PN, et al. The CC domain structure from the wheat stem rust resistance protein Sr33 challenges paradigms for dimerization in plant NLR proteins. Proc Natl Acad Sci U S A. 2016;113(45):12856–12861. doi: 10.1073/pnas.1609922113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesari S. Multiple strategies for pathogen perception by plant immune receptors. New Phytol. 2018;219(1):17–24. doi: 10.1111/nph.14877. [DOI] [PubMed] [Google Scholar]
- Cesari S, Moore J, Chen C, Webb D, Periyannan S, Mago R, Bernoux M, Lagudah ES, Dodds PN. Cytosolic activation of cell death and stem rust resistance by cereal MLA-family CC-NLR proteins. Proc Natl Acad Sci U S A. 2016;113(36):10204–10209. doi: 10.1073/pnas.1605483113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Yu D, Jiao J, Jing S, Schulze-Lefert P, Shen Q-H. Barley MLA immune receptors directly interfere with antagonistically acting transcription factors to initiate disease resistance signaling. Plant Cell. 2013;25(3):1158–1173. doi: 10.1105/tpc.113.109942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman A, Elmore JM, McReynolds M, Walley J, Wise RP. SGT1-specific domain mutations impair interactions with the barley MLA6 immune receptor in association with loss of NLR protein. Mol Plant Microbe Interact. 2022;35(3):274–289. doi: 10.1094/MPMI-08-21-0217-R. [DOI] [PubMed] [Google Scholar]
- Chapman AVE, Hunt M, Surana P, Velásquez-Zapata V, Xu W, Fuerst G, Wise RP. Disruption of barley immunity to powdery mildew by an in-frame Lys-Leu deletion in the essential protein SGT1. Genetics. 2021;217(2):iyaa026. doi: 10.1093/genetics/iyaa026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coego A, Ramirez V, Gil MJ, Flors V, Mauch-Mani B, Vera P. An Arabidopsis homeodomain transcription factor, OVEREXPRESSOR OF CATIONIC PEROXIDASE 3, mediates resistance to infection by necrotrophic pathogens. Plant Cell. 2005;17(7):2123–2137. doi: 10.1105/tpc.105.032375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins NC, Thordal-Christensen H, Lipka V, Bau S, Kombrink E, Qiu J-L, Hückelhoven R, Stein M, Freialdenhoven A, Somerville SC, et al. SNARE-protein-mediated disease resistance at the plant cell wall. Lett to Nat. 2003;425(6961):973–977. doi: 10.1038/nature02076. [DOI] [PubMed] [Google Scholar]
- Csardi G, Nepusz T. The igraph software package for complex network research. InterJournal. Complex Sytems. 2006;1695.https://igraph.org. [Google Scholar]
- Cui L, Yang G, Yan J, Pan Y, Nie X. Genome-wide identification, expression profiles and regulatory network of MAPK cascade gene family in barley. BMC Genomics. 2019;20(1):750.doi:10.1186/s12864-019–6144-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl JL, Jones JDG. A pentangular plant inflammasome. Science. 2019;364(6435):31–32. doi: 10.1126/science.aax0174. [DOI] [PubMed] [Google Scholar]
- Dean R, Van Kan JAL, Pretorius ZA, Hammond-Kosack KE, Di Pietro A, Spanu PD, Rudd JJ, Dickman M, Kahmann R, Ellis J, et al. The top 10 fungal pathogens in molecular plant pathology. Mol Plant Pathol. 2012;13(4):414–430. doi: 10.1111/j.1364-3703.2011.00783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Ning Y, Yang D-L, Zhai K, Wang G-L, He Z. Molecular basis of disease resistance and perspectives on breeding strategies for resistance improvement in crops. Mol Plant. 2020;13(10):1402–1419. doi: 10.1016/j.molp.2020.09.018. [DOI] [PubMed] [Google Scholar]
- Dong L, Liu H, Zhang J, Yang S, Kong G, Chu JSC, Chen N, Wang D. Single-molecule real-time transcript sequencing facilitates common wheat genome annotation and grain transcriptome research. BMC Genomics. 2015;16:1039.doi: 10.1186/s12864-015-2257-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreze M, Monachello D, Lurin C, Cusick ME, Hill DE, Vidal M, Braun P. High-Quality Binary Interactome Mapping, 2nd edn. New York: Elsevier Inc.; 2010. [DOI] [PubMed] [Google Scholar]
