Abstract
Recent findings have revealed the important roles of microRNAs (miRNAs) in the secondary responses to oxidative damage caused by iron (Fe) excess. However, the functional importance of miRNAs in plant responses to Fe deficiency remains to be explored. Here, we show that the expression level of miR164 in Arabidopsis (Arabidopsis thaliana) roots was repressed by Fe deficiency. Primary root length, lateral root number, ferric reductase activity, and mRNA abundance of IRON-REGULATED TRANSPORTER1 (IRT1) and FERRIC REDUCTION OXIDASE2 (FRO2) were higher in the mir164b mutant than in the wild-type (WT) under Fe-deficient conditions. Analysis of the Fe concentrations and ferric reductase activities in the roots of miR164 knockdown transgenic plants showed that members of the miR164 family had different functions in Fe-deficiency responses. Promoter::GUS analysis showed that NAM/ATAF/CUC (NAC) domain transcription factor5 (NAC5) is regulated at both transcriptional and posttranscriptional levels under Fe-deficient conditions. Transgenic Arabidopsis plants overexpressing NAC5 were more tolerant of Fe deficiency than the WT. NAC5 has transactivation activity and directly transactivates the expression of Nuclear Factor Y, Subunit A8 (NFYA8), as demonstrated by chromatin immunoprecipitation followed by quantitative polymerase chain reaction, electrophoretic mobility shift assay (EMSA), and dual-luciferase reporter assay. Like overexpression of NAC5, overexpression of NFYA8 increases primary root length, lateral root number, ferric reductase activity, and mRNA abundance of IRT1 and FRO2 under Fe-deficient conditions. Thus, MIR164b is important for Fe-deficiency responses by its regulation of the NAC5–NFYA8 module.
The NAC domain transcription factor5 protein plays an important role in Arabidopsis tolerance to iron deficiency by promoting the expression of Nuclear Factor Y, Subunit A8.
Introduction
As the cofactor for many important enzymatic reactions, iron (Fe) is essential for all organisms. The fixation of insoluble Fe in the field, however, reduces the availability of Fe to plants, especially in calcareous soils with high pH and high bicarbonate content. Fe deficiency has become a major factor limiting crop yield worldwide (Briat et al., 2015). Because limited Fe availability in soil also reduces the Fe content of plant-derived food, Fe deficiency-induced anemia is a common nutritional disorder for humans in developing countries (Grillet et al., 2018; Narayanan et al., 2019). An improved understanding of the genetic mechanisms of Fe homeostasis in plants could help increase the Fe content in crops.
To cope with Fe-deficient environments, plants have evolved two responsive strategies. Graminaceous species use a chelation-based Fe acquisition mechanism (Strategy II) in that they secret phytosiderophores and uptake Fe(III)-phytosiderophores via the Yellow Stripe Like transporters (Römheld and Marschner, 1986; Curie et al., 2001). With the exception of graminaceous species, most plants, including Arabidopsis (Arabidopsis thaliana), employ a reduction-based Fe-acquisition mechanism (Strategy I). They first acidify the rhizosphere by excreting protons and phenolic compounds to increase the solubility of Fe (Kobayashi and Nishizawa, 2012). The Fe(III)-chelates is then reduced to Fe2+ by the ferric-chelate reductase FERRIC REDUCTION OXIDASE2 (FRO2), which is the rate-limiting step for mobilizing Fe from soil (Robinson et al., 1999; Satbhai et al., 2017). Finally, the reduced Fe2+ is transported across the root plasma membrane by the metal transporter IRON-REGULATED TRANSPORTER1 (IRT1) (Eide et al., 1996; Vert et al., 2002).
Although recent research has demonstrated the importance of posttranslational modifications in plant Fe-nutritional responses (Shin et al., 2013; Dubeaux et al., 2018; Gratz et al., 2019), the transcriptional regulation of gene expression plays a crucial role in Fe deficiency responses in plants (Riaz and Guerinot, 2021; Sun et al., 2021). The basic helix–loop–helix (bHLH) transcription factors FERLIKE IRON DEFICIENCY-INDUCED TRANSCRIPTION FACTOR (FIT) and POPEYE (PYE) are the key regulators of the Fe-deficiency response in Arabidopsis (Colangelo and Guerinot, 2004; Yuan et al., 2005; Long et al., 2010). The nonoverlapping locations of the target genes of FIT and PYE indicate that the two transcription factors are involved in separate gene regulons. FIT physically interacts with four subgroup Ib bHLH proteins, bHLH038, bHLH039, bHLH100, and bHLH101, to upregulate its target genes (Yuan et al., 2008; Wang et al., 2013; Cai et al., 2021). PYE acts as a transcriptional repressor and is involved in the internal mobilization of Fe in the root and its transport to the shoot (Long et al., 2010). To date, 17 of 133 members in the bHLH family has been determined to be involved in Fe homeostasis in Arabidopsis (Gao et al., 2015; Cui et al., 2018; Lei et al., 2020), indicating the importance of the bHLH family in plant adaptation to Fe deficiency.
NAM, ATAF1/2, and CUC1/2 (NAC) proteins constitute a large plant-specific family of transcription factors, with over 100 members in Arabidopsis (Olsen et al., 2005). Several reports indicate that the NAC family could be associated with Fe homeostasis in Strategy II plants. In wheat (Triticum aestivum L.), a NAC transcription factor, NAM-B1, accelerated senescence and increased grain protein, zinc, and Fe content (Uauy et al., 2006). In rice (Oryza sativa), IDE-binding factor 2 (IDEF2), which belongs to an uncharacterized branch of the NAC transcription factor family, regulates the genes involved in Fe homeostasis by recognizing Fe deficiency-responsive element 2 in their promoters (Ogo et al., 2008). However, an understanding of NAC in Fe-deficiency responses in Strategy I plants is lacking.
A subset of NAC transcription factors, including CUP-SHAPED COTYLEDON1 (CUC1), CUC2, NAC domain transcription factor1 (NAC1), NAC4, NAC5 (At5g61430), and ORE1 are posttranscriptionally regulated by miRNA164 (Laufs et al., 2004; Mallory et al., 2004; Guo et al., 2005; Kim et al., 2009). Plant miRNAs are involved in various developmental processes. Recent findings have revealed the important roles of miRNAs in plant adaptation to nutrient deficiency or excess, such as copper deficiency (Yamasaki et al., 2009; Pilon, 2017), phosphorus deficiency (Kuo and Chiou 2011), and copper/Fe excess (Sunkar et al., 2006). The expression and phenotypic analyses of transgenic plants over-expressing Fe and other heavy metal-related miRNAs have suggested that the miRNAs mainly function in the secondary responses to oxidative damage caused by heavy metal stress. The functional importance of miRNAs in Fe-deficiency responses in plants remains to be explored.
Here, we report that the expression of miR164 in Arabidopsis roots is repressed by Fe deficiency. Analysis of mir164b knockout plants, miR164 knock-down transgenic plants, and NAC5-overexpressing lines showed that the posttranscriptional regulation of NAC5 by miR164 is important for Fe uptake in Arabidopsis. We also demonstrated that NAC5 could directly activate the expression level of Nuclear Factor Y, Subunit A8 (NFYA8). NFYA8-overexpressing Arabidopsis was tolerant of Fe deficiency. Taken together, our results reveal the importance of the MIR164b–NAC5–NFYA8 module in Fe homeostasis in Arabidopsis.
Results
miR164 expression is repressed by Fe deficiency in Arabidopsis roots
To identify miRNAs and other endogenous small RNAs that respond to Fe deficiency, we used Solexa high-throughput sequencing technology to generate and sequence two small RNA libraries from Fe-sufficient and -deficient Arabidopsis roots. The Fe-deficient treatment was effective because the expression of the marker gene, AtIRT1 (At4G19690), was significantly induced by Fe deficiency as reported previously (Eide et al., 1996; Vert et al., 2002; Supplemental Figure S1). The two small RNA libraries yielded a total of >23 million raw reads, and after 5′ and 3′ adaptor trimming, >87% of the remaining raw reads ranged in size from 18 to 26 nt (Supplemental Table S1). Of these, >16 million reads corresponding to 3.1 million unique sequences could be perfectly mapped to Arabidopsis genome (The Arabidopsis Information Resource 9). Sequences that could not be mapped to Arabidopsis genome were discarded and only those perfectly mapped were analyzed further. The genome-matched sequences were assigned to several RNA groups, including known miRNAs, rRNAs, tRNAs, repeats, snRNAs, snoRNAs, and others (Supplemental Table S2). The known miRNAs accounted for 8.34% and 7.89% of all sequence reads for Fe-sufficient and -deficient small RNA libraries, respectively. Based on the difference in numbers of normalized sequence reads between Fe-deficient and -sufficient roots, 38 conserved miRNAs with >1.5-fold relative change were identified (Supplemental Table S3).
