Abstract
Gene targeting (GT) for precise gene insertion or swap into pre-defined genomic location has been a bottleneck for expedited soybean precision breeding. We report a robust selectable marker-free GT system in soybean, one of the most economically important crops. An efficient Oh H1-8 (Ochrobactrum haywardense H1-8)-mediated embryonic axis transformation method was used for the delivery of CRISPR-Cas9 components and donor template to regenerate T0 plants 6–8 weeks after transformation. This approach generated up to 3.4% targeted insertion of the donor sequence into the target locus in T0 plants, with ∼ 90% mutation rate observed at the genomic target site. The GT was demonstrated in two genomic sites using two different donor DNA templates without the need for a selectable marker within the template. High-resolution Southern-by-Sequencing analysis identified T1 plants with precise targeted insertion and without unintended plasmid DNA. Unlike previous low-frequency GT reports in soybean that involved particle bombardment–mediated delivery and extensive selection, the method described here is fast, efficient, reproducible, does not require a selectable marker within the donor DNA, and generates nonchimeric plants with heritable GT.
A gene targeting (GT) method shows 2%–3.4% targeted insertion of the selectable marker-free donor in two different soybean genomic sites.
Introduction
Soybean (Glycine max) has become one of the most economically important legume seed crops. In addition to being a major oilseed crop, it also provides more than a quarter of the total protein for the world’s food and animal feed (Graham and Vance, 2003). Due to population growth along with increased societal interest in plant-based protein diets, the demand for soybean is gradually expanding (Ray et al., 2013). However, unlike considerable progress made in increasing the yields of rice, wheat (Triticum aestivum), and maize (Zea mays) through the Green Revolution, only limited improvements have been made for soybean (Liu et al., 2020). The yield and other trait improvement in soybean will require complex and precise genetic manipulations to obtain desired plant height, node number, internode length, branch number, disease resistance, and seed size supplemented with high oil and protein contents. Conventional plant breeding has played a major role in the genetic improvement of soybean and will continue to do so in the future. However, genome editing technology could expedite and revolutionize traditional breeding processes and enable markedly improved precision in soybean breeding. Clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9 (Jinek et al., 2012) is a robust and versatile genome editing tool to make targeted DNA double-strand breaks (DSBs) (Kim et al., 1996; Epinat et al., 2003; Christian et al., 2010; Gasiunas et al., 2012; Jinek et al., 2012). Genomic DSBs in somatic cells are repaired predominantly by nonhomologous end joining (NHEJ), which is an error-prone repair pathway often resulting in small insertions or deletions (indels) at the DSB site. Homologous recombination (HR) (Wood, 1996), in contrast, is a precise repair pathway that requires a DNA repair template with flanking sequences homologous to those flanking the genomic DSB. Unlike NHEJ, HR is a very low frequency process in somatic cells used for plant transformation. Consequently, NHEJ-mediated gene mutations, which can be recovered in 30%–100% of regenerated plants, have become routine (Puchta, 2017), whereas precise gene targeting (GT) via HR remains inefficient and challenging. Some monogenic simple traits that depend on endogenous gene mutation can be generated through NHEJ-mediated DNA repair (Li et al., 2016a, 2016b, 2018; Liu et al., 2017; Nonaka et al., 2017). Complex yield traits require precise GT via HR to make large modifications in the genome to expedite the breeding process (Shi et al., 2017; Yu et al., 2017). Two recent technologies, base editing (Komor et al., 2016; Gaudelli et al., 2017) and prime editing (Marzec et al., 2020) can make precise changes in the desired genomic location without making any DSBs or needing donor DNA. Compared to HR, these technologies are more efficient and could potentially overcome the limitations of HR-mediated GT for making small changes (Molla et al., 2021). However, GT via HR would still be required for making large and complex genomic modifications,
The majority of genome editing work reported in soybean describes successful NHEJ-mediated targeted mutagenesis (Al Amin et al., 2019; Bao et al., 2019; Campbell et al., 2019; Cheng et al., 2019; Di et al., 2019; Do et al., 2019; Han et al., 2019; Li et al., 2019; Cai et al., 2020; Michno et al., 2020; Zheng et al., 2020). So far, very limited success has been obtained in HR-mediated GT in soybean, with the majority of regenerated plants being chimeric and mutations not inherited to the next generation (Li et al., 2015).
Here, we report an Oh H1-8 (Ochrobactrum haywardense H1-8)-mediated embryonic axis transformation system for high-frequency GT in soybean, where the resultant HR-modified target locus is selectable-marker-free. Using two different donor designs 2%–3.4% GT was achieved in two soybean genomic sites. Analyses of T1 progeny confirmed Mendelian inheritance for the majority of GT events, while randomly integrated T-DNA segregated independently of the GT locus resulting in T-DNA-free GT-positive T1 progeny. The method being simple, robust, highly efficient, and reproducible, could expedite soybean precision breeding for yield and other traits.