- Dwivedi SK, Tjärnberg A, Tegnér J, Gustafsson M. Deriving disease modules from the compressed transcriptional space embedded in a deep autoencoder. Nat Commun. 2020;11(1):856.doi: 10.1038/s41467-020-14666-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forde BG, Roberts MR. Glutamate receptor-like channels in plants: a role as amino acid sensors in plant defence? F1000Prime Rep. 2014;6:37.doi: 10.12703/P6-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frantzeskakis L, Kracher B, Kusch S, Yoshikawa-Maekawa M, Bauer S, Pedersen C, Spanu PD, Maekawa T, Schulze-Lefert P, Panstruga R, et al. Signatures of host specialization and a recent transposable element burst in the dynamic one-speed genome of the fungal barley powdery mildew pathogen. BMC Genomics. 2018;19(1):381. doi: 10.1186/s12864-018-4750-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao D, Appiano M, Huibers RP, Chen X, Loonen AEHM, Visser RGF, Wolters A-MA, Bai Y. Activation tagging of ATHB13 in Arabidopsis thaliana confers broad-spectrum disease resistance. Plant Mol Biol. 2014;86(6):641–653. doi: 10.1007/s11103-014-0253-2. [DOI] [PubMed] [Google Scholar]
- Geisler-Lee J, O'Toole N, Ammar R, Provart NJ, Millar AH, Geisler M. A predicted interactome for Arabidopsis. Plant Physiol. 2007;145(2):317–329. doi: 10.1104/pp.107.103465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H, Zhu P, Jiao Y, Meng Y, Chen M. PRIN: a predicted rice interactome network. BMC Bioinformatics. 2011;12:161.doi:10.1186/1471–2105-12–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustavsen JA, Pai S, Isserlin R, Demchak B, Pico AR. Rcy3: network biology using cytoscape from within R. F1000Res. 2019;8:1774.doi: 10.12688/f1000research.20887.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halterman D, Zhou F, Wei F, Wise RP, Schulze-Lefert P. The MLA6 coiled-coil, NBS-LRR protein confers AvrMla6-dependent resistance specificity to Blumeria graminis f. sp. hordei in barley and wheat. Plant J. 2001;25(3):335–348. doi:10.1046/j.1365-313X.2001.00982.x. [DOI] [PubMed] [Google Scholar]
- Halterman DA, Wei F, Wise RP. Powdery mildew-induced Mla mRNAs are alternatively spliced and contain multiple upstream open reading frames. Plant Physiol. 2003;131(2):558–567. doi:10.1104/pp. 014407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halterman DA, Wise RP. A single-amino acid substitution in the sixth leucine-rich repeat of barley MLA6 and MLA13 alleviates dependence on RAR1 for disease resistance signaling. Plant J. 2004;38(2):215–226. doi: 10.1111/j.1365-313X.2004.02032.x. [DOI] [PubMed] [Google Scholar]
- Heese M, Gansel X, Sticher L, Wick P, Grebe M, Granier F, Jurgens G. Functional characterization of the KNOLLE-interacting t-SNARE AtSNAP33 and its role in plant cytokinesis. J Cell Biol. 2001;155(2):239–249. doi: 10.1083/jcb.200107126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho C-L, Wu Y, Shen HH, Provart NNJ, Geisler M. A predicted protein interactome for rice. Rice (NY). 2012;5(1):15.doi:10.1186/1939–8433-5–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbaert A, Zhang C, Tiwari M, Wang K, de Marcos Serrano A, Savatin DV, Urs MJ, Zhiponova MK, Gudesblat GE, Vanhoutte I, et al. POLAR-guided signalling complex assembly and localization drive asymmetric cell division. Nature. 2018;563(7732):574–578. doi: 10.1038/s41586-018-0714-x. [DOI] [PubMed] [Google Scholar]
- Howe KL, Contreras-Moreira B, De Silva N, Maslen G, Akanni W, Allen J, Alvarez-Jarreta J, Barba M, Bolser DM, Cambell L, et al. Ensembl Genomes 2020-enabling non-vertebrate genomic research. Nucleic Acids Res. 2020;48(D1):D689–D695. doi: 10.1093/nar/gkz890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt M, Banerjee S, Surana P, Liu M, Fuerst G, Mathioni S, Meyers BC, Nettleton D, Wise RP. Small RNA discovery in the interaction between barley and the powdery mildew pathogen. BMC Genomics. 2019;20(1):610.doi: 10.1186/s12864-019-5947-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z, Dong X, Zhang Z. Network-based comparative analysis of Arabidopsis immune responses to Golovinomyces orontii and Botrytis cinerea infections. Sci Rep. 2016;6:19149.doi: 10.1038/srep19149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Tian F, Yang D-C, Meng Y-Q, Kong L, Luo J, Gao G. PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017;45(D1):D1040–D1045. doi: 10.1093/nar/gkw982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan T, Seeholzer S, Schwizer S, Töller A, Somssich IE, Keller B. The wheat Mla homologue TmMla1 exhibits an evolutionarily conserved function against powdery mildew in both wheat and barley. Plant J. 2011;65(4):610–621. doi:10.1111/j.1365-313X.2010.04445.x. [DOI] [PubMed] [Google Scholar]