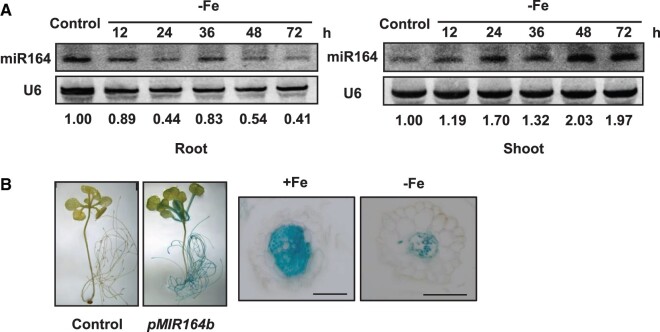
miR164 attracted our attention because of its high expression levels in roots (Mallory et al., 2004; Guo et al., 2005; Supplemental Table S3). Fe deficiency significantly reduced the expression levels of MIR164a and MIR164b in Arabidopsis roots (Supplemental Table S3). To confirm the results obtained with Solexa high-throughput sequencing technology, we conducted small RNA northern blot analysis. Consistent with the Solexa high-throughput sequencing results, the expression level of miR164 in roots reduced by ∼60% (relative to unstressed plants) when plants were subjected to Fe deficiency for 3 d (Figure 1A). Interestingly, the expression level of miR164 in shoots was upregulated by Fe deficiency (Figure 1A). We further collected xylem sap of Arabidopsis and found that miR164 existed in the xylem sap under both Fe-sufficient and -deficient conditions (Supplemental Figure S2), which indicated that miR164 could be moved from root-to-shoot. In addition, the expression level of miR164 was enhanced by Fe excess in both shoots and roots of Arabidopsis (Supplemental Figure S3).
Figure 1.
Regulation of miR164 by Fe deficiency. A, Assay of miR164 abundance in Arabidopsis roots and shoots by small RNA northern blot. U6 was used as the loading control. Numbers below each lane indicate the expression level of miR164 relative to U6. B, MIR164bpro::GUS expression pattern in transgenic Arabidopsis roots. Scale bars = 50 µm.
The miR164 family in Arabidopsis has three members, among which miR164b is the most abundant (Raman et al., 2008). We therefore selected miR164b for further study. We first used the promoter::GUS transgenic lines to examine the expression pattern of MIR164b. Glucuronidase (GUS) staining patterns in several transgenic lines showed that the promoter was active mainly in petiole and root vascular systems of 14-d-old Arabidopsis under Fe-sufficient conditions (Figure 1B). In agreement with the small RNA northern blot analysis, Fe deficiency caused a substantial decrease in the GUS activity in Arabidopsis roots (Figure 1B).
The mir164b mutant is tolerant of Fe-deficient stress
The downregulation of miR164 by Fe deficiency and its strong expression in roots prompted us to analyze its potential role in Arabidopsis responses to Fe deficiency. To investigate the function of miR164b, we searched the publicly available T-DNA collections and obtained a T-DNA insertion mutant (SALK_136105C in the Columbia background) from the Arabidopsis Biological Resource Center. Plants homozygous for the T-DNA insertion were identified by PCR and sequencing of the T-DNA flanking region confirmed the insertion site in loop of the predicted hairpin precursor of MIR164b (Supplemental Figure S4A). Reverse transcription PCR (RT-PCR) showed that MIR164b transcripts were absent in the T-DNA line designated as mir164b (Supplemental Figure S4B). The hybridization signal from the miR164 probe was much weaker in the mir164b lines than in wild-type (WT) plants (Supplemental Figure S4C), indicating that mir164b represented a null allele as described by Mallory et al. (2004).
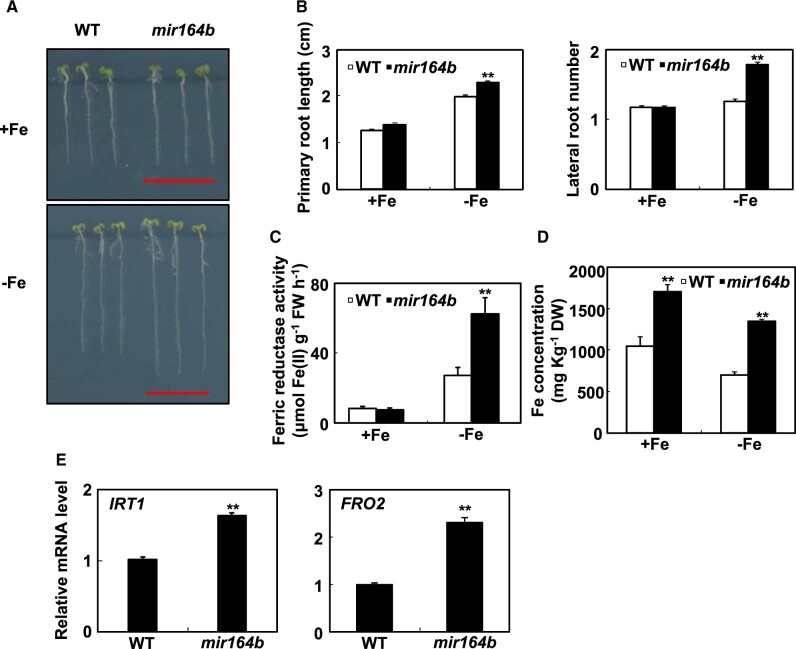
In a previous study, the mir164b mutant promoted lateral root emergence when grown on Murashige and Skoog (MS) medium (Guo et al., 2005). When we grew the mir164b mutant on 0.5 MS medium, we observed no difference in root morphology between the mir164b mutant and WT Arabidopsis (Figure 2, A and B). When the mir164b loss-of-function mutant was germinated and grown on Fe-free 0.5 MS medium for 7 d, however, primary root length and lateral root number were significantly higher for the mir164b mutant than for the WT (Figure 2, A and B). We also generated transgenic Arabidopsis plants overexpressing MIR164b, and found that primary root length was much shorter for 35S::MIR164b transgenic plants than for the WT under Fe-deficient conditions (Supplemental Figure S5). In parallel, the rate of leaf discoloration was higher for 35S::MIR164b transgenic plants than for the WT (Supplemental Figure S5). Interestingly, the ferric reductase activity was 2.3 times greater for mir164b mutant plants than for WT plants under Fe-deficient conditions (Figure 2C). These results indicated that the root system is more developed for mir164b mutant plants than for WT plants and that Fe acquisition might be more efficient in mir164b mutant plants than in the WT plants under Fe-deficient conditions.
Figure 2.
mir164b mutants have increased tolerance to Fe deficiency. A, Representative images of mir164b mutant seedlings grown on Fe-deficient medium for 7 d. Scale bars = 1 cm. B, The primary root length and lateral root number of mir164b mutant seedlings grown on Fe-deficient medium for 7 d. C, The ferric reductase activity in the roots of mir164b mutant seedlings grown on Fe-deficient medium for 7 d. D, The Fe concentration in the roots of 18-d-old hydroponically grown Arabidopsis, either supplied with Fe or transferred to a Fe-free solution for 36 h. E, The effects of Fe deficiency on the expression levels of IRT1 and FRO2 in 18-d-old hydroponically grown WT and mir164b mutant plants. RT-qPCR quantifications were normalized to the expression of TUB2. Error bars represent standard errors of three independent experiments, with 20–30 plants per experiment (B–D). Error bars represent standard errors of three biological replicates (E). Asterisks indicate significant differences between Fe-sufficient and Fe-deficient treatments at P < 0.01 according to t tests.
To test the latter hypothesis, we used inductively coupled plasma mass spectrometer (ICP-MS) to measure the Fe concentrations in the roots of the mir164b mutant and the WT. Given the small root biomass of Arabidopsis seedlings, we used hydroponically grown 18-d-old Arabidopsis plants. Under both Fe-sufficient and -deficient conditions, the Fe concentrations in the roots were much higher in the mir164b mutant than in the WT (Figure 2D). After the plants were subjected to Fe deficiency for 36 h, the Fe concentration in the roots of the mir164b mutant was 1,352 mg kg−1 DW, which was ∼two times greater than that in WT plants (Figure 2D). Consistent with these results, the mRNA abundance of IRT1 and FRO2 in the roots under Fe-deficient conditions was much higher for the mir164b mutant than for the WT (Figure 2E). These results supported our hypothesis that mir164b mutant has increased tolerance to Fe deficiency.