Results
GT experiment design
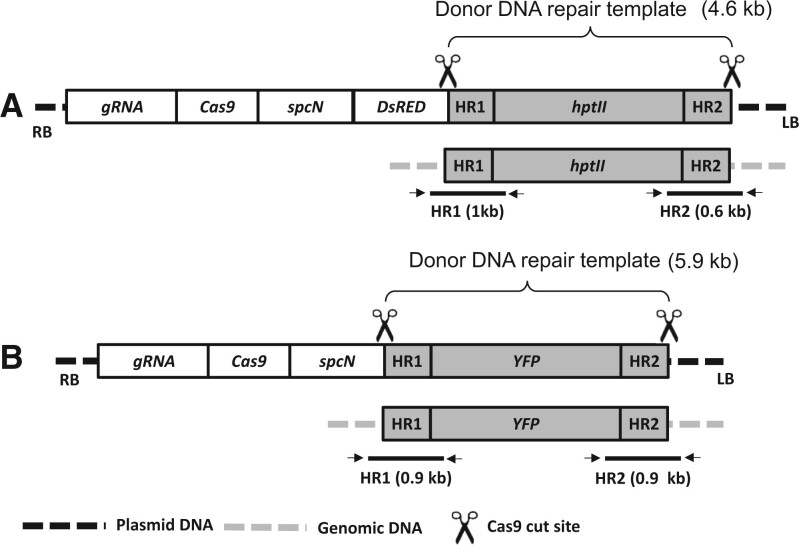
An overview of the GT construct design used in this study is described in Figure 1. The T-DNA construct (Figure 1A and Supplemental Figure S1A) targeting the soybean DD38 (Cigan et al., 2016) genomic site contained five expression cassettes. The expression cassette nearest the LB encoded a hygromycin-resistance gene (hptII) donor DNA repair template flanked by homology arms, HR1 and HR2, which were in turn flanked by two Cas9 cut sites matching the genomic target site. The T-DNA construct also contained in order between the RB and hptII cassette a single guide RNA (gRNA) expression cassette, a Cas9 expression cassette, a spectinomycin-resistance gene (spcN), and a DsRED color marker cassette. Following T-DNA transfer, transient Cas9 expression results in two DSBs at the Cas9 cut sites next to the two homology arms flanking hptII, leading to release of the donor DNA repair template. The T-DNA containing spcN as the plant selectable marker, which is randomly inserted in the genome, is used for positive selection and regeneration of transformed cells. In addition, a DSB is generated at the genomic target locus, which could then be repaired via HR using the donor template released from T-DNA, resulting in GT. The T-DNA could also be initially randomly integrated into the genome, with Cas9 expression leading to the release of the donor DNA repair template resulting in intragenomic GT as described previously (Barone et al., 2020). The donor DNA repair template in the T-DNA construct (Figure 1B and Supplemental Figure S1B) targeting the soybean DD51 (Cigan et al., 2016) genomic site contained the yellow fluorescent protein (YFP) gene.
Figure 1.
Schematic description of the pDD38 and pDD51 constructs containing donor template for GT at soybean DD38 and DD51 genomic sites, respectively. Both constructs contain gRNA and Cas9 expression cassettes for genome editing, and a spectinomycin (spcN) cassette as a plant selectable marker. Construct A for the DD38 site also contained the DsRED color marker. The donor DNA repair template contained a Hygromycin (hptII, A) or YFP (B) gene cassette flanked by homology arms (HR1 and HR2) and Cas9 cut sites (shown as scissors) matching the DD38 or DD51 genomic site. Following Cas9 and gRNA expression, the donor template will be released and used as template for repair at the DD38 or DD51 target site for GT (Bottom panel). HR1 and HR2 diagnostic PCRs were conducted to detect GT as described in Figure 2. RB and LB: right and left T-DNA border, respectively. The component sizes are not to the scale.
GT in soybean
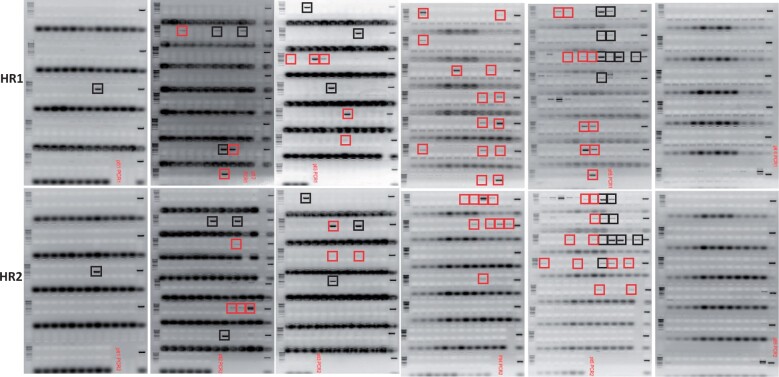
A plant transformation construct pDD38 containing constitutively expressed Cas9 was used for GT at the soybean DD38 site (Figure 1A and Supplemental Figure S1A). Oh H1-8-mediated embryonic axis transformation was carried out to generate stably transformed shoots ∼1.5 cm in size using spectinomycin selection as described in “Materials and methods.” In 6–8 weeks after transformation, a total of 466 shoots were regenerated and analyzed for GT and targeted NHEJ-mediated mutation at the DD38 site. HR-mediated GT was detected by HR1 and HR2 junction PCR as depicted in Figure 1A. The junction PCR was designed for quick screening of putative GT positive plants, further analyses for perfect GT was performed in the T1 generation. Out of 466 shoots analyzed, an HR1 junction-PCR positive product was observed in 48 (10%) shoots, while 43 (9.2%) shoots were found positive for HR2 junction-PCR (Figure 2 and Table 1). Positive PCR products for both HR1 and HR2 junctions were obtained in 16 plants indicating 3.4% putative GT at the DD38 site. Targeted insertion mutation analysis of the target site revealed 93.3% of shoots containing mutations in at least one allele, while >7% of the plants were observed with bi-allelic mutations (Table 1).
Figure 2.
Quick PCR screening to detect GT. HR1 (upper panel) and HR2 PCR (lower panel) of leaf samples from 466 T0 shoots regenerated for the DD38 GT experiment. Positive PCR products of expected sizes obtained both for HR1 and HR2 junctions are shown in black frames. PCR products amplified only for HR1 or HR2 are indicated in red frames. The PCR results are summarized in Table 1.
Table 1.
Summary of GT and mutation analyses at soybean DD38 genomic site
| Plants analyzed | GT |
Mutationa |
||||
|---|---|---|---|---|---|---|
| HR1 | HR2 | HR1 and HR2 | Mutated plants | Mono-allelic | Bi-allelic | |
| 466 | 48 (10%) | 43 (9.2%) | 16 (3.4%) | 435 (93.3%) | 401 (86%) | 34 (7.3%) |
qPCR read from 0 to 0.099: bi-allelic; 0.1–0.7: mono-allelic; 0.7–1.0: no mutation (WT).
After having successfully demonstrated GT at the DD38 site, we created another donor construct pDD51 (Figure 1B and Supplemental Figure S1B) targeting the DD51 site in the soybean genome. The construct pDD51 contained a YFP cassette as a donor repair template while DsRed was not included in the T-DNA, the rest of the construct design was similar to pDD38. Using Oh H1-8-mediated embryonic axis transformation, 690 shoots were generated on spectinomycin selection. HR1 and HR2 junction PCR revealed 2.8% and 2% positive plants (Table 2), respectively. Fourteen plants were observed positive for both HR1 and HR2 junctions demonstrating 2% GT at the DD51 site. Similar to the DD38 site, a high frequency (89%) of NHEJ-mediated mutations was detected at the DD51 site, with 9.5% of plants showing bi-allelic mutations.