- Kayum MA, Park J-I, Nath UK, Saha G, Biswas MK, Kim H-T, Nou I-S. Genome-wide characterization and expression profiling of PDI family gene reveals function as abiotic and biotic stress tolerance in Chinese cabbage (Brassica rapa ssp. pekinensis). BMC Genomics. 2017;18(1):885.doi: 10.1186/s12864-017-4277-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingenberg H, Meinicke P. How to normalize metatranscriptomic count data for differential expression analysis. PeerJ. 2017;5:e3859.doi: 10.7717/peerj.3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koschützki D, Schreiber F. Centrality analysis methods for biological networks and their application to gene regulatory networks. Gene Regul Syst Bio. 2008;2:193–201. doi: 10.4137/grsb.s702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krattinger SG, Keller B. Molecular genetics and evolution of disease resistance in cereals. New Phytol. 2016;212(2):320–332. doi: 10.1111/nph.14097. [DOI] [PubMed] [Google Scholar]
- Kwon Y, Shen J, Lee MH, Geem KR, Jiang L, Hwang I. AtCAP2 is crucial for lytic vacuole biogenesis during germination by positively regulating vacuolar protein trafficking. Proc Natl Acad Sci U S A. 2018;115(7):E1675–E1683. doi: 10.1073/pnas.1717204115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambertucci S, Orman KM, Das Gupta S, Fisher JP, Gazal S, Williamson RJ, Cramer R, Bindschedler LV. Analysis of barley leaf epidermis and extrahaustorial proteomes during powdery mildew infection reveals that the PR5 Thaumatin-like protein TLP5 is required for susceptibility towards Blumeria graminis f. sp. hordei. Front Plant Sci. 2019;10:1138.doi: 10.3389/fpls.2019.01138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landi S, Capasso G, Ben Azaiez FE, Jallouli S, Ayadi S, Trifa Y, Esposito S. Different roles of heat shock proteins (70 kDa) during abiotic stresses in barley (Hordeum vulgare) genotypes. Plants. 2019;8(8):248.doi: 10.3390/plants8080248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latijnhouwers M, Gillespie T, Boevink P, Kriechbaumer V, Hawes C, Carvalho CM. Localization and domain characterization of Arabidopsis golgin candidates. J Exp Bot. 2007;58(15–16):4373–4386. doi: 10.1093/jxb/erm304. [DOI] [PubMed] [Google Scholar]
- Lee H-Y, Seo Y-E, Lee JH, Lee SE, Oh S, Kim J, Jung S, Kim H, Park H, Kim S, et al. Plasma membrane‐localized plant immune receptor targets H+ ‐ATPase for membrane depolarization to regulate cell death. New Phytol. 2022;233(2):934–947. doi: 10.1111/nph.17789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Eschen-Lippold L, Lassowskat I, Böttcher C, Scheel D. Cellular reprogramming through mitogen-activated protein kinases. Front Plant Sci. 2015;6:940.doi: 10.3389/fpls.2015.00940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JD, Wu R, Guttman DS, Desveaux D. Allele-specific virulence attenuation of the Pseudomonas syringae HopZ1a type III effector via the Arabidopsis ZAR1 resistance protein. PLoS Genet. 2010;6(4):e1000894.doi: 10.1371/journal.pgen.1000894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo-Presti L, Lanver D, Schweizer G, Tanaka S, Liang L, Tollot M, Zuccaro A, Reissmann S, Kahmann R. Fungal effectors and plant susceptibility. Annu Rev Plant Biol. 2015;66(1):513–545. doi: 10.1146/annurev-arplant-043014-114623. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550.doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Kracher B, Saur IML, Bauer S, Ellwood SR, Wise R, Yaeno T, Maekawa T, Schulze-Lefert P. Allelic barley MLA immune receptors recognize sequence-unrelated avirulence effectors of the powdery mildew pathogen. Proc Natl Acad Sci U S A. 2016;113(42):E6486–E6495. doi: 10.1073/pnas.1612947113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa T, Cheng W, Spiridon LN, Töller A, Lukasik E, Saijo Y, Liu P, Shen Q-H, Micluta MA, Somssich IE, et al. Coiled-coil domain-dependent homodimerization of intracellular barley immune receptors defines a minimal functional module for triggering cell death. Cell Host Microbe. 2011;9(3):187–199. doi:10.1016/j.chom.2011.02.008. [DOI] [PubMed] [Google Scholar]