Functional specialization in Fe-deficiency responses of miR164 members
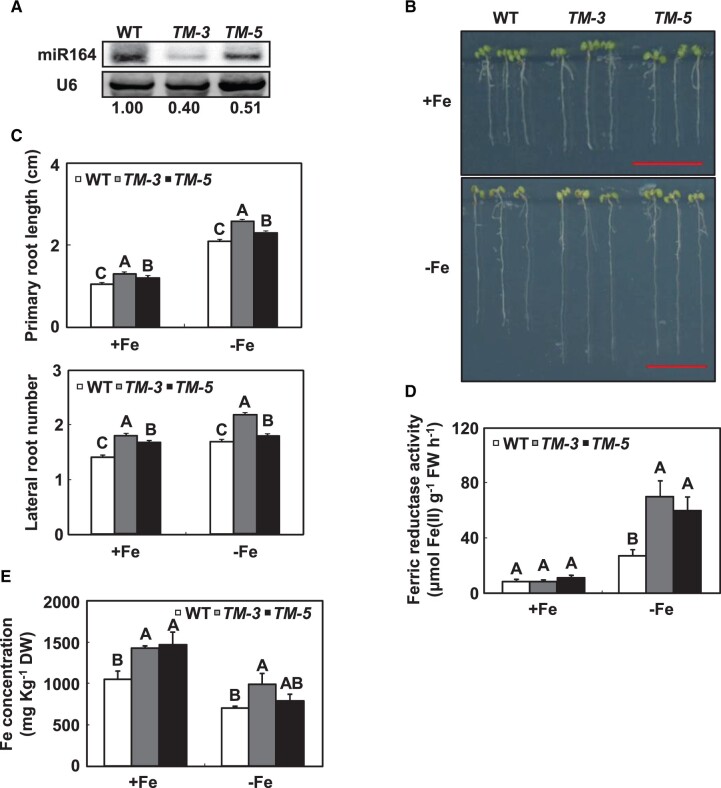
Analysis of mir164abc triple-mutant plants revealed that miR164a, miR164b, and miR164c control flower development in a redundant manner (Sieber et al., 2007). However, different expression patterns of the individual miR164 genes suggested that these genes may be redundant for some functions but not for others (Sieber et al., 2007). mir164a-4 had deepened serrations of the leaf margins (Nikovics et al., 2006), while the phenotype of mir164b-1 was the same as that of the WT (Mallory et al., 2004). These results suggested that miR164 family members have different functions during plant development. To check whether functional redundancy existed in Fe-deficient response among the members in miR164 family, we designed the short tandem target mimic transgenic lines targeting MIR164a, MIR164b, and MIR164c as described by Yan et al. (2012). The destruction of MIR164a, MIR164b, and MIR164c was verified by small RNA northern blot, and two independent knock-down lines (TM-3 and TM-5) were chosen for analysis of their expression levels (Figure 3A). Under both Fe-sufficient and -deficient conditions, TM transgenic seedlings had longer primary roots and more lateral roots than WT seedlings (Figure 3, B and C). Consistent with the observation in the mir164b mutant, the ferric reductase activities in roots were higher for TM transgenic seedlings than for WT seedlings under Fe-deficient conditions (Figure 3D).
Figure 3.
The responses of miR164 knock-known transgenic plants (TM) to Fe deficiency. A, miR164 levels in TM transgenic plants as indicated by small RNA northern blot. U6 was used as the loading control. Numbers below each lane indicate the expression level of miR164 relative to U6. B, Representative images of TM transgenic seedlings grown on Fe-deficient medium for 7 d. Scale bars = 1 cm. C, The primary root length and lateral root number of TM transgenic seedlings grown on Fe-deficient medium for 7 d. D, The ferric reductase activity in the roots of TM transgenic seedlings grown on Fe-deficient medium for 7 d. E, The Fe concentration in the roots of 18-d-old hydroponically grown Arabidopsis, either supplied with Fe or transferred to a Fe-free solution for 36 h. Error bars represent standard errors of three independent experiments, with 20–30 plants per experiment (C–E). Means with the same letter are not significantly different at P < 0.01 according to one-way ANOVA followed by Tukey’s multiple comparison test.
TM transgenic plants were similar to WT plants during vegetative development, except that the leaf margins of TM transgenic plants developed deepened serrations (Supplemental Figure S6). After the plants were subjected to Fe deficiency for 36 h, the Fe concentrations in the roots of 18-d-old hydroponically plants were higher for TM transgenic plants than for WT plants (Figure 3E). However, the increased ratios of Fe concentrations and ferric reductase activities in the roots of TM transgenic plants versus the WT were comparable to those of mir164b mutants versus the WT. These results suggested that miR164 family members have at least partial functional specialization in their Fe-deficiency responses.
NAC5 is regulated at both transcriptional and posttranscriptional levels under Fe-deficient conditions
miR164 was previously verified to posttranscriptionally regulate a subset of NAC-domain transcription factors, including CUC1, CUC2, NAC1, NAC4, NAC5 (At5g61430), and ORE1 (Laufs et al., 2004; Mallory et al., 2004; Guo et al., 2005; Kim et al., 2009). Of the six targets directly cleaved by miR164, only NAC1 and NAC5 were highly expressed in roots (Mallory et al., 2004). NAC1 was mainly observed in root tips and lateral root initiation sites (Xie et al., 2000), which was inconsistent with the expression patterns of MIR164b. In addition, Fe deficiency did not alter the mRNA abundance of NAC1 in Arabidopsis roots (Supplemental Figure S7). We, therefore, investigated the function of NAC5.
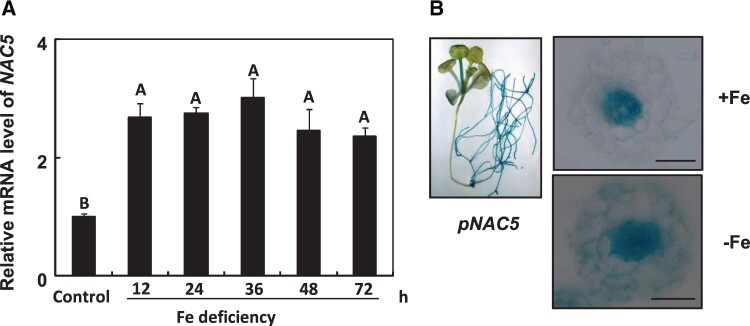
The mRNA abundance of NAC5 was higher in the mir164b mutant than in the WT (Supplemental Figure S8), further demonstrating the posttranscriptional regulation of NAC5 by MIR164b. In contrast to the downregulation of miR164 expression by Fe deficiency, NAC5 was strongly induced by Fe deficiency in roots (Figure 4A). To determine whether the induction of NAC5 in response to Fe deficiency was caused by increased promoter activity or altered RNA stability, we constructed NAC5 promoter::GUS transgenic lines. In agreement with the expression patterns of MIR164b, NAC5 expression was high in the vascular tissues of petioles and roots during vegetative development under Fe-sufficient conditions (Figure 4B). Fe deficiency enhanced the GUS activity of NAC5 (Figure 4B). Unlike the expression pattern of MIR164b under Fe-deficient conditions, NAC5 was expressed in both root vascular tissue and root cortical tissue under Fe-deficient conditions (Figure 4B), indicating that Fe deficiency also induced NAC5 mRNA abundance at transcriptional level.
Figure 4.
Regulation of NAC5 by Fe deficiency. A, Relative quantification of NAC5 mRNA abundance in Arabidopsis roots in response to Fe deficiency by RT-qPCR. The expression levels were normalized to that of TUB2. Error bars represent standard errors of three biological replicates. Means with the same letter are not significantly different at P < 0.01 according to one-way ANOVA followed by Tukey’s multiple comparison test. B, NAC5pro::GUS expression pattern in transgenic Arabidopsis roots. Scale bars = 50 µm.
35S::NAC5 transgenic plants, like the mir164b mutant, are tolerant of Fe deficiency
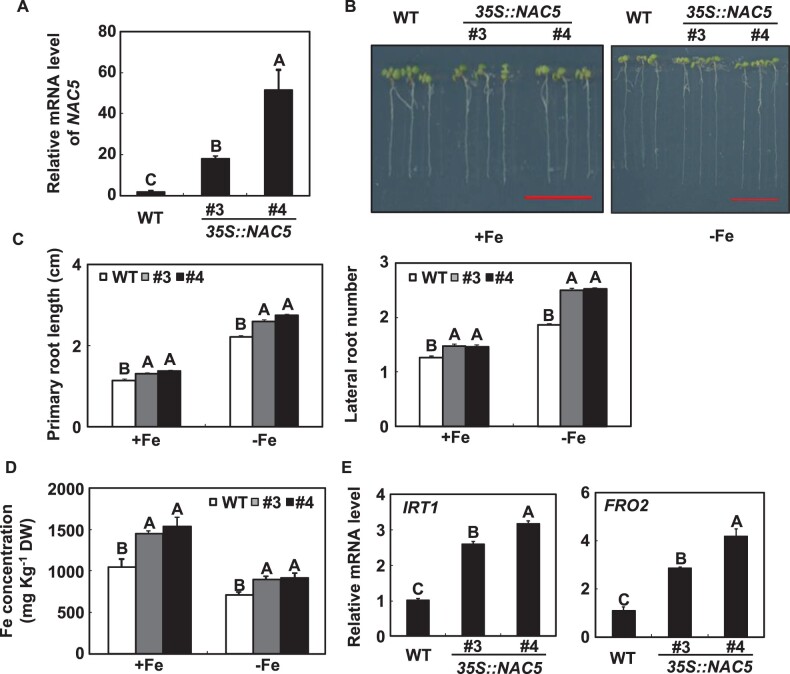
To characterize the potential roles of NAC5 in Fe-deficiency tolerance, we generated transgenic Arabidopsis plants overexpressing the full-length coding sequence (CDS) of NAC5. Two transgenic lines (#3 and #4) were chosen for further analysis based on their high level of NAC5 (Figure 5A). Under both Fe-sufficient and -deficient conditions, the primary root length and lateral root number of 35S::NAC5 transgenic Arabidopsis seedlings, and the Fe concentrations in the roots of 18-d-old hydroponically grown 35S::NAC5 transgenic Arabidopsis plants, were significantly higher than those of WT plants (Figure 5, B–D). Correspondingly, the mRNA abundances of IRT1 and FRO2 in roots were much higher in 35S::NAC5 transgenic plants than in WT plants under Fe-deficient conditions (Figure 5E).