Table 2.
Summary of GT and mutation analyses at soybean DD51 genomic site
| Plants analyzed | GT |
Mutationa |
||||
|---|---|---|---|---|---|---|
| HR1 | HR2 | HR1 and HR2 | Mutated plants | Mono-allelic | Bi-allelic | |
| 690 | 22 (3.2%) | 19 (2.8%) | 14 (2%) | 614 (89%) | 548 (78.5%) | 66 (9.5%) |
qPCR read from 0 to 0.099: bi-allelic; 0.1–0.7: mono-allelic; 0.7–1.0: no mutation (WT).
Inheritance and segregation analysis of GT plants
To study inheritance and segregation of GT, four randomly selected GT positive T0 plants at the DD38 site (designated as P1–P4) were self-pollinated to generate T1 progeny. Approximately 90 T1 plants from each of the 4 GT-positive T0 plants were analyzed for the presence of GT and T-DNA components. Dsred though present within T-DNA was not used for prescreening. To confirm GT and validate the integrity of the insertion, HR1/HR2 junction PCR data were supplemented with Southern-by-Sequencing (SbS) (Zastrow-Hayes et al., 2015; Table 3). Approximately 75% of progeny from 3 GT-positive T0 plants (P2, P3, and P4) were observed positive for both HR1 and HR2 junction PCR, and conformed to expected Mendelian inheritance of the GT insertion at the DD38 locus. These plants contained 1–2 copies of randomly inserted T-DNA components that segregated 1:2:1 (homo:hemi:null) as expected for a single locus insertion. For two of the four DD38 insertion lines, the number of T1 plants positive for both the HR1 and HR2 junctions and also null for the randomly inserted T-DNA, conformed to the expected Mendelian ratios of ∼25% (16 nulls out of 67 for P3, 22 nulls out of 71 for P4). Only one T-DNA-free GT T1 plant was observed from the P2 plant. The T1 progeny from the P1 T0 plant did not show expected Mendelian inheritance, only seven plants (∼7%) were observed GT positive and no T-DNA-free GT T1 plant was obtained (Table 3).
Table 3.
Inheritance and segregation of GT in T1 generation
| T0 plant | Total T1 plants | T-DNA |
GT |
|||||
|---|---|---|---|---|---|---|---|---|
| Nulls | Homo | Hemi | Copy# (PSB1) | HR1/2 positive T1 plants |
SBSa | |||
| Total | Transgene free | |||||||
| P1 | 91 | 31 | 21 | 39 | 3 | 7 | 1b | Deletion |
| P2 | 93 | 19 | 16 | 58 | 1 | 71 | 1 | Clean |
| P3 | 86 | 19 | 26 | 41 | 2 | 67 | 16 | Rearrangement |
| P4 | 95 | 26 | 20 | 49 | 2 | 71 | 22 | Clean |
Maximum five plants analyzed.
HR2 only.
Further molecular characterization of T-DNA component-free GT plants was done using SbS, which utilizes in-solution sequence capture coupled with NGS (Zastrow-Hayes et al., 2015). The method is highly sensitive in detection of construct-to-genome and construct-to-construct novel junctions that could have been created during the transformation process. The junction sequence data are then used to detect unintentionally inserted sequences from the transformation plasmid and help identify small T-DNA fragments, truncations of the intended donor DNA for GT, or the presence of transformation plasmid backbone sequences. One T1 plant each from P1 and P2 and five T1 plants each from P3 and P4 T0 plants were sampled for SbS (Table 3). The P1 plant although PCR-negative for HR1 was included to confirm sensitivity of SbS. In addition, samples from the wild-type negative control and the plasmid-spiked positive control were also included.
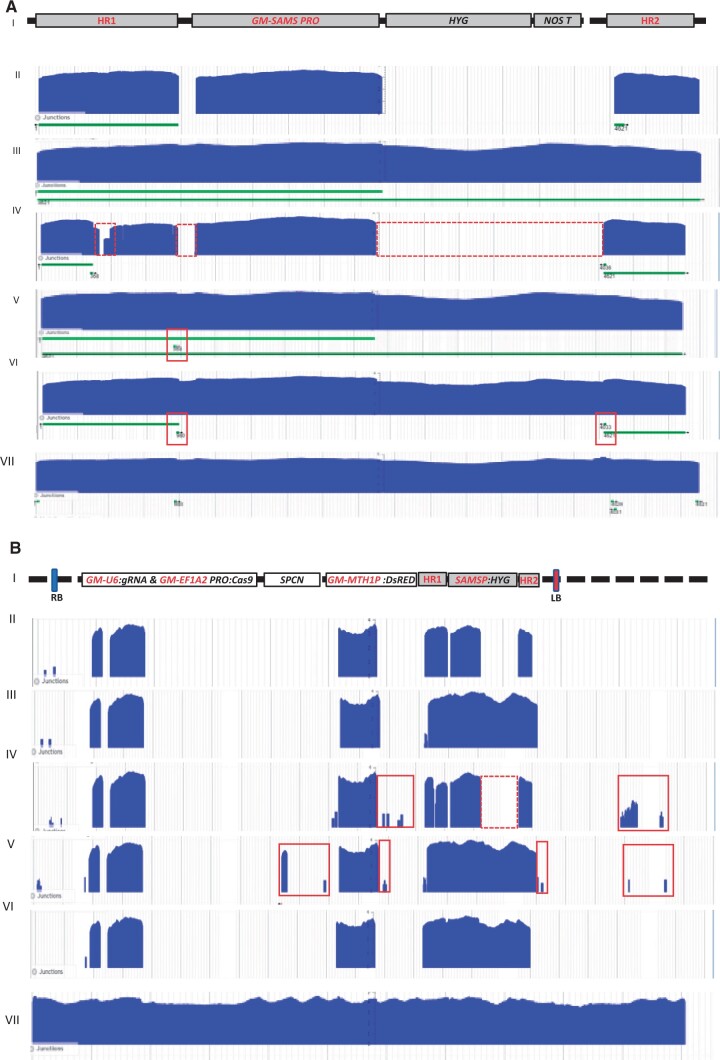
The sequence coverage graphs of the GT plants and controls were mapped to the expected schematic of the precise donor insertion at the DD38 genomic site (Figure 3A) and to the schematic of the construct used (Figure 3B) to confirm precise donor DNA insertion and absence of unintentional randomly inserted construct sequences. The SbS analyses revealed T-DNA-free precise GT in P2 and four P4 plants. A representative result is shown in Figure 3, AIII and BIII. One T1 plant from P4 was observed to contain an unexpected unique junction at the DD38 target site indicating insertion of multiple copies of rearranged donor DNA (Figure 3A VI). Similarly, all five P3 progeny plants were found to contain rearranged donor DNA at the DD38 target site (Figure 3A V). In addition, these plants also contained small T-DNA and backbone fragments (Figure 3B V) that were undetected by quick PCR screening conducted in the T0 generation. As expected from the HR1 PCR analysis, one P1 progeny plant was observed to contain a partial donor DNA insertion lacking majority of intended donor DNA sequence (Figure 3A IV).