- Maekawa T, Kracher B, Vernaldi S, Van Themaat EVL, Schulze-Lefert P. Conservation of NLR-triggered immunity across plant lineages. Proc Natl Acad Sci U S A. 2012;109(49):20119–20123. doi: 10.1073/pnas.1218059109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mago R, Zhang P, Vautrin S, Šimková H, Bansal U, Luo M-C, Rouse M, Karaoglu H, Periyannan S, Kolmer J, et al. The wheat Sr50 gene reveals rich diversity at a cereal disease resistance locus. Nat Plants. 2015;1(12):15186. doi: 10.1038/nplants.2015.186. [DOI] [PubMed] [Google Scholar]
- Manzoor H, Kelloniemi J, Chiltz A, Wendehenne D, Pugin A, Poinssot B, Garcia-Brugger A. Involvement of the glutamate receptor AtGLR3.3 in plant defense signaling and resistance to Hyaloperonospora arabidopsidis. Plant J. 2013;76(3):466–480. doi: 10.1111/tpj.12311. [DOI] [PubMed] [Google Scholar]
- Martin GB, Bogdanove AJ, Sessa G. Understanding the functions of plant disease resistance proteins. Annu Rev Plant Biol. 2003;54:23–61. doi: 10.1146/annurev.arplant.54.031902.135035. [DOI] [PubMed] [Google Scholar]
- Matthews LR, Vaglio P, Reboul J, Ge H, Davis BP, Garrels J, Vincent S, Vidal M. Identification of potential interaction networks using sequence-based searches for conserved protein-protein interactions or “interologs”. Genome Res. 2001;11(12):2120–2126. doi: 10.1101/gr.205301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34(Suppl 1):108–110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWhite CD, Papoulas O, Drew K, Cox RM, June V, Dong OX, Kwon T, Wan C, Salmi ML, Roux SJ, et al. A pan-plant protein complex map reveals deep conservation and novel assemblies. Cell. 2020;181(2):460–474. doi: 10.1016/j.cell.2020.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Y, Moscou MJ, Wise RP. Blufensin1 negatively impacts basal defense in response to barley powdery mildew. Plant Physiol. 2009;149(1):271–285. doi: 10.1104/pp.108.129031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra B, Sun Y, Ahmed H, Liu X, Mukhtar MS. Global temporal dynamic landscape of pathogen-mediated subversion of Arabidopsis innate immunity. Sci Rep. 2017;7(1):7849.doi:10.1038/s41598-017–08073-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistry D, Wise RP, Dickerson JA. DiffSLC: a graph centrality method to detect essential proteins of a protein-protein interaction network. PLoS One. 2017;12(11):e0187091.doi: 10.1371/journal.pone.0187091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monat C, Padmarasu S, Lux T, Wicker T, Gundlach H, Himmelbach A, Ens J, Li C, Muehlbauer GJ, Schulman AH, et al. TRITEX: chromosome-scale sequence assembly of Triticeae genomes with open-source tools. Genome Biol. 2019;20(1):284. doi:10.1186/s13059-019–1899-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro F, Nishimura MT. Structural, functional, and genomic diversity of plant NLR proteins: an evolved resource for rational engineering of plant immunity. Annu Rev Phytopathol. 2018;56:243–267. doi: 10.1146/annurev-phyto-080417-045817. [DOI] [PubMed] [Google Scholar]
- Moscou MJ, Lauter N, Caldo RA, Nettleton D, Wise RP. Quantitative and temporal definition of the Mla transcriptional regulon during barley-powdery mildew interactions. Mol Plant Microbe Interact. 2011;24(6):694–705. doi: 10.1094/MPMI-09-10-0211. [DOI] [PubMed] [Google Scholar]
- Mukhtar MS, Carvunis A-R, Dreze M, Epple P, Steinbrenner J, Moore J, Tasan M, Galli M, Hao T, Nishimura MT, et al. ; European Union Effectoromics Consortium. Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science. 2011;333(6042):596–601. doi: 10.1126/science.1203659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muschalik N, Munro S. Golgins. Curr Biol. 2018;28(8):R374–R376. doi: 10.1016/j.cub.2018.01.006. [DOI] [PubMed] [Google Scholar]