Figure 5.
The responses of 35S::NAC5 transgenic plants to Fe deficiency. A, Detection of NAC5 mRNA in 35S::NAC5 transgenic Arabidopsis. RT-qPCR quantifications were normalized to the expression of TUB2. Error bars represent standard errors of three biological replicates. B, Representative images of 35S::NAC5 transgenic Arabidopsis seedlings grown on Fe-deficient medium for 7 d. Scale bars = 1 cm. C, The primary root length and lateral root number of 35S::NAC5 transgenic Arabidopsis seedlings grown on Fe-deficient medium for 7 d. D, The Fe concentration in the roots of 18-d-old hydroponically grown Arabidopsis, either supplied with Fe or transferred to a Fe-free solution for 36 h. Error bars represent standard errors of three independent experiments, with 20–30 plants per experiment (C and D). E, The effects of Fe deficiency on the expression levels of IRT1 and FRO2 in 18-d-old hydroponically grown WT and 35S::NAC5 transgenic plants. Error bars represent standard errors of three biological replicates. Means with the same letter are not significantly different at P < 0.01 according to one-way ANOVA followed by Tukey’s multiple comparison test.
To further characterize the function of NAC5 in Fe-deficiency responses, we identified a NAC5 loss-of-function mutant and designated as nac5 (Supplemental Figure S9). In contrast with WT plants, nac5 loss-of-function mutant had shorter primary root and higher rates of leaf discoloration under Fe-deficient conditions (Supplemental Figure S9). We also detected the mRNA level of FIT and PYE in 35S::NAC5 transgenic Arabidopsis plants and nac5 loss-of-function mutant under Fe-deficient conditions. Transcriptional levels of NAC5 did not affect FIT and PYE mRNA abundance (Supplemental Figure S10). In addition, knock-out of FIT and PYE did not affect expression level of NAC5 (Colangelo and Guerinot, 2004; Long et al., 2010). These results suggested that NAC5, FIT, and PYE were involved in separate gene regulons of Fe-deficient responses in Arabidopsis.
NAC5 has transactivation activity
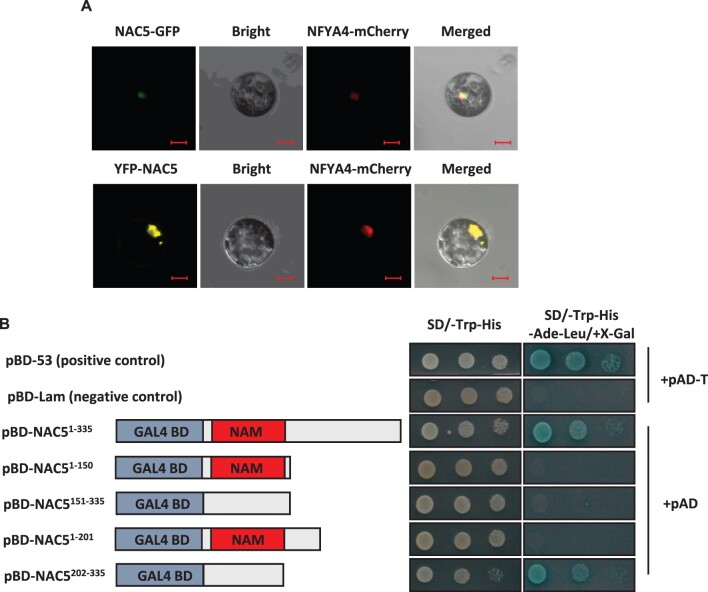
The UniProt program predicted the nuclear localization of NAC5 protein (https://www.uniprot.org/locations). To confirm the subcellular localization of NAC5 protein, we generated NAC5-GFP and YFP-NAC5 constructs, and separately introduced the constructs into Arabidopsis leaf protoplasts. NAC5-GFP and YFP-NAC5 signals were detected only in the nucleus (Figure 6A). The localizations were confirmed by co-localization with the nucleus marker gene, NFYA4 (Figure 6A; Zhou et al., 2015).
Figure 6.
Subcellular localization and transcriptional activation assay of the NAC5 protein. A, NAC5-GFP and YFP-NAC5 constructs were transiently coexpressed with the nuclear marker NFYA4-mCherry in Arabidopsis leaf protoplasts. GFP, YFP, and mCherry signals were imaged by confocal microscopy. Scale bars = 10 μm. B, Transcriptional activation assay of NAC5 protein in yeast. NAC5 cDNAs encoding different fragments of NAC5 were fused to the GAL4 activation domain on the pGBKT7 vector to generate pBD-NAC5 fusion plasmids. pBD-53 and pBD-Lam were used as a positive control and a negative control, respectively.
NAC5 contains an NAC domain, indicating that NAC5 should have transcription activity. To test this hypothesis, NAC5 CDS encoding different fragments of NAC5 were fused to the GAL4 activation domain on the pGBKT7 vector in order to generate pBD-NAC5 fusion plasmids. The pBD-NAC5 fusion plasmids were co-transformed with the pGADT7 AD vector into the yeast strain Y2Hgold. The full-length NAC5 protein and the C-terminal 202–335 amino-acid fusion protein had transactivation activities, leading to the α-galactosidase activity and survival in a medium without Trp/His/Ade/Leu (Figure 6B). Deletion of the C-terminal domain showed that amino acids 1–150, 1–201, and 151–335 abolished the activation of the expression of the LacZ reporter gene (Figure 6B). These results indicated that the C-terminal is essential for the transactivation activities of NAC5 protein and that at least two NAC repression domains (amino acids 1–150 and 151–201) exist in the N-terminal of the NAC5 protein.
NAC5 directly transactivates the expression of NFYA8
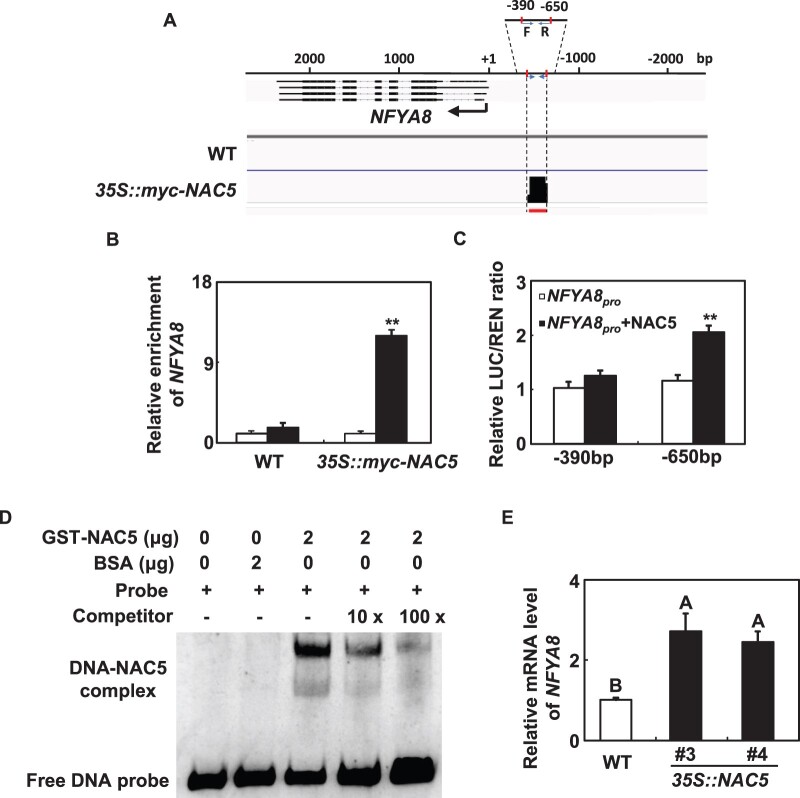
To further characterize the function of NAC5 in Fe homeostasis, we identified NAC5 targets by determining its genome-wide DNA-binding profile. Chromatin immunoprecipitation followed by sequencing (ChIP-seq) was carried out using 35S::myc-NAC5 transgenic plants (Supplemental Figure S11). As a control, the same experiments were performed in parallel with the WT. A total of 7,217 peaks were unique to 35S::myc-NAC5 transgenic plants. Among these peaks, 1,101 were located in the core promoter regions (−1 kb to +100 bp of the transcription start site) of 1,007 genes. NFYA8, a member of the CCAAT-binding transcription factor (CBF-B/NFYA) family attracted our attention because the core promoter regions (1,000 bp) of IRT1 and FRO2 contain two and four CCAAT boxes, respectively (Figure 7A;Supplemental Figure S12), and because NFYA8 expression was consistent with the location of NAC5 (Supplemental Figure S13). To verify the ChIP-seq data, we performed ChIP-qPCR and found that the ChIP-seq detected peak of the NFYA8 promoter was enriched by up to 14-fold in 35S::myc-NAC5 transgenic plants (Figure 7B). In contrast, promoter fragments of NFYA6, the negative control, were not enriched in 35S::myc-NAC5 transgenic plants (Supplemental Figure S14).