Figure 3.
SbS for high-resolution molecular characterization of PCR positive gene-targeted T1 soybean plants. A, Sequence coverage graphs of targeted plants and controls mapped to the expected schematic of the precise donor insertion at the DD38 genomic site. (I) Schematic representation of the precisely targeted soybean DD38 site, which is mapped to the sequence coverage graph of the wild type (WT) plant as a negative control (II), representative precisely targeted P2 T1 plant (III), P2 T1 plant observed with imprecise insertion (IV) due to missing part of the donor (indicted as dotted red boxes), P3 and P4 T1 plants (V and VI, respectively) showing imprecise insertion due to rearrangements at the targeted locus (indicted as red boxes), WT plant DNA with spiked in plasmid DNA used for GT as positive SBS control (VII). B, Sequence coverage graphs of targeted plants and controls mapped to schematic of the transgene used. (I) Schematic representation of plasmid map aligned to sequence coverage graph of plants and controls described in A. I–VII as above. Red box indicates the presence of plasmid DNA, dotted box shows the missing part of the donor. Red font in the maps indicates soybean genomic sequences.
Discussion
Being a major source of plant proteins and oils, soybean is an economically important food and oilseed crop. The recent societal shift toward plant-based meat alternatives (Choudhury et al., 2020) has further increased the demand for soybean, making this crop even more important than ever. While the Green Revolution brought a considerable increase in the yields of rice, wheat, and maize, only modest yield improvements have been made for soybean (Liu et al., 2020). The unique architecture of soybean plants with leaves, inflorescences, and pods growing at each node, and the determined internode number greatly affect the final yield. Therefore, soybean yield being a multifactorial, complex trait, is likely to require simultaneous and precise modification of several components to positively affect the yield. Modern precision breeding tools, including genome editing, present an opportunity for such targeted breeding approaches. In addition, genome editing would be extremely useful for the pathway engineering needed to develop soybean varieties with enhanced protein and oil contents (Aili Bao et al., 2020; Subedi et al., 2020).
Due to its flexibility and ease of design, CRISPR-Cas9 has become an important genome engineering tool for creating targeted DSBs required for precision breeding applications (Wiedenheft et al., 2011; Jinek et al., 2012; Cong et al., 2013; Xing et al., 2014). A DSB at the desired target site triggers induction of a DNA repair pathway. In somatic cells NHEJ remains a dominant pathway, which mainly involves ligation of unrelated sequences (Salomon and Puchta, 1998; Puchta, 1999; Orel et al., 2003). Therefore, CRISPR-Cas9-medaited targeted mutagenesis, which is based on NHEJ repair, is now becoming routine in plants that are amenable for transformation including soybean (Al Amin et al., 2019; Bao et al., 2019; Campbell et al., 2019; Cheng et al., 2019; Di et al., 2019; Do et al., 2019; Han et al., 2019; Li et al., 2019; Cai et al., 2020; Michno et al., 2020; Zheng et al., 2020). In contrast, GT for targeted insertion or allele replacement remains a huge challenge mainly due to low-frequency HR in somatic cells coupled with the complexity of making externally supplied donor template accessible at the DSB site (Mao et al., 2019; Rozov et al., 2019). The complex genetic changes needed to improve soybean yield, oil, and protein traits are likely to require precise insertions and replacements of DNA sequences, which can only be accomplished by HR-mediated GT. The challenge of inefficient GT is further confounded by the need for a selectable marker to recover, enrich and regenerate plants from rare GT cell(s), while the presence of the selectable marker in the final GT plants is undesirable for commercial deployment of the technology. Therefore, development of a GT system which does not have a selectable marker in the final plant is highly desirable for application of GT for commercial product development.
Here we report Oh H1-mediated delivery of CRISPR-Cas9 and donor DNA template to obtain high-frequency GT in soybean. The delivery method utilized an embryonic axis derived from soybean seed to generate stably transformed shoots (Cho et al., 2022). First, we demonstrated efficient GT at the soybean DD38 genomic site. While up to 10% targeted insertion was observed by HR1 or HR2 junction PCR, 3.4% putative true GT was obtained as indicated by positive PCR for both HR1 and HR2 junctions. Second, to establish reproducibility of our GT method we targeted another genomic site (DD51) using a different construct design. Similar to the DD38 site, we successfully generated 2% GT at the DD51 site indicating that the GT method described in this report is reliable and can be applied for different genomic sites. High frequency (∼90%) targeted mutagenesis at both DD38 and DD51 sites further confirmed that our CRISPR-Cas9 genome editing is robust and efficient.
