Abstract
Rice (Oryza sativa) germination and seedling establishment, particularly in increasingly saline soils, are critical to ensure successful crop yields. Seed vigor, which determines germination and seedling growth, is a complex trait affected by exogenous (environmental) and endogenous (hormonal) factors. Here, we used genetic and biochemical analyses to uncover the role of an APETALA2-type transcription factor, SALT AND ABA RESPONSE ERF1 (OsSAE1), as a positive regulator of seed germination and salt tolerance in rice by repressing the expression of ABSCISIC ACID-INSENSITIVE5 (OsABI5). ossae1 knockout lines exhibited delayed seed germination, enhanced sensitivity to abscisic acid (ABA) during germination and in early seedling growth, and reduced seedling salt tolerance. OsSAE1 overexpression lines exhibited the converse phenotype, with increased seed germination and salt tolerance. In vivo and in vitro assays indicated that OsSAE1 binds directly to the promoter of OsABI5, a major downstream component of the ABA signaling pathway and acts as a major regulator of seed germination and stress response. Genetic analyses revealed that OsABI5-mediated ABA signaling functions downstream of OsSAE1. This study provides important insights into OsSAE1 regulation of seed vigor and salt tolerance and facilitates the practical use of OsSAE1 in breeding salt-tolerant varieties suitable for direct seeding cultivation.
An APETALA2 transcription factor positively regulates seed germination, seedling growth, and salt tolerance in rice by repressing the expression of an abscisic acid signaling component.
Introduction
Rice (Oryza sativa) is a staple crop for half of the world’s population, and rice production is critical for ensuring global food security. To ensure grain yield and quality, rice seeds must be vigorous (Gommers and Monte, 2018). In the agricultural production, seeds with high vigor will behave the characteristics of speed and uniform germination and vigorous seedling growth (Chen et al., 2019). As a crucial developmental step for continuing the plant life cycle, seed germination, and seedling establishment are complex processes precisely regulated by environmental stimuli and phytohormones (He et al., 2019, 2020; Xu et al., 2020; Wang et al., 2020a, 2020b). Dissecting the key genes involved in seed vigor and elucidating the underlying mechanism will facilitate rice breeding.
The phytohormone abscisic acid (ABA), which is considered a stress hormone, plays pivotal roles in abiotic stress tolerance and seed germination in plants (Tang et al., 2016; Ma et al., 2018, 2019; Zhao et al., 2020a, 2020b). Mutation or overexpression (OE) of genes involved in ABA biosynthesis or signaling pathway often results in abnormal germination and stress-tolerant phenotypes (Tang et al., 2012; Liu et al., 2014; Zong et al., 2016; Han et al., 2017). Numerous studies in Arabidopsis (Arabidopsis thaliana) have determined a core ABA signaling pathway: ABA is perceived by the PYRABACTIN-RESISTANCE PROTEINS/PYR-LIKE PROTEINS/REGULA TORY COMPONENTS OF ABA RECEPTOR (PYR/PYL/RCAR) receptors, which interact with the subclass A type 2C protein phosphatases (PP2Cs), thus blocking the dephosphorylation of SNF1-RELATED PROTEIN KINASE 2 (SnRK2s). As a result, SnRK2s phosphorylate and activate basic leucine zipper (bZIP) transcription factors to regulate the expression of ABA-responsive genes (Cutler et al., 2010; Raghavendra et al., 2010; Umezawa et al., 2010). ABSCISIC ACID-INSENSITIVE 5 (ABI5), a bZIP transcription factor functioning in ABA signaling, has been reported as a major regulator of seed germination and stress responses (Zou et al., 2008; Kong et al., 2013; Skubacz et al., 2016; Chang et al., 2019).
APETALA2/ethylene response factors (AP2/ERFs) are a large super-family of transcription factors found mainly in plants (Okamuro et al., 1997; Magnani et al., 2004; Nakano et al., 2006). AP2/ERF proteins contain one or two AP2/ERF domain, which consists of ∼60–70 amino acids and is important for DNA binding. The AP2/ERF family can be divided into four subfamilies: ERF, AP2, related to ABI3/VIVIPAROUS 1 (VP1) (RAV), and dehydration-responsive element-binding protein (DREB) (Nakano et al., 2006; Sharoni et al., 2011). Numerous AP2 subfamily genes have been identified and investigated in plants, and shown to regulate various developmental processes, including responses to hormones and stress (Zhang et al., 2013; Zhao et al., 2015; Cheng et al., 2018; Xie et al., 2019; Feng et al., 2020). For example, OsERF3 negatively regulates drought tolerance in rice by repressing ethylene biosynthesis (Zhang et al., 2013); RICE STARCH REGULATOR1 (OsRSR1) negatively regulates starch biosynthesis (Fu and Xue, 2010); SHATTERING ABORTION1 (OsSHAT1) is required for seed shattering through specifying abscission zone development in rice (Zhou et al., 2012); SUPERNUMERARY BRACT (OsSNB) controls rice seed shattering and seed size, and regulates the transition from spikelet to floral meristem development (Lee and An, 2012; Jiang et al., 2019); and INDETERMINATE SPIKELET1 (OsIDS1) regulates salt tolerance, inflorescence architecture, and floral meristem establishment in rice (Lee and An, 2012; Cheng et al., 2018). However, the function of AP2 proteins in seed germination is largely unclear.
Previously, via evaluating the survival rates and yield traits in saline and alkali soil, we identified 20 genes’ transgenic plants that generated by artificial microRNA interfering ethylene-responsive element binding factor-associated amphiphilic repression (EAR) motif-containing transcription factor, including OsERF3 and OsIDS1 (Zhang et al., 2013; Cheng et al., 2018). Here, we have functionally characterized SALT AND ABA RESPONSE ERF 1 (OsSAE1), a member of the AP2 subfamily. Using genetic and biochemical analyses, we reveal it to be a positive regulator of seed germination, seedling growth, and salt tolerance via direct repression of OsABI5. These results deepen our understanding of the regulatory function of AP2 proteins involved in ABA signaling and stress response, and provide avenues to improve seed vigor and seedling tolerance via molecular breeding.
Results
OsSAE1 is a transcriptional repressor and can bind the GCC-box motif
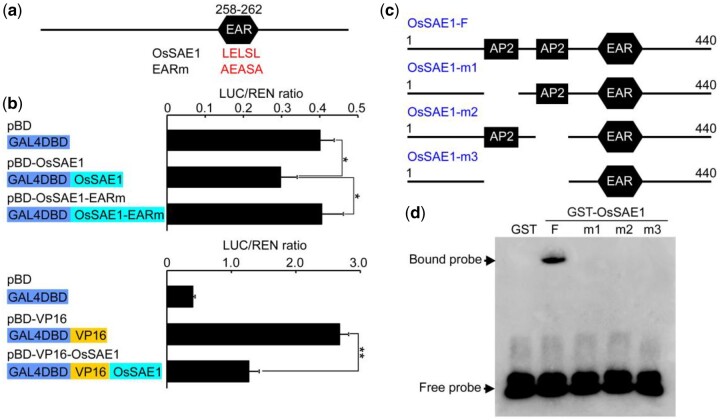
Previously, we identified 20 EAR-motif-containing transcription factors that are associated with salt tolerance in rice, and OsSAE1 is one of them (Cheng et al., 2018). OsSAE1 encodes 440 amino acids and belongs to AP2 subfamily. Sequence alignments showed that OsSAE1 contains two typical AP2 domains and an EAR motif (LELSL; Supplemental Figure S1, a and b). Phylogenetic tree analysis showed that OsSAE1 is closely related to OsSHAT1, OsRSR1, OsSNB, and OsIDS1 (Supplemental Figure S1c). These proteins function as transcriptional repressors that modulate processes such as seed size and shattering, spikelet and floral organ establishment, and stress tolerance (Fu and Xue, 2010; Lee and An, 2012; Zhou et al., 2012; Cheng et al., 2018; Jiang et al., 2019), and are all targets of miR172 (Zhu et al., 2009; Lee et al., 2014). OsSAE1 similarity with these proteins suggests that OsSAE1 may also be a transcriptional repressor with roles in reproductive development and/or stress response.
To examine whether OsSAE1 possesses transcriptional repression activity, we performed a transcription activity assay using an effector-reporter system by comparing luciferase (LUC) expression levels after the accumulation of GAL4 DNA binding domain (BD), BD-OsSAE1 fusion protein, and BD-OsSAE1 with a mutated EAR motif (BD-OsSAE1-EARm; Figure 1A), and a Renilla (REN) LUC reporter driven by the 35S promoter was used as an internal control. As shown in Figure 1B, LUC activity was significantly repressed in BD-OsSAE1 compared with that in BD, whereas the LUC activity in BD-OsSAE1-EARm showed no obvious difference with that in BD (Figure 1B), indicating that OsSAE1 has intrinsic transcriptional repression activity in vivo, and the transcriptional repression activity of OsSAE1 is largely dependent on the EAR motif. To further confirm the repression activity of OsSAE1, the coding region of OsSAE1 was fused with the activator VP16 to generate VP16-OsSAE1. According to our results, VP16-OsSAE1 exhibited much lower relative LUC activity compared to the control VP16 (Figure 1B). These results suggested that OsSAE1 may function as a transcriptional repressor.
Figure 1.
OsSAE1 confers transcriptional repression activity and DNA-binding activity to the GCC-box element. A, Intact and mutated amino acid sequence of the OsSAE1 EAR motif. B, Transient dual-LUC expression assays illustrating the transcriptional repression activity of OsSAE1. The reporters LUC and REN and the effectors (pBD, pBD-OsSAE1, pBD-OsSAE1-EARm, pBD-VP16, and pBD-OsSAE1-VP16) were separately constructed as shown on the left. The activities of LUC and REN were determined 16 h after transformation, and the relative LUC/REN ratios represent the transcriptional activation activities. Error bars represent STDEVs (sds) among three independent replicates. Asterisks indicate significant differences between the compared two samples at *P < 0.05 and **P < 0.01 using a Student’s t test. C, Schematic diagrams of wild-type and deletion mutants of OsSAE1 cloned into the pGEX-6p-1 vector. D, EMSA using GCC-box probe with GST-tagged OsSAE1 (GST-OsSAE1-F) or mutant OsSAE1 (GST-OsSAE1-m1/m2/m3). GST-tag was used in place of GST-OsSAE1 for no-protein controls. Three biological replicates were performed, with similar results.
Previous studies have shown that AP2/ERF superfamily transcription factors preferentially bind a GCC-box upstream of target genes to regulate downstream gene expression (Cai et al., 2014; Mao et al., 2016). To examine the DNA-binding properties of OsSAE1, we conducted an electrophoretic mobility shift assay (EMSA) with the biotin-labeled DNA probes that contained the GCC-box motif (5′-GCCGCC-3′). Our results showed that the intact OsSAE1 protein was able to bind the probe containing the GCC-box motif, whereas no binding was observed for OsSAE1 protein lacking one or two AP2 domains (Figure 1, C and D), indicating that both AP2 domains are involved in DNA binding.