Figure 7.
NFYA8 is the direct target of NAC5. A, NAC5 binding peak in the promoter region of NFYA8 detected by ChIP-seq. F and R indicate the positions of forward and reverse primers used in ChIP-qPCR. B, ChIP-qPCR assays verified the in vivo binding activity of NAC5 to the promoter of NFYA8. TUB2 was used as an internal control. Error bars represent standard errors of three biological replicates. Asterisks indicate significant differences (P < 0.01) between the amount of NFYA8 fragment in the immunoprecipitate and the amount in the input according to t tests. C, Transient transactivation assay of NAC5 protein with the promoter of NFYA8 in Arabidopsis mesophyll protoplasts. Relative REN activity was used as an internal control and the relative LUC/REN ratios are shown. Error bars represent standard errors of three biological replicates. Asterisks indicate significant differences based on t tests (P < 0.01). D, EMSA with NAC5 protein using the probes derived from the NFYA8 promoter. Competition for the labeled sequences was tested by adding an excess of unlabeled probes as indicated. The free probes and DNA-NAC5 are marked. E, Detection of NFYA8 transcripts in 35S::NAC5 transgenic plants by RT-qPCR. Quantifications were normalized to the expression of TUB2. Error bars represent standard errors of three biological replicates. Means with the same letter are not significantly different at P < 0.01 according to one-way ANOVA followed by Tukey’s multiple comparison test.
We also utilize the dual-luciferase (LUC) reporter assay system to confirm the direct regulation of the NFYA8 promoter by NAC5. NAC5 significantly induced the expression of LUC driven by the NFYA8 promoter with the NAC5-binding site (−650 bp), but not by the NFYA8 promoters without the NAC5-binding site (−390 bp) (Figure 7C). To further confirm the direct binding of NAC5 to the NFYA8 promoter, we performed electrophoretic mobility shift assays (EMSAs). The results of the EMSAs showed that NAC5 was bound to the NFYA8 promoter. When unlabeled NAC5 probe was added to the system as a competitor, the signal was suppressed (Figure 7D), further demonstrating the direct binding of the NFYA8 promoter by NAC5. Furthermore, the mRNA abundance of NFYA8 significantly increased in 35S::myc-NAC5 transgenic plants (Figure 7E). These results strongly suggested that NAC5 regulates the expression of NFYA8 in vivo.
35S::NFYA8 transgenic plants are also tolerant of Fe deficiency
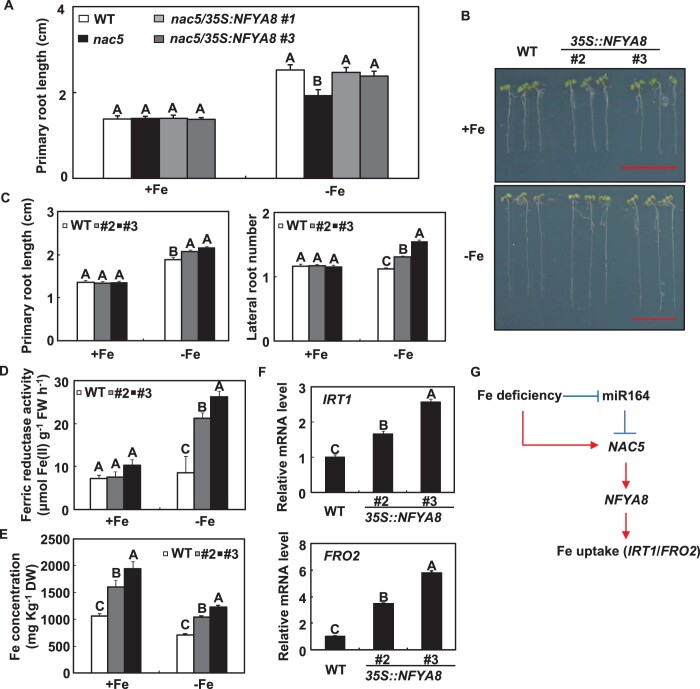
The direct binding of the NFYA8 promoter by NAC5 suggested that NFYA8 was a directly downstream target of NAC5. To test the hypothesis, we overexpressed NFYA8 in nac5 mutant (Supplemental Figure S15). NFYA8 overexpression almost completely rescued the retarded root development and leaf discoloration of nac5 mutant under Fe-deficient conditions (Figure 8A;Supplemental Figure S16). Then, we constructed NFYA8 overexpression lines and tested the responses of the 35S::NFYA8 transgenic plants to Fe deficiency. There are four splicing variants of NFYA8 transcripts (Figure 7A). Fortunately, these splicing variants share a common coding region, and the full-length CDS region of NFYA8 was therefore used for the construct (Supplemental Figure S17). Consistent with mir164b mutant and 35S::NAC5 transgenic plants, the primary root length, lateral root number, ferric reductase activity, Fe content, and mRNA abundance of IRT1 and FRO2 were higher in 35S::NFYA8 transgenic plants than in WT plants under Fe-deficient conditions (Figure 8, B–F). We also used the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated Protein 9 (Cas9) system to generate NFYA8 loss-of-function mutants (Supplemental Figure S18A). Cas9-free transgenic plants were identified for the phenotypic analysis. Compared with the WT, NFYA8 loss-of-function mutants were more sensitive to Fe deficiency (Supplemental Figure S18, B and C). These results indicated that NFYA8 plays important role in plant tolerance to Fe deficiency.
Figure 8.
Responses of 35S::NFYA8 transgenic plants to Fe deficiency. A, Rescue of the nac5 phenotype by NFYA8 overexpression. B, Representative images of 35S::NFYA8 transgenic Arabidopsis seedlings grown on Fe-deficient medium for 7 d. Scale bars = 1 cm. C, The primary root length and lateral root number of 35S::NFYA8 transgenic Arabidopsis seedlings grown on Fe-deficient medium for 7 d. D, The ferric reductase activity in the roots of 35S::NFYA8 transgenic Arabidopsis seedlings grown on Fe-deficient medium for 7 d. E, The Fe concentration in the roots of 18-d-old hydroponically grown Arabidopsis, either supplied with Fe or transferred to a Fe-free solution for 36 h. F, The effects of Fe deficiency on the expression levels of IRT1 and FRO2 in 18-d-old hydroponically grown WT and 35S::NFYA8 transgenic plants. Error bars represent standard errors of three independent experiments, with 20–30 plants per experiment (A, C–E). Error bars represent standard errors of three biological replicates (F). Means with the same letter are not significantly different at P < 0.01 according to one-way ANOVA followed by Tukey’s multiple comparison test. G, A model for the co-function among MIR164b, NAC5, and NFYA8 in Arabidopsis under Fe-deficient conditions. The solid arrows and solid lines with perpendicular lines represent the upregulation and downregulation of corresponding mRNA abundance, respectively.
Discussion
Recent research has clarified the transcriptional responses, especially of bHLH family members, to Fe deficiency in plants (Colangelo and Guerinot, 2004; Yuan et al., 2005, 2008; Long et al., 2010; Wang et al., 2013; Zhang et al., 2015; Li et al., 2016; Liang et al., 2017; Cui et al., 2018). In this study, we identified a transcription factor from the NAC family, NAC5, that is important for Fe homeostasis in Arabidopsis. Although the NAC family was previously determined to be associated with Fe homeostasis in Strategy II plants, the current study demonstrates that NAC protein could be involved in Fe-deficiency responses in Strategy I plants.
NAC5 is a functional transcription factor because (1) both NAC5-GFP and YFP-NAC5 are localized in the nucleus; (2) NAC5 contains a conserved NAC DNA-binding domain at the N terminus (amino acids 17–141); and (3) NAC5 has transcriptional activation activity as demonstrated by autoactivation testing in yeast cells and dual-LUC transient expression assay in Arabidopsis mesophyll protoplasts. In Arabidopsis, FIT and PYE regulons are the two key networks that regulate Fe homeostasis. NAC5 might define another Fe-deficiency-responsive transcriptional cascade. The NAC5 transcriptional cascade is important for Fe-deficiency responses because mir164b knockout plants, miR164 knock-down transgenic plants, and NAC5-overexpressing lines are tolerant of Fe deficiency. Compared with the WT, these mutants and transgenic plants produce longer primary roots and more lateral roots under Fe-deficient conditions. Another related NAC transcription factor, NAC1 increases lateral root emergence by affecting auxin signals (Xie et al., 2000; Guo et al., 2005; D’haeseleer et al., 2011). NAC1 and NAC5 might be involved in separate gene regulons for root development, as indicated by the following observations: (1) under normal conditions, 35S::NAC5 transgenic plants but not 35S::NAC1 transgenic plants had longer primary roots than the WT (Guo et al., 2005); (2) the mRNA abundance of NAC5, but not of NAC1, accumulated under Fe-deficient conditions (this study); and (3) the lateral root number of 35S::NAC5 transgenic plants increased only under Fe-deficient conditions (this study).