While the donor DNA template contained a hptII or YFP gene cassette, they were not used for selection; instead, spcN located on the T-DNA outside the donor DNA sequence was used to identify plants with randomly inserted T-DNA. Such construct design enables the donor DNA to contain only the trait gene(s) and the GT at the genomic target site to be free from a selectable marker. The randomly inserted T-DNA containing the spcN cassette can be segregated out in the next generation to obtain clean GT plants without unintended T-DNA sequences. The unique donor construct design with Cas9 cut sites flanking the donor template (Peterson et al., 2021) enabled such indirect selection using a randomly inserted spcN selection cassette while GT at the genomic target site remains selection free. Recently, we applied a similar construct design for an intra-genomic GT in maize, however, it utilized heat-shock inducible expression of Cas9 (Barone et al., 2020). The GT work on soybean described in this article is based on the constitutive expression of Cas9. While we assume that the selective regeneration advantage for the Cas9-expressing cells with donor DNA released from the T-DNA provided preferential enrichment for GT events, intra-genomic GT with donor released from the T-DNA randomly integrated into the plant genome cannot be ruled out. Therefore, similar to our maize intra-genomic GT (Barone et al., 2020), inducible Cas9 expression coupled with selectable marker activation would be an attractive approach to further enhance GT in soybean.
Lack of a fast, efficient, and reproducible transformation method has been a key bottleneck for soybean genome editing (Altpeter et al., 2016). The common transformation method used to transform soybean utilizes Agrobacterium-mediated DNA delivery to cotyledonary-node explants (Paz et al., 2006), which is not only inefficient (∼4% transformation frequency) but often leads to chimerism and nontransgenic escape plants, making the transformation process labor-intensive and expensive (Zheng et al., 2020). The current status of soybean transformation probably explains the lack of a viable GT system for commercial product development. The only previously published report on GT in soybean utilized particle bombardment-mediated delivery, showing GT in the callus stage with the majority being chimeric and not inherited to the next generation (Li et al., 2015). In contrast, the Oh H1-8 mediated delivery used here is fast, highly efficient (∼35% transformation frequency), reproducible, and generates nonchimeric plants (Cho et al., 2022). The transformation method coupled with the construct design described in this report was instrumental for successful demonstration of an efficient, heritable, and selectable marker free GT system paving the way for next-generation precision breeding in soybean.
After demonstrating a successful GT in T0 plants, the next step was to validate that our GT method produces nonchimeric plants with stable and precise insertion, while unintentionally randomly inserted T-DNA sequences are segregated away to generate transgene-free GT plants in the T1 generation. Among the self-pollinated T1 progeny planted from four GT-positive T0 plants, Mendelian inheritance of GT at the DD38 locus and randomly inserted T-DNA was observed for progeny from three T0 plants. An expected 25% T-DNA-free GT positive T1 plants were obtained from two T0 GT plants. Only one T-DNA-free GT plant was observed from the third plant indicating genetic linkage between the DD38 target site and the T-DNA insertion site in this line. The T1 progeny from one plant showed chimeric GT and no T-DNA-free GT T1 plant was obtained in this line. The next goal was to confirm the fidelity of PCR-positive GT calls and ensure the selected GT T1 plants are free from any unintentionally inserted T-DNA sequences. We employed high-resolution SbS (Zastrow-Hayes et al., 2015) to further characterize selected transgene-free GT positive T1 plants. Compared with whole-genome sequencing, SbS utilizes sequence capture technology, which reduces sequence analysis complexity by enriching the target sequences of interest, in turn decreasing the amount of sequence data generated by NGS technology. Bioinformatic analysis of the targeted sequencing identifies novel plasmid-to-genome or plasmid-to-plasmid junctions providing comprehensive information about the number of unique insertion loci, potential rearrangements of the inserted DNA, and the presence or absence of plasmid backbone sequences. SBS analyses revealed random T-DNA-free precise GT in T1 plants from two T0 lines. Some T1 plants from these lines were also observed with insertion of multiple copies of rearranged donor DNA. All T1 plants screened for one T0 line had rearranged donor DNA at the DD38 target site, in addition to containing small T-DNA and backbone fragments that were undetected by quick PCR screening in the T0 generation. More extensive diagnostic PCR in T0 plants could have detected these imperfect GT or T-DNA fragment-containing events. Given the limited number of plants analyzed for T1 inheritance, more work is needed to confirm the findings reported in this study.
In summary, using an Oh H1-8-mediated delivery, we developed an efficient and reliable GT system in a major dicot crop plant. This work also provides general analytical guidelines for T0 to T1 generations posttransformation for selecting plants with precise GT and void of any unintentionally inserted plasmid DNA sequences. Given GT without a selectable marker gene or any transgene in the final plant is required for nontransgenic applications of genome editing technology, successful demonstration of the system design for a selectable marker-free GT in this report is a major step toward commercial application of CRISPR-Cas9 genome editing in soybean precision breeding.
Materials and methods
Genomic target sites, plasmids, and reagents used for plant transformation
Genomic target sites DD38 and DD51 used in this study were selected using criteria described previously (Gao et al., 2020), which includes no known gene within 2 kb, the site being unique in the genome and conserved among the targeted inbred lines, and unique 200–500 bp sequence flanking the site. Both DD38 and DD51 sites are within chromosome 4 with genetic position (Wm82.a4) 90.91 and 92.40 cM, respectively. The physical location of DD38-CR1 and DD38-CR2 is Gm04:45802466.45802445 and Gm04:45802465.45802486, respectively. The location of DD51-CR1 and DD51-CR2 is Gm04:45937312.45937333 and Gm04:45937298.45937320, respectively.
Standard cloning methods were used to construct DNA plasmids used in this study. All plasmids were quality controlled through deep sequencing and 100% identity was confirmed. Supplemental Table S1 and Supplemental Figure S2 provide the details of expression and other elements used in different constructs described in Figure 1 and Supplemental Figure S1.
Soybean transformation
Corteva Agriscience elite soybean (G. max) variety 93Y21 was used. Mature dry seeds were surface-sterilized for 16 h using chlorine gas as described by Di et al. (1996). Disinfected seeds were imbibed on solid agar medium containing 5 g/L sucrose and 6 g/L agar for 6–8 h and then the seeds were soaked in distilled sterile water for overnight at room temperature in the dark. Intact embryonic axes were isolated using a scalpel blade.