- Musungu B, Bhatnagar D, Brown RL, Fakhoury AM, Geisler M. A predicted protein interactome identifies conserved global networks and disease resistance subnetworks in maize. Front Genet. 2015;6:201.doi: 10.3389/fgene.2015.00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima N, Hayashida M, Jansson J, Maruyama O, Akutsu T. Determining the minimum number of protein-protein interactions required to support known protein complexes. PLoS One. 2018;13(4):e0195545.doi: 10.1371/journal.pone.0195545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngou BPM, Jones JDG, Ding P. Plant immune networks. Feature Review. 2022;27(3):255–273. doi: 10.1016/j.tplants.2021.08.012. [DOI] [PubMed] [Google Scholar]
- Park GT, Frost JM, Park J-S, Kim TH, Lee JS, Oh SA, Twell D, Brooks JS, Fischer RL, Choi Y, et al. Nucleoporin MOS7/Nup88 is required for mitosis in gametogenesis and seed development in Arabidopsis. Proc Natl Acad Sci U S A. 2014;111(51):18393–18398. doi: 10.1073/pnas.1421911112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AH, Bowers JE, Chapman BA. Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc Natl Acad Sci U S A. 2004;101(26):9903–9908. doi: 10.1073/pnas.0307901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017;14(4):417–419. doi:10.0.4.14/nmeth.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennington HG, Gheorghe DM, Damerum A, Pliego C, Spanu PD, Cramer R, Bindschedler LV. Interactions between the powdery mildew effector BEC1054 and barley proteins identify candidate host targets. J Proteome Res. 2016;15(3):826–839. doi: 10.1021/acs.jproteome.5b00732. [DOI] [PubMed] [Google Scholar]
- Periyannan S, Moore J, Ayliffe M, Bansal U, Wang X, Huang L, Deal K, Luo M, Kong X, Bariana H, et al. The gene Sr33, an ortholog of barley Mla genes, encodes resistance to wheat stem rust race Ug99. Science. 2013;341(6147):786–788. [DOI] [PubMed] [Google Scholar]
- Petrey D, Honig B. Structural bioinformatics of the interactome. Annu Rev Biophys. 2014;43(1):193–210. doi: 10.1146/annurev-biophys-051013-022726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pusztahelyi T. Chitin and chitin-related compounds in plant–fungal interactions. Mycology. 2018;9(3):189–201. doi:10.1080/21501203.2018.1473299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y, Xi J, Du L, Poovaiah BW. The function of calreticulin in plant immunity. Plant Signal Behav. 2012;7(8):907–910. doi:10.4161/psb.20721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randhawa V, Pathania S. Advancing from protein interactomes and gene co-expression networks towards multi-omics-based composite networks: approaches for predicting and extracting biological knowledge. Brief Funct Genomics. 2020;19(5–6):364–376. doi: 10.1093/bfgp/elaa015. [DOI] [PubMed] [Google Scholar]
- Ray S, Anderson JM, Urmeev FI, Goodwin SB. Rapid induction of a protein disulfide isomerase and defense-related genes in wheat in response to the hemibiotrophic fungal pathogen Mycosphaerella graminicola. Plant Mol Biol. 2003;53(5):741–714. doi: 10.1023/b:plan.0000019120.74610.52. [DOI] [PubMed] [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing. Vienna (Austria): R Core Team; 2013. http://www.r-project.org. [Google Scholar]
- Ridout CJ, Skamnioti P, Porritt O, Sacristan S, Jones JDG, Brown JKM. Multiple avirulence paralogues in cereal powdery mildew fungi may contribute to parasite fitness and defeat of plant resistance. Plant Cell. 2006;18(9):2402–2414. doi: 10.1105/tpc.106.043307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salanoubat M, Lemcke K, Rieger M, Ansorge W, Unseld M, et al. Sequence and analysis of chromosome 3 of the plant Arabidopsis thaliana. Nature. 2000;402:761–768. [Google Scholar]
- Sánchez-Martín J, Keller B. NLR immune receptors and diverse types of non-NLR proteins control race-specific resistance in Triticeae. Curr Opin Plant Biol. 2021;62:102053.doi:10.1016/j.pbi.2021.102053. [DOI] [PubMed] [Google Scholar]