By determining the genome-wide DNA-binding profile of NAC5, we identified 1,007 cis-acting targets. Gene ontology (GO; http://systemsbiology.cau.edu.cn/agriGOv2/) analysis indicated that these genes were related to various metabolic processes, including cytochrome-c oxidase activity (GO:0004129, P = 0.00019), oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor (GO:0016676, P = 0.00019), NADH dehydrogenase activity (GO:0003954, P = 0.00027), and mitochondrion (GO:0005739, P = 2.3e−24). These results suggest that NAC5 is involved in oxidative stress.
The peak in the core promoter region of NFYA8 (202 bp) contains eight NAC-binding sites (http://jaspar.genereg.net/). The binding of NFYA8 was verified by ChIP-qPCR, dual-LUC reporter assay, EMSAs, and the corresponding mRNA abundance in 35S::myc-NAC5 transgenic plants. NF-Y is a universal transcription factor with high affinity and sequence specificity for the CCAAT box in ∼25% of eukaryotic gene promoters. There are two and four CCAAT boxes located in core promoter regions of IRT1 and FRO2, respectively, and 35S::NFYA8 transgenic plants are as tolerant as 35S::NAC5 plants to Fe deficiency, indicating that NAC5 affected Fe-deficient responses in Arabidopsis by regulation of the expression of NFYA8. The 202-bp sequence was not conserved among other members of the NFYA family, and promoter fragments of NFYA6, another member of the NFYA family, were not enriched in 35S::myc-NAC5 transgenic plants, which suggests that the NAC5-NFYA8 route is specific for Fe-deficiency responses in Arabidopsis.
Transcriptional induction may explain part of the NAC5 transcript accumulation under Fe deficiency. Our results suggest that the posttranscriptional regulation of NAC5 transcripts by miR164 is also important for Fe homeostasis in Arabidopsis. Under Fe-sufficient conditions, long primary roots were produced by MIR164a/b/c TM transgenic plants, but not by mir164b mutant plants. Given the variations of Fe concentrations and ferric reductase activities in the roots of TM transgenic plants and mir164b mutants, we inferred that MIR164b is the member of the miR164 family that is mainly responsible for Fe-deficiency responses in Arabidopsis. GUS staining patterns showed that MIR164b expression was consistent with the location of NAC5, and mir164b plants were as tolerant of Fe deficiency as 35S::NAC5 and 35S::NFYA8 transgenic plants. We, therefore, propose a model for the co-function among MIR164b, NAC5, and NFYA8 in Arabidopsis under Fe-deficient conditions (Figure 8G). Fe deficiency reduces MIR164b abundance and thereby, relieves miR164-guided repression of NAC5 in Arabidopsis roots. NAC5 directly transactivates the expression of NFYA8, which induces the mRNA accumulation of the Fe transporters IRT1 and of the ferric-chelate reductase FRO2. In addition, the expression levels of miR164 were induced by Fe excess in Arabidopsis. This increases the tolerance of Arabidopsis to Fe stress.
Materials and methods
Plant materials and growth conditions
Arabidopsis (A. thaliana) Col-0 ecotype plants were grown hydroponically in a growth chamber with a light intensity of 150 µmol·m−2·s−1, 60%–70% relative humidity, and a 22°C/18°C day/night temperature regime. The hydroponic culture was performed as previously described (Zhao et al., 2011). After they had grown for 18 d, the plants were supplied with Fe free or 500 µM Fe nutrient solutions to simulate Fe-deficient and excess conditions. Nutrient solutions were renewed daily to ensure pH stability. Shoots and roots were separately harvested, flash frozen, and stored at −80°C. Arabidopsis seedlings in 0.5 MS (Murashige and Skoog, 1962) nutrient agar medium were grown under continuous light (70 μmol m−2s−1) at 23°C ± 1°C.
Constructs and generation of transgenic plants
The full-length CDS of NAC5 (AT5G61430) and NFYA8 (AT1G17590) was amplified by PCR to generate 35S::NAC5, 35S::NFYA8 transgenic plants. A fragment surrounding the MIR164b (AT5G01747) sequence including the fold-back structure was amplified from genomic DNA. The amplified fragments were introduced into the pENTR/D-TOPO vector (Invitrogen, Waltham, MA, USA) and cloned into pMDC32 (Curtis and Grossniklaus, 2003) by LR reactions (Invitrogen).
To generate the miR164 knock-down mutant, the short tandem target mimic transgenic lines targeting MIR164a, MIR164b, and MIR164c were designed as described by Yan et al. (2012). To generate 35S::myc-NAC5 transgenic plants, the full-length CDS of NAC5 was amplified. Thereafter, the fragments were cloned into the pCPB vector using the BamH I restriction site by In-Fusion reaction.
The CRISPR/Cas9-mediated NFYA8 editing was performed as described by Xing et al. (2014) with some modifications. In brief, two gRNAs that direct target sequences located at the first and fifth exon of NFYA8 were produced. The fragments were cloned into the pHEC401 vector.
For MIR164b, NAC5, and NFYA8 promoter::GUS constructs, the 906-bp, 831-bp, and 1,955-bp fragments upstream from the initiation codon were amplified and separately cloned into the pMDC164 vector following Gateway recombination.
The fusions of yellow fluorescent protein (YFP) to the C-terminal end and green fluorescent protein (GFP) to the N-terminal end of NAC5 were generated and introduced separately into pAN580 and pCPB vectors using the BamHI restriction site by In-Fusion reaction. The plasmids were transiently expressed in Arabidopsis mesophyll protoplast. YFP/GFP images were collected with a Leica SP2 confocal microscope. YFP and GFP were detected at 488 nm laser excitation, their collection bandwidth was at 515–535 nm and 510–550 nm, respectively (intensity at 3%, Gain <20%).
The plasmids were electroporated into Agrobacterium tumefaciens GV3101, and Arabidopsis plants were transformed using the floral dip method (Clough and Bent, 1998). Transgenic plants were selected with the use of 35 µg mL−1 hygromycin or 45 μM glufosinate ammonium. T3 or T4 homozygous lines were used for all experiments. The sequences of the primer pairs used in the experiments are listed in Supplemental Table S4.
Small RNA isolation, library construction, and sequencing
Total RNA was extracted with Trizol reagent (Invitrogen) from roots subjected to Fe deficiency for 2 d. Small RNA isolation, library construction, and data analysis were performed as previously described (Gao et al., 2015). Solexa sequencing was performed at BGI (Shenzhen, China). The unique sequencing data were aligned to the Arabidopsis genome (The Arabidopsis Information Resource 9). Only perfectly matching sequences were considered for further analysis. The count information was used to determine normalized miRNA expression levels as reads of exon model per million mapped reads (RPM).
Gene expression analysis
For quantitative reverse transcription PCR (RT-qPCR), 1 µg of total RNA isolated with the RNeasy plant mini kit was used for the first-strand cDNA synthesis with the SuperScript III first-strand synthesis supermix (Invitrogen). Specific primers were used to detect the expression levels of NAC5, NFYA8, IRT1 (AT4G19690), and FRO2 (AT1G01580). Primer efficiencies were determined (Ramakers et al., 2003). For stem–loop RT-qPCR, 0.5 µg of total RNA was used for the cDNA synthesis with miR164 stem–loop RT primer according to the protocol of Varkonyi-Gasic et al. (2007). RT-qPCR was carried out in an ABI 7500 system using the SYBR Premix Ex Taq (perfect real time) kit (TaKaRa Biomedicals, San Jose, CA, USA) as previously described (Zhao et al., 2011). Each experiment was replicated three times. The comparative Ct method was applied. The sequences of the specific primers are listed in Supplemental Table S4.
Transactivation assay in yeast cells
The yeast strain Y2Hgold (MATa trp1-901 leu2-3 112 ura3-52 his3-200 gal4Δ gal80Δ LYS2::GAL1UAS-Gal1TATA-His3 GAL2UAS-Gal2TATA-Ade2 URA3::MEL1UAS-Mel1TATA AUR1-C MEL1), which contains HIS3 and lacZ, was used in the assay. Different fragments of NAC5 were obtained by PCR and cloned into the pGBKT7 vector to generate pBD-NAC5 fusion plasmids. The fusion plasmids were transformed into Y2Hgold according to the manufacturer’s instructions (Clontech). The transformants were selected on synthetic medium lacking Leu and His. The positive clones were plated on SD/-Leu-Trp-His-Ade selective dropout medium. The transactivation was further confirmed by chloroform overlay α-galactosidase plate assay without Leu, Trp, His, and Ade.
ChIP assay
Two-weeks-old 35S::myc-NAC5 transgenic plants grown on 0.5 MS medium were used for ChIP assays as described by Huysmans et al. (2018). A 3-g quantity of ground plant tissue was cross-linked in 1% (v/v) formaldehyde. The chromatin complexes were isolated and sonicated with a Bioruptor to obtain 200- to 800-bp DNA fragments. Protein–DNA complexes were immunoprecipitated using an anti-Myc Tag antibody (Millipore, Burlington, MA, USA). The purified DNA fragments were quantified by ChIP-qPCR with the primers listed in Supplemental Table S4. The enrichment was calculated based on the amount of fragment in the immunoprecipitate relative to the amount in the input.