Expression of OsSAE1 in rice tissues and in response to various treatments
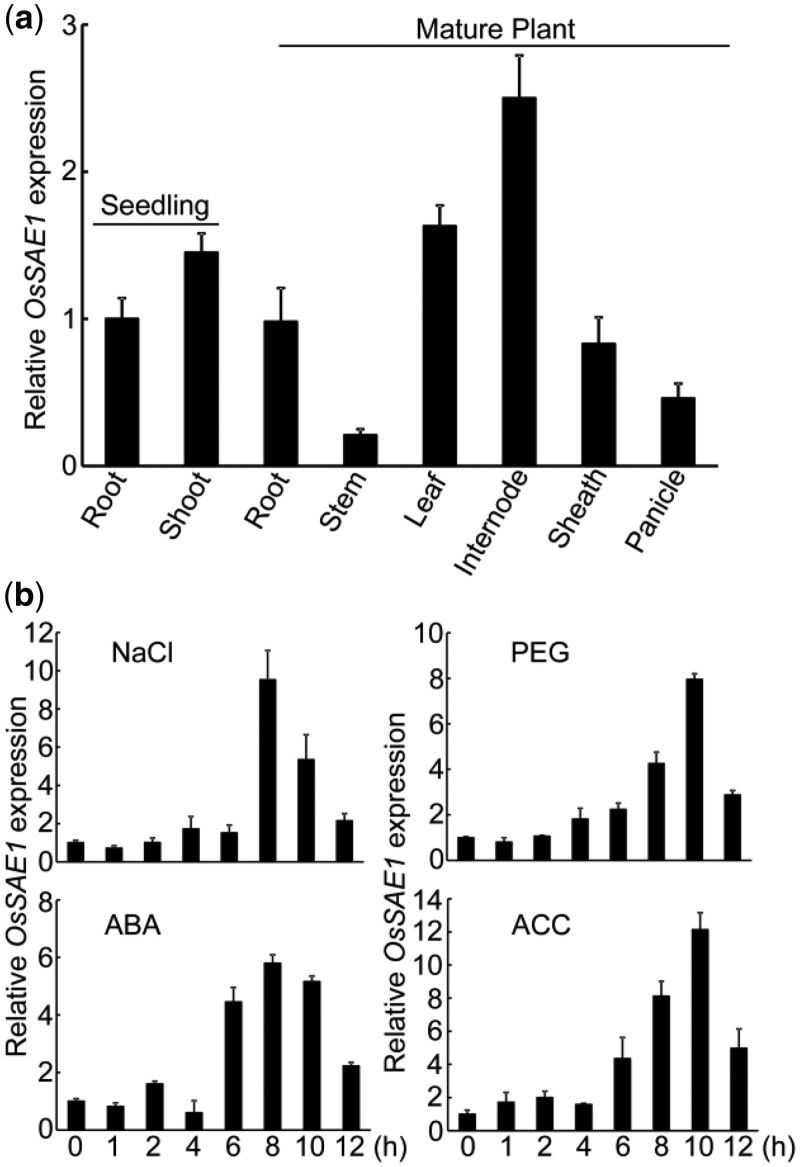
To speculate on the function of OsSAE1, we checked the expression profile of OsSAE1 in different tissues by reverse transcription-quantitative polymerase chain reaction (RT-qPCR). The results showed that OsSAE1 is constitutively expressed in young and mature plant tissues (Figure 2A). In seedling 10 d after imbibition (DAI), OsSAE1 was expressed at similar levels in root and shoot (Figure 2A). In mature plants, its expression was detected in all organs from the vegetative to reproductive stages and was preferentially expressed in leaves and internodes (Figure 2A), suggesting the possible role of OsSAE1 throughout the development process.
Figure 2.
OsSAE1 expression in different plant tissues and under stress and hormone treatments. A, OsSAE1 expression in different seedling and mature rice tissues. B, Expression of OsSAE1 in response to salt (NaCl), PEG, ABA, and ACC. The 10 DAI Nip seedlings were treated with 150-mM NaCl, 10% (w/v) PEG6000, 10-μM ABA, or 10-μM ACC for 0, 1, 2, 4, 6, 8, 10, and 12 h. RNA was isolated from shoots and used for RT-qPCR. Actin1 was used as an internal control. The relative expression levels were represented by fold change relative to the expression levels of root (A) or 0 h (B). The error bars indicate the sds based on three independent replicates.
Next, we investigated the expression pattern of OsSAE1 in response to various abiotic stresses and hormone treatments. Our results showed that the expression of OsSAE1 was substantially induced by salt (NaCl), osmotic stress (PEG), ABA and 1-aminocyclopropane-1-carboxylate (an ethylene precursor), and reached peak at 8 or 10 h after treatment (Figure 2B). These results suggest that OsSAE1 might be associated with ethylene- and ABA-related environmental stimuli in rice.
OsSAE1 negatively regulates ABA sensitivity in seed germination and postgermination growth
To facilitate our analysis of OsSAE1 function, we generated OsSAE1 knockout mutants (ossae1-1 and ossae1-2) using clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated (Cas) protein 9 system. The ossae1-1 mutant contained 2-bp insertion in the coding region of the target gene, resulting in an early generation of termination codon, whereas the ossae1-2 mutant contained 5-bp deletion in the coding region of the target gene, leading to a frame shift in the open reading frame and the generation of a premature stop codon (Supplemental Figure S2, a and b). We also generated the OE lines by overexpressing the coding region of OsSAE1 under the control of the Ubiquitin promoter, the increased expression of the OsSAE1 was confirmed by RT-qPCR (Supplemental Figure S2c).
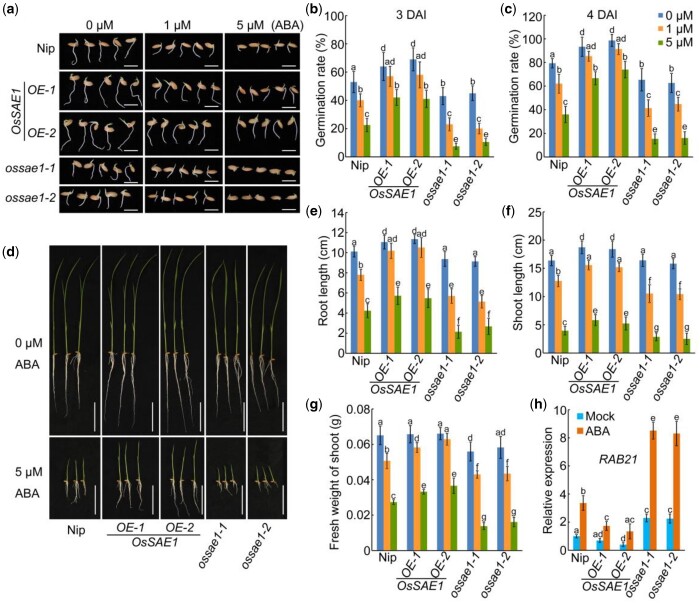
The finding that exogenous ABA treatment induces the expression of OsSAE1 enlightens us that OsSAE1 might be involved in ABA signaling in rice. To confirm this, we assessed the ABA sensitivity of transgenic plants at the germination stage. Seeds of wild-type Nipponbare (Nip), OsSAE1 OE (OsSAE1-OE) lines, and ossae1 mutants were imbibed in water for 48 h at 4°C, and then transferred to ABA solution to germinate at 28°C. Under control conditions, the germination rate of OsSAE1-OE lines reached 65%–70% at 3 DAI and 95% at 4 DAI, compared with the Nip germination rate of 53% at 3 DAI and 80% at 4 DAI. The germination rate of ossae1 mutants was lower than that of Nip, 45% at 3 DAI and 65% at 4 DAI (Figure 3, A–C), indicating that OsSAE1 positively regulates rice seed germination. Under ABA treatments, seeds of all the three genotypes exhibited some delay in germination, with OsSAE1-OE seeds being less sensitive to increasing concentrations of ABA than Nip seeds, which in turn were less sensitive than ossae1 seeds (Figure 3, A–C), indicating that OsSAE1 negatively regulates ABA sensitivity during rice seed germination.
Figure 3.
OsSAE1 negatively regulates ABA sensitivity at germination and in seedlings. A, Germination performance of wild-type (Nip), OsSAE1-OE, and OsSAE1 knockout (ossae1) seeds under 0, 1, or 5 μM ABA treatment at 3 DAI. Bars = 1 cm. B and C, Germination rate of seeds in (A) at 3 DAI (B) and 4 DAI (C). The error bars indicate the sds based on three independent replicates. D, Growth of the 10 DAI Nip, OsSAE1-OE and ossae1 seedlings under 0- or 5-μM ABA treatment. Bars = 5 cm. The images of the plants were digitally extracted for comparison. E–G, Root length (E), shoot length (F), and fresh weight of shoots (G) under 0, 1, or 5 μM ABA treatment of 10 DAI Nip, OsSAE1-OE, and ossae1 seedlings. Error bars indicate sds, n ≥ 10. H, Expression of RAB21 in Nip, OsSAE1-OE, and ossae1 seedlings with or without ABA treatment. The 10 DAI seedlings were treated with or without 1-μM ABA for 6 h. RNA was isolated from shoots and used for RT-qPCR. Actin1 was used as an internal control. The relative expression levels were represented by fold change relative to the expression levels of Nip-mock. The error bars indicate the sds based on three independent replicates. For (B, C, E, F, G, and H), different letters indicate significant differences (P < 0.05, one-way ANOVA with Tukey’s test).
Nip, OsSAE1-OE, and ossae1 seedlings were also assessed after 10 d of ABA treatment. Without ABA, root and shoot lengths of OsSAE1-OE seedlings were significantly longer than those of Nip seedlings, whereas root and shoot lengths of ossae1 seedlings were identical to Nip seedlings (Figure 3, D–F). Further analysis of fresh weight of shoots showed that the fresh weight of ossae1 shoots was slightly lower than Nip and OsSAE1-OE, which were similar to each other (Figure 3G). ABA treatment significantly reduced the root length, shoot length, and fresh weight of shoots in Nip seedlings, with a milder phenotype in OsSAE1-OE seedlings and a more pronounced phenotype in ossae1 seedlings (Figure 3, D–G). We further checked the expression level of RAB21, a commonly used marker gene in ABA response (Tang et al., 2012). The expression level of RAB21 was lower in OsSAE1-OE plants and higher in ossae1 mutants compared with that in Nip under normal growth conditions (Figure 3H). ABA treatment significantly induced the expression of RAB21 in Nip seedlings, and this tendency was weakened in OsSAE1-OE plants, but enhanced in ossae1 mutants (Figure 3H). These results indicate that OsSAE1 may negatively regulate ABA signaling in rice during germination and postgermination growth.
At maturation stage, the plant height and panicle length of the OsSAE1-OE plants were shorter than Nip and ossae1 mutants (Supplemental Figure S2, d–f). Tiller number was not significantly affected in the three genotypes (Supplemental Figure S2g). The grain size was further examined in ossae1 mutants and OsSAE1-OE plants using well-filled grains. In ossae1 mutants, grain length and grain width were slightly increased compared to those of Nip, whereas grain length and grain width of OsSAE1-OE plants were identical to Nip plants (Supplemental Figure S2, h–i). Measurement of 1000-grain weight for well-filled grains showed that OsSAE1-OE plants had significant reductions, whereas ossae1 mutants had slight increases in this parameter (Supplemental Figure S2j). These results indicate that OsSAE1 negatively regulates yield-related traits in rice.