- Saur IM, Bauer S, Kracher B, Lu X, Franzeskakis L, Müller MC, Sabelleck B, Kümmel F, Panstruga R, Maekawa T, et al. Multiple pairs of allelic MLA immune receptor-powdery mildew AVRA effectors argue for a direct recognition mechanism. Elife. 2019;8:e44471. doi: 10.7554/eLife.44471.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeholzer S, Tsuchimatsu T, Jordan T, Bieri S, Pajonk S, Yang W, Jahoor A, Shimizu KK, Keller B, Schulze-Lefert P, et al. Diversity at the Mla powdery mildew resistance locus from cultivated barley reveals sites of positive selection. Mol Plant Microbe Interact. 2010;23(4):497–509. doi: 10.1094/MPMI-23-4-0497. [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303.metabolite. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Menche J, Huang CC, Ort T, Zhou X, Kitsak M, Sahni N, Thibault D, Voung L, Guo F, et al. A disease module in the interactome explains disease heterogeneity, drug response and captures novel pathways and genes in asthma. Hum Mol Genet. 2015;24(11):3005–3020. doi: 10.1093/hmg/ddv001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q-H, Zhou F, Bieri S, Haizel T, Shirasu K, Schulze-Lefert P. Recognition specificity and RAR1/SGT1 dependence in barley Mla disease resistance genes to the powdery mildew fungus. Plant Cell. 2003;15(3):732–744. doi: 10.1105/tpc.009258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q-H, Saijo Y, Mauch S, Biskup C, Bieri S, Keller B, Seki H, Ulker B, Somssich IE, Schulze-Lefert P, et al. Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science. 2007;315(5815):1098–1103. doi: 10.1126/science.1136372. [DOI] [PubMed] [Google Scholar]
- Shirasu K. The HSP90-SGT1 chaperone complex for NLR immune sensors. Annu Rev Plant Biol. 2009;60(1):139–164. doi: 10.1146/annurev.arplant.59.032607.092906. [DOI] [PubMed] [Google Scholar]
- Singh V, Singh G, Singh V. TulsiPIN: an interologous protein interactome of Ocimum tenuiflorum. J Proteome Res. 2020;19(2):884–899. doi: 10.1021/acs.jproteome.9b00683. [DOI] [PubMed] [Google Scholar]
- Smakowska-Luzan E, Mott GA, Parys K, Stegmann M, Howton TC, Layeghifard M, Neuhold J, Lehner A, Kong J, Grünwald K, et al. An extracellular network of Arabidopsis leucine-rich repeat receptor kinases. Nature. 2018;553(7688):342–346. doi: 10.1038/nature25184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammer ELL, Östlund G. InParanoid 8: orthology analysis between 273 proteomes, mostly eukaryotic. Nucleic Acids Res. 2015;43(Database Issue):D234–D239. doi: 10.1093/nar/gku1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanu PD, Abbott JC, Amselem J, Burgis TA, Soanes DM, Stüber K, Ver Loren van Themaat E, Brown JKM, Butcher SA, Gurr SJ, et al. Genome expansion and gene loss in powdery mildew fungi reveal tradeoffs in extreme parasitism. Science. 2010;330(6010):1543–1546. doi: 10.1126/science.1194573. [DOI] [PubMed] [Google Scholar]
- Spies A, Korzun V, Bayles R, Rajaraman J, Himmelbach A, Hedley PE, Schweizer P. Allele mining in barley genetic resources reveals genes of race-non-specific powdery mildew resistance. Front Plant Sci. 2012;2:113.doi: 10.3389/fpls.2011.00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srichumpa P, Brunner S, Keller B, Yahiaoui N. Allelic series of four powdery mildew resistance genes at the Pm3 locus in hexaploid bread wheat. Plant Physiol. 2005;139(2):885–895. doi:10.1104/pp.105.062406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadnik MJ, Buchenauer H. Inhibition of phenylalanine ammonia-lyase suppresses the resistance induced by benzothiadiazole in wheat to Blumeria graminis f. sp. tritici. Physiol Mol Plant Pathol. 2000;57(1):25–34. doi: 10.1006/pmpp.2000.0276. [DOI] [Google Scholar]
- Stark C, Breitkreutz B-J, Reguly T, Boucher L, Breitkreutz A, Tyers M. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34(Database Issue):D535–D539. doi:10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steuernagel B, Periyannan SK, Hernández-Pinzón I, Witek K, Rouse MN, Yu G, Hatta A, Ayliffe M, Bariana H, Jones JDG, et al. Rapid cloning of disease-resistance genes in plants using mutagenesis and sequence capture. Nat Biotechnol. 2016;34(6):652–655. doi: 10.1038/nbt.3543. [DOI] [PubMed] [Google Scholar]
- Sun Y, Zhu YX, Balint-Kurti PJ, Wang GF. Fine-tuning immunity: players and regulators for plant NLRs. Trends Plant Sci. 2020;25(7):695–713. doi: 10.1016/j.tplants.2020.02.008. [DOI] [PubMed] [Google Scholar]
- Surana P. Membrane trafficking in resistance gene-mediated defense against the barley powdery mildew fungus. Iowa State University; 2017.