All O. haywardense H1-8 (Oh H1-8)-mediated soybean transformation was carried out prior to 2020 as described by Cho et al Oh H1-8 lines containing the vectors listed in Figure 1 and Supplemental Figure S1 were used for transformation. Oh H1-8 suspension (OD 0.5 at 600 nm) in infection medium composed of 1/10× Gamborg B5 basal medium, 30 g/L sucrose, 20 mM MES (2-(N-morpholino) ethanesulfonic acid), 0.25 mg/L GA3 (Gibberellic acid), 1.67 mg/L BAP (6-Benzylaminopurine), 200 µM acetosyringone, and 1 mM dithiothreitol in pH 5.4 was added to about 200–300 EAs on a 25 × 100 mm deep petri dish. The plates were sealed with parafilm, then sonicated for 30 s. After sonication, EAs were incubated 2 h at room temperature. After inoculation, about 200–300 embryonic axes were transferred to a single layer of autoclaved sterile filter paper (Cat No. 28320-020, VWR) in a 25 × 100 mm petri dish. The plates were sealed with Micropore tape (1530-0, 3M, St. Paul, MN, USA) and incubated under dim light (1–2 μE/m2/s), using cool white fluorescent lamps for 16 h at 21°C for 3 d. After co-cultivation, the base of each embryonic axis was embedded in shoot induction medium (R7100, PhytoTech Labs) containing 30 g/L sucrose, 6 g/L agar, 25 mg/L spectinomycin as a selectable agent and 250 mg/L cefotaxime (GoldBio, ST Louis, MO, USA) in pH5.7. Shoot induction was carried out in a growth room at 26°C with a photoperiod of 16 h and a light intensity of 60–100 μE/m2/s.
After 4–6 weeks in selection medium, the spectinomycin-resistant shoots were cut and transferred to 0.5 strength MS rooting medium (M404, PhytoTech Labs) containing 15 g/L sucrose, 6 g/L agar, 10 mg/L spectinomycin, and 250 mg/L cefotaxime for further shoot and root elongations. Transformation efficiency was calculated based on the number of positive transgenic soybean T0 plants divided by the total number of EAs. Transgenic soybean plantlets were transferred to moistened Berger BM2 soil (Berger, Saint-Modeste, QC, Canada), and hardened plantlets were potted in 2-gallon pots containing moistened SunGro 702 and grown to maturity for harvest in a greenhouse.
Molecular analysis
Plants were sampled by leaf punching at 7 d after transplantation for qPCR to confirm the presence of the selectable marker (spcN), CRISPR-Cas9 (PSN1), the left (PSB1), and right border (PSA2) regions. The spcN, PSN1, PSB1, and PSA2 regions for qPCR are shown in T-DNA maps in Supplemental Figure S1 and PCR conditions are given by Wu et al. (2014). The primers and probes used for qPCR assays are described in Supplemental Table S2. DNA was extracted from the 7 mm leaf disk with sbeadex chemistry (LGC Biosearch, Middlesex, UK) as per vendor recommendations and resulting DNA was analyzed for quality and quantity via DropSense (Unchained Labs, Pleasanton, CA, USA). PCR to detect mutations and HR junction PCR have been described previously (Barone et al., 2020). Mutation frequencies at genomic target sites were analyzed by qPCR using primer pair and probes given in Supplemental Table S3. HR Junctions were analyzed by using HR spanning primers (F1 and R1) described in in Supplemental Table S3. All PCR positive samples were cloned (TOPO) and sequenced (Sanger sequencing), and only sequence confirmed samples were counted positive for putative GT.
SbS data were generated according to Zastrow-Hayes et al. (2015). Briefly, genomic DNA was isolated from 4-mm leaf punches using the Sbeadex Maxi Plant kit (LGC Genomics) then randomly sheared to an average size of ∼400 bps with a Covaris E210 ultrasonicator (Covaris, Woburn, MA, USA). Indexed Illumina genomic libraries were generated with the KAPA HTP Library Preparation Kit (Roche, Basel, Switzerland). Adapter-ligated DNA fragments were subsequently PCR-amplified for eight cycles, pooled in equal molar ratios then normalized in nuclease-free water to 5 ng/μL. Following library construction, pooling and normalization, and target enrichment was performed using a modified Roche Nimblegen double capture protocol (Zastrow-Hayes et al., 2015), with biotinylated DNA probes designed from the entire plasmid sequences. Enriched DNA fragments were sequenced on an Illumina NextSeq sequencer, according to the manufacturer’s instructions. The resulting DNA sequences were quality-trimmed and aligned to the plasmid sequence for coverage analysis and characterization of the event. Positive SbS controls were derived from wild-type genomic DNA combined with spiked-in plasmid DNA for Illumina genomic library construction and enrichment.
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers given in Supplemental Table S1.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Plasmid maps of the constructs pDD38 (A) and pDD51 (B) containing donor template for GT at soybean DD38 and DD51 genomic sites, respectively.
Supplemental Figure S2. gRNA spacer and homology sequences used in pDD38 and pDD51 plasmids.
Supplemental Table S1. Description of vector components.
Supplemental Table S2. Primers and probes used for qPCR detection of transgene.
Supplemental Table S3. Primers and probes used for GT and mutation analyses.
Supplementary Material
Acknowledgments
We thank Lijuan Wang and Super Vector team for vector construction, Shujun Chang and Sam Ellis for soybean transformation support, Joel Van Wyk and Becca Vickroy for greenhouse care, and Marv Taylor and Balaji Boovaraghan and the PCR Analysis and Characterization team for high-throughput PCR support. We also thank Scott Betts, Maria Fedorova, and Sendil Devadas for critical review of the manuscript. Finally, we acknowledge Clara Alarcon, Todd Jones, and Doane Chilcoat for providing resources and support for this work.
Data availability
The data supporting the findings of this study are available within the paper and its supplementary information files.
Conflict of interest statement. Authors are employees of Corteva Agriscience. Some of the authors are inventors on pending applications on this work.
S.K., Z.-B.L., and A.A designed the experiment. N.S.-D, J.B., and H.-J.C. performed the soybean transformation. B.L., A.W., and A.R. designed and performed the DNA analysis. S.K. led the project and wrote the manuscript. All authors reviewed the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Sandeep Kumar (sandeep.kumar@corteva.com).