OsSAE1 positively regulates salt tolerance in rice seedlings
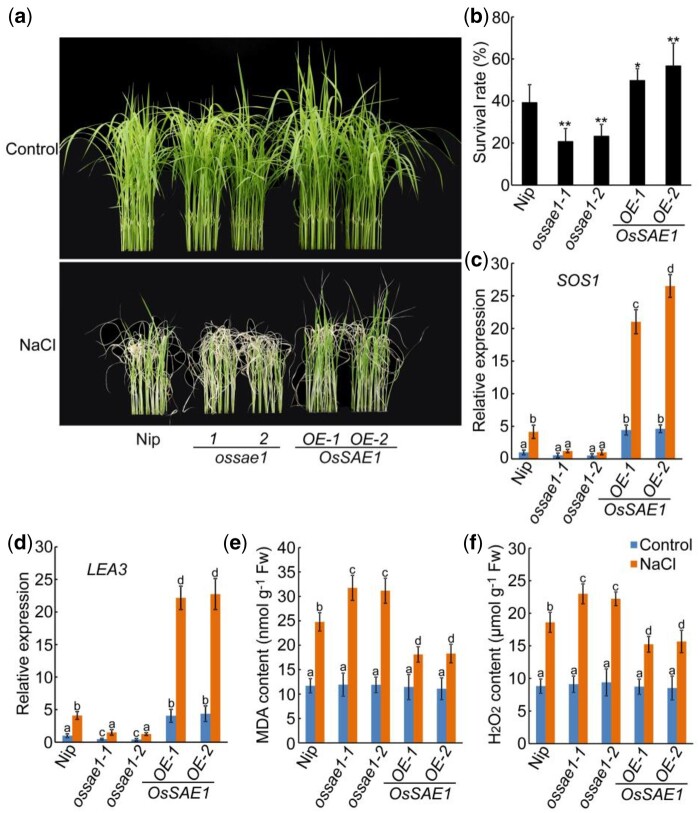
Regulators of ABA signaling have also been shown to contribute to plant tolerance to abiotic stress (Xiang et al., 2008; Joo et al., 2019; Liu et al., 2019). As OsSAE1 was obtained by salt tolerance analysis of rice ethylene-responsive element binding factor-associated amphiphilic repression (EAR)-containing transcription factors and salt induces OsSAE1 expression, we hypothesized that OsSAE1 is involved in regulating salt tolerance in rice. To verify this, we treated Nip, OsSAE1-OE, and ossae1 seedlings with NaCl. After 7 d of treatment with 150-mM NaCl, OsSAE1-OE seedlings were obviously less affected than ossae1 and Nip seedlings by salt stress (Figure 4A). After recovery for 7 d in nonsalt conditions, the survival rate of ossae1 seedlings was significantly lower, and that of OsSAE1-OE seedlings was significantly higher, than that of the Nip seedlings (Figure 4B). The expression of salt stress-responsive marker genes, SALT OVERLY SENSITIVE 1 (SOS1), and LATE EMBRYOGENESIS ABUNDANT PROTEIN 3 (LEA3; Cheng et al., 2018), was further checked. The results showed that salt treatment significantly induced the expression of SOS1 and LEA3 in Nip seedlings, this tendency was weakened in ossae1 mutants, but enhanced in OsSAE1-OE lines (Figure 4, C and D). These results indicate that OsSAE1 positively regulates the response of rice to salt stress at seedling stage.
Figure 4.
OsSAE1 positively regulates salt tolerance in rice seedlings. A, Phenotypes of Nip, OsSAE1-OE, and OsSAE1 knockout (ossae1) seedlings under salt stress. The images of the plants were digitally extracted for comparison. B, Survival rates of the plants shown in (A), 7 d after stopping salt stress. Approximately 50–60 seedlings were used per experiment. Data show mean ± sd of three independent replicates. Asterisks indicate significant differences compared to Nip at *P < 0.05 and **P < 0.01 by Student’s t test. C and D, Expression of SOS1 (C) and LEA3 (D) in Nip, OsSAE1-OE, and ossae1 seedlings with or without salt treatment. Ten DAI seedlings were treated with or without 150-mM NaCl for 6 h. RNA was isolated from shoots and used for RT-qPCR. Actin1 was used as an internal control. The relative expression levels were represented by fold change relative to the expression levels of Nip-control. E and F, MDA (E) and H2O2 (F) contents in leaves of 10 DAI seedlings with or without 150-mM NaCl treatment for 3 d. For (C, D, E, and F), the error bars indicate the sds based on three independent replicates, different letters indicate significant differences (P < 0.05, one-way ANOVA with Tukey’s test).
Salt stress leads to accumulation of reactive oxygen species (ROS) and malondialdehyde (MDA), which have detrimental effects on plant tissues (Zhao et al., 2020a, 2020b). Thus, both ROS and MDA contents serve as important indicators to evaluate abiotic stress tolerance. Although the MDA and hydrogen peroxide (H2O2) contents increased in all genotypes following a 3-d salt treatment, these indicators were significantly lower in OsSAE1-OE plants compared to the Nip and ossae1 mutants (Figure 4, E and F). These results suggest that OsSAE1 positively regulates salt tolerance in rice by reducing the accumulation of MDA and H2O2 under salt stress.
OsSAE1 directly binds to the promoter of OsABI5 and represses its expression
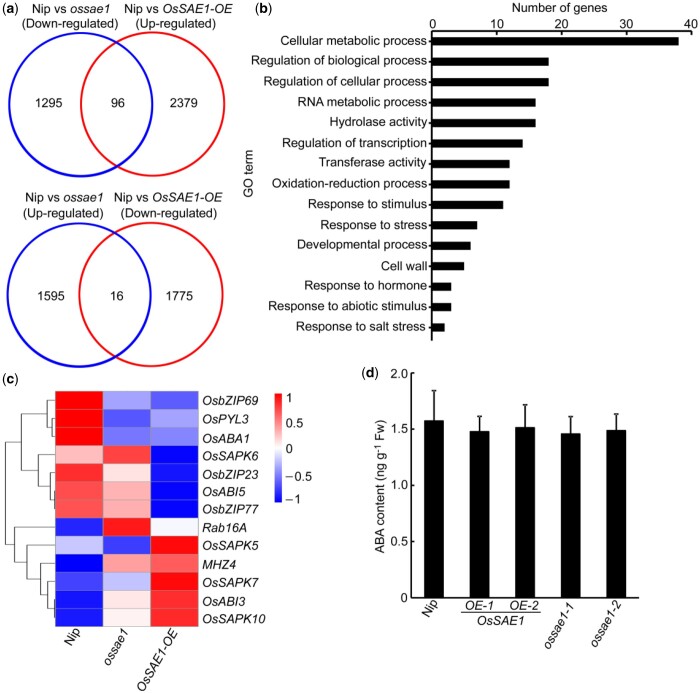
To elucidate the molecular network underlying OsSAE1-regulated seed germination and salt tolerance, we examined changes in gene expression in Nip, OsSAE1-OE, and ossae1 plants using transcriptome deep sequencing (RNA-seq). We identified 3,002 differentially expressed genes (DEGs; 1,611 upregulated and 1,391 downregulated) in ossae1 plants compared to the Nip plants, and 4,266 DEGs (2,475 upregulated and 1,791 downregulated) in OsSAE1-OE plants compared to the Nip plants (Figure 5A; Supplemental Dataset 1). Venn diagram analysis showed that 96 genes were upregulated in OsSAE1-OE and downregulated in ossae1, and 16 genes were downregulated in OsSAE1-OE and upregulated in ossae1 (Figure 5A; Supplemental Dataset 1). Gene ontology (GO) enrichment analysis showed that these genes from the intersection of the Venn diagram were involved in cellular metabolic process, regulation of biological process, regulation of cellular process, hydrolase activity, oxidation–reduction process, and stress response (Figure 5B). These results indicate that OsSAE1 is involved in a diverse range of biological processes and molecular functions, including those related to abiotic stress responses.
Figure 5.
Transcriptome analysis of DEGs regulated by OsSAE1. A, Venn diagrams of the DEGs upregulated and downregulated in OsSAE1-OE and OsSAE1 knockout (ossae1) seedlings compared with Nip. B, GO term analysis of OsSAE1-regulated genes. C, Heat map of microarray expression profiles for ABA biosynthesis and signaling-related genes. D, ABA contents in 5 DAI Nip, OsSAE1-OE, and ossae1 seedlings. The error bars indicate the sds based on three independent replicates. Asterisks indicate significant differences compared to Nip at *P < 0.05 and **P < 0.01 by Student’s t test.
Considering that OsSAE1 negatively regulates ABA sensitivity during rice seed germination and postgermination growth. We analyzed genes associated with ABA signal transduction in the transcriptome data and found that several genes in the ABA signaling core (OsSAPKs, OsbZIPs, OsABIs, and OsPYL) were regulated by OsSAE1 (Figure 5C). Further determination of ABA content showed that no significant differences in ABA content between wild-type, ossae1 mutants and OsSAE1-OE plants (Figure 5D), indicating that disrupting OsSAE1 does not affect ABA biosynthesis, and implying that OsSAE1 might inhibit ABA signaling to reduce ABA sensitivity during rice seed germination and postgermination growth.
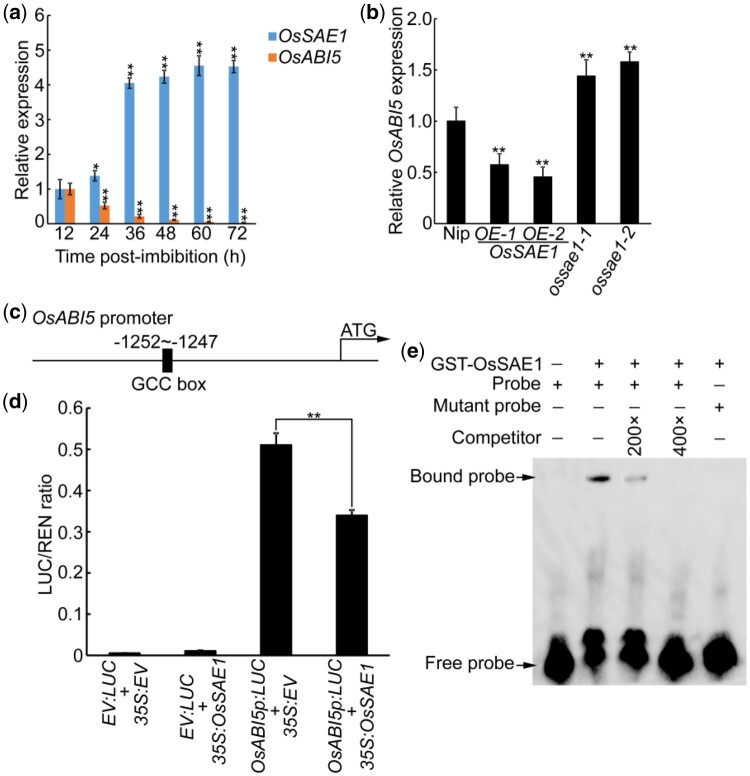
ABIs are downstream factors in ABA signaling pathways, and play key roles in seed germination and stress response (Skubacz et al., 2016; Lin et al., 2020; Zhao et al., 2020a, 2020b). Among the genes involved in the ABA signaling pathway regulated by OsSAE1, we found that OsABI5 was significantly downregulated in OsSAE1-OE plant (Figure 5C). Previous studies have shown that OsABI5 is involved in stress response and ABA signal transduction (Zou et al., 2007, 2008). RT-qPCR analysis showed that the expression of OsSAE1 significantly increased, while the expression of OsABI5 significantly decreased, during wild-type seed germination (Figure 6A), suggesting that OsSAE1 and OsABI5 play opposite roles during seed germination. Further analysis in OsSAE1-OE lines and ossae1 mutants revealed that OsABI5 expression was significantly lower in OsSAE1-OE lines, but higher in ossae1 mutants, compared with Nip (Figure 6B), suggesting that OsSAE1 negatively regulates OsABI5 expression and OsABI5 is a potential target of OsSAE1.