- Surana P, Xu R, Fuerst G, Chapman AVE, Nettleton D, Wise RP. Interchromosomal transfer of immune regulation during infection of barley with the powdery mildew pathogen. G3 (Bethesda). 2017;7(10):3317–3329. doi: 10.1534/g3.117.300125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Székely GJ, Rizzo ML, Bakirov NK. Measuring and testing dependence by correlation of distances. Ann Stat. 2007;35(6):2769–2794. doi: 10.1214/009053607000000505. [DOI] [Google Scholar]
- Tamborski J, Krasileva KV. Evolution of plant NLRs: from natural history to precise modifications. Annu Rev Plant Biol. 2020;71:355–378. doi: 10.1146/annurev-arplant-081519-035901. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Ade J, Frye CA, Innes RW. A mutation in the GTP hydrolysis site of Arabidopsis dynamin-related protein 1E confers enhanced cell death in response to powdery mildew infection. Plant J. 2006;47(1):75–84. doi: 10.1111/j.1365-313X.2006.02769.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toruño TY, Stergiopoulos I, Coaker G. Plant-pathogen effectors: cellular probes interfering with plant defenses in spatial and temporal manners. Annu Rev Phytopathol. 2016;54(1):419–441. doi: 10.1146/annurev-phyto-080615-100204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trigg SA, Garza RM, MacWilliams A, Nery JR, Bartlett A, Castanon R, Goubil A, Feeney J, O'Malley R, Huang S-SC, et al. CrY2H-seq: a massively multiplexed assay for deep-coverage interactome mapping. Nat Methods. 2017;14(8):819–825. doi:10.1038/nmeth.4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van-Wersch S, Tian L, Hoy R, Li X. Plant NLRs: the whistleblowers of plant immunity. Plant Commun. 2020;1(1):100016.doi:10.1016/j.xplc.2019.100016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velásquez-Zapata V, Elmore JM, Banerjee S, Dorman KS, Wise RP. Next-generation yeast-two-hybrid analysis with Y2H-SCORES identifies novel interactors of the MLA immune receptor. PLoS Comput Biol. 2021;17(4):e1008890.doi:10.1371/journal.pcbi.1008890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Hu M, Wang J, Qi J, Han Z, Wang G, Qi Y, Wang H-W, Zhou J-M, Chai J, et al. Reconstitution and structure of a plant NLR resistosome conferring immunity. Science. 2019;364(6435):44. doi: 10.1126/science.aav5870. [DOI] [PubMed] [Google Scholar]
- Wang T, Chang C, Gu C, Tang S, Xie Q, Shen Q-H. An E3 ligase affects the NLR receptor stability and immunity to powdery mildew. Plant Physiol. 2016;172(4):2504–2515. doi: 10.1104/pp.16.01520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Peng Q, Liu B, Liu Y, Wang Y. Disease module identification based on representation learning of complex networks integrated from GWAS, eQTL summaries, and human interactome. Front Bioeng Biotechnol. 2020;8:418.doi: 10.3389/fbioe.2020.00418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts DJ, Strogatz SH. Collective dynamics of ‘small-world’ networks. Nature. 1998;393(6684):440–442. [DOI] [PubMed] [Google Scholar]
- Wei F, Wing RA, Wise RP. Genome dynamics and evolution of the Mla (powdery mildew) resistance locus in barley. Plant Cell. 2002;14(8):1903–1917. doi: 10.1105/tpc.002238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weirauch MT, Yang A, Albu M, Cote AG, Montenegro-Montero A, Drewe P, Najafabadi HS, Lambert SA, Mann I, Cook K, et al. Determination and inference of eukaryotic transcription factor sequence specificity. Cell. 2014;158(6):1431–1443. doi:10.1016/j.cell.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weßling R, Epple P, Altmann S, He Y, Yang L, Henz SR, McDonald N, Wiley K, Bader KC, Gläßer C, et al. Convergent targeting of a common host protein-network by pathogen effectors from three kingdoms of life. Cell Host Microbe. 2014;16(3):364–375. doi:10.1016/j.chom.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierbowski SD, Vo TV, Falter-Braun P, Jobe TO, Kruse LH, Wei X, Liang J, Meyer MJ, Akturk N, Rivera-Erick CA, et al. A massively parallel barcoded sequencing pipeline enables generation of the first ORFeome and interactome map for rice. Proc Natl Acad Sci U S A. 2020;117(21):11836–11842. doi: 10.1073/pnas.1918068117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiermer M, Germain H, Cheng YT, García AV, Parker JE, Li X. Nucleoporin MOS7/Nup88 contributes to plant immunity and nuclear accumulation of defense regulators. Nucleus. 