References
- Aili Bao CZ, Huang Y, Chen H, Zhou X, Cao D (2020) Genome editing technology and application in soybean improvement. Oil Crop Sci 5: 31–40 [Google Scholar]
- Al Amin N, Ahmad N, Wu N, Pu X, Ma T, Du Y, Bo X, Wang N, Sharif R, Wang P (2019) CRISPR-Cas9 mediated targeted disruption of FAD2-2 microsomal omega-6 desaturase in soybean (Glycine max L.). BMC Biotechnol 19: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altpeter F, Springer NM, Bartley LE, Blechl AE, Brutnell TP, Citovsky V, Conrad LJ, Gelvin SB, Jackson DP, Kausch AP, et al. (2016) Advancing crop transformation in the era of genome editing. Plant Cell 28: 1510–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao A, Chen H, Chen L, Chen S, Hao Q, Guo W, Qiu D, Shan Z, Yang Z, Yuan S, et al. (2019) CRISPR/Cas9-mediated targeted mutagenesis of GmSPL9 genes alters plant architecture in soybean. BMC Plant Biol 19: 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barone P, Wu E, Lenderts B, Anand A, Gordon-Kamm W, Svitashev S, Kumar S (2020) Efficient gene targeting in maize using inducible CRISPR-Cas9 and marker-free donor template. Mol Plant 13: 1219–1227 [DOI] [PubMed] [Google Scholar]
- Cai Y, Wang L, Chen L, Wu T, Liu L, Sun S, Wu C, Yao W, Jiang B, Yuan S, et al. (2020) Mutagenesis of GmFT2a and GmFT5a mediated by CRISPR/Cas9 contributes for expanding the regional adaptability of soybean. Plant Biotechnol J 18: 298–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell BW, Hoyle JW, Bucciarelli B, Stec AO, Samac DA, Parrott WA, Stupar RM (2019) Functional analysis and development of a CRISPR/Cas9 allelic series for a CPR5 ortholog necessary for proper growth of soybean trichomes. Sci Rep 9: 14757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Q, Dong L, Su T, Li T, Gan Z, Nan H, Lu S, Fang C, Kong L, Li H, et al. (2019) CRISPR/Cas9-mediated targeted mutagenesis of GmLHY genes alters plant height and internode length in soybean. BMC Plant Biol 19: 562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HJ, Moy Y, Rudnick NA, Klein TM, Yin J, Bolar J, Hendrick C, Beatty M, Castaneda L, Kinney AJ, et al. (2022) Development of an efficient marker-free soybean transformation method using the novel bacterium Ochrobactrum haywardense H1. Plant Biotechnol J https://doi.org/10.1111/pbi.13777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury D, Singh S, Seah JSH, Yeo DCL, Tan LP (2020) Commercialization of plant-based meat alternatives. Trends Plant Sci 25: 1055–1058 [DOI] [PubMed] [Google Scholar]
- Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, Bogdanove AJ, Voytas DF (2010) Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 186: 757–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cigan M, Gao H, Liu Z-B, Mutti J, Podlich D, Scelonge C (2016) Generation of site-specific-integration sites for complex trait loci in corn and soybean, and methods of use. https://patents.google.com/patent/WO2016040030A1/en
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di R, Purcell V, Collins GB, Ghabrial SA (1996) Production of transgenic soybean lines expressing the bean pod mottle virus coat protein precursor gene. Plant Cell Rep 15: 746–750 [DOI] [PubMed] [Google Scholar]
- Di YH, Sun XJ, Hu Z, Jiang QY, Song GH, Zhang B, Zhao SS, Zhang H (2019) Enhancing the CRISPR/Cas9 system based on multiple GmU6 promoters in soybean. Biochem Biophys Res Commun 519: 819–823 [DOI] [PubMed] [Google Scholar]
- Do PT, Nguyen CX, Bui HT, Tran LTN, Stacey G, Gillman JD, Zhang ZJ, Stacey MG (2019) Demonstration of highly efficient dual gRNA CRISPR/Cas9 editing of the homeologous GmFAD2-1A and GmFAD2-1B genes to yield a high oleic, low linoleic and alpha-linolenic acid phenotype in soybean. BMC Plant Biol 19: 311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epinat JC, Arnould S, Chames P, Rochaix P, Desfontaines D, Puzin C, Patin A, Zanghellini A, Paques F, Lacroix E (2003) A novel engineered meganuclease induces homologous recombination in yeast and mammalian cells. Nucleic Acids Res 31: 2952–2962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Mutti J, Young JK, Yang M, Schroder M, Lenderts B, Wang L, Peterson D, St. Clair G, Jones S, et al. (2020) Complex trait loci in maize enabled by CRISPR-Cas9 mediated gene insertion. Front Plant Sci 11: 535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasiunas G, Barrangou R, Horvath P, Siksnys V (2012) Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci USA 109: E2579–2586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudelli NM, Komor AC, Rees HA, Packer MS, Badran AH, Bryson DI, Liu DR (2017) Programmable base editing of AT to GC in genomic DNA without DNA cleavage. Nature 551: 464–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham PH, Vance CP (2003) Legumes: importance and constraints to greater use. Plant Physiol 131: 872–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Guo B, Guo Y, Zhang B, Wang X, Qiu LJ (2019) Creation of early flowering germplasm of soybean by CRISPR/Cas9 technology. Front Plant Sci 10: 1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YG, Cha J, Chandrasegaran S (1996) Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci USA 93: 1156–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR (2016) Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533: 420–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Gao F, Xie K, Zeng X, Cao Y, Zeng J, He Z, Ren Y, Li W, Deng Q, et al. (2016b) The osmiR396c-OsGRF4-OsGIF1 regulatory module determines grain size and yield in rice. Plant Biotechnol J 14: 2134–2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Li X, Zhou Z, Wu P, Fang M, Pan X, Lin Q, Luo W, Wu G, Li H (2016a) Reassessment of the four yield-related genes Gn1a, DEP1, GS3, and IPA1 in rice using a CRISPR/Cas9 system. Front Plant Sci 7: 377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Liu ZB, Xing A, Moon BP, Koellhoffer JP, Huang L, Ward RT, Clifton E, Falco SC, Cigan AM (2015) Cas9-guide RNA directed genome editing in soybean. Plant Physiol 169: 960–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Nguyen V, Liu J, Fu W, Chen C, Yu K, Cui Y (2019) Mutagenesis of seed storage protein genes in Soybean using CRISPR/Cas9. BMC Res Notes 12: 176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Wang Y, Chen S, Tian H, Fu D, Zhu B, Luo Y, Zhu H (2018) Lycopene is enriched in tomato fruit by CRISPR/Cas9-mediated multiplex genome editing. Front Plant Sci 9: 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Chen J, Zheng X, Wu F, Lin Q, Heng Y, Tian P, Cheng Z, Yu X, Zhou K, et al. (2017) GW5 acts in the brassinosteroid signalling pathway to regulate grain width and weight in rice. Nat Plants 3: 17043. [DOI] [PubMed] [Google Scholar]