Figure 6.
OsSAE1 directly binds to the OsABI5 promoter to repress its expression. A, Expression of OsSAE1 and OsABI5 in germinating Nip seeds. Actin1 was used as an internal control. The relative expression levels were represented by fold change relative to the expression levels of 12 h. B, Expression of OsABI5 in Nip, OsSAE1-OE, and OsSAE1 knockout (ossae1) seeds at 24-h postimbibition. Actin1 was used as an internal control. The relative expression levels were represented by fold change relative to the expression levels of Nip. For (A and B), the error bars indicate the sds based on three independent replicates. Asterisks indicate significant differences from control values (12 h and Nip, respectively) using a Student’s t test at *P < 0.05; **P < 0.01. C, Schematic diagram of the OsABI5 promoter, showing the location of the GCC-box (GCCGCC). D, Dual-LUC assay in rice protoplasts using constructs constitutively expressing OsSAE1 and/or the LUC reporter gene under control of the OsABI5 promoter. The error bars indicate the sds based on three independent replicates. Asterisks indicate significant differences between the compared two samples using a Student’s t test at **P < 0.01. E, EMSA using normal (GCCGCC) and mutated (AAAAAA) OsABI5 promoter GCC-box probes with GST-tagged OsSAE1 (GST-OsSAE1). GST-tag was used in place of GST-OsSAE1 for no-protein controls. Protein was incubated with biotin-labeled DNA fragments (Probe), tested for competition by adding an excess of unlabeled probe (Competitor), and for specificity with labeled mutant probe. Three biological replicates were performed, with similar results.
To determine whether OsSAE1 can bind to the OsABI5 promoter and inhibit the promoter activity, we analyzed the promoter sequence of OsABI5 and identified one GCC-box, approximately 1.25-kb upstream of the transcriptional start, in the OsABI5 promoter (Figure 6C). Hence, we performed a transient, dual-LUC expression assay using the 2-kb OsABI5 promoter to direct expression of the LUC reporter gene (OsABI5p:LUC) in rice protoplasts cotransfected with or without the effector plasmid (35S:OsSAE1). The presence of OsSAE1 significantly decreased LUC activity driven by the OsABI5 promoter (Figure 6D). Subsequently, we conducted an EMSA using the GST-OsSAE1 fusion protein, which was able to bind directly to DNA probes containing the 5′-GCCGCC-3′ motif in the OsABI5 promoter (Figure 6E). The binding was specific, as demonstrated by competition using unlabeled (competitor) and mutant probes (Figure 6E). These results indicate that OsSAE1 binds directly to the OsABI5 promoter to repress its expression.
OsABI5 is a negative regulator of seed germination and salt tolerance
OsABI5 is a member of the third subfamily of bZIP transcription factors and exhibits a high sequence similarity to OsbZIP23 and OsbZIP46, two transcriptional factors that have been characterized as having an essential role in ABA signaling and stress tolerance in rice (Xiang et al., 2008; Tang et al., 2012). Previous studies have shown that the OsABI5 gene presents only one copy in the rice genome, which contains two splicing variants, OsABI5-1 and OsABI5-2. Both OsABI5-1 and OsABI5-2 had transactivation capacity, but only OsABI5-2 could specifically bind to G-box element (5′-CACGTG-3′), and constitutive expression of OsABI5-1 and OsABI5-2 can rescue ABA-insensitivity of abi5-1 and confer ABA hypersensitivity to wild-type transgenic plants in Arabidopsis (Zou et al., 2007, 2008), suggesting that OsABI5-1 and OsABI5-2 have similar functions to AtABI5. Studies show that OE of OsABI5-2 confers ABA hypersensitivity and salt sensitivity, and the gene has been designated as OsABI5 (Zou et al., 2007, 2008). To investigate the role of OsABI5 in seed germination and salt tolerance, we generated OsABI5 OE lines (OsABI5-OE-1 and OsABI5-OE-2) and OsABI5 loss-of-function mutants (osabi5-1 and osabi5-2). The increased expression of the target gene was confirmed by RT-qPCR and mutant form was identified by nucleotide sequences (Supplemental Figure S3, a–c). The osabi5-1 and osabi5-2 mutants harbor a 77-bp and 8-bp deletion in the coding region, respectively, resulting in the early generation of stop codons (Supplemental Figure S3, a and b), indicating that the osabi5-1 and osabi5-2 mutants lacked OsABI5. The progeny of these homozygous mutants were used in subsequent experiments.
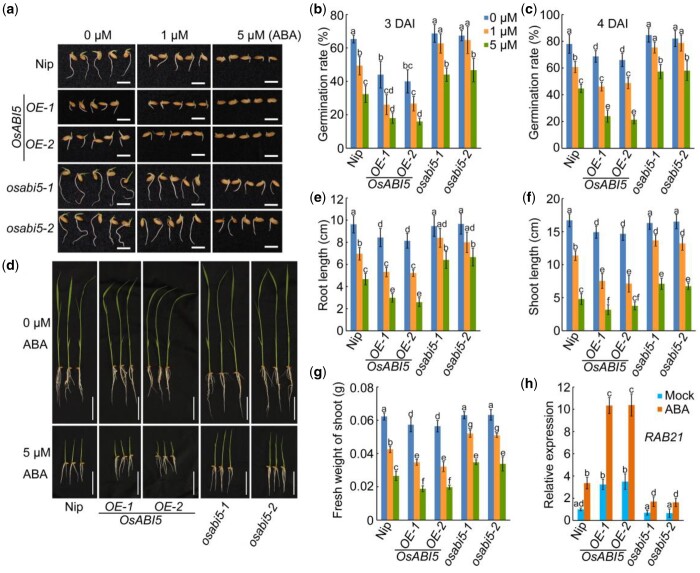
Next, we investigated the seed germination of Nip, OsABI5-OE lines, and osabi5 mutants. Our results showed that the germination rate of OsABI5-OE seeds under normal conditions was significantly lower than that of Nip, whereas the osabi5 seeds exhibited similar germination rate to Nip seeds (Figure 7, A–C). When subject to ABA treatment, the OsABI5-OE seeds were more sensitive, and osabi5 seeds less sensitive, to ABA compared with Nip seeds (Figure 7, A–C). Consistent with ABA sensitivity during germination, the root length, shoot length, and fresh weight of shoots of OsABI5-OE seedlings was significantly lower, whereas osabi5 seedlings was significantly higher, than those of Nip seedlings in the presence of ABA (Figure 7, D–G). Under normal growth conditions, the expression level of RAB21 was higher in OsABI5-OE plants than this in Nip, whereas no obvious difference between osabi5 mutants and Nip (Figure 7H). ABA treatment significantly induced the expression of RAB21 in Nip seedlings, and this tendency was enhanced in OsABI5-OE plants, but weakened in osabi5 mutants (Figure 7h). These results indicate that OsABI5 is a positive regulator of ABA signaling, and plays a negative role in seed germination and postgermination growth.
Figure 7.
OsABI5 positively regulates ABA sensitivity at germination and in seedlings. a, Germination performance of wild-type (Nip), OsABI5 OE (OsABI5-OE), and OsABI5 knockout (osabi5) seeds under 0-, 1-, or 5-μM ABA treatment at 3 DAI. Bars = 1 cm. B and C, Germination rate of seeds in (A) at 3 DAI (B) and 4 DAI (C). The error bars indicate the sds based on three independent replicates. D, Growth of 10 DAI Nip, OsABI5-OE, and osabi5 seedlings under 0- or 5-μM ABA treatment. Bars = 5 cm. The images of the plants were digitally extracted for comparison. E–G, Root length (E), shoot length (F), and fresh weight of shoots (G) under 0-, 1-, or 5-μM ABA treatment of 10 DAI Nip, OsABI5-OE, and osabi5 seedlings. Error bars indicate sds, n ≥ 10. H, Expression of RAB21 in Nip, OsABI5-OE, and osabi5 seedlings with or without ABA treatment. Ten DAI seedlings were treated with or without 1-μM ABA for 6 h. RNA was isolated from shoots and used for RT-qPCR. Actin1 was used as an internal control. The relative expression levels were represented by fold change relative to the expression levels of Nip-mock. The error bars indicate the sds based on three independent replicates. For (B, C, E, F, G, and H), different letters indicate significant differences (P < 0.05, one-way ANOVA with Tukey’s test).
Previous studies showed that OsABI5 is involved in the stress tolerance of rice and acts as a negative regulator (Zou et al., 2008). To further identify the function of OsABI5 in salt tolerance, we examined the salt tolerance of OsABI5-OE lines and osabi5 mutants at seedling stage. Ten DAI seedlings were treated with 150-mM NaCl for 7 d until the phenotype was observed. Then the plants were transferred to water without salt for additional 7 d before the survival rate was investigated. The results showed that survival rate of osabi5 mutants (∼45%–55%) were significantly higher than that of wild-type (∼38%) and OsABI5 OE lines (∼20%–25%; Supplemental Figure S3, d and e). Further detection of the expression of SOS1 and LEA3 showed that salt-induced expression of SOS1 and LEA3 was weakened in OsABI5-OE plants, whereas enhanced in osabi5 mutants, compared to that in Nip plants (Supplemental Figure S3, f and g). Correspondingly, OsABI5-OE plants accumulated more, and osabi5 mutants accumulated less, MDA and H2O2 under salt stress (Supplemental Figure S3, h and i). All these results suggest that OsABI5 plays a negative role in salt tolerance in rice.