2010;1(4):332–336. doi: 10.4161/nucl.1.4.12109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wróblewski T, Spiridon L, Martin EC, Petrescu A-J, Cavanaugh K, Truco MJ, Xu H, Gozdowski D, Pawłowski K, Michelmore RW, et al. Genome-wide functional analyses of plant coiled–coil NLR-type pathogen receptors reveal essential roles of their N-terminal domain in oligomerization, networking, and immunity. PLoS Biol. 2018;16(12):e2005821. doi: 10.1371/journal.pbio.2005821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Kapos P, Cheng YT, Li M, Zhang Y, Li X. NLR-associating transcription factor bHLH84 and its paralogs function redundantly in plant immunity. PLoS Pathog. 2014;10(8):e1004312.doi: 10.1371/journal.ppat.1004312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Meng Y, Surana P, Fuerst G, Nettleton D, Wise RP. The knottin-like Blufensin family regulates genes involved in nuclear import and the secretory pathway in barley-powdery mildew interactions. Front Plant Sci. 2015;6:409.doi:10.3389/fpls.2015.00409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Smith DK, Zhu H, Guan Y, Lam TT-Y. GGTREE: An R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol Evol. 2017;8(1):28–36. doi: 10.1111/2041-210X.12628. [DOI] [Google Scholar]
- Yu G, Wang L-G, Han Y, He Q-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M, Jiang Z, Bi G, Nomura K, Liu M, et al. Pattern-recognition receptors are required for NLR-mediated plant immunity. Nature. 2020;592:105–112. doi: 10.1101/2020.04.10.031294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng LR, Vega-Sánchez ME, Zhu T, Wang GL. Ubiquitination-mediated protein degradation and modification: an emerging theme in plant-microbe interactions. Cell Res. 2006;16(5):413–426. doi: 10.1038/sj.cr.7310053. [DOI] [PubMed] [Google Scholar]
- Zhou F, Kurth J, Wei F, Elliott C, Valè G, Yahiaoui N, Keller B, Somerville S, Wise R, Schulze-Lefert P, et al. Cell-autonomous expression of barley Mla1 confers race-specific resistance to the powdery mildew fungus via a Rar1-independent signaling pathway. Plant Cell. 2001;13(2):337–350. doi: 10.1105/tpc.13.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G, Wu A, Xu X-J, Xiao P-P, Lu L, Liu J, Cao Y, Chen L, Wu J, Zhao X-M, et al. PPIM: a protein-protein interaction database for maize. Plant Physiol. 2016;170(2):618–626. doi: 10.1104/pp.15.01821. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains and plasmids are available upon request. Supplemental files available at figshare: https://doi.org/10.25386/genetics.19089992. Supplementary Data 1 contain the predicted barley interactome (HvInt) and experimentally validated Arabidopsis interactome (AtInt). Supplementary Data 2 contain node properties of HvInt and enrichment of essential proteins across clusters. Supplementary Data 3 contain DMs and DM DE genes at Bgh penetration and haustorial development. Supplementary Data 4 contain resistant and susceptible interactomes. Supplementary Data 5 contain the MLA interactome (MLAInt). Supplementary Figure 1 contains core and unique DM GO terms associated with DE genes by timepoint. Supplementary Figure 2 contains complete time–course expression patterns DM DE genes by type of response. Supplementary Figure 3 contains complete Y2H validation of interactions for the CC + NB domains of Mla alleles, orthologs and outgroup NLRs. Supplementary Text 1 contains bait and prey sequences used for Y2H binary validation among Mla alleles, orthologs, and outgroup NLRs. Supplementary Text 2 contains the protein alignment of the Mla alleles, orthologs, and outgroup used for Y2H tests. Code used to support the main findings of this manuscript are at the GitHub page https://github.com/Wiselab2/Barley_Interactome [accessed 15 April 2022]. MIAME-compliant Barley1 GeneChip profiling data (Affymetrix part number 900515) from the Q21861 × SM89010 eQTL analysis are available as accession GSE68963 at NCBI’s Gene Expression Omnibus (GEO). Conversion of the GeneChip Probe sets to gene IDs was done using Ensemble and Biomart. Infection–time–course RNA-Seq datasets are available in NCBI-GEO under accession number GSE101304 (https://www.ncbi.nlm.nih.gov/gds/?term=GSE101304). R code and the ReadMe file for the NGPINT and Y2H-SCORES software used to identify MLA interactors are provided at the GitHub page https://github.com/Wiselab2/ [accessed 15 April 2022]. Raw Y2H-Seq reads are at NCBI-GEO under the accession number GSE164815 (MLA6CC+NB).
Su pplemental material is available at figshare DOI: https://doi.org/10.25386/genetics.19089992