The dual-LUC transient expression assay
NFYA8 promoters with and without NAC5 binding site were amplified by PCR. The amplified fragments were cloned into the pGreen0800-LUC double reporter vector using the BamH I restriction site to generate reporters. The coding sequence of NAC5 was amplified and introduced into the pRI101-AN vector to generate effectors. The constructs with various combinations were infiltrated into Arabidopsis mesophyll protoplasts as previously described (Yoo et al., 2007). The activities of Firefly LUC and Renilla LUC (REN) were measured using the Dual-LUC Reporter Assay System (Promega, Madison, WI, USA). The relative reporter ratio (LUC/REN) was normalized with respect to the average values of the NFYA8 promoter.
EMSAs
EMSAs were performed as previously described (Liu et al., 2014). Affinity-purified GST-NAC5 recombinant protein was used for EMSAs. For the generation of GST-NAC5 recombinant protein, full length coding region of NAC5 was amplified and cloned into pGEX-4T vector using the BamH I restriction site by In-Fusion reaction. GST-NAC5 construct was expressed in Escherichia coli BL21 and purified by glutathione-Sepharose 4B (GE Healthcare, Stockholm, Sweden). The DNA probe from NFYA8 promoter was amplified by PCR and labeled DIG-ddUTP using DIG Gel Shift Kit (Roche, Basel, Switzerland). The same unlabeled fragment was used as competitor probe and the bovine serum albumin (BSA) alone was used as the negative control.
Determination of Fe concentration
After heating to 120°C for 30 min, the roots of hydroponic Arabidopsis plants were dried at 65°C for 72 h and milled to a fine powder. The samples were weighed into PTFE digestion tubes and digested in HNO3 using a microwave digester (CEM Mars6 Xpress). The Fe concentration in the samples was determined by ICP-MS (Agilent 7700x; Agilent Technologies, Santa Clara, CA, USA).
Assay of Fe(III)-chelate reductase activity
The activity of Fe(III)-chelate reductase was assayed according to the procedure of Connolly et al. (2003) and Satbhai et al. (2017). Eight 7-d-old Arabidopsis plants were pooled together and whole roots of the plants were submerged in a medium containing 0.1 mM Fe(III)-ethylenediaminetetraacetic acid (EDTA) and 0.3 mM ferrozine. After incubation for 30 min in darkness at 25°C, the transmittance of Fe(II)-ferrozine at 562 nm was determined.
GO analysis
GO enrichment was performed using AgriGO version 2.0 (http://systemsbiology.cau.edu.cn/agriGOv2/) with default parameters. GO terms were considered significantly enriched at false discovery rates <0.05.
Statistical analysis
In this study, data were presented as means ± standard errors. Number of replicates can be found in the corresponding figure legends. The Student’s t test was used to determine statistical significance between two groups, asterisk indicate significant difference at P < 0.01. For multiple group comparisons, the data were analyzed by one-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison test and means with the same letter are not significantly different at P < 0.01.
Accession numbers
Arabidopsis Genome Initiative numbers for the genes discussed in this article are as follows: MIR164b, AT5G01747; NAC1, AT1G56010; NAC5, AT5G61430; NFYA8, AT1G17590; IRT1, AT4G19690; FRO2, AT1G01580; NFYA6, AT3G14020; NFYA4, AT2G34720; PYE, AT3G47640; FIT1, AT2G28160; TUB2, AT5G62690.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Marker gene expression is significantly induced by Fe deficiency in Arabidopsis.
Supplemental Figure S2. Detection of mature miR164 expression by stem–loop RT-qPCR in the xylem sap of Arabidopsis under Fe-sufficient and -deficient conditions.
Supplemental Figure S3. Regulation of miR164 by Fe excess.
Supplemental Figure S4. Diagram of the T-DNA insertion site in the MIR164b locus and detection of MIR164b abundance.
Supplemental Figure S5. The responses of 35S::MIR164b transgenic plants to Fe deficiency.
Supplemental Figure S6. Leaf phenotype of WT and mir164b mutant and TM transgenic plants.
Supplemental Figure S7. Relative quantification of NAC1 mRNA abundance in Arabidopsis roots in response to Fe deficiency by RT-qPCR.
Supplemental Figure S8. The mRNA abundance of NAC5 in mir164b.
Supplemental Figure S9. The responses of nac5 to Fe deficiency.
Supplemental Figure S10. The relative mRNA level of FIT1 and PYE in WT, 35S::NAC5 transgenic plants, and nac5 mutants.
Supplemental Figure S11. NAC5 protein abundance in 35S::myc-NAC5 transgenic plants.
Supplemental Figure S12. Schematic diagram of CCAAT box distribution in the core promoter regions of IRT1 and FRO2 genes.
Supplemental Figure S13. NAC5pro::GUS and NFYA8pro::GUS expression pattern in transgenic Arabidopsis roots.
Supplemental Figure S14. NAC5 did not bind to the promoter of NFYA6.
Supplemental Figure S15. Detection of NFYA8 mRNA in nac5/35S::NFYA8 transgenic Arabidopsis.
Supplemental Figure S16. The effects of Fe deficiency on leaf color of WT, nac5 mutant, and nac5/35S::NFYA8 transgenic Arabidopsis.
Supplemental Figure S17. Detection of NFYA8 mRNA in 35S::NFYA8 transgenic Arabidopsis.
Supplemental Figure S18. The responses of nfya8 to Fe deficiency.
Supplemental Table S1. Statistics of small RNA sequences from Arabidopsis roots with or without Fe.
Supplemental Table S2. Different categories of small RNAs in two small RNA libraries.
Supplemental Table S3. Expression profiles of conserved miRNAs in response to Fe deficiency in Arabidopsis roots.
Supplemental Table S4. Oligos used as primers in the experiment.
Funding
This work was supported by grants to W.-X.L. from the National Science Foundation of China (grant number 31870224) and the Agricultural Science and Technology Innovation Program of CAAS.
Conflict of interest statement. We declare that we have no conflict of interest.
Supplementary Material
These authors contributed equally (Q.D., W.L., and Y.G.).
W.-X.L. designed the research, analyzed the data, and wrote the article. Q.D., W.L., Y.G., J.Y., and S.W. performed the research.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Wen-Xue Li (liwenxue@caas.cn).
References
- Briat JF, Dubos C, Gaymard F (2015) Iron nutrition, biomass production, and plant product quality. Trends Plant Sci 20: 33–40 [DOI] [PubMed] [Google Scholar]
- Cai Y, Li Y, Liang G (2021) FIT and bHLH Ib transcription factors modulate iron and copper crosstalk in Arabidopsis. Plant Cell Environ 44: 1679–1691 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Colangelo EP, Guerinot ML (2004) The essential basic helix–loop–helix protein FIT1 is required for the iron deficiency response. Plant Cell 16: 3400–3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly EL, Campbell NH, Grotz N, Prichard CL, Guerinot ML (2003) Overexpression of the FRO2 ferric chelate reductase confers tolerance to growth on low iron and uncovers posttranscriptional control. Plant Physiol 133: 1102–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y, Chen CL, Cui M, Zhou WJ, Wu HL, Ling HQ (2018) Four IVa bHLH transcription factors are novel interactors of FIT and mediate JA inhibition of iron uptake in Arabidopsis. Mol Plant 11: 1166–1183 [DOI] [PubMed] [Google Scholar]
- Curie C, Panaviene Z, Loulergue C, Dellaporta SL, Briat JF, Walker EL (2001) Maize yellow stripe1 encodes a membrane protein directly involved in Fe(III) uptake. Nature 409: 346–349 [DOI] [PubMed] [Google Scholar]
- Curtis M D, Grossniklaus U (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Haeseleer K, Den Herder G, Laffont C, Plet J, Mortier V, Lelandais-Brière C, De Bodt S, De Keyser A, Crespi M, Holsters M, et al. (2011) Transcriptional and post-transcriptional regulation of a NAC1 transcription factor in Medicago truncatula roots. New Phytol 191: 647–661 [DOI] [PubMed] [Google Scholar]
- Dubeaux G, Neveu J, Zelazny E, Vert G (2018) Metal sensing by the IRT1 transporter-receptor orchestrates its own degradation and plant metal nutrition. Mol Cell 69: 953–964 [DOI] [PubMed] [Google Scholar]
- Eide DJ, Broderius M, Fett J, Guerinot ML (1996) A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc Natl Acad Sci USA 93: 5624–5628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W, Liu W, Zhao M, Li WX (2015) NERF encodes a RING E3 ligase important for drought resistance and enhances the expression of its antisense gene NFYA5 in Arabidopsis. Nucleic Acids Res 46: 607–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz R, Manishankar P, Ivanov R, Köster P, Mohr I, Trofimov K, Steinhorst L, Meiser J, Mai HJ, Drerup M, et al. (2019) CIPK11-dependent phosphorylation modulates FIT activity to promote Arabidopsis iron acquisition in response to calcium signaling. Dev Cell 48: 726–740 [DOI] [PubMed] [Google Scholar]