OsABI5 acts downstream of OsSAE1 to regulate seed germination and salt tolerance
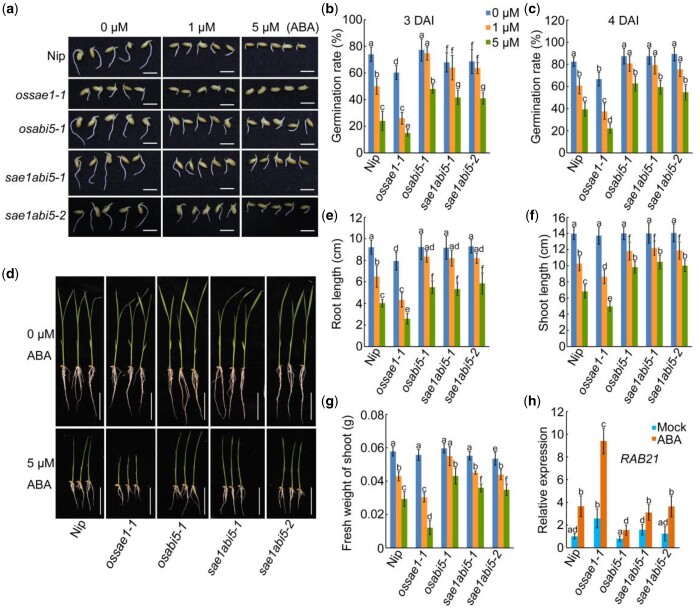
To examine the genetic relationship between OsABI5 and OsSAE1, we generated double mutant lines, sae1abi5-1 and sae1abi5-2. These lines contained a 2-bp insertion and a 5-bp deletion in OsSAE1, respectively, and both contained a 1-bp deletion in OsABI5; all mutations generated premature stop codons in the protein sequence (Supplemental Figure S4). The germination rates of sae1abi5-1 and sae1abi5-2 seeds (∼68%) were slightly higher than that of ossae1-1 (∼60%) but lower than that of osabi5-1 (∼77%) and Nip (∼74%) seeds at 3 DAI under normal conditions (Figure 8, A and B). In the presence of ABA, seeds of all genotypes exhibited some delay in germination, but sae1abi5-1 and sae1abi5-2 seeds were similar to osabi5 seeds, and less sensitive than Nip and ossae1 seeds (Figure 8, A–C). Similarly, root length, shoot length, and fresh weight of shoots showed no significant difference between the sae1abi5-1, sae1abi5-2, and osabi5 seedlings under ABA treatment, whereas root length, shoot length, and fresh weight of shoots of Nip and ossae1 seedlings were significantly lower than osabi5 seedlings (Figure 8, D–G). Analysis of the expression of RAB21 showed that the expression level of RAB21 in sae1abi5 plants was similar to that in osabi5 mutant, which was significantly lower than that in ossae1 mutant in the presence or absence of ABA treatment (Figure 8H). These results indicate that OsABI5 acts downstream of OsSAE1 to regulate ABA sensitivity during seed germination and postgermination growth.
Figure 8.
OsABI5 functions downstream of OsSAE1 to regulate ABA sensitivity at germination and in seedlings. A, Germination performance of Nip, ossae1, osabi5, and sae1abi5 double mutant seeds under 0-, 1-, or 5-μM ABA treatment at 3 DAI. Bars = 1 cm. B and C, Germination rate of seeds in (A) at 3 DAI (B) and 4 DAI (C). The error bars indicate the sds based on three independent replicates. D, Growth of 10 DAI Nip, ossae1, osabi5, and sae1abi5 double mutant seedlings under 0- or 5-μM ABA treatment. Bars = 5 cm. The images of the plants were digitally extracted for comparison. E–G, Root length (E), shoot length (F), and fresh weight of shoots (G) under 0-, 1-, or 5-μM ABA treatment of 10 DAI Nip, ossae1, osabi5, and sae1abi5 double mutant seedlings. Error bars indicate sds, n ≥ 10. H, Expression of RAB21 in Nip, ossae1, osabi5, and sae1abi5 double mutant seedlings with or without ABA treatment. Ten DAI seedlings were treated with or without 1-μM ABA for 6 h. RNA was isolated from shoots and used for RT-qPCR. Actin1 was used as an internal control. The relative expression levels were represented by fold change relative to the expression levels of Nip-mock. The error bars indicate the sds based on three independent replicates. For (B, C, E, F, G, and H), different letters indicate significant differences (P < 0.05, one-way ANOVA with Tukey’s test).
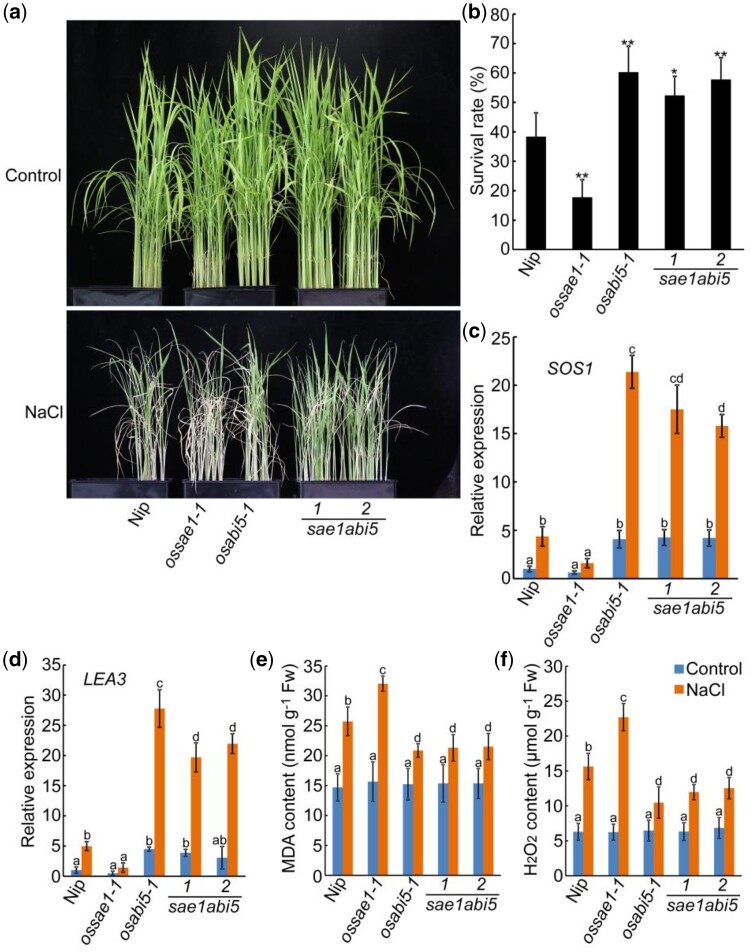
To investigate the above genetic relationship in salt tolerance, we subject 10 DAI seedlings to salt stress (150-mM NaCl). After exposure to NaCl for 7 d, the sae1abi5-1, sae1abi5-2, and osabi5 seedlings showed increased tolerance to salt stress compared to Nip seedlings, which in turn was higher than ossae1 seedlings (Figure 9A). After recovery for 7 d, the survival rate of ossae1 seedlings was significantly lower, and that of sae1abi5-1, sae1abi5-2, and osabi5 seedlings significantly higher, than that of Nip seedlings (Figure 9B). The expression levels of SOS1 and LEA3 in sae1abi5-1, sae1abi5-2, and osabi5 seedlings were significantly higher than that of ossae1 and Nip seedlings in the presence or absence of salt treatment (Figure 9, C and D). Determination of MDA and H2O2 content showed that the sae1abi5-1, sae1abi5-2, and osabi5 seedlings accumulated similar levels of MDA and H2O2 under salt stress, which were significantly lower than that of ossae1 and Nip seedlings (Figure 9, E and F). These results indicate that OsABI5-mediated pathway is required for OsSAE1 to regulate salt tolerance in rice seedlings.
Figure 9.
OsABI5 acts downstream of OsSAE1 to regulate salt tolerance in rice seedlings. A, Phenotypes of Nip, ossae1, osabi5, and sae1abi5 double mutants under salt stress. The images of the plants were digitally extracted for comparison. B, Survival rates of the plants shown in (A), 7 d after stopping salt stress. Approximately 50–60 seedlings were used per experiment. Data show mean ± sd of three independent replicates. Asterisks indicate significant differences compared to Nip at *P < 0.05 and **P < 0.01 by Student’s t test. C and D, Expression of SOS1 (C) and LEA3 (D) in Nip, ossae1, osabi5, and sae1abi5 double mutant seedlings with or without salt treatment. Ten DAI seedlings were treated with or without 150-mM NaCl for 6 h. RNA was isolated from shoots and used for RT-qPCR. Actin1 was used as an internal control. The relative expression levels were represented by fold change relative to the expression levels of Nip-control. E and F, MDA (E) and H2O2 (F) contents in leaves of 10 DAI seedlings with or without 150-mM NaCl treatment for 3 d. For (C, D, E, and F), the error bars indicate the sds based on three independent replicates, different letters indicate significant differences (P < 0.05, one-way ANOVA with Tukey’s test).
To further elucidate the relationship between OsSAE1- and OsABI5-mediated pathways, we compared the downstream DEGs regulated by OsSAE1 or OsABI5 in rice seedlings with or without salt treatment through transcriptome analysis. In the absence of salt stress, 34% (1,004) of DEGs in ossae1 mutant existed in OsABI5-OE plant, and 36% (1,561) of DEGs in OsSAE1-OE plant existed in osabi5 mutant (Supplemental Figure S5a; Supplemental Dataset 1). Further analysis of these DEGs showed that many genes involved in ROS-related pathway were regulated by OsABI5 or OsSAE1 (Supplemental Figure S5b). After exposure to salt treatment, 96 overlapping DEGs were identified in OsSAE1-OE plant (3,561 DEGs) and osabi5 mutant (140 DEGs), and 423 overlapping DEGs were identified in ossae1 mutant (3,650 DEGs) and OsABI5-OE plant (838 DEGs; Supplemental Figure S5c; Supplemental Dataset 2). These results suggest that OsSAE1 and OsABI5 are differential in terms of gene expression, and OsABI5 plays a partial role downstream of OsSAE1.
Discussion
Seed germination is a complex process, controlled by multiple genes and easily affected by environmental cues (Han and Yang, 2015; Penfield and MacGregor, 2017). Multiple phytohormones are involved in regulating seed germination; ABA has been recognized as a key inhibitor of seed germination (Han and Yang, 2015; Shu et al., 2016). ABA sensitivity is an important aspect of ABA signaling-dependent regulation. In this study, we identified an AP2 transcription factor, OsSAE1, which functions as a positive regulator of seed germination and salt tolerance in rice. OsSAE1-OE promotes seed germination and reduces ABA sensitivity, whereas OsSAE1 knockout retards seed germination and enhances ABA sensitivity, demonstrating that OsSAE1 is an important regulator in ABA signaling-dependent germination. Moreover, OsSAE1 directly binds the OsABI5 promoter to repress gene transcription during seed germination, resulting in a decrease in ABA sensitivity in seed germination and postgermination growth. The OsABI5-mediated pathway is required for OsSAE1-regulated seed germination and ABA sensitivity, as OsABI5 mutation rescues the seed germination and ABA sensitivity in ossae1 lines, revealing that OsSAE1 is a key factor involved in seed vigor that might be useful to genetic improvements of seed vigor for direct seeding of rice.
AP2/ERF transcription factors are a large group of factors that are known to regulate diverse processes of plant development and play very important roles in hormonal regulation and stress responses (Licausi et al., 2013; Xie et al., 2019; Feng et al., 2020). OsSAE1 encodes an AP2-type transcription factor and exhibits a close evolutionary relationship with OsSHAT1, OsRSR1, OsSNB, and OsIDS1. These four proteins are known to function as transcriptional repressors and, together with OsSAE1, are miR172 target genes (Zhu et al., 2009; Lee et al., 2014); miR172 has been shown to play a role in floral and seed developmental and salt tolerance in rice (Zhu et al., 2009; Cheng et al., 2021). This study deepens our understanding of the role of AP2 transcription factors, by determining that OsSAE1 acts as a transcriptional repressor involved in regulating seed germination and ABA sensitivity during seed germination and in young seedlings, through ABA signaling pathways mediated by OsABI5. Further investigation will be required to determine how miR172 may be involved in coordinating these developmental processes via OsSAE1 and OsABI5.