- Liu S, Zhang M, Feng F, Tian Z (2020) Toward a “green revolution” for soybean. Mol Plant 13: 688–697 [DOI] [PubMed] [Google Scholar]
- Mao Y, Botella JR, Liu Y, Zhu JK (2019) Gene editing in plants: progress and challenges. Natl Sci Rev 6: 421–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzec M, Braszewska-Zalewska A, Hensel G (2020) Prime editing: A new way for genome editing. Trends Cell Biol 30: 257–259. [DOI] [PubMed] [Google Scholar]
- Michno JM, Virdi K, Stec AO, Liu J, Wang X, Xiong Y, Stupar RM (2020) Integration, abundance, and transmission of mutations and transgenes in a series of CRISPR/Cas9 soybean lines. BMC Biotechnol 20: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molla KA, Sretenovic S, Bansal KC, Qi Y (2021) Precise plant genome editing using base editors and prime editors. Nat Plants 7: 1166–1187 [DOI] [PubMed] [Google Scholar]
- Nonaka S, Arai C, Takayama M, Matsukura C, Ezura H (2017) Efficient increase of aminobutyric acid (GABA) content in tomato fruits by targeted mutagenesis. Sci Rep 7: 7057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orel N, Kyryk A, Puchta H (2003) Different pathways of homologous recombination are used for the repair of double-strand breaks within tandemly arranged sequences in the plant genome. Plant J 35: 604–612 [DOI] [PubMed] [Google Scholar]
- Paz MM, Martinez JC, Kalvig AB, Fonger TM, Wang K (2006) Improved cotyledonary node method using an alternative explant derived from mature seed for efficient Agrobacterium-mediated soybean transformation. Plant Cell Rep 25: 206–213 [DOI] [PubMed] [Google Scholar]
- Peterson D, Barone P, Lenderts B, Schwartz C, Feigenbutz L, St Clair G, Jones S, Svitashev S (2021) Advances in Agrobacterium transformation and vector design result in high frequency targeted gene insertion in maize. Plant Biotechnol J 19: 2000–2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puchta H (1999) Double-strand break-induced recombination between ectopic homologous sequences in somatic plant cells. Genetics 152: 1173–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puchta H (2017) Applying CRISPR/Cas for genome engineering in plants: The best is yet to come. Curr Opin Plant Biol 36: 1–8 [DOI] [PubMed] [Google Scholar]
- Ray DK, Mueller ND, West PC, Foley JA (2013) Yield trends are insufficient to double global crop production by 2050. PLoS ONE 8: e66428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozov SM, Permyakova NV, Deineko EV (2019) The problem of the low rates of CRISPR/Cas9-mediated knock-ins in plants: Approaches and solutions. Int J Mol Sci 20: 3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomon S, Puchta H (1998) Capture of genomic and T-DNA sequences during double-strand break repair in somatic plant cells. EMBO J 17: 6086–6095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Gao H, Wang H, Lafitte HR, Archibald RL, Yang M, Hakimi SM, Mo H, Habben JE (2017) ARGOS8 variants generated by CRISPR-Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnol J 15: 207–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subedi U, Jayawardhane KN, Pan X, Ozga J, Chen G, Foroud NA, Singer SD (2020) The potential of genome editing for improving seed oil content and fatty acid composition in oilseed crops. Lipids 55: 495–512 [DOI] [PubMed] [Google Scholar]
- Wiedenheft B, van Duijn E, Bultema JB, Waghmare SP, Zhou K, Barendregt A, Westphal W, Heck AJ, Boekema EJ, Dickman MJ, et al. (2011) RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. Proc Natl Acad Sci USA 108: 10092–10097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood RD (1996) DNA repair in eukaryotes. Annu Rev Biochem 65: 135–167 [DOI] [PubMed] [Google Scholar]
- Wu E, Lenderts B, Glassman K, Berezowska-Kaniewska M, Christensen H, Asmus T, Zhen S, Chu U, Cho MJ, Zhao ZY (2014) Optimized Agrobacterium-mediated sorghum transformation protocol and molecular data of transgenic sorghum plants. In Vitro Cell Dev Biol Plant 50: 9–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing HL, Dong L, Wang ZP, Zhang HY, Han CY, Liu B, Wang XC, Chen QJ (2014) A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol 14: 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu QH, Wang B, Li N, Tang Y, Yang S, Yang T, Xu J, Guo C, Yan P, Wang Q, et al. (2017) CRISPR/Cas9-induced targeted mutagenesis and gene replacement to generate long-shelf life tomato lines. Sci Rep 7: 11874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zastrow-Hayes GM, Lin H, Sigmund AL, Hoffman JL, Alarcon CM, Hayes KR, Richmond TA, Jeddeloh JA, May GD, Beatty MK (2015) Southern-by-sequencing: A robust screening approach for molecular characterization of genetically modified crops. Plant Genome 8: eplantgenome2014.08.0037 [DOI] [PubMed] [Google Scholar]
- Zheng N, Li T, Dittman JD, Su J, Li R, Gassmann W, Peng D, Whitham SA, Liu S, Yang B (2020) CRISPR/Cas9-based gene editing using egg cell-specific promoters in Arabidopsis and soybean. Front Plant Sci 11: 800. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the findings of this study are available within the paper and its supplementary information files.
Conflict of interest statement. Authors are employees of Corteva Agriscience. Some of the authors are inventors on pending applications on this work.