- Grillet L, Lan P, Li W, Mokkapati G, Schmidt W (2018) IRON MAN is a ubiquitous family of peptides that control iron transport in plants. Nat Plants 4: 953–963 [DOI] [PubMed] [Google Scholar]
- Guo HS, Xie Q, Fei JF, Chua NH (2005) MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for Arabidopsis lateral root development. Plant Cell 17: 1376–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huysmans M, Buono RA, Skorzinski N, Radio MC, De Winter F, Parizot B, Mertens J, Karimi M, Fendrych M, Nowack MK (2018) NAC transcription factors ANAC087 and ANAC046 control distinct aspects of programmed cell death in the Arabidopsis columella and lateral root cap. Plant Cell 30: 2197–2213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Woo HR, Kim J, Lim PO, Lee IC, Choi SH, Hwang D, Nam HG (2009) Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis. Science 323: 1053–1057 [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Nishizawa NK (2012) Iron uptake, translocation, and regulation in higher plants. Annu Rev Plant Biol 63: 131–152 [DOI] [PubMed] [Google Scholar]
- Kuo HF, Chiou TJ (2011) The role of microRNAs in phosphorus deficiency signaling. Plant Physiol 156: 1016–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufs P, Peaucelle A, Morin H, Traas J (2004) MicroRNA regulation of the CUC genes is required for boundary size control in Arabidopsis meristems. Development 131: 4311–4322 [DOI] [PubMed] [Google Scholar]
- Lei R, Li Y, Cai Y, Li C, Pu M, Lu C, Yang Y, Liang G (2020) bHLH121 functions as a direct link that facilitates the activation of FIT by bHLH IVc transcription factors for maintaining Fe homeostasis in Arabidopsis. Mol Plant 13: 634–649 [DOI] [PubMed] [Google Scholar]
- Li X, Zhang H, Ai Q, Liang G, Yu D (2016) Two bHLH transcription factors, bHLH34 and bHLH104, regulate iron homeostasis in Arabidopsis thaliana. Plant Physiol 170: 2478–2493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G, Zhang H, Li X, Ai Q, Yu D. (2017) bHLH transcription factor bHLH115 regulates iron homeostasis in Arabidopsis thaliana. J Exp Bot 68: 1743–1755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Tai H, Li S, Gao W, Zhao M, Xie C, Li WX (2014) bHLH122 is important for drought and osmotic stress resistance in Arabidopsis and in the repression of ABA catabolism. New Phytol 201: 1192–1204 [DOI] [PubMed] [Google Scholar]
- Long TA, Tsukagoshi H, Busch W, Lahner B, Salt DE, Benfey PN (2010) The bHLH transcription factor POPEYE regulates response to iron deficiency in Arabidopsis roots. Plant Cell 22: 2219–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory AC, Dugas DV, Bartel DP, Bartel B (2004) MicroRNA regulation of NAC-domain targets is required for proper formation and separation of adjacent embryonic, vegetative, and floral organs. Curr Biol 14: 1035–1046 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Narayanan N, Beyene G, Chauhan RD, Gaitán-Solís E, Gehan J, Butts P, Siritunga D, Okwuonu I, Woll A, Jiménez-Aguilar DM, et al. (2019) Biofortification of field-grown cassava by engineering expression of an iron transporter and ferritin. Nat Biotechnol 37: 144–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikovics K, Blein T, Peaucelle A, Ishida T, Morin H, Aida M, Laufs P (2006) The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell 8: 2929–2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogo Y, Kobayashi T, Nakanishi Itai R, Nakanishi H, Kakei Y, Takahashi M, Toki S, Mori S, Nishizawa NK (2008) A novel NAC transcription factor, IDEF2, that recognizes the iron deficiency-responsive element 2 regulates the genes involved in iron homeostasis in plants. J Biol Chem 283: 3407–13417 [DOI] [PubMed] [Google Scholar]
- Olsen AN, Ernst HA, Leggio LL, Skriver K (2005) NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci 10: 79–87 [DOI] [PubMed] [Google Scholar]
- Pilon M (2017) The copper microRNAs. New Phytol 213: 1030–1035 [DOI] [PubMed] [Google Scholar]
- Ramakers C, Ruijter JM, Deprez RH, Moorman AF (2003) Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett 339: 62–66 [DOI] [PubMed] [Google Scholar]
- Raman S, Greb T, Peaucelle A, Blein T, Laufs P, Theres K (2008) Interplay of miR164, CUP-SHAPED COTYLEDON genes and LATERAL SUPPRESSOR controls axillary meristem formation in Arabidopsis thaliana. Plant J 55: 65–76 [DOI] [PubMed] [Google Scholar]
- Riaz N, Guerinot ML (2021) All together now: regulation of the iron deficiency response. J Exp Bot 72: 2045–2055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson NJ, Procter CM, Connolly EL, Guerinot ML (1999) A ferric-chelate reductase for iron uptake from soils. Nature 397: 694–697 [DOI] [PubMed] [Google Scholar]
- Römheld V, Marschner H (1986) Evidence for a specific uptake system for iron phytosiderophores in roots of grasses. Plant Physiol 80: 175–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satbhai SB, Setzer C, Freynschlag F, Slovak R, Kerdaffrec E, Busch W (2017) Natural allelic variation of FRO2 modulates Arabidopsis root growth under iron deficiency. Nat Commun 8: 15603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin L, Lo J, Chen G, Callis J, Fu H, Yeh KC (2013) IRT1 degradation factor1, a RING E3 ubiquitin ligase, regulates the degradation of iron-regulated transporter1 in Arabidopsis. Plant Cell 25: 3039–3051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieber P, Wellmer F, Gheyselinck J, Riechmann JL, Meyerowitz EM (2007) Redundancy and specialization among plant microRNAs: role of the MIR164 family in developmental robustness. Development 134: 1051–1060 [DOI] [PubMed] [Google Scholar]
- Sun L, Wei YQ, Wu KH, Yan JY, Xu JN, Wu YR, Li GX, Xu JM, Harberd NP, Ding ZJ, et al. (2021) Restriction of iron loading into developing seeds by a YABBY transcription factor safeguards successful reproduction in Arabidopsis. Mol Plant 14: 1624–1639 [DOI] [PubMed] [Google Scholar]
- Sunkar R, Kapoor A, Zhu JK (2006) Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell 18: 2051–2065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uauy C, Distelfeld A, Fahima T, Blechl A, Dubcovsky J (2006) A NAC gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science 314: 1298–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varkonyi-Gasic E, Wu R, Wood M, Walton EF, Hellens RP (2007) Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 3: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert G, Grotz N, Dédaldéchamp F, Gaymard F, Guerinot ML, Briat JF, Curie C (2002) IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell 14: 1223–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Cui Y, Liu Y, Fan H, Du J, Huang Z, Yuan Y, Wu H, Ling HQ (2013) Requirement and functional redundancy of Ib subgroup bHLH proteins for iron deficiency responses and uptake in Arabidopsis thaliana. Mol Plant 6: 503–513 [DOI] [PubMed] [Google Scholar]
- Xie Q, Frugis G, Colgan D, Chua NH (2000) Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev 14: 3024–3036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing HL, Dong L, Wang ZP, Zhang HY, Han CY, Liu B, Wang XC, Chen QJ (2014) A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol 14: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki H, Hayashi M, Fukazawa M, Kobayashi Y, Shikanai T (2009) SQUAMOSA promoter binding protein-like7 is a central regulator for copper homeostasis in Arabidopsis. Plant Cell 21: 347–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Gu Y, Jia X, Kang W, Pan S, Tang X, Chen X, Tang G (2012) Effective small RNA destruction by the expression of a short tandem target mimic in Arabidopsis. Plant Cell 24: 415–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Yuan Y, Wu H, Wang N, Li J, Zhao W, Du J, Wang D, Ling HQ (2008) FIT interacts with AtbHLH38 and AtbHLH39 in regulating iron uptake gene expression for iron homeostasis in Arabidopsis. Cell Res 18: 385–397 [DOI] [PubMed] [Google Scholar]
- Yuan YX, Zhang J, Wang DW, Ling HQ (2005) AtbHLH29 of Arabidopsis thaliana is a functional ortholog of tomato FER involved in controlling iron acquisition in strategy I plants. Cell Res 15: 613–621 [DOI] [PubMed] [Google Scholar]
- Zhang J, Liu B, Li M, Feng D, Jin H, Wang P, Liu J, Xiong F, Wang J, Wang HB (2015) The bHLH transcription factor bHLH104 interacts with IAA-LEUCINE RESISTANT3 and modulates iron homeostasis in Arabidopsis. Plant Cell 27: 787–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Ding H, Zhu JK, Zhang F, Li WX (2011) Involvement of miR169 in the nitrogen-starvation responses in Arabidopsis. New Phytol 190: 906–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou SF, Sun L, Valdés AE, Engström P, Song ZT, Lu SJ, Liu JX (2015) Membrane-associated transcription factor peptidase, site-2 protease, antagonizes ABA signaling in Arabidopsis. New Phytol 208: 188–197 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.