ABA-inhibited seed germination and postgermination growth is regulated by a core regulatory mechanism involving the following core components: PYR1/PYL/RCAR receptors, PP2C co-receptors, and SnRK2 kinases (Ma et al., 2009; Park et al., 2009; Nishimura et al., 2010; Soon et al., 2012). ABA binds to PYR1/PYL/RCAR to enable the interaction between PYR1/PYL/RCAR and PP2Cs, thus leading to the activation of SnRK2 protein kinases from inhibition by the PP2Cs. Activated SnRK2 kinases thereby phosphorylate downstream transcription factors and activate the expression of ABA-responsive genes (Ma et al., 2009; Park et al., 2009). In this study, we show that OsSAE1, a negative regulator of ABA signaling pathway, negatively regulates ABA-inhibited seed germination and postgermination growth by repressing the expression of OsABI5. Transcriptome analysis showed that several genes in the ABA signaling core were obviously regulated by OsSAE1, such as OsSAPKs, OsbZIPs, OsABIs, and OsPYL, indicating that the effect of OsSAE1 on seed germination and postgermination growth occurs through multiple pathways, of which the OsABI5-mediated pathway is one. Further studies should focus on the interaction of OsSAE1 with the upstream components of the ABA signaling pathway.
ABA-responsive element (ABRE) binding factor/ABRE binding protein/ABI5 is a small subfamily of bZIP transcription factors that function as key regulators of ABA and abiotic stress responses of plants. Previous reports indicate that OsABI5 possesses transactivation activity and the N-terminal 9-amino acids transactivation domain is critical for transactivation (Zou et al., 2007, 2008; Hong et al., 2011). Moreover, OsABI5 acts as a negative regulator of salt stress response and can be phosphorylated by SnRK2 family member OSRK1 (Chae et al., 2007; Zou et al., 2008), together with PYL5, PP2C30, and SAPK2, constitutes the minimal ABA signaling unit that modulates seed germination and early seedling growth (Kim et al., 2012). Constitutive expression of OsABI5 can rescue ABA-insensitivity of abi5-1 and confer ABA hypersensitivity to wild-type transgenic plants in Arabidopsis (Zou et al., 2007), suggesting that OsABI5 has similar functions as AtABI5, and OsABI5 is involved in ABA signal transduction. In this study, we indicate that OsABI5 is a positive regulator of ABA signaling and negatively regulates seed germination, postgermination growth, and salt tolerance in rice. Further investigations showed that OsABI5 is a downstream target of OsSAE1. Thus, our results enrich the regulatory network of OsABI5. Several studies have shown that OsABI5 could interact with other factors to regulate plant growth and response to external stimuli (Zou et al., 2007; Kim et al., 2012; Chen et al., 2018), it will be interesting to know whether the OsSAE1 and OsABI5 could interact physically in the future.
Salinity is a major environmental stress that constraints rice cultivation worldwide. Improving salt tolerance in rice is considered to be the most economic and effective options to ensure food security and ameliorate the constraints of increasingly saline soils in rice-cultivating regions. Rice is highly susceptible to salinity during early seedling growth stage (Nam et al., 2015), so improving the salt tolerance of rice seedlings is critical for seedling establishment and yield under salinity stress. ABA is a major abiotic stress-responsive hormone and plays pivotal roles in the stress responses by modulating the gene expression and cellular processes (Tang et al., 2016; Xu et al., 2018). The role of ABA in drought tolerance has been extensively investigated and it is generally believed that enhancing ABA biosynthesis or signaling transduction could improve the drought tolerance of plants (Sah et al., 2016; Zhao et al., 2016; Chang et al., 2017). Literature focusing on ABA-mediated responses under salt stress is less consistent, and both positive and negative effects of ABA on salt tolerance have been reported (Xiang et al., 2008; Liu et al., 2014; Sah et al., 2016; Kakan et al., 2021). In this study, we have shown that OsSAE1 positively regulates salt tolerance in rice seedlings by repressing the expression of OsABI5, a positive regulator of ABA signaling that increases susceptibility of rice to salt stress. OsABI5 exhibits high sequence similarity to OsbZIP23 and OsbZIP46 (Tang et al., 2012), which have both been characterized with essential roles in ABA signaling and stress tolerance in rice (Xiang et al., 2008; Tang et al., 2012).
Taken together, our results demonstrate that OsSAE1 may exert its functions through repression of ABI5-mediated ABA signaling for seed germination and salt stress tolerance. Thus, our research presents insights into the roles of OsSAE1 in improving seed vigor and salt tolerance in rice, and provides further evidence for the role of ABA signaling via OsABI5 in driving the rice salt response. These results offer targets and strategies for the breeding and development of new, direct seeding rice varieties suitable for saline–alkali land.
Materials and methods
Plant materials and grown conditions
Rice (O. sativa) variety Nip (O. sativa ssp. japonica) was used as a wild-type plant and recipient for genetic transformation in this study. Rice plants were cultivated at the Experimental Station of the Chinese Academy of Agricultural Sciences in Beijing during the natural growing season. In general, germinated seeds were sown in seed beds over the period 25–30 April, and planted in the fields 20–30 d later.
Generation of transgenic plants
Knockout ossae1 mutants, osabi5 mutants, and sae1abi5 double mutants were generated using CRISPR/Cas9 (Xu et al., 2017), in which the target regions 5′-AAGGGTCTGGGAGGGTCTTT-3′ of OsSAE1 and 5′-CAGGAGGAAGCGGTTTATGT-3′ of OsABI5 were introduced into the pHUN4c12 vector backbone, and the recombinant vector was transformed into Agrobacterium strain EHA105-pSOUP for rice transformation. To generate OE lines, the OsSAE1 or OsABI5 coding sequence was amplified from Nip cDNA and cloned into the binary vector pCAMBIA3301 (Wang et al., 2020a, 2020b) under control of the Ubiquitin promoter using the PstI and BamHI sites. The resulting plasmids were introduced separately into Nip via Agrobacterium-mediated transformation. Primers used for gene editing and plasmid construction are listed in Supplemental Table S1.
ABA and salt treatments
The effect of ABA on seed germination was assessed in triplicate as previously described (Song et al., 2020) with minor modifications. Briefly, 50 seeds per replicate of Nip and transgenic lines were imbibed in Petri dishes with 10-mL sterile distill water at 4°C for 48 h. Water was drained and replaced with ABA solution (0, 1, and 5 μM), and seeds were transferred to a growth chamber under a 14-h light/10-h dark cycle at 28°C. Germination was defined as the emergence of the radical, and the number of germinated seeds was counted every 24 h. The germination rate was calculated as the number of germinated seeds divided by 50 (total seeds assayed).
To determine the ABA sensitivity of postgermination growth, germinated rice seeds (∼30 seeds) were placed on a stainless steel sieve, which was placed in an air-tight 10-L plastic box. Seeds were treated with 6 L of Yoshida’s culture solution (Cui et al., 2015) containing 0, 1, or 5 μM ABA. The seedlings were grown under a 14-h light/10-h dark cycle at 28°C for 10 d. Root and shoot lengths were measured via ImageJ software; shoot weight was assessed using the whole shoot. Three replications were performed.
To asses salt-tolerance of rice seedlings, ∼30 germinated seeds (per replicate) were sown in a bottomless 96-well plate in a container of Yoshida’s culture solution (Cui et al., 2015) and grown under a 14-h light/10-h dark cycle at 28°C for 10 d. Treated plants were then transferred to Yoshida’s culture solution with 150- mM NaCl for another 7 d, then back to salt-free solution for 7 d of recovery, after which surviving seedlings were counted. Experiments were repeated at least 3 times with similar results.
RT-qPCR
Total RNA was extracted from seeds using a Plant Total RNA Purification Kit (GeneMark, Taichung, Taiwan) or from other tissues using an Ultrapure RNA Kit (CWBIO, Jiangsu, China), according to manufacturer’s instructions. Approximately 2-μg total RNA was reverse transcribed to cDNA with HiScript II Q RT SuperMix (Vazyme, Nanjing, China) according to manufacturer’s instructions. RT-qPCR was performed as previously described (Zhang et al., 2012), using the rice Actin1 gene as an internal standard to normalize gene expression. Primers used for RT-qPCR are listed in Supplemental Table S1.
Measurement of H2O2 and MDA contents
H2O2 content was determined using the method described previously (Wang et al., 2019). Briefly, 0.1 g of normal or salt-stressed rice leaves were harvested and homogenized in 1-mL cold acetone. Then, H2O2 content was determined using an H2O2 assay kit (Solarbio, Beijing, China) according to the manufacturer’s instructions.
To measure MDA, 0.1 g of normal or salt-stressed rice leaves was homogenized in 1 mL 0.1% (w/v) trichloroacetic acid (TCA) followed by centrifugation at 8,000g for 10 min at 4°C. Four volumes of 0.5% (w/v) thiobarbituric acid in 20% (w/v) TCA were added to one volume of supernatant; the mixture was incubated at 100°C for 1 h. The reaction was terminated by incubating the mixture on ice for 15 min, after which the absorbance was measured spectrophotometrically at 450, 532, and 600 nm. The amount of MDA was calculated from the formula provided with the MDA assay kit (Solarbio, Beijing, China).
Transcriptome analysis
Total RNA was isolated in triplicate from 5 DAI Nip, OsSAE1-OE, OsSAE1 knockout (ossae1), OsABI5 OE (OsABI5-OE), and OsABI5 knockout (osabi5) seedlings treated with 150-mM NaCl for 0 h (as control) and 3 h. Whole seedling was collected, immediately frozen in liquid nitrogen, and stored at −80°C. Total mRNA was isolated using the TRIzol method (TIANGEN BIOTECH, Beijing, China), quality checked on a Bioanalyzer (Agilent 2100), and used for RNA-seq library construction with the NEBNext Ultra RNA Library Prep Kilt for Illumina (NEB, Ipswich, MA, USA), according to manufacturer’s instructions. Multiplex paired-end adapters were used for multiplex libraries, which were sequenced (paired-end, 150 bp each) by the Illumina genome analyzer (Hiseq TM2500/4000). After removing adaptor and low-quality reads, clean reads were mapped to rice genome MSU version 7.0 using Tophat2, and analyzed using Cufflinks (Trapnell et al., 2012). The Poisson-dispersion model of fragments was used to conduct statistical analysis (false discovery rate < 0.05) and gene expression levels were estimated by fragments per kilobase of transcript per million fragments mapped. Differential expression analysis of two samples was performed using the DESeq R package and genes with an adjusted P < 0.05 found by DESeq were assigned as DEGs (Anders and Huber, 2010). Repeatability and correlation were evaluated by Pearson’s correlation coefficient (Schulze et al., 2012). GO enrichment analysis of the DEGs was implemented by the GOseq R packages based Wallenius noncentral hyper-geometric distribution (Young et al., 2010).
LUC transient expression assay
The transient dual-LUC expression assays were performed using rice protoplasts. To test the transcriptional repression activity of OsSAE1, a dual-LUC reporter assay was conducted. For the effectors generation, OsSAE1 coding region was fused with the GAL4 DBD to generate pBD-OsSAE1. OsSAE1 containing a transactivating domain VP16 in its N terminus was also fused with the GAL4 DBD to generate a pBD-VP16-OsSAE1 effector. The GAL4 DBD with or without VP16 (pBD and pBD-VP16) was used as the positive and the negative controls, respectively.
To evaluate the transcriptional regulatory activity of OsSAE1 on the OsABI5 promoter, the reporter (OsABI5p:LUC) and effector (35S:OsSAE1) plasmids or empty vector (EV) controls were transformed into rice protoplasts via polyethylene glycol-mediated transfection as previously described (Bart et al., 2006).
Firefly LUC and REN activities were measured with a dual-LUC reporting assay kit (Promega, Madison, WI, USA), according to manufacturer’s instructions. For each replication, three independent transformations were conducted. Primers used for plasmid construction are listed in Supplemental Table S1.
EMSA
Glutathione S-transferase-tagged OsSAE1 fusion protein (GST-OsSAE1) was expressed in Escherichia coli BL21 (DE3) and purified with ProteinIso GST Resin (Transgen, Beijing, China), according to manufacturer’s instructions. Oligonucleotides were synthesized and labeled with biotin. Unlabeled oligonucleotides were used as competitors. To generate double-stranded probes, an equal quantity of the complementary single-strand oligos was mixed, heated to 95°C for 2 min, and annealed by gradually cooling to 25°C. Probe sequences are given in Supplemental Table S1.
EMSA was performed using a LightShift Chemiluminescent EMSA Kit (Thermo Fisher, Waltham, MA, USA) according to manufacturer’s instructions. Reaction solutions were incubated for 20 min at room temperature. The protein–probe mixture was separated on a 5% (v/v) native polyacrylamide gel and transferred to a nylon membrane (GE, UK). Following crosslinking under UV light, DNA on the membrane was detected using a Chemiluminescent Nucleic Acid Detection Module (Thermo Fisher, Waltham, MA, USA), according to manufacturer’s instructions.
Phylogenetic analysis
All sequences of the AP2/ERF proteins were available at the National Center for Biotechnology Information (NCBI) website (http://www.ncbi.nlm.nih.gov). The nucleotide sequences and deduced amino acid sequences were aligned using the Multiple Sequence Alignment program of the DNAMAN software (version 7.0, LynnonBiosoft, Quebec, Canada). A phylogenetic tree was also constructed from the alignment these sequences using DNAMAN software.
Statistical analysis
Student’s t test was used for significant difference analysis between two samples. One-way analysis of variance (ANOVA) followed with Tukey’s test (P < 0.05) was used for pairwise multiple comparisons. All the analyses were performed with GraphPad Prism version 5 software.
Accession numbers
Sequence data from this article can be found in the Rice Genome Annotation Project website (http://rice.plantbiology.msu.edu/) under the following accession numbers: OsActin1, LOC_Os03g50885; OsSAE1, LOC_Os06g43220; OsABI5, LOC_Os01g64000; RAB21, LOC_Os11g26790; SOS1, LOC_Os12g44360; LEA3, LOC_Os05g46480. The transcriptome data from this study can be found in the National Center for Biotechnology Information Sequence Read Archive (NCBI SRA) under the accession number PRJNA793282.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Sequence and phylogenetic analysis of OsSAE1.
Supplemental Figure S2. Agronomic traits of OsSAE1 knockout and OE lines.
Supplemental Figure S3. OsABI5 negatively regulates salt tolerance in rice seedlings.
Supplemental Figure S4. Identification of sae1abi5 double mutants.
Supplemental Figure S5. DEGs in OsSAE1-OE, ossae1, OsABI5-OE, and osabi5 seedlings with or without salt treatment.
Supplemental Table S1. Primers used in this study.
Supplemental Dataset 1. DEGs in OsSAE1-OE, ossae1, OsABI5-OE, and osabi5 seedlings under normal conditions.
Supplemental Dataset 2. DEGs in response to NaCl in Nip, OsSAE1-OE, ossae1, OsABI5-OE, and osabi5 seedlings.
Funding
This work was funded by National Natural Science Foundation of China grants 31771706, 32030079, 31801445, and 31871551 to R.Q., R.H., and H.Q.; National Key R&D Program of China grant 2020YFE0202300 to R.Q.; and the Agricultural Science and Technology Innovation Program of the Chinese Academy of Agricultural Sciences to J.W. and R.H.
Conflict of interest statement. None declared.
Supplementary Material
These authors contributed equally (Y.L., J.Z., and Z.L.).
R.H., R.Q., and H.Q. conceived the projects. H.Q., Y.L., J.Z., Z.L., J.Q., J.W., and R.Q. performed the experiments and analyzed the data. H.Q. and R.H. wrote the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Hua Qin (qinhua@caas.cn).
References
- Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bart R, Chern M, Park CJ, Bartley L, Ronald PC (2006) A novel system for gene silencing using siRNAs in rice leaf and stem-derived protoplasts. Plant Methods 2: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai XT, Xu P, Zhao PX, Liu R, Yu LH, Xiang CB (2014) Arabidopsis ERF109 mediates cross-talk between jasmonic acid and auxin biosynthesis during lateral root formation. Nat Commun 5: 5833. [DOI] [PubMed] [Google Scholar]
- Chae MJ, Lee JS, Nam MH, Cho K, Hong JY, Yi SA, Suh SC, Yoon IS (2007) A rice dehydration-inducible SNF1-related protein kinase 2 phosphorylates an abscisic acid responsive element-binding factor and associates with ABA signaling. Plant Mol Biol 63: 151–169 [DOI] [PubMed] [Google Scholar]
- Chang HC, Tsai MC, Wu SS, Chang IF (2019) Regulation of ABI5 expression by ABF3 during salt stress responses in Arabidopsis thaliana. Bot Stud 60: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y, Nguyen BH, Xie Y, Xiao B, Tang N, Zhu W, Mou T, Xiong L (2017) Co-overexpression of the constitutively active form of OsbZIP46 and ABA-activated protein kinase SAPK6 improves drought and temperature stress resistance in rice. Front Plant Sci 8: 1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HL, Xu N, Wu Q, Yu B, Chu YL, Li XX, Huang JL, Jin L (2018) OsMADS27 regulates the root development in a NO3--Dependent manner and modulates the salt tolerance in rice (Oryza sativa L.). Plant Sci 277: 20–32 [DOI] [PubMed] [Google Scholar]
- Chen K, Zhang Q, Wang CC, Liu ZX, Jiang YJ, Zhai LY, Zheng TQ, Xu JL, Li ZK (2019) Genetic dissection of seedling vigour in a diverse panel from the 3,000 Rice (Oryza sativa L.) Genome Project. Sci Rep 9: 4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, He Q, Tang S, Wang H, Zhang X, Lv M, Liu H, Gao Q, Zhou Y, Wang Q, et al. (2021) The miR172/IDS1 signaling module confers salt tolerance through maintaining ROS homeostasis in cereal crops. New Phytol 230: 1017–1033 [DOI] [PubMed] [Google Scholar]
- Cheng X, Zhang S, Tao W, Zhang X, Liu J, Sun J, Zhang H, Pu L, Huang R, Chen T (2018) INDETERMINATE SPIKELET1 recruits histone deacetylase and a transcriptional repression complex to regulate rice salt tolerance. Plant Physiol 178: 824–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui LG, Shan JX, Shi M, Gao JP, Lin HX (2015) DCA1 acts as a transcriptional co-activator of DST and contributes to drought and salt tolerance in rice. PLoS Genet 11: e1005617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR (2010) Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol 61: 651–679 [DOI] [PubMed] [Google Scholar]
- Feng K, Hou XL, Xing GM, Liu JX, Duan AQ, Xu ZS, Li MY, Zhuang J, Xiong AS (2020) Advances in AP2/ERF super-family transcription factors in plant. Crit Rev Biotechnol 40: 750–776 [DOI] [PubMed] [Google Scholar]
- Fu FF, Xue HW (2010) Coexpression analysis identifies rice starch regulator1, a rice AP2/EREBP family transcription factor, as a novel rice starch biosynthesis regulator. Plant Physiol 154: 927–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gommers CMM, Monte E (2018) Seedling establishment: a dimmer switch-regulated process between dark and light signaling. Plant Physiol 176: 1061–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han C, Yang P (2015) Studies on the molecular mechanisms of seed germination. Proteomics 15: 1671–1679 [DOI] [PubMed] [Google Scholar]
- Han S, Min MK, Lee SY, Lim CW, Bhatnagar N, Lee Y, Shin D, Chung KY, Lee SC, Kim BG, Lee S (2017) Modulation of ABA signaling by altering VxGΦL motif of PP2Cs in Oryza sativa. Mol Plant 10: 1190–1205 [DOI] [PubMed] [Google Scholar]
- He Y, Yang B, He Y, Zhan C, Cheng Y, Zhang J, Zhang H, Cheng J, Wang Z (2019) A quantitative trait locus, qSE3, promotes seed germination and seedling establishment under salinity stress in rice. Plant J 97: 1089–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Zhao J, Yang B, Sun S, Peng L, Wang Z (2020) Indole-3-acetate beta-glucosyltransferase OsIAGLU regulates seed vigour through mediating crosstalk between auxin and abscisic acid in rice. Plant Biotechnol J 18: 1933–1945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong JY, Chae MJ, Lee IS, Lee YN, Nam MH, Kim DY, Byun MO, Yoon IS (2011) Phosphorylation-mediated regulation of a rice ABA responsive element binding factor. Phytochemistry 72: 27–36 [DOI] [PubMed] [Google Scholar]
- Jiang L, Ma X, Zhao S, Tang Y, Liu F, Gu P, Fu Y, Zhu Z, Cai H, Sun C, et al. (2019) The APETALA2-like transcription factor SUPERNUMERARY BRACT controls rice seed shattering and seed size. Plant Cell 31: 17–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo J, Lee YH, Song SI (2019) OsbZIP42 is a positive regulator of ABA signaling and confers drought tolerance to rice. Planta 249: 1521–1533 [DOI] [PubMed] [Google Scholar]
- Kakan X, Yu Y, Li S, Li X, Huang R, Wang J (2021) Ascorbic acid modulation by ABI4 transcriptional repression of VTC2 in the salt tolerance of Arabidopsis. BMC Plant Biol 21: 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Hwang H, Hong JW, Lee YN, Ahn IP, Yoon IS, Yoo SD, Lee S, Lee SC, Kim BG (2012) A rice orthologue of the ABA receptor, OsPYL/RCAR5, is a positive regulator of the ABA signal transduction pathway in seed germination and early seedling growth. J Exp Bot 63: 1013–1024 [DOI] [PubMed] [Google Scholar]
- Kong Y, Chen S, Yang Y, An C (2013) ABA-insensitive (ABI) 4 and ABI5 synergistically regulate DGAT1 expression in Arabidopsis seedlings under stress. FEBS Lett 587: 3076–3082 [DOI] [PubMed] [Google Scholar]
- Lee DY, An G (2012) Two AP2 family genes, supernumerary bract (SNB) and osindeterminate spikelet 1 (OsIDS1), synergistically control inflorescence architecture and floral meristem establishment in rice. Plant J 69: 445–461 [DOI] [PubMed] [Google Scholar]
- Lee YS, Lee DY, Cho LH, An G (2014) Rice miR172 induces flowering by suppressing OsIDS1 and SNB, two AP2 genes that negatively regulate expression of Ehd1 and florigens. Rice 7: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licausi F, Ohme-Takagi M, Perata P (2013) APETALA2/Ethylene Responsive Factor (AP2/ERF) transcription factors: mediators of stress responses and developmental programs. New Phytol 199: 639–649 [DOI] [PubMed] [Google Scholar]
- Lin JH, Yu LH, Xiang CB (2020) ARABIDOPSIS NITRATE REGULATED 1 acts as a negative modulator of seed germination by activating ABI3 expression. New Phytol 225: 835–847 [DOI] [PubMed] [Google Scholar]
- Liu C, Mao B, Ou S, Wang W, Liu L, Wu Y, Chu C, Wang X (2014) OsbZIP71, a bZIP transcription factor, confers salinity and drought tolerance in rice. Plant Mol Biol 84: 19–36 [DOI] [PubMed] [Google Scholar]
- Liu CT, Schlappi MR, Mao BG, Wang W, Wang AJ, Chu CC (2019) The bZIP73 transcription factor controls rice cold tolerance at the reproductive stage. Plant Biotechnol J 17: 1834–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, Tang N, Li X, Xie Y, Xiang D, Fu J, Shen J, Yang J, Tu H, Li X, et al. (2019) Reversible histone H2B monoubiquitination fine-tunes abscisic acid signaling and drought response in rice. Mol Plant 12: 263–277 [DOI] [PubMed] [Google Scholar]
- Ma Y, Cao J, He J, Chen Q, Li X, Yang Y (2018) Molecular mechanism for the regulation of ABA homeostasis during plant development and stress responses. Int J Mol Sci 19: 3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E (2009) Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324: 1064–1068 [DOI] [PubMed] [Google Scholar]
- Magnani E, Sjolander K, Hake S (2004) From endonucleases to transcription factors: evolution of the AP2 DNA binding domain in plants. Plant Cell 16: 2265–2277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao JL, Miao ZQ, Wang Z, Yu LH, Cai XT, Xiang CB (2016) Arabidopsis ERF1 mediates cross-talk between ethylene and auxin biosynthesis during primary root elongation by regulating ASA1 expression. PLoS Genet 12: e1006076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T, Suzuki K, Fujimura T, Shinshi H (2006) Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol 140: 411–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam MH, Bang E, Kwon TY, Kim Y, Kim EH, Cho K, Park WJ, Kim BG, Yoon IS (2015) Metabolite profiling of diverse rice germplasm and identification of conserved metabolic markers of rice roots in response to long-term mild salinity stress. Int J Mol Sci 16: 21959–21974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura N, Sarkeshik A, Nito K, Park SY, Wang A, Carvalho PC, Lee S, Caddell DF, Cutler SR, Chory J, et al. (2010) PYR/PYL/RCAR family members are major in-vivo ABI1 protein phosphatase 2C-interacting proteins in Arabidopsis. Plant J 61: 290–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamuro JK, Caster B, Villarroel R, VanMontagu M, Jofuku KD (1997) The AP2 domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis. Proc Natl Acad Sci USA 94: 7076–7081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TF, et al. (2009) Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324: 1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S, MacGregor DR (2017) Effects of environmental variation during seed production on seed dormancy and germination. J Exp Bot 68: 819–825 [DOI] [PubMed] [Google Scholar]
- Raghavendra AS, Gonugunta VK, Christmann A, Grill E (2010) ABA perception and signalling. Trends Plant Sci 15: 395–401 [DOI] [PubMed] [Google Scholar]
- Sah SK, Reddy KR, Li J (2016) Abscisic acid and abiotic stress tolerance in crop plants. Front Plant Sci 7: 571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze SK, Kanwar R, Golzenleuchter M, Therneau TM, Beutler AS (2012) SERE: single-parameter quality control and sample comparison for RNA-Seq. BMC Genomics 13: 524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharoni AM, Nuruzzaman M, Satoh K, Shimizu T, Kondoh H, Sasaya T, Choi IR, Omura T, Kikuchi S (2011) Gene structures, classification and expression models of the AP2/EREBP transcription factor family in rice. Plant Cell Physiol 52: 344–360 [DOI] [PubMed] [Google Scholar]
- Shu K, Liu XD, Xie Q, He ZH (2016) Two faces of one seed: hormonal regulation of dormancy and germination. Mol Plant 9: 34–45 [DOI] [PubMed] [Google Scholar]
- Skubacz A, Daszkowska-Golec A, Szarejko I (2016) The role and regulation of ABI5 (ABA-Insensitive 5) in plant development, abiotic stress responses and phytohormone crosstalk. Front Plant Sci 7: 1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Wang G, Wu H, Fan X, Liang L, Zhao H, Li S, Hu Y, Liu H, Ayaad M, Xing Y (2020) OsMFT2 is involved in the regulation of ABA signaling-mediated seed germination through interacting with OsbZIP23/66/72 in rice. Plant J 103: 532–546 [DOI] [PubMed] [Google Scholar]
- Soon FF, Ng LM, Zhou XE, West GM, Kovach A, Tan MH, Suino-Powell KM, He Y, Xu Y, Chalmers MJ, et al. (2012) Molecular mimicry regulates ABA signaling by SnRK2 kinases and PP2C phosphatases. Science 335: 85–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang N, Ma S, Zong W, Yang N, Lv Y, Yan C, Guo Z, Li J, Li X, Xiang Y, et al. (2016) MODD mediates deactivation and degradation of OsbZIP46 to negatively regulate ABA signaling and drought resistance in rice. Plant Cell 28: 2161–2177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang N, Zhang H, Li X, Xiao J, Xiong L (2012) Constitutive activation of transcription factor OsbZIP46 improves drought tolerance in rice. Plant Physiol 158: 1755–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7: 562–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa T, Nakashima K, Miyakawa T, Kuromori T, Tanokura M, Shinozaki K, Yamaguchi-Shinozaki K (2010) Molecular basis of the core regulatory network in ABA responses: sensing, signaling and transport. Plant Cell Physiol 51: 1821–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Qin H, Zhou S, Wei P, Zhang H, Zhou Y, Miao Y, Huang R (2020a) The ubiquitin-binding protein OsDSK2a mediates seedling growth and salt responses by regulating gibberellin metabolism in rice. Plant Cell 32: 414–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Hou Y, Qiu J, Wang H, Wang S, Tang L, Tong X, Zhang J (2020b) Abscisic acid promotes jasmonic acid biosynthesis via a ‘SAPK10-bZIP72-AOC’ pathway to synergistically inhibit seed germination in rice (Oryza sativa). New Phytol 228: 1336–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Liang C, Meng Z, Li Y, Abid MA, Askari M, Wang P, Wang Y, Sun G, Cai Y (2019) Leveraging atriplex hortensis choline monooxygenase to improve chilling tolerance in cotton. Environ Exp Bot 162: 364–373 [Google Scholar]
- Xiang Y, Tang N, Du H, Ye HY, Xiong LZ (2008) Characterization of OsbZIP23 as a key player of the basic leucine zipper transcription factor family for conferring abscisic acid sensitivity and salinity and drought tolerance in rice. Plant Physiol 148: 1938–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Nolan TM, Jiang H, Yin Y (2019) AP2/ERF transcription factor regulatory networks in hormone and abiotic stress responses in Arabidopsis. Front Plant Sci 10: 228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Lantzouni O, Bruggink T, Benjamins R, Lanfermeijer F, Denby K, Schwechheimer C, Bassel GW (2020) A molecular signal integration network underpinning Arabidopsis seed germination. Curr Biol 30: 3703–3712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N, Chu Y, Chen H, Li X, Wu Q, Jin L, Wang G, Huang J (2018) Rice transcription factor OsMADS25 modulates root growth and confers salinity tolerance via the ABA-mediated regulatory pathway and ROS scavenging. PLoS Genet 14: e1007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R, Wei P, Yang J (2017) Use of CRISPR/Cas genome editing technology for targeted mutagenesis in rice. Methods Mol Biol 1498: 33–40 [DOI] [PubMed] [Google Scholar]
- Young MD, Wakefield MJ, Smyth GK, Oshlack A (2010) Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 11: R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zhang J, Quan R, Pan X, Wan L, Huang R (2013) EAR motif mutation of rice OsERF3 alters the regulation of ethylene biosynthesis and drought tolerance. Planta 237: 1443–1451 [DOI] [PubMed] [Google Scholar]
- Zhang ZJ, Wang J, Zhang RX, Huang RF (2012) The ethylene response factor AtERF98 enhances tolerance to salt through the transcriptional activation of ascorbic acid synthesis in Arabidopsis. Plant J 71: 273–287 [DOI] [PubMed] [Google Scholar]
- Zhao C, Zhang H, Song C, Zhu JK, Shabala S (2020a) Mechanisms of plant responses and adaptation to soil salinity. Innovation (N Y) 1: 100017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Nie K, Zhou H, Yan X, Zhan Q, Zheng Y, Song CP (2020b) ABI5 modulates seed germination via feedback regulation of the expression of the PYR/PYL/RCAR ABA receptor genes. New Phytol 228: 596–608 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Chan Z, Gao J, Xing L, Cao M, Yu C, Hu Y, You J, Shi H, Zhu Y, et al. (2016) ABA receptor PYL9 promotes drought resistance and leaf senescence. Proc Natl Acad Sci USA 113: 1949–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Cheng SF, Song YL, Huang YL, Zhou SL, Liu XY, Zhou DX (2015) The interaction between rice ERF3 and WOX11 promotes crown root development by regulating gene expression involved in cytokinin signaling. Plant Cell 27: 2469–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Lu D, Li C, Luo J, Zhu BF, Zhu J, Shangguan Y, Wang Z, Sang T, Zhou B, et al. (2012) Genetic control of seed shattering in rice by the APETALA2 transcription factor shattering abortion1. Plant Cell 24: 1034–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu QH, Upadhyaya NM, Gubler F, Helliwell CA (2009) Over-expression of miR172 causes loss of spikelet determinacy and floral organ abnormalities in rice (Oryza sativa). BMC Plant Biol 9: 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong W, Tang N, Yang J, Peng L, Ma SQ, Xu Y, Li GL, Xiong LZ (2016) Feedback regulation of ABA signaling and biosynthesis by a bZIP transcription factor targets drought-resistance-related genes. Plant Physiol 171: 2810–2825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou M, Guan Y, Ren H, Zhang F, Chen F (2007) Characterization of alternative splicing products of bZIP transcription factors OsABI5. Biochem Biophys Res Commun 360: 307–313 [DOI] [PubMed] [Google Scholar]
- Zou M, Guan Y, Ren H, Zhang F, Chen F (2008) A bZIP transcription factor, OsABI5, is involved in rice fertility and stress tolerance. Plant Mol Biol 66: 675–683 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.